
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:UVRAG-ZMAT4 (FusionGDB2 ID:97822) |
Fusion Gene Summary for UVRAG-ZMAT4 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: UVRAG-ZMAT4 | Fusion gene ID: 97822 | Hgene | Tgene | Gene symbol | UVRAG | ZMAT4 | Gene ID | 7405 | 79698 |
| Gene name | UV radiation resistance associated | zinc finger matrin-type 4 | |
| Synonyms | DHTX|VPS38|p63 | - | |
| Cytomap | 11q13.5 | 8p11.21 | |
| Type of gene | protein-coding | protein-coding | |
| Description | UV radiation resistance-associated gene proteinbeclin 1 binding proteindisrupted in heterotaxy | zinc finger matrin-type protein 4 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000356136, ENST00000528420, ENST00000531818, ENST00000532130, ENST00000533454, ENST00000539288, ENST00000525872, ENST00000538870, | ENST00000523823, ENST00000297737, ENST00000315769, | |
| Fusion gene scores | * DoF score | 41 X 18 X 15=11070 | 15 X 12 X 7=1260 |
| # samples | 49 | 17 | |
| ** MAII score | log2(49/11070*10)=-4.49772966266634 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(17/1260*10)=-2.88981708224958 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: UVRAG [Title/Abstract] AND ZMAT4 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | UVRAG(75728024)-ZMAT4(40389757), # samples:4 | ||
| Anticipated loss of major functional domain due to fusion event. | UVRAG-ZMAT4 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. UVRAG-ZMAT4 seems lost the major protein functional domain in Hgene partner, which is a tumor suppressor due to the frame-shifted ORF. UVRAG-ZMAT4 seems lost the major protein functional domain in Tgene partner, which is a transcription factor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | UVRAG | GO:0071900 | regulation of protein serine/threonine kinase activity | 22542840 |
| Hgene | UVRAG | GO:0097352 | autophagosome maturation | 28306502 |
| Hgene | UVRAG | GO:0097680 | double-strand break repair via classical nonhomologous end joining | 22542840 |
 Fusion gene breakpoints across UVRAG (5'-gene) Fusion gene breakpoints across UVRAG (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
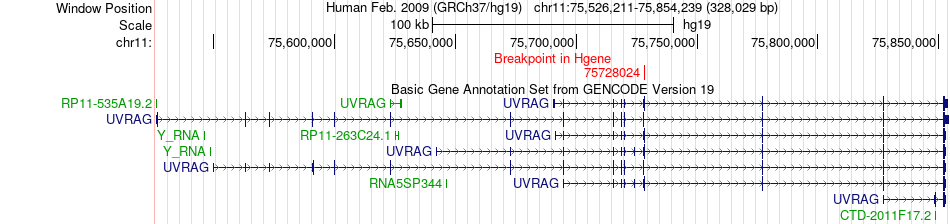 |
 Fusion gene breakpoints across ZMAT4 (3'-gene) Fusion gene breakpoints across ZMAT4 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
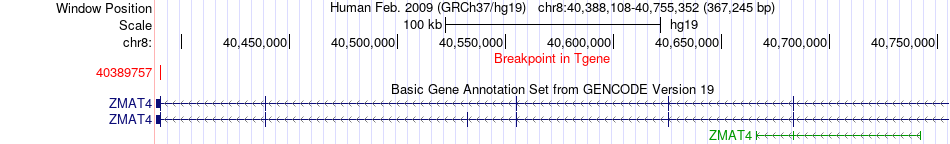 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-A2-A0ES-01A | UVRAG | chr11 | 75728024 | - | ZMAT4 | chr8 | 40389757 | - |
| ChimerDB4 | BRCA | TCGA-A2-A0ES-01A | UVRAG | chr11 | 75728024 | + | ZMAT4 | chr8 | 40389757 | - |
| ChimerDB4 | BRCA | TCGA-A2-A0ES | UVRAG | chr11 | 75728024 | + | ZMAT4 | chr8 | 40389757 | - |
Top |
Fusion Gene ORF analysis for UVRAG-ZMAT4 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000356136 | ENST00000523823 | UVRAG | chr11 | 75728024 | + | ZMAT4 | chr8 | 40389757 | - |
| 5CDS-intron | ENST00000528420 | ENST00000523823 | UVRAG | chr11 | 75728024 | + | ZMAT4 | chr8 | 40389757 | - |
| 5CDS-intron | ENST00000531818 | ENST00000523823 | UVRAG | chr11 | 75728024 | + | ZMAT4 | chr8 | 40389757 | - |
| 5CDS-intron | ENST00000532130 | ENST00000523823 | UVRAG | chr11 | 75728024 | + | ZMAT4 | chr8 | 40389757 | - |
| 5CDS-intron | ENST00000533454 | ENST00000523823 | UVRAG | chr11 | 75728024 | + | ZMAT4 | chr8 | 40389757 | - |
| 5CDS-intron | ENST00000539288 | ENST00000523823 | UVRAG | chr11 | 75728024 | + | ZMAT4 | chr8 | 40389757 | - |
