
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:WDR48-HERC2 (FusionGDB2 ID:99012) |
Fusion Gene Summary for WDR48-HERC2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: WDR48-HERC2 | Fusion gene ID: 99012 | Hgene | Tgene | Gene symbol | WDR48 | HERC2 | Gene ID | 57599 | 8924 |
| Gene name | WD repeat domain 48 | HECT and RLD domain containing E3 ubiquitin protein ligase 2 | |
| Synonyms | P80|SPG60|UAF1 | D15F37S1|MRT38|SHEP1|jdf2|p528 | |
| Cytomap | 3p22.2 | 15q13.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | WD repeat-containing protein 48USP1 associated factor 1WD repeat endosomal proteintesticular tissue protein Li 224 | E3 ubiquitin-protein ligase HERC2HECT-type E3 ubiquitin transferase HERC2hect domain and RCC1-like domain-containing protein 2hect domain and RLD 2probable E3 ubiquitin-protein ligase HERC2 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | O95714 | |
| Ensembl transtripts involved in fusion gene | ENST00000302313, ENST00000544962, ENST00000396258, ENST00000418020, ENST00000466405, | ENST00000563945, ENST00000261609, | |
| Fusion gene scores | * DoF score | 5 X 4 X 2=40 | 7 X 8 X 4=224 |
| # samples | 5 | 8 | |
| ** MAII score | log2(5/40*10)=0.321928094887362 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(8/224*10)=-1.48542682717024 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: WDR48 [Title/Abstract] AND HERC2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | WDR48(39093564)-HERC2(28397976), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | WDR48 | GO:0016579 | protein deubiquitination | 18082604|19075014|24145035 |
| Tgene | HERC2 | GO:0006974 | cellular response to DNA damage stimulus | 22508508 |
| Tgene | HERC2 | GO:0016567 | protein ubiquitination | 20304803 |
 Fusion gene breakpoints across WDR48 (5'-gene) Fusion gene breakpoints across WDR48 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
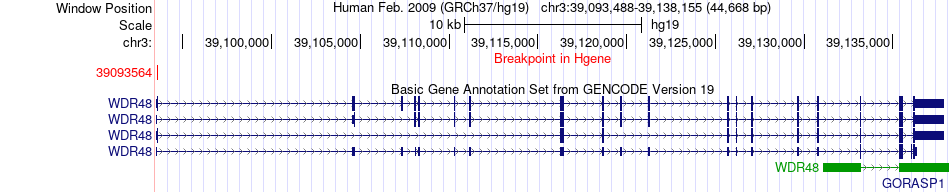 |
 Fusion gene breakpoints across HERC2 (3'-gene) Fusion gene breakpoints across HERC2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
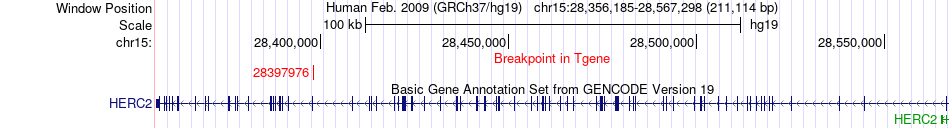 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BLCA | TCGA-XF-AAMG-01A | WDR48 | chr3 | 39093564 | + | HERC2 | chr15 | 28397976 | - |
| ChimerDB4 | BLCA | TCGA-XF-AAMG | WDR48 | chr3 | 39093564 | + | HERC2 | chr15 | 28397976 | - |
Top |
Fusion Gene ORF analysis for WDR48-HERC2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000302313 | ENST00000563945 | WDR48 | chr3 | 39093564 | + | HERC2 | chr15 | 28397976 | - |
| 5CDS-intron | ENST00000544962 | ENST00000563945 | WDR48 | chr3 | 39093564 | + | HERC2 | chr15 | 28397976 | - |
