
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:USP9X-DDX3X (FusionGDB2 ID:97653) |
Fusion Gene Summary for USP9X-DDX3X |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: USP9X-DDX3X | Fusion gene ID: 97653 | Hgene | Tgene | Gene symbol | USP9X | DDX3X | Gene ID | 8239 | 1654 |
| Gene name | ubiquitin specific peptidase 9 X-linked | DEAD-box helicase 3 X-linked | |
| Synonyms | DFFRX|FAF|FAM|MRX99|MRXS99F | CAP-Rf|DBX|DDX14|DDX3|HLP2|MRX102 | |
| Cytomap | Xp11.4 | Xp11.4 | |
| Type of gene | protein-coding | protein-coding | |
| Description | probable ubiquitin carboxyl-terminal hydrolase FAF-XDrosophila fat facets related, X-linkeddeubiquitinating enzyme FAF-Xfat facets in mammalsfat facets protein related, X-linkedhFAMubiquitin specific protease 9, X chromosome (fat facets-like Drosoph | ATP-dependent RNA helicase DDX3XDEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linkedDEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linkedDEAD box protein 3, X-chromosomalDEAD box, X isoformDEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 3DEAD/H box-3helicase- | |
| Modification date | 20200313 | 20200329 | |
| UniProtAcc | . | O00571 | |
| Ensembl transtripts involved in fusion gene | ENST00000324545, ENST00000378308, | ENST00000478993, ENST00000399959, ENST00000441189, ENST00000457138, ENST00000542215, | |
| Fusion gene scores | * DoF score | 19 X 12 X 12=2736 | 7 X 9 X 4=252 |
| # samples | 23 | 8 | |
| ** MAII score | log2(23/2736*10)=-3.57236246394152 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(8/252*10)=-1.65535182861255 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: USP9X [Title/Abstract] AND DDX3X [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | USP9X(40945362)-DDX3X(41223527), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | USP9X | GO:0016579 | protein deubiquitination | 19135894 |
| Hgene | USP9X | GO:0030509 | BMP signaling pathway | 19135894 |
| Hgene | USP9X | GO:1901537 | positive regulation of DNA demethylation | 25944111 |
| Tgene | DDX3X | GO:0009615 | response to virus | 18636090 |
| Tgene | DDX3X | GO:0010501 | RNA secondary structure unwinding | 22872150 |
| Tgene | DDX3X | GO:0010628 | positive regulation of gene expression | 10074132 |
| Tgene | DDX3X | GO:0030308 | negative regulation of cell growth | 16818630 |
| Tgene | DDX3X | GO:0031333 | negative regulation of protein complex assembly | 17667941 |
| Tgene | DDX3X | GO:0031954 | positive regulation of protein autophosphorylation | 30341167 |
| Tgene | DDX3X | GO:0032727 | positive regulation of interferon-alpha production | 30341167 |
| Tgene | DDX3X | GO:0032728 | positive regulation of interferon-beta production | 27980081 |
| Tgene | DDX3X | GO:0034063 | stress granule assembly | 21883093 |
| Tgene | DDX3X | GO:0034157 | positive regulation of toll-like receptor 7 signaling pathway | 30341167 |
| Tgene | DDX3X | GO:0034161 | positive regulation of toll-like receptor 8 signaling pathway | 30341167 |
| Tgene | DDX3X | GO:0035556 | intracellular signal transduction | 18636090 |
| Tgene | DDX3X | GO:0045727 | positive regulation of translation | 18596238|22872150 |
| Tgene | DDX3X | GO:0045944 | positive regulation of transcription by RNA polymerase II | 16818630|18636090|28128295 |
| Tgene | DDX3X | GO:0071243 | cellular response to arsenic-containing substance | 21883093 |
| Tgene | DDX3X | GO:0071470 | cellular response to osmotic stress | 21883093 |