| Frame-shift | ENST00000356136 | ENST00000297737 | UVRAG | chr11 | 75728024 | + | ZMAT4 | chr8 | 40389757 | - |
| Frame-shift | ENST00000528420 | ENST00000297737 | UVRAG | chr11 | 75728024 | + | ZMAT4 | chr8 | 40389757 | - |
| Frame-shift | ENST00000531818 | ENST00000297737 | UVRAG | chr11 | 75728024 | + | ZMAT4 | chr8 | 40389757 | - |
| Frame-shift | ENST00000532130 | ENST00000297737 | UVRAG | chr11 | 75728024 | + | ZMAT4 | chr8 | 40389757 | - |
| Frame-shift | ENST00000533454 | ENST00000297737 | UVRAG | chr11 | 75728024 | + | ZMAT4 | chr8 | 40389757 | - |
| Frame-shift | ENST00000539288 | ENST00000297737 | UVRAG | chr11 | 75728024 | + | ZMAT4 | chr8 | 40389757 | - |
| In-frame | ENST00000356136 | ENST00000315769 | UVRAG | chr11 | 75728024 | + | ZMAT4 | chr8 | 40389757 | - |
| In-frame | ENST00000528420 | ENST00000315769 | UVRAG | chr11 | 75728024 | + | ZMAT4 | chr8 | 40389757 | - |
| In-frame | ENST00000531818 | ENST00000315769 | UVRAG | chr11 | 75728024 | + | ZMAT4 | chr8 | 40389757 | - |
| In-frame | ENST00000532130 | ENST00000315769 | UVRAG | chr11 | 75728024 | + | ZMAT4 | chr8 | 40389757 | - |
| In-frame | ENST00000533454 | ENST00000315769 | UVRAG | chr11 | 75728024 | + | ZMAT4 | chr8 | 40389757 | - |
| In-frame | ENST00000539288 | ENST00000315769 | UVRAG | chr11 | 75728024 | + | ZMAT4 | chr8 | 40389757 | - |
| intron-3CDS | ENST00000525872 | ENST00000297737 | UVRAG | chr11 | 75728024 | + | ZMAT4 | chr8 | 40389757 | - |
| intron-3CDS | ENST00000525872 | ENST00000315769 | UVRAG | chr11 | 75728024 | + | ZMAT4 | chr8 | 40389757 | - |
| intron-3CDS | ENST00000538870 | ENST00000297737 | UVRAG | chr11 | 75728024 | + | ZMAT4 | chr8 | 40389757 | - |
| intron-3CDS | ENST00000538870 | ENST00000315769 | UVRAG | chr11 | 75728024 | + | ZMAT4 | chr8 | 40389757 | - |
| intron-intron | ENST00000525872 | ENST00000523823 | UVRAG | chr11 | 75728024 | + | ZMAT4 | chr8 | 40389757 | - |
| intron-intron | ENST00000538870 | ENST00000523823 | UVRAG | chr11 | 75728024 | + | ZMAT4 | chr8 | 40389757 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000356136 | UVRAG | chr11 | 75728024 | + | ENST00000315769 | ZMAT4 | chr8 | 40389757 | - | 3116 | 1467 | 163 | 1482 | 439 |
| ENST00000528420 | UVRAG | chr11 | 75728024 | + | ENST00000315769 | ZMAT4 | chr8 | 40389757 | - | 2857 | 1208 | 96 | 1223 | 375 |
| ENST00000533454 | UVRAG | chr11 | 75728024 | + | ENST00000315769 | ZMAT4 | chr8 | 40389757 | - | 2511 | 862 | 2188 | 1733 | 151 |
| ENST00000531818 | UVRAG | chr11 | 75728024 | + | ENST00000315769 | ZMAT4 | chr8 | 40389757 | - | 2689 | 1040 | 447 | 1055 | 202 |
| ENST00000532130 | UVRAG | chr11 | 75728024 | + | ENST00000315769 | ZMAT4 | chr8 | 40389757 | - | 2236 | 587 | 48 | 602 | 184 |
| ENST00000539288 | UVRAG | chr11 | 75728024 | + | ENST00000315769 | ZMAT4 | chr8 | 40389757 | - | 2303 | 654 | 1980 | 1525 | 151 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000356136 | ENST00000315769 | UVRAG | chr11 | 75728024 | + | ZMAT4 | chr8 | 40389757 | - | 0.000602721 | 0.9993973 |
| ENST00000528420 | ENST00000315769 | UVRAG | chr11 | 75728024 | + | ZMAT4 | chr8 | 40389757 | - | 0.000741677 | 0.99925834 |
| ENST00000533454 | ENST00000315769 | UVRAG | chr11 | 75728024 | + | ZMAT4 | chr8 | 40389757 | - | 0.6957949 | 0.30420512 |
| ENST00000531818 | ENST00000315769 | UVRAG | chr11 | 75728024 | + | ZMAT4 | chr8 | 40389757 | - | 0.7220262 | 0.2779738 |
| ENST00000532130 | ENST00000315769 | UVRAG | chr11 | 75728024 | + | ZMAT4 | chr8 | 40389757 | - | 0.7216378 | 0.27836218 |