| 5UTR-3CDS | ENST00000396258 | ENST00000261609 | WDR48 | chr3 | 39093564 | + | HERC2 | chr15 | 28397976 | - |
| 5UTR-3CDS | ENST00000418020 | ENST00000261609 | WDR48 | chr3 | 39093564 | + | HERC2 | chr15 | 28397976 | - |
| 5UTR-intron | ENST00000396258 | ENST00000563945 | WDR48 | chr3 | 39093564 | + | HERC2 | chr15 | 28397976 | - |
| 5UTR-intron | ENST00000418020 | ENST00000563945 | WDR48 | chr3 | 39093564 | + | HERC2 | chr15 | 28397976 | - |
| In-frame | ENST00000302313 | ENST00000261609 | WDR48 | chr3 | 39093564 | + | HERC2 | chr15 | 28397976 | - |
| In-frame | ENST00000544962 | ENST00000261609 | WDR48 | chr3 | 39093564 | + | HERC2 | chr15 | 28397976 | - |
| intron-3CDS | ENST00000466405 | ENST00000261609 | WDR48 | chr3 | 39093564 | + | HERC2 | chr15 | 28397976 | - |
| intron-intron | ENST00000466405 | ENST00000563945 | WDR48 | chr3 | 39093564 | + | HERC2 | chr15 | 28397976 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000302313 | WDR48 | chr3 | 39093564 | + | ENST00000261609 | HERC2 | chr15 | 28397976 | - | 4558 | 76 | 28 | 3834 | 1268 |
| ENST00000544962 | WDR48 | chr3 | 39093564 | + | ENST00000261609 | HERC2 | chr15 | 28397976 | - | 4540 | 58 | 10 | 3816 | 1268 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000302313 | ENST00000261609 | WDR48 | chr3 | 39093564 | + | HERC2 | chr15 | 28397976 | - | 0.000716921 | 0.99928313 |
| ENST00000544962 | ENST00000261609 | WDR48 | chr3 | 39093564 | + | HERC2 | chr15 | 28397976 | - | 0.000721562 | 0.9992785 |
Top |
Fusion Genomic Features for WDR48-HERC2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
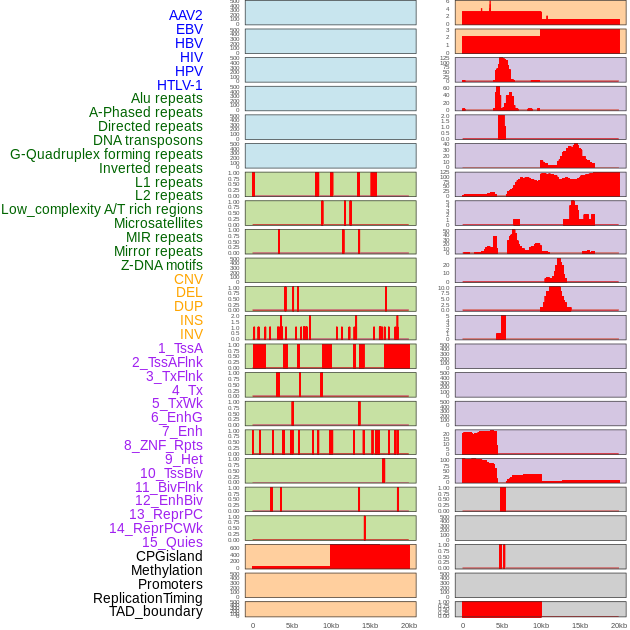 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for WDR48-HERC2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:39093564/chr15:28397976) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | HERC2 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: E3 ubiquitin-protein ligase that regulates ubiquitin-dependent retention of repair proteins on damaged chromosomes. Recruited to sites of DNA damage in response to ionizing radiation (IR) and facilitates the assembly of UBE2N and RNF8 promoting DNA damage-induced formation of 'Lys-63'-linked ubiquitin chains. Acts as a mediator of binding specificity between UBE2N and RNF8. Involved in the maintenance of RNF168 levels. E3 ubiquitin-protein ligase that promotes the ubiquitination and proteasomal degradation of XPA which influences the circadian oscillation of DNA excision repair activity. By controlling the steady-state expression of the IGF1R receptor, indirectly regulates the insulin-like growth factor receptor signaling pathway (PubMed:26692333). {ECO:0000269|PubMed:20023648, ECO:0000269|PubMed:20304803, ECO:0000269|PubMed:22508508, ECO:0000269|PubMed:26692333}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | HERC2 | chr3:39093564 | chr15:28397976 | ENST00000261609 | 68 | 93 | 4457_4794 | 3582 | 4835.0 | Domain | HECT | |
