
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:BIRC6-MATN3 (FusionGDB2 ID:9760) |
Fusion Gene Summary for BIRC6-MATN3 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: BIRC6-MATN3 | Fusion gene ID: 9760 | Hgene | Tgene | Gene symbol | BIRC6 | MATN3 | Gene ID | 57448 | 4148 |
| Gene name | baculoviral IAP repeat containing 6 | matrilin 3 | |
| Synonyms | APOLLON|BRUCE | DIPOA|EDM5|HOA|OADIP|OS2 | |
| Cytomap | 2p22.3 | 2p24.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | baculoviral IAP repeat-containing protein 6BIR repeat-containing ubiquitin-conjugating enzymeRING-type E3 ubiquitin transferase BIRC6ubiquitin-conjugating BIR-domain enzyme apollon | matrilin-3 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q9NR09 | O15232 | |
| Ensembl transtripts involved in fusion gene | ENST00000421745, | ENST00000407540, ENST00000421259, | |
| Fusion gene scores | * DoF score | 21 X 23 X 16=7728 | 3 X 3 X 2=18 |
| # samples | 33 | 3 | |
| ** MAII score | log2(33/7728*10)=-4.54955716458996 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(3/18*10)=0.736965594166206 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: BIRC6 [Title/Abstract] AND MATN3 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | BIRC6(32641231)-MATN3(20206071), # samples:4 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across BIRC6 (5'-gene) Fusion gene breakpoints across BIRC6 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
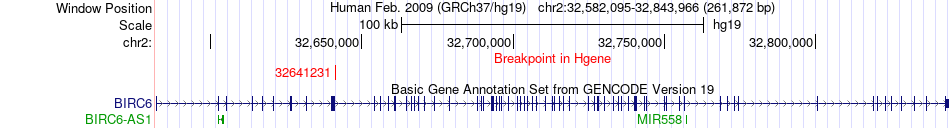 |
 Fusion gene breakpoints across MATN3 (3'-gene) Fusion gene breakpoints across MATN3 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
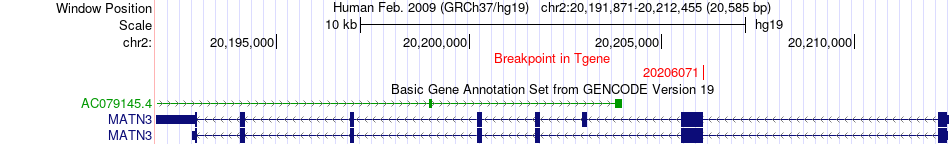 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | UCEC | TCGA-EO-A3KW-01A | BIRC6 | chr2 | 32641231 | - | MATN3 | chr2 | 20206071 | - |
| ChimerDB4 | UCEC | TCGA-EO-A3KW-01A | BIRC6 | chr2 | 32641231 | + | MATN3 | chr2 | 20206071 | - |
| ChimerDB4 | UCEC | TCGA-EO-A3KW | BIRC6 | chr2 | 32641231 | + | MATN3 | chr2 | 20206071 | - |
Top |
Fusion Gene ORF analysis for BIRC6-MATN3 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000421745 | ENST00000407540 | BIRC6 | chr2 | 32641231 | + | MATN3 | chr2 | 20206071 | - |