| Tgene | DDX3X | GO:0071902 | positive regulation of protein serine/threonine kinase activity | 23413191 |
| Tgene | DDX3X | GO:1902523 | positive regulation of protein K63-linked ubiquitination | 27980081 |
 Fusion gene breakpoints across USP9X (5'-gene) Fusion gene breakpoints across USP9X (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 Fusion gene breakpoints across DDX3X (3'-gene) Fusion gene breakpoints across DDX3X (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-23-2077-01A | USP9X | chrX | 40945362 | + | DDX3X | chrX | 41217086 | + |
| ChimerDB4 | OV | TCGA-23-2077-01A | USP9X | chrX | 40945362 | + | DDX3X | chrX | 41223527 | + |
| ChimerKB4 | . | . | USP9X | chrX | 41060312 | + | DDX3X | chrX | 41060312 | + |
Top |
Fusion Gene ORF analysis for USP9X-DDX3X |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5UTR-3UTR | ENST00000324545 | ENST00000478993 | USP9X | chrX | 40945362 | + | DDX3X | chrX | 41217086 | + |
| 5UTR-3UTR | ENST00000324545 | ENST00000478993 | USP9X | chrX | 40945362 | + | DDX3X | chrX | 41223527 | + |
| 5UTR-3UTR | ENST00000378308 | ENST00000478993 | USP9X | chrX | 40945362 | + | DDX3X | chrX | 41217086 | + |
| 5UTR-3UTR | ENST00000378308 | ENST00000478993 | USP9X | chrX | 40945362 | + | DDX3X | chrX | 41223527 | + |
| 5UTR-intron | ENST00000324545 | ENST00000399959 | USP9X | chrX | 40945362 | + | DDX3X | chrX | 41217086 | + |
| 5UTR-intron | ENST00000324545 | ENST00000399959 | USP9X | chrX | 40945362 | + | DDX3X | chrX | 41223527 | + |
| 5UTR-intron | ENST00000324545 | ENST00000441189 | USP9X | chrX | 40945362 | + | DDX3X | chrX | 41217086 | + |
| 5UTR-intron | ENST00000324545 | ENST00000441189 | USP9X | chrX | 40945362 | + | DDX3X | chrX | 41223527 | + |
| 5UTR-intron | ENST00000324545 | ENST00000457138 | USP9X | chrX | 40945362 | + | DDX3X | chrX | 41217086 | + |
| 5UTR-intron | ENST00000324545 | ENST00000457138 | USP9X | chrX | 40945362 | + | DDX3X | chrX | 41223527 | + |
| 5UTR-intron | ENST00000324545 | ENST00000542215 | USP9X | chrX | 40945362 | + | DDX3X | chrX | 41217086 | + |
| 5UTR-intron | ENST00000324545 | ENST00000542215 | USP9X | chrX | 40945362 | + | DDX3X | chrX | 41223527 | + |
| 5UTR-intron | ENST00000378308 | ENST00000399959 | USP9X | chrX | 40945362 | + | DDX3X | chrX | 41217086 | + |
| 5UTR-intron | ENST00000378308 | ENST00000399959 | USP9X | chrX | 40945362 | + | DDX3X | chrX | 41223527 | + |
| 5UTR-intron | ENST00000378308 | ENST00000441189 | USP9X | chrX | 40945362 | + | DDX3X | chrX | 41217086 | + |
| 5UTR-intron | ENST00000378308 | ENST00000441189 | USP9X | chrX | 40945362 | + | DDX3X | chrX | 41223527 | + |
| 5UTR-intron | ENST00000378308 | ENST00000457138 | USP9X | chrX | 40945362 | + | DDX3X | chrX | 41217086 | + |
| 5UTR-intron | ENST00000378308 | ENST00000457138 | USP9X | chrX | 40945362 | + | DDX3X | chrX | 41223527 | + |
| 5UTR-intron | ENST00000378308 | ENST00000542215 | USP9X | chrX | 40945362 | + | DDX3X | chrX | 41217086 | + |
| 5UTR-intron | ENST00000378308 | ENST00000542215 | USP9X | chrX | 40945362 | + | DDX3X | chrX | 41223527 | + |
| intron-intron | ENST00000324545 | ENST00000399959 | USP9X | chrX | 41060312 | + | DDX3X | chrX | 41060312 | + |
| intron-intron | ENST00000324545 | ENST00000441189 | USP9X | chrX | 41060312 | + | DDX3X | chrX | 41060312 | + |
| intron-intron | ENST00000324545 | ENST00000457138 | USP9X | chrX | 41060312 | + | DDX3X | chrX | 41060312 | + |
| intron-intron | ENST00000324545 | ENST00000478993 | USP9X | chrX | 41060312 | + | DDX3X | chrX | 41060312 | + |