| ENST00000539288 | ENST00000315769 | UVRAG | chr11 | 75728024 | + | ZMAT4 | chr8 | 40389757 | - | 0.4546522 | 0.5453478 |
Top |
Fusion Genomic Features for UVRAG-ZMAT4 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
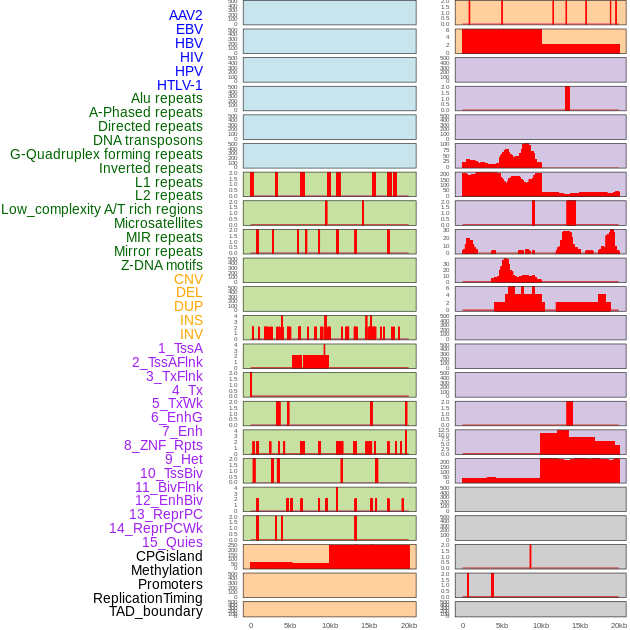 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for UVRAG-ZMAT4 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr11:75728024/chr8:40389757) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | UVRAG | chr11:75728024 | chr8:40389757 | ENST00000356136 | + | 12 | 15 | 224_305 | 408 | 700.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | UVRAG | chr11:75728024 | chr8:40389757 | ENST00000356136 | + | 12 | 15 | 23_149 | 408 | 700.0 | Domain | C2 |
| Tgene | ZMAT4 | chr11:75728024 | chr8:40389757 | ENST00000315769 | 4 | 6 | 198_228 | 148 | 154.0 | Zinc finger | Note=Matrin-type 4 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | ZMAT4 | chr11:75728024 | chr8:40389757 | ENST00000297737 | 5 | 7 | 145_175 | 224 | 230.0 | Zinc finger | Note=Matrin-type 3 | |
| Tgene | ZMAT4 | chr11:75728024 | chr8:40389757 | ENST00000297737 | 5 | 7 | 14_44 | 224 | 230.0 | Zinc finger | Note=Matrin-type 1 | |
| Tgene | ZMAT4 | chr11:75728024 | chr8:40389757 | ENST00000297737 | 5 | 7 | 198_228 | 224 | 230.0 | Zinc finger | Note=Matrin-type 4 | |
| Tgene | ZMAT4 | chr11:75728024 | chr8:40389757 | ENST00000297737 | 5 | 7 | 72_106 | 224 | 230.0 | Zinc finger | Note=Matrin-type 2 | |
| Tgene | ZMAT4 | chr11:75728024 | chr8:40389757 | ENST00000315769 | 4 | 6 | 145_175 | 148 | 154.0 | Zinc finger | Note=Matrin-type 3 | |
| Tgene | ZMAT4 | chr11:75728024 | chr8:40389757 | ENST00000315769 | 4 | 6 | 14_44 | 148 | 154.0 | Zinc finger | Note=Matrin-type 1 | |
| Tgene | ZMAT4 | chr11:75728024 | chr8:40389757 | ENST00000315769 | 4 | 6 | 72_106 | 148 | 154.0 | Zinc finger | Note=Matrin-type 2 |
Top |
Fusion Gene Sequence for UVRAG-ZMAT4 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >97822_97822_1_UVRAG-ZMAT4_UVRAG_chr11_75728024_ENST00000356136_ZMAT4_chr8_40389757_ENST00000315769_length(transcript)=3116nt_BP=1467nt AATGAGCGCTCCGGGTCTCAGCGGGGTGGAGGGGTTGCACTGCGGTAATATGGCTCTTCCTTAGCCAGCGGCGGCAACGGCGGCAGCGGC GGCAGCGGCGGCGGCTACTGTCTGGGCTGAGCAGTAGTGCCTCTCGGGTGGCGGGTTTCTAGGCTGCAGGGGCTTGGTAGGTGGTGGCAA GGGGGCGGCGGCGGATGCCGGAAGAGTGCCCGCCCCGCCGCTTGGCGGCCCCTGGATCGAGATGAGCGCCTCCGCGTCGGTCGGGGGCCC CGTCCCCCAGCCACCCCCGGGCCCGGCCGCTGCTCTGCCTCCCGGTTCTGCCGCGCGGGCCCTGCATGTGGAGCTGCCGTCTCAGCAGCG GCGTCTTCGACATCTTCGGAACATTGCTGCCCGGAACATTGTTAATAGAAATGGCCATCAGCTCCTTGATACCTACTTTACACTTCACTT GTGTAGTACTGAAAAGATATATAAAGAATTTTATAGAAGTGAAGTGATTAAGAATTCCTTGAATCCCACGTGGCGAAGTCTCGATTTTGG AATTATGCCAGACCGTCTTGATACATCTGTGTCTTGTTTCGTGGTGAAGATATGGGGTGGAAAGGAGAACATCTACCAGCTGTTGATTGA ATGGAAAGTCTGTTTGGATGGGCTGAAATACTTGGGTCAGCAGATTCATGCCCGAAACCAAAATGAAATAATTTTTGGGCTGAATGATGG ATACTATGGTGCTCCATTTGAACATAAGGGTTATTCAAATGCTCAGAAGACTATTCTTCTGCAGGTGGATCAGAACTGTGTTCGCAATTC TTACGATGTCTTCTCTTTGCTACGGCTTCATAGAGCCCAGTGTGCAATTAAACAGACTCAGGTAACTGTTCAGAAAATTGGAAAGGAAAT TGAAGAAAAACTAAGACTCACATCTACAAGCAATGAACTGAAAAAAAAAAGTGAATGCCTGCAGTTAAAAATTTTGGTGCTTCAGAATGA ACTGGAACGGCAGAAGAAAGCTTTGGGACGGGAGGTGGCATTACTGCATAAGCAACAAATTGCATTACAAGACAAAGGAAGTGCATTTTC AGCTGAGCACCTCAAACTTCAACTCCAGAAGGAATCCCTAAATGAGCTGAGGAAGGAGTGCACTGCAAAAAGAGAACTCTTCTTGAAGAC TAATGCTCAGTTGACAATTCGTTGCAGGCAGTTACTCTCTGAGCTTTCCTACATTTACCCTATTGATTTGAATGAACATAAGGATTACTT TGTATGCGGTGTCAAGTTGCCTAATTCTGAGGACTTCCAAGCAAAAGATGATGGAAGCATTGCTGTTGCCCTTGGTTATACTGCACATCT