| In-frame | ENST00000421745 | ENST00000421259 | BIRC6 | chr2 | 32641231 | + | MATN3 | chr2 | 20206071 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000421745 | BIRC6 | chr2 | 32641231 | + | ENST00000407540 | MATN3 | chr2 | 20206071 | - | 5244 | 3006 | 134 | 4243 | 1369 |
| ENST00000421745 | BIRC6 | chr2 | 32641231 | + | ENST00000421259 | MATN3 | chr2 | 20206071 | - | 4192 | 3006 | 134 | 4117 | 1327 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000421745 | ENST00000407540 | BIRC6 | chr2 | 32641231 | + | MATN3 | chr2 | 20206071 | - | 0.000217172 | 0.99978286 |
| ENST00000421745 | ENST00000421259 | BIRC6 | chr2 | 32641231 | + | MATN3 | chr2 | 20206071 | - | 0.000473035 | 0.999527 |
Top |
Fusion Genomic Features for BIRC6-MATN3 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
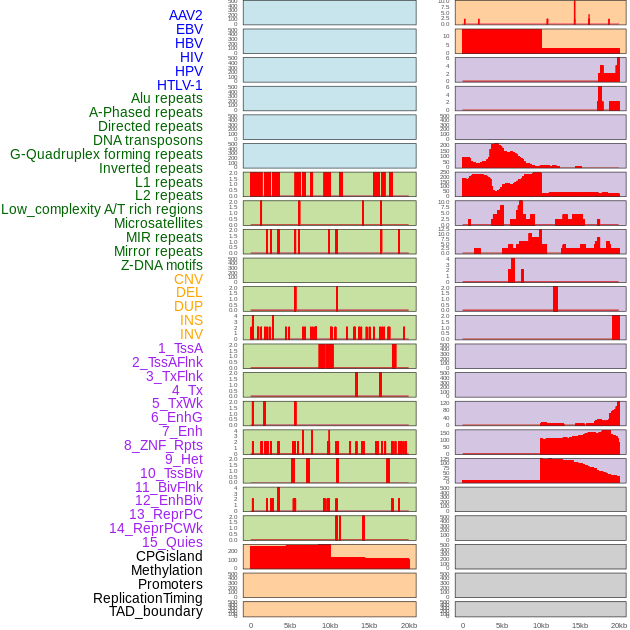 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for BIRC6-MATN3 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr2:32641231/chr2:20206071) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| BIRC6 | MATN3 |
| FUNCTION: Anti-apoptotic protein which can regulate cell death by controlling caspases and by acting as an E3 ubiquitin-protein ligase. Has an unusual ubiquitin conjugation system in that it could combine in a single polypeptide, ubiquitin conjugating (E2) with ubiquitin ligase (E3) activity, forming a chimeric E2/E3 ubiquitin ligase. Its tragets include CASP9 and DIABLO/SMAC. Acts as an inhibitor of CASP3, CASP7 and CASP9. Important regulator for the final stages of cytokinesis. Crucial for normal vesicle targeting to the site of abscission, but also for the integrity of the midbody and the midbody ring, and its striking ubiquitin modification. {ECO:0000269|PubMed:14765125, ECO:0000269|PubMed:15200957, ECO:0000269|PubMed:18329369}. | FUNCTION: Major component of the extracellular matrix of cartilage and may play a role in the formation of extracellular filamentous networks. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | BIRC6 | chr2:32641231 | chr2:20206071 | ENST00000421745 | + | 10 | 74 | 30_36 | 957 | 4858.0 | Compositional bias | Note=Poly-Ala |
| Hgene | BIRC6 | chr2:32641231 | chr2:20206071 | ENST00000421745 | + | 10 | 74 | 284_358 | 957 | 4858.0 | Repeat | Note=BIR |
| Tgene | MATN3 | chr2:32641231 | chr2:20206071 | ENST00000407540 | 0 | 8 | 456_480 | 74 | 487.0 | Coiled coil | Ontology_term=ECO:0000250 | |