| intron-intron | ENST00000324545 | ENST00000542215 | USP9X | chrX | 41060312 | + | DDX3X | chrX | 41060312 | + |
| intron-intron | ENST00000378308 | ENST00000399959 | USP9X | chrX | 41060312 | + | DDX3X | chrX | 41060312 | + |
| intron-intron | ENST00000378308 | ENST00000441189 | USP9X | chrX | 41060312 | + | DDX3X | chrX | 41060312 | + |
| intron-intron | ENST00000378308 | ENST00000457138 | USP9X | chrX | 41060312 | + | DDX3X | chrX | 41060312 | + |
| intron-intron | ENST00000378308 | ENST00000478993 | USP9X | chrX | 41060312 | + | DDX3X | chrX | 41060312 | + |
| intron-intron | ENST00000378308 | ENST00000542215 | USP9X | chrX | 41060312 | + | DDX3X | chrX | 41060312 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
Top |
Fusion Genomic Features for USP9X-DDX3X |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| USP9X | chrX | 40945362 | + | DDX3X | chrX | 41223526 | + | 7.89E-06 | 0.99999213 |
| USP9X | chrX | 40945362 | + | DDX3X | chrX | 41217085 | + | 2.77E-07 | 0.99999976 |
| USP9X | chrX | 40945362 | + | DDX3X | chrX | 41223526 | + | 7.89E-06 | 0.99999213 |
| USP9X | chrX | 40945362 | + | DDX3X | chrX | 41217085 | + | 2.77E-07 | 0.99999976 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
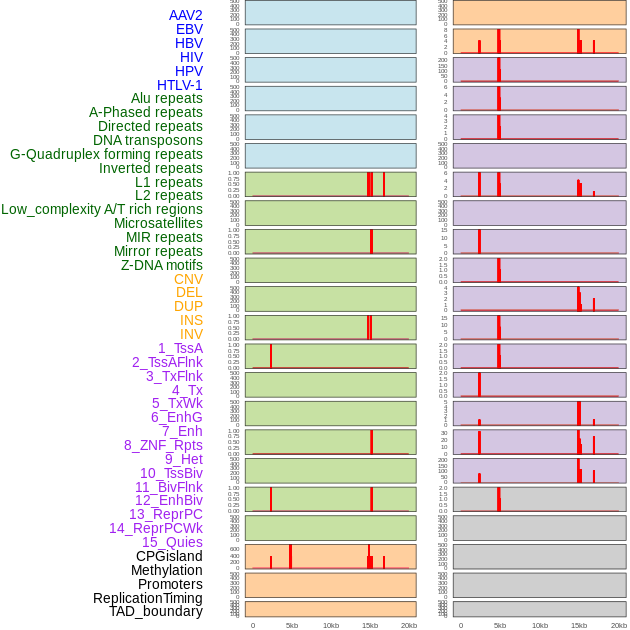 |
Top |
Fusion Protein Features for USP9X-DDX3X |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/:40945362/:41223527) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | DDX3X |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Multifunctional ATP-dependent RNA helicase (PubMed:17357160, PubMed:21589879, PubMed:31575075). The ATPase activity can be stimulated by various ribo-and deoxynucleic acids indicative for a relaxed substrate specificity (PubMed:29222110). In vitro can unwind partially double-stranded DNA with a preference for 5'-single-stranded DNA overhangs (PubMed:17357160, PubMed:21589879). Binds RNA G-quadruplex (rG4s) structures, including those located in the 5'-UTR of NRAS mRNA (PubMed:30256975). Involved in many cellular processes, which do not necessarily require its ATPase/helicase catalytic activities (Probable). Involved in transcription regulation (PubMed:16818630, PubMed:18264132). Positively regulates CDKN1A/WAF1/CIP1 transcription in an SP1-dependent manner, hence inhibits cell growth. This function requires its ATPase, but not helicase activity (PubMed:16818630, PubMed:18264132). CDKN1A up-regulation may be cell-type specific (PubMed:18264132). Binds CDH1/E-cadherin promoter and represses its transcription (PubMed:18264132). Potentiates HNF4A-mediated MTTP transcriptional activation; this function requires ATPase, but not helicase activity. Facilitates HNF4A acetylation, possibly catalyzed by CREBBP/EP300, thereby increasing the DNA-binding affinity of HNF4 to its response element. In addition, disrupts the interaction between HNF4 and SHP that forms inactive heterodimers and enhances the formation of active HNF4 homodimers. By promoting HNF4A-induced MTTP expression, may play a role in lipid homeostasis (PubMed:28128295). May positively regulate TP53 transcription (PubMed:28842590). Associates with mRNPs, predominantly with spliced mRNAs carrying an exon junction complex (EJC) (PubMed:17095540, PubMed:18596238). Involved in the regulation of translation initiation (PubMed:18628297, PubMed:17667941, PubMed:22872150). Not involved in the general process of translation, but promotes efficient translation of selected complex