GGTCTCCATGATTTCCTTTTTCCTACAAGTGCCCCTCAGATATCCTATAATTCATAAGGGGTCTAGATCAACAATCAAAGACAATATCAA TGACAAACTGACGGAAAAGGAGAGAGACCTGAAGAATAAGTAGTGAAAGCATCAATCAAGACATAAGAACAAAACATTAGCATTTCTCTG CCGTGGAGAATTGCTTATCAACCACCAGAGGAGGCTTCTTTCTTGAACAATAAACATTTCTTATAAGGATTCACAGATTCACATACGACT GATCTTGATTTTTGGAAATGAATGAGGTTTCTTTTTTCTTTTTCCTTTTTTTAATTTTGGGGTAAGTTATGATATTTGGATGGATTTTTA AATTCTTTCCTGATAACATATTTAGCACATGTTCTAAATTATAATCCTATAGCAAACAGTTGGAGCATTATTCAAACTGAAAGTGGAAAA ATTTAAATTTCCAATTTATTCTAGATTTCCTCAGAGCATAATTATTCTGTTAAATCCTCAATGAGTGTGATGTAAACCACCTCTATCCAG AAATATACATTCTTTTCTCATCATGTTGGACACAGTTGAGGGTGACATGCACAGAACTGGAACAGATCACTATTAGTGGAAAATACCAAA TGGACAAATAAATACCAGTCGTTTTCTCCGTTCTCCAAGCACAGGAGCCAGGTTTACCATCTGAACAATGAAGACGAAGGGAGTAAATAA AGGAAGAATTCTCATCTTTTTTCCTGATCATTCAAAGAACAGTTTCTCAAGGTTAAGCCAAGTCCTCCTTGCAAGTTGCCAAATAATAGC TTAGGAAAAGAATTAGTCTGCCTGCATGATGATCTTCTTAGGCAAAAACGTCTTCACAGCCCTTGACCTTGGTGAATTTTTTTCCCCAAA AGCATCCAAAAGAAGAATTATAAACCCCAGAATGAGATGGAAATAAACAAGTATTTTTTTTTATGATGTTTGGCCTGAACTGTGGGCTTT AATTGGGGGATACTGATCGTTTGGAAAGAAGTGAGAAAATTCTGAAGAAATGGCGGCCTTGGGCTAGGCGGGGTCCCCTATTTCTTCTGT TTCTCACTGAAGTCCTACTGCTGAGCCAAGACTCAGTCACTCTGGAAAGAGCATGACCGATAAAGAAAACAGTTCCTTTCTGATGGGGAG CGTCTGAGTGCAGATCATGAGGCTCTTTCTCTAGGTTTAATTCTTTTCCATGGTGACCGGACTTGGTGTCTTGTAGCCTGGTTACGAAGT GGGACGTTGAGCTTCTACTGACGATGCCCTGCATGGACCAGCTGGGATCTGGCTGGGGCTGCCCTGTGTCCCTAACGACCATAGGCAATC CATCTTCTTGTGTCAGCAATTTCTGGACACCCACTGTTTTCCACCAAGAGCTGAGGTGGCAACAACTCAGTGAGCAATAAACAAAATGAC ACAGAAATGCACAGTGTTGTTATGAAGGAGCCTGTTTACCTGTGTTCAAAATCTGGCACCATTCCCTTGAGCAGGGCCCGCTCAGGAGGG ACCAGGTCTGCCAGTTTCTGTGCCTGCAGAGAGACGAAGCCCCACGAGCCACACCCTACTCTACAAGAGGAAAGGGGGTTGGATGGGAAG AATCTATTTTGCTGTTTTGGAAAGCACACAGCCGACCTACAAACCTCCTGTGATGGTGTTTCTTCGGATGTGTAAAATAAGGCTTTATTT >97822_97822_1_UVRAG-ZMAT4_UVRAG_chr11_75728024_ENST00000356136_ZMAT4_chr8_40389757_ENST00000315769_length(amino acids)=439AA_BP= MVGGGKGAAADAGRVPAPPLGGPWIEMSASASVGGPVPQPPPGPAAALPPGSAARALHVELPSQQRRLRHLRNIAARNIVNRNGHQLLDT YFTLHLCSTEKIYKEFYRSEVIKNSLNPTWRSLDFGIMPDRLDTSVSCFVVKIWGGKENIYQLLIEWKVCLDGLKYLGQQIHARNQNEII FGLNDGYYGAPFEHKGYSNAQKTILLQVDQNCVRNSYDVFSLLRLHRAQCAIKQTQVTVQKIGKEIEEKLRLTSTSNELKKKSECLQLKI LVLQNELERQKKALGREVALLHKQQIALQDKGSAFSAEHLKLQLQKESLNELRKECTAKRELFLKTNAQLTIRCRQLLSELSYIYPIDLN -------------------------------------------------------------- >97822_97822_2_UVRAG-ZMAT4_UVRAG_chr11_75728024_ENST00000528420_ZMAT4_chr8_40389757_ENST00000315769_length(transcript)=2857nt_BP=1208nt AGTGCTCAGCTCCCTTGCCATGCGATACCCTGCAAGTCCTCACCAGCAAAAAGGCTCTCACCAGAGGCTGCCCCTCGGCCTGTGTTATCA GATTTTTTGCGGCGTCTTCGACATCTTCGGAACATTGCTGCCCGGAACATTGTTAATAGAAATGGCCATCAGCTCCTTGATACCTACTTT ACACTTCACTTGTGTAGTACTGAAAAGATATATAAAGAATTTTATAGAAGTGAAGTGATTAAGAATTCCTTGAATCCCACGTGGCGAAGT CTCGATTTTGGAATTATGCCAGACCGTCTTGATACATCTGTGTCTTGTTTCGTGGTGAAGATATGGGGTGGAAAGGAGAACATCTACCAG CTGTTGATTGAATGGAAAGTCTGTTTGGATGGGCTGAAATACTTGGGTCAGCAGATTCATGCCCGAAACCAAAATGAAATAATTTTTGGG CTGAATGATGGATACTATGGTGCTCCATTTGAACATAAGGGTTATTCAAATGCTCAGAAGACTATTCTTCTGCAGGTGGATCAGAACTGT GTTCGCAATTCTTACGATGTCTTCTCTTTGCTACGGCTTCATAGAGCCCAGTGTGCAATTAAACAGACTCAGGTAACTGTTCAGAAAATT GGAAAGGAAATTGAAGAAAAACTAAGACTCACATCTACAAGCAATGAACTGAAAAAAAAAAGTGAATGCCTGCAGTTAAAAATTTTGGTG CTTCAGAATGAACTGGAACGGCAGAAGAAAGCTTTGGGACGGGAGGTGGCATTACTGCATAAGCAACAAATTGCATTACAAGACAAAGGA AGTGCATTTTCAGCTGAGCACCTCAAACTTCAACTCCAGAAGGAATCCCTAAATGAGCTGAGGAAGGAGTGCACTGCAAAAAGAGAACTC TTCTTGAAGACTAATGCTCAGTTGACAATTCGTTGCAGGCAGTTACTCTCTGAGCTTTCCTACATTTACCCTATTGATTTGAATGAACAT AAGGATTACTTTGTATGCGGTGTCAAGTTGCCTAATTCTGAGGACTTCCAAGCAAAAGATGATGGAAGCATTGCTGTTGCCCTTGGTTAT