| Tgene | MATN3 | chr2:32641231 | chr2:20206071 | ENST00000407540 | 0 | 8 | 264_305 | 74 | 487.0 | Domain | EGF-like 1 | |
| Tgene | MATN3 | chr2:32641231 | chr2:20206071 | ENST00000407540 | 0 | 8 | 306_347 | 74 | 487.0 | Domain | EGF-like 2 | |
| Tgene | MATN3 | chr2:32641231 | chr2:20206071 | ENST00000407540 | 0 | 8 | 348_389 | 74 | 487.0 | Domain | EGF-like 3 | |
| Tgene | MATN3 | chr2:32641231 | chr2:20206071 | ENST00000407540 | 0 | 8 | 390_431 | 74 | 487.0 | Domain | EGF-like 4 | |
| Tgene | MATN3 | chr2:32641231 | chr2:20206071 | ENST00000407540 | 0 | 8 | 83_258 | 74 | 487.0 | Domain | VWFA |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | BIRC6 | chr2:32641231 | chr2:20206071 | ENST00000421745 | + | 10 | 74 | 1660_1668 | 957 | 4858.0 | Compositional bias | Note=Poly-Ala |
| Hgene | BIRC6 | chr2:32641231 | chr2:20206071 | ENST00000421745 | + | 10 | 74 | 4573_4740 | 957 | 4858.0 | Domain | UBC core |
Top |
Fusion Gene Sequence for BIRC6-MATN3 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >9760_9760_1_BIRC6-MATN3_BIRC6_chr2_32641231_ENST00000421745_MATN3_chr2_20206071_ENST00000407540_length(transcript)=5244nt_BP=3006nt TCCCTGCTTCTCCCCCTCTCCCGTCAGCCTCCCTCCGAGTTTGGCCCCTCCGGCCGGGCGATCGACGTTCCGCGTGCGTGCGGGCGCCTG ACTTCACTTCCGGCTAACGCGCTCGGCTTGCCCCCTGGCCCCGGATGGTGACTGGTGGTGGTGCTGCACCTCCCGGGACTGTCACTGAGC CGCTTCCCAGTGTGATTGTGCTGAGCGCAGGCCGGAAGATGGCGGCTGCGGCTGCGGCGGCCTCGGGCCCCGGCTGCTCCTCGGCGGCGG GGGCGGGGGCGGCCGGGGTCTCAGAGTGGCTGGTGCTGCGGGACGGCTGCATGCACTGCGACGCCGACGGGCTGCACAGCCTGTCCTACC ACCCTGCGCTCAACGCCATCCTGGCCGTCACTAGCCGCGGGACCATCAAAGTCATCGACGGCACCTCGGGGGCCACACTGCAGGCCTCCG CGCTCAGTGCTAAACCAGGTGGACAGGTGAAATGTCAGTATATCTCTGCTGTGGATAAAGTTATATTTGTGGATGATTATGCAGTAGGGT GTAGGAAGGACCTTAATGGAATCTTGTTGTTAGACACTGCTCTGCAAACTCCAGTTTCAAAGCAGGATGATGTGGTTCAGCTTGAATTAC CCGTTACAGAGGCACAGCAGCTCTTATCAGCATGTTTAGAAAAGGTAGATATTTCTAGTACAGAGGGTTATGATTTGTTCATCACACAGC TCAAAGATGGTTTAAAAAATACATCTCATGAGACTGCAGCAAACCACAAAGTTGCTAAGTGGGCCACAGTTACATTTCATCTTCCTCATC ATGTGTTGAAGTCCATTGCCAGTGCCATTGTAAATGAACTCAAGAAAATAAATCAAAATGTTGCTGCCTTACCTGTGGCGTCCTCAGTGA TGGACAGATTGTCTTACCTCTTACCTAGTGCACGTCCAGAACTCGGAGTGGGGCCAGGCCGTTCTGTAGACAGATCACTGATGTATAGTG AAGCTAACAGACGGGAGACATTTACCTCATGGCCTCATGTAGGCTATAGGTGGGCACAACCAGATCCCATGGCTCAAGCTGGATTTTATC ATCAGCCTGCCTCATCTGGAGATGATAGAGCCATGTGTTTTACTTGTAGTGTATGCCTCGTTTGTTGGGAACCTACTGATGAACCTTGGT