| Tgene | HERC2 | chr3:39093564 | chr15:28397976 | ENST00000261609 | 68 | 93 | 3951_4002 | 3582 | 4835.0 | Repeat | RCC1 3-1 | |
| Tgene | HERC2 | chr3:39093564 | chr15:28397976 | ENST00000261609 | 68 | 93 | 4004_4056 | 3582 | 4835.0 | Repeat | RCC1 3-2 | |
| Tgene | HERC2 | chr3:39093564 | chr15:28397976 | ENST00000261609 | 68 | 93 | 4058_4108 | 3582 | 4835.0 | Repeat | RCC1 3-3 | |
| Tgene | HERC2 | chr3:39093564 | chr15:28397976 | ENST00000261609 | 68 | 93 | 4110_4162 | 3582 | 4835.0 | Repeat | RCC1 3-4 | |
| Tgene | HERC2 | chr3:39093564 | chr15:28397976 | ENST00000261609 | 68 | 93 | 4164_4214 | 3582 | 4835.0 | Repeat | RCC1 3-5 | |
| Tgene | HERC2 | chr3:39093564 | chr15:28397976 | ENST00000261609 | 68 | 93 | 4216_4266 | 3582 | 4835.0 | Repeat | RCC1 3-6 | |
| Tgene | HERC2 | chr3:39093564 | chr15:28397976 | ENST00000261609 | 68 | 93 | 4268_4318 | 3582 | 4835.0 | Repeat | RCC1 3-7 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | WDR48 | chr3:39093564 | chr15:28397976 | ENST00000302313 | + | 1 | 19 | 115_154 | 16 | 678.0 | Repeat | Note=WD 3 |
| Hgene | WDR48 | chr3:39093564 | chr15:28397976 | ENST00000302313 | + | 1 | 19 | 166_205 | 16 | 678.0 | Repeat | Note=WD 4 |
| Hgene | WDR48 | chr3:39093564 | chr15:28397976 | ENST00000302313 | + | 1 | 19 | 208_247 | 16 | 678.0 | Repeat | Note=WD 5 |
| Hgene | WDR48 | chr3:39093564 | chr15:28397976 | ENST00000302313 | + | 1 | 19 | 250_289 | 16 | 678.0 | Repeat | Note=WD 6 |
| Hgene | WDR48 | chr3:39093564 | chr15:28397976 | ENST00000302313 | + | 1 | 19 | 28_67 | 16 | 678.0 | Repeat | Note=WD 1 |
| Hgene | WDR48 | chr3:39093564 | chr15:28397976 | ENST00000302313 | + | 1 | 19 | 292_334 | 16 | 678.0 | Repeat | Note=WD 7 |
| Hgene | WDR48 | chr3:39093564 | chr15:28397976 | ENST00000302313 | + | 1 | 19 | 358_397 | 16 | 678.0 | Repeat | Note=WD 8 |
| Hgene | WDR48 | chr3:39093564 | chr15:28397976 | ENST00000302313 | + | 1 | 19 | 73_112 | 16 | 678.0 | Repeat | Note=WD 2 |
| Hgene | WDR48 | chr3:39093564 | chr15:28397976 | ENST00000396258 | + | 1 | 18 | 115_154 | 0 | 596.0 | Repeat | Note=WD 3 |
| Hgene | WDR48 | chr3:39093564 | chr15:28397976 | ENST00000396258 | + | 1 | 18 | 166_205 | 0 | 596.0 | Repeat | Note=WD 4 |