ACTGCACATCTGGTCTCCATGATTTCCTTTTTCCTACAAGTGCCCCTCAGATATCCTATAATTCATAAGGGGTCTAGATCAACAATCAAA GACAATATCAATGACAAACTGACGGAAAAGGAGAGAGACCTGAAGAATAAGTAGTGAAAGCATCAATCAAGACATAAGAACAAAACATTA GCATTTCTCTGCCGTGGAGAATTGCTTATCAACCACCAGAGGAGGCTTCTTTCTTGAACAATAAACATTTCTTATAAGGATTCACAGATT CACATACGACTGATCTTGATTTTTGGAAATGAATGAGGTTTCTTTTTTCTTTTTCCTTTTTTTAATTTTGGGGTAAGTTATGATATTTGG ATGGATTTTTAAATTCTTTCCTGATAACATATTTAGCACATGTTCTAAATTATAATCCTATAGCAAACAGTTGGAGCATTATTCAAACTG AAAGTGGAAAAATTTAAATTTCCAATTTATTCTAGATTTCCTCAGAGCATAATTATTCTGTTAAATCCTCAATGAGTGTGATGTAAACCA CCTCTATCCAGAAATATACATTCTTTTCTCATCATGTTGGACACAGTTGAGGGTGACATGCACAGAACTGGAACAGATCACTATTAGTGG AAAATACCAAATGGACAAATAAATACCAGTCGTTTTCTCCGTTCTCCAAGCACAGGAGCCAGGTTTACCATCTGAACAATGAAGACGAAG GGAGTAAATAAAGGAAGAATTCTCATCTTTTTTCCTGATCATTCAAAGAACAGTTTCTCAAGGTTAAGCCAAGTCCTCCTTGCAAGTTGC CAAATAATAGCTTAGGAAAAGAATTAGTCTGCCTGCATGATGATCTTCTTAGGCAAAAACGTCTTCACAGCCCTTGACCTTGGTGAATTT TTTTCCCCAAAAGCATCCAAAAGAAGAATTATAAACCCCAGAATGAGATGGAAATAAACAAGTATTTTTTTTTATGATGTTTGGCCTGAA CTGTGGGCTTTAATTGGGGGATACTGATCGTTTGGAAAGAAGTGAGAAAATTCTGAAGAAATGGCGGCCTTGGGCTAGGCGGGGTCCCCT ATTTCTTCTGTTTCTCACTGAAGTCCTACTGCTGAGCCAAGACTCAGTCACTCTGGAAAGAGCATGACCGATAAAGAAAACAGTTCCTTT CTGATGGGGAGCGTCTGAGTGCAGATCATGAGGCTCTTTCTCTAGGTTTAATTCTTTTCCATGGTGACCGGACTTGGTGTCTTGTAGCCT GGTTACGAAGTGGGACGTTGAGCTTCTACTGACGATGCCCTGCATGGACCAGCTGGGATCTGGCTGGGGCTGCCCTGTGTCCCTAACGAC CATAGGCAATCCATCTTCTTGTGTCAGCAATTTCTGGACACCCACTGTTTTCCACCAAGAGCTGAGGTGGCAACAACTCAGTGAGCAATA AACAAAATGACACAGAAATGCACAGTGTTGTTATGAAGGAGCCTGTTTACCTGTGTTCAAAATCTGGCACCATTCCCTTGAGCAGGGCCC GCTCAGGAGGGACCAGGTCTGCCAGTTTCTGTGCCTGCAGAGAGACGAAGCCCCACGAGCCACACCCTACTCTACAAGAGGAAAGGGGGT TGGATGGGAAGAATCTATTTTGCTGTTTTGGAAAGCACACAGCCGACCTACAAACCTCCTGTGATGGTGTTTCTTCGGATGTGTAAAATA >97822_97822_2_UVRAG-ZMAT4_UVRAG_chr11_75728024_ENST00000528420_ZMAT4_chr8_40389757_ENST00000315769_length(amino acids)=375AA_BP= MRRLRHLRNIAARNIVNRNGHQLLDTYFTLHLCSTEKIYKEFYRSEVIKNSLNPTWRSLDFGIMPDRLDTSVSCFVVKIWGGKENIYQLL IEWKVCLDGLKYLGQQIHARNQNEIIFGLNDGYYGAPFEHKGYSNAQKTILLQVDQNCVRNSYDVFSLLRLHRAQCAIKQTQVTVQKIGK EIEEKLRLTSTSNELKKKSECLQLKILVLQNELERQKKALGREVALLHKQQIALQDKGSAFSAEHLKLQLQKESLNELRKECTAKRELFL KTNAQLTIRCRQLLSELSYIYPIDLNEHKDYFVCGVKLPNSEDFQAKDDGSIAVALGYTAHLVSMISFFLQVPLRYPIIHKGSRSTIKDN -------------------------------------------------------------- >97822_97822_3_UVRAG-ZMAT4_UVRAG_chr11_75728024_ENST00000531818_ZMAT4_chr8_40389757_ENST00000315769_length(transcript)=2689nt_BP=1040nt CAGTTCGAGCTTCTGGGCAGCTTTGTTTACCTGCTCAAGCCTCAGCATTGGTGGGCGCCCCTCCCCCAGCCTCGCTGCCGCCTTGCAGTT TGATCTCAGACTGCTGTGCTAGCAGTGAGCAAGGCTCCGTGGGCGTGGGACCCTCTGAGCCAGGCGCGGGATATAATCTTCTGGTATGCT GTTTGCTAAGACCTTTGGAAAAGCACAGTATTAGGGTGGGAGTGACCTGATTTTCCGGGTGCGATTTTCCAGGTGCCGTCTGTCGTGGCT TCCCTTGGCTGGGAAAGGGAGTTCCCCGACCCCTTGTGCTTCCCTGGTGAGGCGATGCCTTGCCCTGCTTTGGCTCACGGTCCGTGGGCT GCACCCACTGTCCTACAAGCCCCAGTGAGATGAACCCGGCACCTCAATTGGAAATGCAGAAATCATCCATCTTCTGCGTCGCTCACGCTG GAAGCTGTAGACTGGAGCTATTCCTATTCGGCCATCTTGGAACCTCCCTGCCTCTTTTTTCTGAAAAAAAAAAGTGAATGCCTGCAGTTA AAAATTTTGGTGCTTCAGAATGAACTGGAACGGCAGAAGAAAGCTTTGGGACGGGAGGTGGCATTACTGCATAAGCAACAAATTGCATTA CAAGACAAAGGAAGTGCATTTTCAGCTGAGCACCTCAAACTTCAACTCCAGAAGGAATCCCTAAATGAGCTGAGGAAGGAGTGCACTGCA AAAAGAGAACTCTTCTTGAAGACTAATGCTCAGTTGACAATTCGTTGCAGGCAGTTACTCTCTGAGCTTTCCTACATTTACCCTATTGAT TTGAATGAACATAAGGATTACTTTGTATGCGGTGTCAAGTTGCCTAATTCTGAGGACTTCCAAGCAAAAGATGATGGAAGCATTGCTGTT GCCCTTGGTTATACTGCACATCTGGTCTCCATGATTTCCTTTTTCCTACAAGTGCCCCTCAGATATCCTATAATTCATAAGGGGTCTAGA