CTGAACACGAAAGACATTCCCCAAACTGCCCATTTGTGAAAGGTGAGCACACACAGAATGTGCCATTGTCAGTCACTCTTGCAACAAGTC CTGCACAGTTTCCTTGTACGGATGGAACTGACAGAATATCTTGCTTTGGGTCGGGGAGCTGCCCTCATTTTCTAGCTGCTGCAACTAAAC GAGGAAAGATCTGCATATGGGATGTTTCCAAACTTATGAAGGTGCACTTAAAGTTTGAAATTAATGCCTATGATCCAGCAATTGTACAAC AGCTTATTCTATCAGGAGACCCAAGCTCAGGAGTTGATTCAAGGAGACCAACTTTGGCGTGGCTGGAGGACTCCTCTAGTTGCTCAGATA TACCAAAATTGGAAGGAGATAGTGATGATTTACTGGAGGATTCAGACAGTGAAGAGCATTCCAGATCAGATTCTGTGACAGGGCATACAT CACAGAAGGAAGCCATGGAAGTAAGCCTTGATATAACAGCACTCAGCATTCTCCAACAGCCAGAAAAACTTCAGTGGGAGATTGTTGCAA ATGTGCTTGAAGATACTGTTAAGGATCTTGAAGAACTTGGGGCAAATCCTTGTTTAACAAACTCTAAGAGTGAAAAGACAAAGGAAAAGC ACCAGGAGCAACACAACATTCCTTTTCCATGTTTATTAGCTGGAGGTTTATTAACATATAAATCTCCTGCTACCTCACCCATTAGTAGTA ATTCTCACAGGTCACTGGATGGTTTAAGCAGAACTCAGGGTGAAAGTATATCAGAACAAGGGTCAACTGACAATGAATCCTGCACTAATT CAGAACTAAATTCTCCTCTGGTAAGGAGGACTTTACCGGTTTTGCTTCTTTATAGCATCAAGGAATCTGATGAGAAAGCAGGAAAGATCT TTTCACAGATGAACAATATTATGAGTAAAAGTTTGCATGATGATGGTTTTACTGTTCCACAGATTATTGAAATGGAGCTGGATAGTCAGG AGCAGTTGTTATTGCAGGATCCTCCTGTGACTTACATTCAGCAATTTGCAGATGCAGCAGCCAACCTTACCTCTCCGGATTCTGAGAAGT GGAACTCTGTGTTTCCCAAGCCTGGGACTTTGGTTCAGTGCTTGAGGCTGCCAAAGTTTGCAGAGGAGGAGAATCTTTGTATAGACTCAA TAACTCCTTGTGCTGACGGAATTCATTTGTTGGTAGGACTGCGGACATGCCCTGTTGAATCCTTGAGTGCAATAAATCAAGTAGAGGCCT TGAATAATTTAAATAAATTAAACTCTGCACTATGTAATAGACGGAAAGGTGAGCTGGAATCAAATCTTGCTGTAGTGAATGGTGCAAATA TTAGTGTAATCCAACATGAATCACCAGCAGATGTACAGACTCCTTTAATAATTCAGCCTGAGCAGAGGAATGTTAGTGGTGGATATTTAG TGCTTTATAAAATGAATTATGCCACTCGGATAGTGACTTTAGAAGAGGAGCCAATAAAAATACAACATATCAAAGATCCCCAGGACACAA TTACCTCGCTCATTTTGCTTCCACCCGATATATTGGATAATCGAGAGGATGACTGTGAGGAACCTATTGAGGACATGCAGTTAACCTCAA AGAATGGTTTTGAGAGAGAAAAAACGTCTGACATTTCTACTCTTGGACACCTGGTAATAACCACTCAGGGAGGATATGTAAAAATACTAG ATCTTTCAAACTTTGAAATTTTGGCCAAAGTGGAGCCTCCCAAAAAGGAGGGCACTGAGGAACAGGACACATTTGTTTCTGTGATTTACT GTTCTGGCACAGACAGGCTGTGTGCATGCACCAAAGGTGTTTGCAAGAGCAGACCCTTGGACCTGGTGTTTATCATTGATAGTTCTCGTA GCGTACGGCCCCTGGAATTCACCAAAGTGAAAACTTTTGTCTCCCGGATAATCGACACTCTGGACATTGGGCCAGCCGACACGCGGGTGG CAGTGGTGAACTATGCTAGCACTGTGAAGATCGAGTTCCAACTCCAGGCCTACACAGATAAGCAGTCCCTGAAGCAGGCCGTGGGTCGAA TCACACCCTTGTCAACAGGCACCATGTCAGGCCTAGCCATCCAGACAGCAATGGACGAAGCCTTCACAGTGGAGGCAGGGGCTCGAGAGC CCTCTTCTAACATCCCTAAGGTGGCCATCATTGTTACAGATGGGAGGCCCCAGGACCAGGTGAATGAGGTGGCGGCTCGGGCCCAAGCAT CTGGTATTGAGCTCTATGCTGTGGGCGTGGACCGGGCAGACATGGCGTCCCTCAAGATGATGGCCAGTGAGCCCCTAGAGGAGCATGTTT