| Hgene | WDR48 | chr3:39093564 | chr15:28397976 | ENST00000396258 | + | 1 | 18 | 208_247 | 0 | 596.0 | Repeat | Note=WD 5 |
| Hgene | WDR48 | chr3:39093564 | chr15:28397976 | ENST00000396258 | + | 1 | 18 | 250_289 | 0 | 596.0 | Repeat | Note=WD 6 |
| Hgene | WDR48 | chr3:39093564 | chr15:28397976 | ENST00000396258 | + | 1 | 18 | 28_67 | 0 | 596.0 | Repeat | Note=WD 1 |
| Hgene | WDR48 | chr3:39093564 | chr15:28397976 | ENST00000396258 | + | 1 | 18 | 292_334 | 0 | 596.0 | Repeat | Note=WD 7 |
| Hgene | WDR48 | chr3:39093564 | chr15:28397976 | ENST00000396258 | + | 1 | 18 | 358_397 | 0 | 596.0 | Repeat | Note=WD 8 |
| Hgene | WDR48 | chr3:39093564 | chr15:28397976 | ENST00000396258 | + | 1 | 18 | 73_112 | 0 | 596.0 | Repeat | Note=WD 2 |
| Hgene | WDR48 | chr3:39093564 | chr15:28397976 | ENST00000544962 | + | 1 | 11 | 115_154 | 16 | 403.0 | Repeat | Note=WD 3 |
| Hgene | WDR48 | chr3:39093564 | chr15:28397976 | ENST00000544962 | + | 1 | 11 | 166_205 | 16 | 403.0 | Repeat | Note=WD 4 |
| Hgene | WDR48 | chr3:39093564 | chr15:28397976 | ENST00000544962 | + | 1 | 11 | 208_247 | 16 | 403.0 | Repeat | Note=WD 5 |
| Hgene | WDR48 | chr3:39093564 | chr15:28397976 | ENST00000544962 | + | 1 | 11 | 250_289 | 16 | 403.0 | Repeat | Note=WD 6 |
| Hgene | WDR48 | chr3:39093564 | chr15:28397976 | ENST00000544962 | + | 1 | 11 | 28_67 | 16 | 403.0 | Repeat | Note=WD 1 |
| Hgene | WDR48 | chr3:39093564 | chr15:28397976 | ENST00000544962 | + | 1 | 11 | 292_334 | 16 | 403.0 | Repeat | Note=WD 7 |
| Hgene | WDR48 | chr3:39093564 | chr15:28397976 | ENST00000544962 | + | 1 | 11 | 358_397 | 16 | 403.0 | Repeat | Note=WD 8 |
| Hgene | WDR48 | chr3:39093564 | chr15:28397976 | ENST00000544962 | + | 1 | 11 | 73_112 | 16 | 403.0 | Repeat | Note=WD 2 |
| Tgene | HERC2 | chr3:39093564 | chr15:28397976 | ENST00000261609 | 68 | 93 | 948_980 | 3582 | 4835.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | HERC2 | chr3:39093564 | chr15:28397976 | ENST00000261609 | 68 | 93 | 1207_1283 | 3582 | 4835.0 | Domain | Cytochrome b5 heme-binding | |
| Tgene | HERC2 | chr3:39093564 | chr15:28397976 | ENST00000261609 | 68 | 93 | 1859_1932 | 3582 | 4835.0 | Domain | MIB/HERC2 | |
| Tgene | HERC2 | chr3:39093564 | chr15:28397976 | ENST00000261609 | 68 | 93 | 2759_2936 | 3582 | 4835.0 | Domain | DOC | |
| Tgene | HERC2 | chr3:39093564 | chr15:28397976 | ENST00000261609 | 68 | 93 | 2958_3009 | 3582 | 4835.0 | Repeat | RCC1 2-1 | |
| Tgene | HERC2 | chr3:39093564 | chr15:28397976 | ENST00000261609 | 68 | 93 | 3010_3064 | 3582 | 4835.0 | Repeat | RCC1 2-2 | |
| Tgene | HERC2 | chr3:39093564 | chr15:28397976 | ENST00000261609 | 68 | 93 | 3065_3116 | 3582 | 4835.0 | Repeat | RCC1 2-3 | |