TCAACAATCAAAGACAATATCAATGACAAACTGACGGAAAAGGAGAGAGACCTGAAGAATAAGTAGTGAAAGCATCAATCAAGACATAAG AACAAAACATTAGCATTTCTCTGCCGTGGAGAATTGCTTATCAACCACCAGAGGAGGCTTCTTTCTTGAACAATAAACATTTCTTATAAG GATTCACAGATTCACATACGACTGATCTTGATTTTTGGAAATGAATGAGGTTTCTTTTTTCTTTTTCCTTTTTTTAATTTTGGGGTAAGT TATGATATTTGGATGGATTTTTAAATTCTTTCCTGATAACATATTTAGCACATGTTCTAAATTATAATCCTATAGCAAACAGTTGGAGCA TTATTCAAACTGAAAGTGGAAAAATTTAAATTTCCAATTTATTCTAGATTTCCTCAGAGCATAATTATTCTGTTAAATCCTCAATGAGTG TGATGTAAACCACCTCTATCCAGAAATATACATTCTTTTCTCATCATGTTGGACACAGTTGAGGGTGACATGCACAGAACTGGAACAGAT CACTATTAGTGGAAAATACCAAATGGACAAATAAATACCAGTCGTTTTCTCCGTTCTCCAAGCACAGGAGCCAGGTTTACCATCTGAACA ATGAAGACGAAGGGAGTAAATAAAGGAAGAATTCTCATCTTTTTTCCTGATCATTCAAAGAACAGTTTCTCAAGGTTAAGCCAAGTCCTC CTTGCAAGTTGCCAAATAATAGCTTAGGAAAAGAATTAGTCTGCCTGCATGATGATCTTCTTAGGCAAAAACGTCTTCACAGCCCTTGAC CTTGGTGAATTTTTTTCCCCAAAAGCATCCAAAAGAAGAATTATAAACCCCAGAATGAGATGGAAATAAACAAGTATTTTTTTTTATGAT GTTTGGCCTGAACTGTGGGCTTTAATTGGGGGATACTGATCGTTTGGAAAGAAGTGAGAAAATTCTGAAGAAATGGCGGCCTTGGGCTAG GCGGGGTCCCCTATTTCTTCTGTTTCTCACTGAAGTCCTACTGCTGAGCCAAGACTCAGTCACTCTGGAAAGAGCATGACCGATAAAGAA AACAGTTCCTTTCTGATGGGGAGCGTCTGAGTGCAGATCATGAGGCTCTTTCTCTAGGTTTAATTCTTTTCCATGGTGACCGGACTTGGT GTCTTGTAGCCTGGTTACGAAGTGGGACGTTGAGCTTCTACTGACGATGCCCTGCATGGACCAGCTGGGATCTGGCTGGGGCTGCCCTGT GTCCCTAACGACCATAGGCAATCCATCTTCTTGTGTCAGCAATTTCTGGACACCCACTGTTTTCCACCAAGAGCTGAGGTGGCAACAACT CAGTGAGCAATAAACAAAATGACACAGAAATGCACAGTGTTGTTATGAAGGAGCCTGTTTACCTGTGTTCAAAATCTGGCACCATTCCCT TGAGCAGGGCCCGCTCAGGAGGGACCAGGTCTGCCAGTTTCTGTGCCTGCAGAGAGACGAAGCCCCACGAGCCACACCCTACTCTACAAG AGGAAAGGGGGTTGGATGGGAAGAATCTATTTTGCTGTTTTGGAAAGCACACAGCCGACCTACAAACCTCCTGTGATGGTGTTTCTTCGG >97822_97822_3_UVRAG-ZMAT4_UVRAG_chr11_75728024_ENST00000531818_ZMAT4_chr8_40389757_ENST00000315769_length(amino acids)=202AA_BP= MEAVDWSYSYSAILEPPCLFFLKKKSECLQLKILVLQNELERQKKALGREVALLHKQQIALQDKGSAFSAEHLKLQLQKESLNELRKECT AKRELFLKTNAQLTIRCRQLLSELSYIYPIDLNEHKDYFVCGVKLPNSEDFQAKDDGSIAVALGYTAHLVSMISFFLQVPLRYPIIHKGS -------------------------------------------------------------- >97822_97822_4_UVRAG-ZMAT4_UVRAG_chr11_75728024_ENST00000532130_ZMAT4_chr8_40389757_ENST00000315769_length(transcript)=2236nt_BP=587nt TTTAGTAGAATTCACTGTGAAACCAACTAGACCTTTTTTTTTTCTCCCCTGTTTCTGAAGAAAAAAAAAAGTGAATGCCTGCAGTTAAAA ATTTTGGTGCTTCAGAATGAACTGGAACGGCAGAAGAAAGCTTTGGGACGGGAGGTGGCATTACTGCATAAGCAACAAATTGCATTACAA GACAAAGGAAGTGCATTTTCAGCTGAGCACCTCAAACTTCAACTCCAGAAGGAATCCCTAAATGAGCTGAGGAAGGAGTGCACTGCAAAA AGAGAACTCTTCTTGAAGACTAATGCTCAGTTGACAATTCGTTGCAGGCAGTTACTCTCTGAGCTTTCCTACATTTACCCTATTGATTTG AATGAACATAAGGATTACTTTGTATGCGGTGTCAAGTTGCCTAATTCTGAGGACTTCCAAGCAAAAGATGATGGAAGCATTGCTGTTGCC CTTGGTTATACTGCACATCTGGTCTCCATGATTTCCTTTTTCCTACAAGTGCCCCTCAGATATCCTATAATTCATAAGGGGTCTAGATCA ACAATCAAAGACAATATCAATGACAAACTGACGGAAAAGGAGAGAGACCTGAAGAATAAGTAGTGAAAGCATCAATCAAGACATAAGAAC AAAACATTAGCATTTCTCTGCCGTGGAGAATTGCTTATCAACCACCAGAGGAGGCTTCTTTCTTGAACAATAAACATTTCTTATAAGGAT TCACAGATTCACATACGACTGATCTTGATTTTTGGAAATGAATGAGGTTTCTTTTTTCTTTTTCCTTTTTTTAATTTTGGGGTAAGTTAT GATATTTGGATGGATTTTTAAATTCTTTCCTGATAACATATTTAGCACATGTTCTAAATTATAATCCTATAGCAAACAGTTGGAGCATTA TTCAAACTGAAAGTGGAAAAATTTAAATTTCCAATTTATTCTAGATTTCCTCAGAGCATAATTATTCTGTTAAATCCTCAATGAGTGTGA TGTAAACCACCTCTATCCAGAAATATACATTCTTTTCTCATCATGTTGGACACAGTTGAGGGTGACATGCACAGAACTGGAACAGATCAC TATTAGTGGAAAATACCAAATGGACAAATAAATACCAGTCGTTTTCTCCGTTCTCCAAGCACAGGAGCCAGGTTTACCATCTGAACAATG AAGACGAAGGGAGTAAATAAAGGAAGAATTCTCATCTTTTTTCCTGATCATTCAAAGAACAGTTTCTCAAGGTTAAGCCAAGTCCTCCTT GCAAGTTGCCAAATAATAGCTTAGGAAAAGAATTAGTCTGCCTGCATGATGATCTTCTTAGGCAAAAACGTCTTCACAGCCCTTGACCTT