TCTACGTGGAGACCTATGGGGTCATTGAGAAACTTTCCTCTAGATTCCAGGAAACCTTCTGTGCGCTGGACCCCTGTGTGCTTGGAACAC ACCAGTGCCAGCACGTCTGCATCAGTGATGGGGAAGGCAAGCACCACTGTGAGTGTAGCCAAGGATACACCTTGAATGCCGACAAGAAAA CGTGTTCAGCTCTTGATAGGTGTGCTCTTAACACCCACGGATGTGAGCACATCTGTGTGAATGACAGAAGTGGCTCTTATCATTGTGAGT GCTATGAAGGTTATACCTTGAATGAAGACAGGAAAACTTGTTCAGCTCAAGATAAATGTGCTTTGGGTACCCATGGGTGTCAGCACATTT GTGTGAATGACAGAACAGGGTCCCATCATTGTGAATGCTATGAGGGCTACACTCTGAATGCAGATAAAAAAACATGTTCAGTCCGTGACA AGTGTGCCCTAGGCTCTCATGGTTGCCAGCACATTTGTGTGAGTGATGGGGCCGCATCCTACCACTGTGATTGCTATCCTGGCTACACCT TAAATGAGGACAAGAAAACATGTTCAGCCACTGAGGAAGCACGAAGACTTGTTTCCACTGAAGATGCTTGTGGATGTGAAGCTACACTGG CATTCCAGGACAAGGTCAGCTCGTATCTTCAAAGACTGAACACTAAACTTGATGACATTTTGGAGAAGTTGAAAATAAATGAATATGGAC AAATACATCGTTAAATTGCTCCAATTTCTCACCTGAAAATGTGGACAGCTTGGTGTACTTAATACTCATGCATTCTTTTGCACACCTGTT ATTGCCAATGTTCCTGCTAATAATTTGCCATTATCTGTATTAATGCTTGAATATTACTGGATAAATTGTATGAAGATCTTCTGCAGAATC AGCATGATTCTTCCAAGGAAATACATATGCAGATACTTATTAAGAGCAAACTTTAGTGTCTCTAAGTTATGACTGTGAAATGATTGGTAG GAAATAGAATGAAAAGTTTAGTGTTTCTTTATCTACTAATTGAGCCATTTAATTTTTAAATGTTTATATTAGATAACCATATTCACAATG GAAACTTTAGGTCTAGTTTCTTTTGATAGTATTTATAATATAAATCAATCTTATTACTGAGAGTGCAAATTGTACAAGGTATTTACACAT ACAACTTCATATAACTGAGATGAATGTAATTTTGAACTGTTTAACACTTTTTGTTTTTTGCTTATTTTGTTGGAGTATTATTGAAGATGT GATCAATAGATTGTAATACACATATCTAAAAATAGTTAACACAGATCAAGTGAACATTACATTGCCATTTTTAATTCATTCTGGTCTTTG AAAGAAATGTACTACTAAAGAGCACTAGTTGTGAATTTAGGGTGTTAAACTTTTTACCAAGTACAAAAATCCCAAATTCACTTTATTATT TTGCTTCAGGATCCAAGTGACAAAGTTATATATTTATAAAATTGCTATAAATCGACAAAATCTAATGTTGTCTTTTTAATGTTAGTGATC CACCTGCCTCAGCCTCCCAAAGTGCTGGGATTACAGGCTTGAAAGTCTAACTTTTTTTTACTTATATATTTGATACATATAATTCTTTTG GCTTTGAAACTTGCAACTTTGAGAACAAAACAGTCCTTTAAATTTTGCACTGCTCAATTCTGTTTTTCGTTTGCATTGTCTTTAATATAA >9760_9760_1_BIRC6-MATN3_BIRC6_chr2_32641231_ENST00000421745_MATN3_chr2_20206071_ENST00000407540_length(amino acids)=1369AA_BP=957 MVTGGGAAPPGTVTEPLPSVIVLSAGRKMAAAAAAASGPGCSSAAGAGAAGVSEWLVLRDGCMHCDADGLHSLSYHPALNAILAVTSRGT IKVIDGTSGATLQASALSAKPGGQVKCQYISAVDKVIFVDDYAVGCRKDLNGILLLDTALQTPVSKQDDVVQLELPVTEAQQLLSACLEK VDISSTEGYDLFITQLKDGLKNTSHETAANHKVAKWATVTFHLPHHVLKSIASAIVNELKKINQNVAALPVASSVMDRLSYLLPSARPEL GVGPGRSVDRSLMYSEANRRETFTSWPHVGYRWAQPDPMAQAGFYHQPASSGDDRAMCFTCSVCLVCWEPTDEPWSEHERHSPNCPFVKG EHTQNVPLSVTLATSPAQFPCTDGTDRISCFGSGSCPHFLAAATKRGKICIWDVSKLMKVHLKFEINAYDPAIVQQLILSGDPSSGVDSR