mRNAs, containing highly structured 5'-untranslated regions (UTR) (PubMed:20837705, PubMed:22872150). This function depends on helicase activity (PubMed:20837705, PubMed:22872150). Might facilitate translation by resolving secondary structures of 5'-UTRs during ribosome scanning (PubMed:20837705). Alternatively, may act prior to 43S ribosomal scanning and promote 43S pre-initiation complex entry to mRNAs exhibiting specific RNA motifs, by performing local remodeling of transcript structures located close to the cap moiety (PubMed:22872150). Independently of its ATPase activity, promotes the assembly of functional 80S ribosomes and disassembles from ribosomes prior to the translation elongation process (PubMed:22323517). Positively regulates the translation of cyclin E1/CCNE1 mRNA and consequently promotes G1/S-phase transition during the cell cycle (PubMed:20837705). May activate TP53 translation (PubMed:28842590). Required for endoplasmic reticulum stress-induced ATF4 mRNA translation (PubMed:29062139). Independently of its ATPase/helicase activity, enhances IRES-mediated translation; this activity requires interaction with EIF4E (PubMed:17667941, PubMed:22323517). Independently of its ATPase/helicase activity, has also been shown specifically repress cap-dependent translation, possibly by acting on translation initiation factor EIF4E (PubMed:17667941). Involved in innate immunity, acting as a viral RNA sensor. Binds viral RNAs and promotes the production of type I interferon (IFN-alpha and IFN-beta) (PubMed:31575075, PubMed:20127681, PubMed:21170385). Potentiate MAVS/DDX58-mediated induction of IFNB in early stages of infection (PubMed:20127681, PubMed:21170385). Enhances IFNB1 expression via IRF3/IRF7 pathway and participates in NFKB activation in the presence of MAVS and TBK1 (PubMed:18583960, PubMed:18636090, PubMed:21170385, PubMed:27980081, PubMed:19913487). Involved in TBK1 and IKBKE-dependent IRF3 activation leading to IFNB induction, acts as a scaffolding adapter that links IKBKE and IRF3 and coordinates their activation (PubMed:23478265). Involved in the TLR7/TLR8 signaling pathway leading to type I interferon induction, including IFNA4 production. In this context, acts as an upstream regulator of IRF7 activation by MAP3K14/NIK and CHUK/IKKA. Stimulates CHUK autophosphorylation and activation following physiological activation of the TLR7 and TLR8 pathways, leading to MAP3K14/CHUK-mediated activatory phosphorylation of IRF7 (PubMed:30341167). Also stimulates MAP3K14/CHUK-dependent NF-kappa-B signaling (PubMed:30341167). Negatively regulates TNF-induced IL6 and IL8 expression, via the NF-kappa-B pathway. May act by interacting with RELA/p65 and trapping it in the cytoplasm (PubMed:27736973). May also bind IFNB promoter; the function is independent of IRF3 (PubMed:18583960). Involved in both stress and inflammatory responses (By similarity). Independently of its ATPase/helicase activity, required for efficient stress granule assembly through its interaction with EIF4E, hence promotes survival in stressed cells (PubMed:21883093). Independently of its helicase activity, regulates NLRP3 inflammasome assembly through interaction with NLRP3 and hence promotes cell death by pyroptosis during inflammation. This function is independent of helicase activity (By similarity). Therefore DDX3X availability may be used to interpret stress signals and choose between pro-survival stress granules and pyroptotic NLRP3 