GGTGAATTTTTTTCCCCAAAAGCATCCAAAAGAAGAATTATAAACCCCAGAATGAGATGGAAATAAACAAGTATTTTTTTTTATGATGTT TGGCCTGAACTGTGGGCTTTAATTGGGGGATACTGATCGTTTGGAAAGAAGTGAGAAAATTCTGAAGAAATGGCGGCCTTGGGCTAGGCG GGGTCCCCTATTTCTTCTGTTTCTCACTGAAGTCCTACTGCTGAGCCAAGACTCAGTCACTCTGGAAAGAGCATGACCGATAAAGAAAAC AGTTCCTTTCTGATGGGGAGCGTCTGAGTGCAGATCATGAGGCTCTTTCTCTAGGTTTAATTCTTTTCCATGGTGACCGGACTTGGTGTC TTGTAGCCTGGTTACGAAGTGGGACGTTGAGCTTCTACTGACGATGCCCTGCATGGACCAGCTGGGATCTGGCTGGGGCTGCCCTGTGTC CCTAACGACCATAGGCAATCCATCTTCTTGTGTCAGCAATTTCTGGACACCCACTGTTTTCCACCAAGAGCTGAGGTGGCAACAACTCAG TGAGCAATAAACAAAATGACACAGAAATGCACAGTGTTGTTATGAAGGAGCCTGTTTACCTGTGTTCAAAATCTGGCACCATTCCCTTGA GCAGGGCCCGCTCAGGAGGGACCAGGTCTGCCAGTTTCTGTGCCTGCAGAGAGACGAAGCCCCACGAGCCACACCCTACTCTACAAGAGG AAAGGGGGTTGGATGGGAAGAATCTATTTTGCTGTTTTGGAAAGCACACAGCCGACCTACAAACCTCCTGTGATGGTGTTTCTTCGGATG >97822_97822_4_UVRAG-ZMAT4_UVRAG_chr11_75728024_ENST00000532130_ZMAT4_chr8_40389757_ENST00000315769_length(amino acids)=184AA_BP= MFLKKKKSECLQLKILVLQNELERQKKALGREVALLHKQQIALQDKGSAFSAEHLKLQLQKESLNELRKECTAKRELFLKTNAQLTIRCR QLLSELSYIYPIDLNEHKDYFVCGVKLPNSEDFQAKDDGSIAVALGYTAHLVSMISFFLQVPLRYPIIHKGSRSTIKDNINDKLTEKERD -------------------------------------------------------------- >97822_97822_5_UVRAG-ZMAT4_UVRAG_chr11_75728024_ENST00000533454_ZMAT4_chr8_40389757_ENST00000315769_length(transcript)=2511nt_BP=862nt ACTACCCACAAGAGGTGGTGAAAACGATCTCTGGTATAAAGCAAATGGAAAAGGCTTCATAGAGCCCAGTGTGCAATTAAACAGACTCAG GTAACTGTTCAGAAAATTGGAAAGGAAATTGAAGAAAAACTAAGACTCACATCTACAAGCAATGAACTGAAAAAAAAAAGTGAATGCCTG CAGTTAAAAATTTTGGTGCTTCAGAATGAACTGGAACGGCAGAAGAAAGCTTTGGGACGGGAGGTGGCATTACTGCATAAGCAACAAATT GCATTACAAGACAAAGGAAGTGCATTTTCAGCTGAGCACCTCAAACTTCAACTCCAGAAGGAATCCCTAAATGAGCTGAGGAAGGAGTGC ACTGCAAAAAGAGAACTCTTCTTGAAGACTAATGCTCAGTTGACAATTCGTTGCAGGCAGTTACTCTCTGAGCTTTCCTACATTTACCCT ATTGATTTGAATGAACATAAGGATTACTTTGTATGCGGTGTCAAGTTGCCTAATTCTGAGGACTTCCAAGGCATGATTTCTTGGAAAGGA GAGTCCATATGTCAAACAATGTCTCATTGAGAACATGGAATGATTCCATAGATGCTGTGGCAGTGATTAGAGTCTTAAATATTTTGTCCT GTCTGGATCAGTGGGTGCTTGGTTCCTTAACAAATCAGCTGCCTCAGTGATTGGGAAGGCAGGATGCAAAAGATGATGGAAGCATTGCTG TTGCCCTTGGTTATACTGCACATCTGGTCTCCATGATTTCCTTTTTCCTACAAGTGCCCCTCAGATATCCTATAATTCATAAGGGGTCTA GATCAACAATCAAAGACAATATCAATGACAAACTGACGGAAAAGGAGAGAGACCTGAAGAATAAGTAGTGAAAGCATCAATCAAGACATA AGAACAAAACATTAGCATTTCTCTGCCGTGGAGAATTGCTTATCAACCACCAGAGGAGGCTTCTTTCTTGAACAATAAACATTTCTTATA AGGATTCACAGATTCACATACGACTGATCTTGATTTTTGGAAATGAATGAGGTTTCTTTTTTCTTTTTCCTTTTTTTAATTTTGGGGTAA GTTATGATATTTGGATGGATTTTTAAATTCTTTCCTGATAACATATTTAGCACATGTTCTAAATTATAATCCTATAGCAAACAGTTGGAG CATTATTCAAACTGAAAGTGGAAAAATTTAAATTTCCAATTTATTCTAGATTTCCTCAGAGCATAATTATTCTGTTAAATCCTCAATGAG TGTGATGTAAACCACCTCTATCCAGAAATATACATTCTTTTCTCATCATGTTGGACACAGTTGAGGGTGACATGCACAGAACTGGAACAG ATCACTATTAGTGGAAAATACCAAATGGACAAATAAATACCAGTCGTTTTCTCCGTTCTCCAAGCACAGGAGCCAGGTTTACCATCTGAA CAATGAAGACGAAGGGAGTAAATAAAGGAAGAATTCTCATCTTTTTTCCTGATCATTCAAAGAACAGTTTCTCAAGGTTAAGCCAAGTCC TCCTTGCAAGTTGCCAAATAATAGCTTAGGAAAAGAATTAGTCTGCCTGCATGATGATCTTCTTAGGCAAAAACGTCTTCACAGCCCTTG ACCTTGGTGAATTTTTTTCCCCAAAAGCATCCAAAAGAAGAATTATAAACCCCAGAATGAGATGGAAATAAACAAGTATTTTTTTTTATG ATGTTTGGCCTGAACTGTGGGCTTTAATTGGGGGATACTGATCGTTTGGAAAGAAGTGAGAAAATTCTGAAGAAATGGCGGCCTTGGGCT AGGCGGGGTCCCCTATTTCTTCTGTTTCTCACTGAAGTCCTACTGCTGAGCCAAGACTCAGTCACTCTGGAAAGAGCATGACCGATAAAG AAAACAGTTCCTTTCTGATGGGGAGCGTCTGAGTGCAGATCATGAGGCTCTTTCTCTAGGTTTAATTCTTTTCCATGGTGACCGGACTTG GTGTCTTGTAGCCTGGTTACGAAGTGGGACGTTGAGCTTCTACTGACGATGCCCTGCATGGACCAGCTGGGATCTGGCTGGGGCTGCCCT GTGTCCCTAACGACCATAGGCAATCCATCTTCTTGTGTCAGCAATTTCTGGACACCCACTGTTTTCCACCAAGAGCTGAGGTGGCAACAA