RPTLAWLEDSSSCSDIPKLEGDSDDLLEDSDSEEHSRSDSVTGHTSQKEAMEVSLDITALSILQQPEKLQWEIVANVLEDTVKDLEELGA NPCLTNSKSEKTKEKHQEQHNIPFPCLLAGGLLTYKSPATSPISSNSHRSLDGLSRTQGESISEQGSTDNESCTNSELNSPLVRRTLPVL LLYSIKESDEKAGKIFSQMNNIMSKSLHDDGFTVPQIIEMELDSQEQLLLQDPPVTYIQQFADAAANLTSPDSEKWNSVFPKPGTLVQCL RLPKFAEEENLCIDSITPCADGIHLLVGLRTCPVESLSAINQVEALNNLNKLNSALCNRRKGELESNLAVVNGANISVIQHESPADVQTP LIIQPEQRNVSGGYLVLYKMNYATRIVTLEEEPIKIQHIKDPQDTITSLILLPPDILDNREDDCEEPIEDMQLTSKNGFEREKTSDISTL GHLVITTQGGYVKILDLSNFEILAKVEPPKKEGTEEQDTFVSVIYCSGTDRLCACTKGVCKSRPLDLVFIIDSSRSVRPLEFTKVKTFVS RIIDTLDIGPADTRVAVVNYASTVKIEFQLQAYTDKQSLKQAVGRITPLSTGTMSGLAIQTAMDEAFTVEAGAREPSSNIPKVAIIVTDG RPQDQVNEVAARAQASGIELYAVGVDRADMASLKMMASEPLEEHVFYVETYGVIEKLSSRFQETFCALDPCVLGTHQCQHVCISDGEGKH HCECSQGYTLNADKKTCSALDRCALNTHGCEHICVNDRSGSYHCECYEGYTLNEDRKTCSAQDKCALGTHGCQHICVNDRTGSHHCECYE GYTLNADKKTCSVRDKCALGSHGCQHICVSDGAASYHCDCYPGYTLNEDKKTCSATEEARRLVSTEDACGCEATLAFQDKVSSYLQRLNT -------------------------------------------------------------- >9760_9760_2_BIRC6-MATN3_BIRC6_chr2_32641231_ENST00000421745_MATN3_chr2_20206071_ENST00000421259_length(transcript)=4192nt_BP=3006nt TCCCTGCTTCTCCCCCTCTCCCGTCAGCCTCCCTCCGAGTTTGGCCCCTCCGGCCGGGCGATCGACGTTCCGCGTGCGTGCGGGCGCCTG ACTTCACTTCCGGCTAACGCGCTCGGCTTGCCCCCTGGCCCCGGATGGTGACTGGTGGTGGTGCTGCACCTCCCGGGACTGTCACTGAGC CGCTTCCCAGTGTGATTGTGCTGAGCGCAGGCCGGAAGATGGCGGCTGCGGCTGCGGCGGCCTCGGGCCCCGGCTGCTCCTCGGCGGCGG GGGCGGGGGCGGCCGGGGTCTCAGAGTGGCTGGTGCTGCGGGACGGCTGCATGCACTGCGACGCCGACGGGCTGCACAGCCTGTCCTACC ACCCTGCGCTCAACGCCATCCTGGCCGTCACTAGCCGCGGGACCATCAAAGTCATCGACGGCACCTCGGGGGCCACACTGCAGGCCTCCG CGCTCAGTGCTAAACCAGGTGGACAGGTGAAATGTCAGTATATCTCTGCTGTGGATAAAGTTATATTTGTGGATGATTATGCAGTAGGGT GTAGGAAGGACCTTAATGGAATCTTGTTGTTAGACACTGCTCTGCAAACTCCAGTTTCAAAGCAGGATGATGTGGTTCAGCTTGAATTAC CCGTTACAGAGGCACAGCAGCTCTTATCAGCATGTTTAGAAAAGGTAGATATTTCTAGTACAGAGGGTTATGATTTGTTCATCACACAGC TCAAAGATGGTTTAAAAAATACATCTCATGAGACTGCAGCAAACCACAAAGTTGCTAAGTGGGCCACAGTTACATTTCATCTTCCTCATC ATGTGTTGAAGTCCATTGCCAGTGCCATTGTAAATGAACTCAAGAAAATAAATCAAAATGTTGCTGCCTTACCTGTGGCGTCCTCAGTGA TGGACAGATTGTCTTACCTCTTACCTAGTGCACGTCCAGAACTCGGAGTGGGGCCAGGCCGTTCTGTAGACAGATCACTGATGTATAGTG AAGCTAACAGACGGGAGACATTTACCTCATGGCCTCATGTAGGCTATAGGTGGGCACAACCAGATCCCATGGCTCAAGCTGGATTTTATC ATCAGCCTGCCTCATCTGGAGATGATAGAGCCATGTGTTTTACTTGTAGTGTATGCCTCGTTTGTTGGGAACCTACTGATGAACCTTGGT