inflammasomes and serve as a live-or-die checkpoint in stressed cells (By similarity). In association with GSK3A/B, negatively regulates extrinsic apoptotic signaling pathway via death domain receptors, including TNFRSF10B, slowing down the rate of CASP3 activation following death receptor stimulation (PubMed:18846110). Cleavage by caspases may inactivate DDX3X and relieve the inhibition (PubMed:18846110). Independently of its ATPase/helicase activity, allosteric activator of CSNK1E. Stimulates CSNK1E-mediated phosphorylation of DVL2, thereby involved in the positive regulation of Wnt/beta-catenin signaling pathway. Also activates CSNK1A1 and CSNK1D in vitro, but it is uncertain if these targets are physiologically relevant (PubMed:23413191, PubMed:29222110). ATPase and casein kinase-activating functions are mutually exclusive (PubMed:29222110). May be involved in mitotic chromosome segregation (PubMed:21730191). {ECO:0000250|UniProtKB:Q62167, ECO:0000269|PubMed:16818630, ECO:0000269|PubMed:17095540, ECO:0000269|PubMed:17357160, ECO:0000269|PubMed:17667941, ECO:0000269|PubMed:18264132, ECO:0000269|PubMed:18583960, ECO:0000269|PubMed:18596238, ECO:0000269|PubMed:18628297, ECO:0000269|PubMed:18636090, ECO:0000269|PubMed:18846110, ECO:0000269|PubMed:19913487, ECO:0000269|PubMed:20127681, ECO:0000269|PubMed:20837705, ECO:0000269|PubMed:21170385, ECO:0000269|PubMed:21589879, ECO:0000269|PubMed:21730191, ECO:0000269|PubMed:21883093, ECO:0000269|PubMed:22323517, ECO:0000269|PubMed:22872150, ECO:0000269|PubMed:23413191, ECO:0000269|PubMed:23478265, ECO:0000269|PubMed:27736973, ECO:0000269|PubMed:27980081, ECO:0000269|PubMed:28128295, ECO:0000269|PubMed:28842590, ECO:0000269|PubMed:29062139, ECO:0000269|PubMed:29222110, ECO:0000269|PubMed:30256975, ECO:0000269|PubMed:30341167, ECO:0000269|PubMed:31575075, ECO:0000305}.; FUNCTION: (Microbial infection) Facilitates hepatitis C virus (HCV) replication (PubMed:29899501). During infection, HCV core protein inhibits the interaction between MAVS and DDX3X and therefore impairs MAVS-dependent INFB induction and might recruit DDX3X to HCV replication complex (PubMed:21170385). {ECO:0000269|PubMed:21170385, ECO:0000269|PubMed:29899501}.; FUNCTION: (Microbial infection) Facilitates HIV-1 replication (PubMed:15507209, PubMed:18583960, PubMed:21589879, PubMed:22872150, PubMed:29899501). Acts as a cofactor for XPO1-mediated nuclear export of HIV-1 Rev RNAs (PubMed:15507209, PubMed:18583960, PubMed:29899501). This function is strongly stimulated in the presence of TBK1 and requires DDX3X ATPase activity (PubMed:18583960). {ECO:0000269|PubMed:15507209, ECO:0000269|PubMed:18583960, ECO:0000269|PubMed:21589879, ECO:0000269|PubMed:22872150, ECO:0000269|PubMed:29899501}.; FUNCTION: (Microbial infection) Facilitates Zika virus (ZIKV) replication. {ECO:0000269|PubMed:29899501}.; FUNCTION: (Microbial infection) Facilitates Dengue virus (DENV) replication. {ECO:0000269|PubMed:29899501}.; FUNCTION: (Microbial infection) Facilitates Venezuelan equine encephalitis virus (VEEV) replication. {ECO:0000269|PubMed:27105836}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
Top |
Fusion Gene Sequence for USP9X-DDX3X |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
Top |
Fusion Gene PPI Analysis for USP9X-DDX3X |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for USP9X-DDX3X |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for USP9X-DDX3X |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