CTCAGTGAGCAATAAACAAAATGACACAGAAATGCACAGTGTTGTTATGAAGGAGCCTGTTTACCTGTGTTCAAAATCTGGCACCATTCC CTTGAGCAGGGCCCGCTCAGGAGGGACCAGGTCTGCCAGTTTCTGTGCCTGCAGAGAGACGAAGCCCCACGAGCCACACCCTACTCTACA AGAGGAAAGGGGGTTGGATGGGAAGAATCTATTTTGCTGTTTTGGAAAGCACACAGCCGACCTACAAACCTCCTGTGATGGTGTTTCTTC >97822_97822_5_UVRAG-ZMAT4_UVRAG_chr11_75728024_ENST00000533454_ZMAT4_chr8_40389757_ENST00000315769_length(amino acids)=151AA_BP= MCHFVYCSLSCCHLSSWWKTVGVQKLLTQEDGLPMVVRDTGQPQPDPSWSMQGIVSRSSTSHFVTRLQDTKSGHHGKELNLEKEPHDLHS -------------------------------------------------------------- >97822_97822_6_UVRAG-ZMAT4_UVRAG_chr11_75728024_ENST00000539288_ZMAT4_chr8_40389757_ENST00000315769_length(transcript)=2303nt_BP=654nt AACTGGAACGGCAGAAGAAAGCTTTGGGACGGGAGGTGGCATTACTGCATAAGCAACAAATTGCATTACAAGACAAAGGAAGTGCATTTT CAGCTGAGCACCTCAAACTTCAACTCCAGAAGGAATCCCTAAATGAGCTGAGGAAGGAGTGCACTGCAAAAAGAGAACTCTTCTTGAAGA CTAATGCTCAGTTGACAATTCGTTGCAGGCAGTTACTCTCTGAGCTTTCCTACATTTACCCTATTGATTTGAATGAACATAAGGATTACT TTGTATGCGGTGTCAAGTTGCCTAATTCTGAGGACTTCCAAGGCATGATTTCTTGGAAAGGAGAGTCCATATGTCAAACAATGTCTCATT GAGAACATGGAATGATTCCATAGATGCTGTGGCAGTGATTAGAGTCTTAAATATTTTGTCCTGTCTGGATCAGTGGGTGCTTGGTTCCTT AACAAATCAGCTGCCTCAGTGATTGGGAAGGCAGGATGCAAAAGATGATGGAAGCATTGCTGTTGCCCTTGGTTATACTGCACATCTGGT CTCCATGATTTCCTTTTTCCTACAAGTGCCCCTCAGATATCCTATAATTCATAAGGGGTCTAGATCAACAATCAAAGACAATATCAATGA CAAACTGACGGAAAAGGAGAGAGACCTGAAGAATAAGTAGTGAAAGCATCAATCAAGACATAAGAACAAAACATTAGCATTTCTCTGCCG TGGAGAATTGCTTATCAACCACCAGAGGAGGCTTCTTTCTTGAACAATAAACATTTCTTATAAGGATTCACAGATTCACATACGACTGAT CTTGATTTTTGGAAATGAATGAGGTTTCTTTTTTCTTTTTCCTTTTTTTAATTTTGGGGTAAGTTATGATATTTGGATGGATTTTTAAAT TCTTTCCTGATAACATATTTAGCACATGTTCTAAATTATAATCCTATAGCAAACAGTTGGAGCATTATTCAAACTGAAAGTGGAAAAATT TAAATTTCCAATTTATTCTAGATTTCCTCAGAGCATAATTATTCTGTTAAATCCTCAATGAGTGTGATGTAAACCACCTCTATCCAGAAA TATACATTCTTTTCTCATCATGTTGGACACAGTTGAGGGTGACATGCACAGAACTGGAACAGATCACTATTAGTGGAAAATACCAAATGG ACAAATAAATACCAGTCGTTTTCTCCGTTCTCCAAGCACAGGAGCCAGGTTTACCATCTGAACAATGAAGACGAAGGGAGTAAATAAAGG AAGAATTCTCATCTTTTTTCCTGATCATTCAAAGAACAGTTTCTCAAGGTTAAGCCAAGTCCTCCTTGCAAGTTGCCAAATAATAGCTTA GGAAAAGAATTAGTCTGCCTGCATGATGATCTTCTTAGGCAAAAACGTCTTCACAGCCCTTGACCTTGGTGAATTTTTTTCCCCAAAAGC ATCCAAAAGAAGAATTATAAACCCCAGAATGAGATGGAAATAAACAAGTATTTTTTTTTATGATGTTTGGCCTGAACTGTGGGCTTTAAT TGGGGGATACTGATCGTTTGGAAAGAAGTGAGAAAATTCTGAAGAAATGGCGGCCTTGGGCTAGGCGGGGTCCCCTATTTCTTCTGTTTC TCACTGAAGTCCTACTGCTGAGCCAAGACTCAGTCACTCTGGAAAGAGCATGACCGATAAAGAAAACAGTTCCTTTCTGATGGGGAGCGT CTGAGTGCAGATCATGAGGCTCTTTCTCTAGGTTTAATTCTTTTCCATGGTGACCGGACTTGGTGTCTTGTAGCCTGGTTACGAAGTGGG ACGTTGAGCTTCTACTGACGATGCCCTGCATGGACCAGCTGGGATCTGGCTGGGGCTGCCCTGTGTCCCTAACGACCATAGGCAATCCAT CTTCTTGTGTCAGCAATTTCTGGACACCCACTGTTTTCCACCAAGAGCTGAGGTGGCAACAACTCAGTGAGCAATAAACAAAATGACACA GAAATGCACAGTGTTGTTATGAAGGAGCCTGTTTACCTGTGTTCAAAATCTGGCACCATTCCCTTGAGCAGGGCCCGCTCAGGAGGGACC AGGTCTGCCAGTTTCTGTGCCTGCAGAGAGACGAAGCCCCACGAGCCACACCCTACTCTACAAGAGGAAAGGGGGTTGGATGGGAAGAAT CTATTTTGCTGTTTTGGAAAGCACACAGCCGACCTACAAACCTCCTGTGATGGTGTTTCTTCGGATGTGTAAAATAAGGCTTTATTTGTC >97822_97822_6_UVRAG-ZMAT4_UVRAG_chr11_75728024_ENST00000539288_ZMAT4_chr8_40389757_ENST00000315769_length(amino acids)=151AA_BP= MCHFVYCSLSCCHLSSWWKTVGVQKLLTQEDGLPMVVRDTGQPQPDPSWSMQGIVSRSSTSHFVTRLQDTKSGHHGKELNLEKEPHDLHS -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for UVRAG-ZMAT4 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for UVRAG-ZMAT4 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for UVRAG-ZMAT4 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