CTGAACACGAAAGACATTCCCCAAACTGCCCATTTGTGAAAGGTGAGCACACACAGAATGTGCCATTGTCAGTCACTCTTGCAACAAGTC CTGCACAGTTTCCTTGTACGGATGGAACTGACAGAATATCTTGCTTTGGGTCGGGGAGCTGCCCTCATTTTCTAGCTGCTGCAACTAAAC GAGGAAAGATCTGCATATGGGATGTTTCCAAACTTATGAAGGTGCACTTAAAGTTTGAAATTAATGCCTATGATCCAGCAATTGTACAAC AGCTTATTCTATCAGGAGACCCAAGCTCAGGAGTTGATTCAAGGAGACCAACTTTGGCGTGGCTGGAGGACTCCTCTAGTTGCTCAGATA TACCAAAATTGGAAGGAGATAGTGATGATTTACTGGAGGATTCAGACAGTGAAGAGCATTCCAGATCAGATTCTGTGACAGGGCATACAT CACAGAAGGAAGCCATGGAAGTAAGCCTTGATATAACAGCACTCAGCATTCTCCAACAGCCAGAAAAACTTCAGTGGGAGATTGTTGCAA ATGTGCTTGAAGATACTGTTAAGGATCTTGAAGAACTTGGGGCAAATCCTTGTTTAACAAACTCTAAGAGTGAAAAGACAAAGGAAAAGC ACCAGGAGCAACACAACATTCCTTTTCCATGTTTATTAGCTGGAGGTTTATTAACATATAAATCTCCTGCTACCTCACCCATTAGTAGTA ATTCTCACAGGTCACTGGATGGTTTAAGCAGAACTCAGGGTGAAAGTATATCAGAACAAGGGTCAACTGACAATGAATCCTGCACTAATT CAGAACTAAATTCTCCTCTGGTAAGGAGGACTTTACCGGTTTTGCTTCTTTATAGCATCAAGGAATCTGATGAGAAAGCAGGAAAGATCT TTTCACAGATGAACAATATTATGAGTAAAAGTTTGCATGATGATGGTTTTACTGTTCCACAGATTATTGAAATGGAGCTGGATAGTCAGG AGCAGTTGTTATTGCAGGATCCTCCTGTGACTTACATTCAGCAATTTGCAGATGCAGCAGCCAACCTTACCTCTCCGGATTCTGAGAAGT GGAACTCTGTGTTTCCCAAGCCTGGGACTTTGGTTCAGTGCTTGAGGCTGCCAAAGTTTGCAGAGGAGGAGAATCTTTGTATAGACTCAA TAACTCCTTGTGCTGACGGAATTCATTTGTTGGTAGGACTGCGGACATGCCCTGTTGAATCCTTGAGTGCAATAAATCAAGTAGAGGCCT TGAATAATTTAAATAAATTAAACTCTGCACTATGTAATAGACGGAAAGGTGAGCTGGAATCAAATCTTGCTGTAGTGAATGGTGCAAATA TTAGTGTAATCCAACATGAATCACCAGCAGATGTACAGACTCCTTTAATAATTCAGCCTGAGCAGAGGAATGTTAGTGGTGGATATTTAG TGCTTTATAAAATGAATTATGCCACTCGGATAGTGACTTTAGAAGAGGAGCCAATAAAAATACAACATATCAAAGATCCCCAGGACACAA TTACCTCGCTCATTTTGCTTCCACCCGATATATTGGATAATCGAGAGGATGACTGTGAGGAACCTATTGAGGACATGCAGTTAACCTCAA AGAATGGTTTTGAGAGAGAAAAAACGTCTGACATTTCTACTCTTGGACACCTGGTAATAACCACTCAGGGAGGATATGTAAAAATACTAG ATCTTTCAAACTTTGAAATTTTGGCCAAAGTGGAGCCTCCCAAAAAGGAGGGCACTGAGGAACAGGACACATTTGTTTCTGTGATTTACT GTTCTGGCACAGACAGGCTGTGTGCATGCACCAAAGGTGTTTGCAAGAGCAGACCCTTGGACCTGGTGTTTATCATTGATAGTTCTCGTA GCGTACGGCCCCTGGAATTCACCAAAGTGAAAACTTTTGTCTCCCGGATAATCGACACTCTGGACATTGGGCCAGCCGACACGCGGGTGG CAGTGGTGAACTATGCTAGCACTGTGAAGATCGAGTTCCAACTCCAGGCCTACACAGATAAGCAGTCCCTGAAGCAGGCCGTGGGTCGAA TCACACCCTTGTCAACAGGCACCATGTCAGGCCTAGCCATCCAGACAGCAATGGACGAAGCCTTCACAGTGGAGGCAGGGGCTCGAGAGC CCTCTTCTAACATCCCTAAGGTGGCCATCATTGTTACAGATGGGAGGCCCCAGGACCAGGTGAATGAGGTGGCGGCTCGGGCCCAAGCAT CTGGTATTGAGCTCTATGCTGTGGGCGTGGACCGGGCAGACATGGCGTCCCTCAAGATGATGGCCAGTGAGCCCCTAGAGGAGCATGTTT TCTACGTGGAGACCTATGGGGTCATTGAGAAACTTTCCTCTAGATTCCAGGAAACCTTCTGTGCTCTTGATAGGTGTGCTCTTAACACCC ACGGATGTGAGCACATCTGTGTGAATGACAGAAGTGGCTCTTATCATTGTGAGTGCTATGAAGGTTATACCTTGAATGAAGACAGGAAAA CTTGTTCAGCTCAAGATAAATGTGCTTTGGGTACCCATGGGTGTCAGCACATTTGTGTGAATGACAGAACAGGGTCCCATCATTGTGAAT GCTATGAGGGCTACACTCTGAATGCAGATAAAAAAACATGTTCAGTCCGTGACAAGTGTGCCCTAGGCTCTCATGGTTGCCAGCACATTT GTGTGAGTGATGGGGCCGCATCCTACCACTGTGATTGCTATCCTGGCTACACCTTAAATGAGGACAAGAAAACATGTTCAGCCACTGAGG AAGCACGAAGACTTGTTTCCACTGAAGATGCTTGTGGATGTGAAGCTACACTGGCATTCCAGGACAAGGTCAGCTCGTATCTTCAAAGAC TGAACACTAAACTTGATGACATTTTGGAGAAGTTGAAAATAAATGAATATGGACAAATACATCGTTAAATTGCTCCAATTTCTCACCTGA >9760_9760_2_BIRC6-MATN3_BIRC6_chr2_32641231_ENST00000421745_MATN3_chr2_20206071_ENST00000421259_length(amino acids)=1327AA_BP=957 MVTGGGAAPPGTVTEPLPSVIVLSAGRKMAAAAAAASGPGCSSAAGAGAAGVSEWLVLRDGCMHCDADGLHSLSYHPALNAILAVTSRGT IKVIDGTSGATLQASALSAKPGGQVKCQYISAVDKVIFVDDYAVGCRKDLNGILLLDTALQTPVSKQDDVVQLELPVTEAQQLLSACLEK VDISSTEGYDLFITQLKDGLKNTSHETAANHKVAKWATVTFHLPHHVLKSIASAIVNELKKINQNVAALPVASSVMDRLSYLLPSARPEL GVGPGRSVDRSLMYSEANRRETFTSWPHVGYRWAQPDPMAQAGFYHQPASSGDDRAMCFTCSVCLVCWEPTDEPWSEHERHSPNCPFVKG EHTQNVPLSVTLATSPAQFPCTDGTDRISCFGSGSCPHFLAAATKRGKICIWDVSKLMKVHLKFEINAYDPAIVQQLILSGDPSSGVDSR RPTLAWLEDSSSCSDIPKLEGDSDDLLEDSDSEEHSRSDSVTGHTSQKEAMEVSLDITALSILQQPEKLQWEIVANVLEDTVKDLEELGA NPCLTNSKSEKTKEKHQEQHNIPFPCLLAGGLLTYKSPATSPISSNSHRSLDGLSRTQGESISEQGSTDNESCTNSELNSPLVRRTLPVL LLYSIKESDEKAGKIFSQMNNIMSKSLHDDGFTVPQIIEMELDSQEQLLLQDPPVTYIQQFADAAANLTSPDSEKWNSVFPKPGTLVQCL RLPKFAEEENLCIDSITPCADGIHLLVGLRTCPVESLSAINQVEALNNLNKLNSALCNRRKGELESNLAVVNGANISVIQHESPADVQTP LIIQPEQRNVSGGYLVLYKMNYATRIVTLEEEPIKIQHIKDPQDTITSLILLPPDILDNREDDCEEPIEDMQLTSKNGFEREKTSDISTL GHLVITTQGGYVKILDLSNFEILAKVEPPKKEGTEEQDTFVSVIYCSGTDRLCACTKGVCKSRPLDLVFIIDSSRSVRPLEFTKVKTFVS RIIDTLDIGPADTRVAVVNYASTVKIEFQLQAYTDKQSLKQAVGRITPLSTGTMSGLAIQTAMDEAFTVEAGAREPSSNIPKVAIIVTDG RPQDQVNEVAARAQASGIELYAVGVDRADMASLKMMASEPLEEHVFYVETYGVIEKLSSRFQETFCALDRCALNTHGCEHICVNDRSGSY HCECYEGYTLNEDRKTCSAQDKCALGTHGCQHICVNDRTGSHHCECYEGYTLNADKKTCSVRDKCALGSHGCQHICVSDGAASYHCDCYP -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for BIRC6-MATN3 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for BIRC6-MATN3 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for BIRC6-MATN3 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