| Tgene | HERC2 | chr3:39093564 | chr15:28397976 | ENST00000261609 | 68 | 93 | 3118_3168 | 3582 | 4835.0 | Repeat | RCC1 2-4 | |
| Tgene | HERC2 | chr3:39093564 | chr15:28397976 | ENST00000261609 | 68 | 93 | 3171_3222 | 3582 | 4835.0 | Repeat | RCC1 2-5 | |
| Tgene | HERC2 | chr3:39093564 | chr15:28397976 | ENST00000261609 | 68 | 93 | 3224_3274 | 3582 | 4835.0 | Repeat | RCC1 2-6 | |
| Tgene | HERC2 | chr3:39093564 | chr15:28397976 | ENST00000261609 | 68 | 93 | 3275_3326 | 3582 | 4835.0 | Repeat | RCC1 2-7 | |
| Tgene | HERC2 | chr3:39093564 | chr15:28397976 | ENST00000261609 | 68 | 93 | 415_461 | 3582 | 4835.0 | Repeat | RCC1 1-1 | |
| Tgene | HERC2 | chr3:39093564 | chr15:28397976 | ENST00000261609 | 68 | 93 | 462_512 | 3582 | 4835.0 | Repeat | RCC1 1-2 | |
| Tgene | HERC2 | chr3:39093564 | chr15:28397976 | ENST00000261609 | 68 | 93 | 513_568 | 3582 | 4835.0 | Repeat | RCC1 1-3 | |
| Tgene | HERC2 | chr3:39093564 | chr15:28397976 | ENST00000261609 | 68 | 93 | 569_620 | 3582 | 4835.0 | Repeat | RCC1 1-4 | |
| Tgene | HERC2 | chr3:39093564 | chr15:28397976 | ENST00000261609 | 68 | 93 | 623_674 | 3582 | 4835.0 | Repeat | RCC1 1-5 | |
| Tgene | HERC2 | chr3:39093564 | chr15:28397976 | ENST00000261609 | 68 | 93 | 675_726 | 3582 | 4835.0 | Repeat | RCC1 1-6 | |
| Tgene | HERC2 | chr3:39093564 | chr15:28397976 | ENST00000261609 | 68 | 93 | 728_778 | 3582 | 4835.0 | Repeat | RCC1 1-7 | |
| Tgene | HERC2 | chr3:39093564 | chr15:28397976 | ENST00000261609 | 68 | 93 | 2702_2749 | 3582 | 4835.0 | Zinc finger | ZZ-type |
Top |
Fusion Gene Sequence for WDR48-HERC2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >99012_99012_1_WDR48-HERC2_WDR48_chr3_39093564_ENST00000302313_HERC2_chr15_28397976_ENST00000261609_length(transcript)=4558nt_BP=76nt AAGTGACGTCGGAGTGTCAACATGCAAGATGGCGGCCCATCACCGGCAGAACACAGCAGGGCGGAGGAAAGTGCAGGTGGCAGATATGCT GTTGGAGCTCTGTGTCACCGAGTTGGAGGATGTGGCCACAGACTCGCAGAGCGGCCGCCTCTCTTCTCAGCCTGTGGTGGTGGAGAGTAG CCACCCTTACACCGACGACACCTCCACCAGTGGCACAGTGAAGATACCAGGTGCAGAAGGACTCAGGGTAGAATTTGACCGGCAGTGCTC CACAGAGAGGCGCCACGACCCTCTCACAGTCATGGACGGCGTCAACAGGATCGTCTCCGTGCGGTCAGGCCGAGAGTGGTCCGACTGGTC CAGCGAGCTGCGCATCCCAGGGGATGAGTTAAAGTGGAAGTTCATCAGCGATGGGTCTGTGAATGGCTGGGGCTGGCGCTTCACCGTCTA TCCCATCATGCCAGCTGCTGGCCCTAAAGAACTCCTCTCTGACCGCTGCGTCCTCTCCTGTCCATCCATGGACTTGGTGACGTGTCTGTT AGACTTCCGACTCAACCTTGCCTCTAACAGAAGCATCGTCCCTCGCCTTGCGGCCTCGCTGGCAGCTTGTGCACAGCTGAGTGCCCTAGC TGCCAGTCACAGAATGTGGGCCCTTCAGAGACTGAGGAAGCTGCTTACAACTGAATTTGGGCAGTCAATTAACATAAATAGGCTGCTTGG AGAAAATGATGGGGAAACAAGAGCTTTGAGTTTTACAGGTAGTGCTCTTGCTGCTTTGGTGAAAGGTCTTCCAGAAGCTTTGCAAAGGCA GTTTGAATATGAAGATCCTATTGTGAGGGGTGGCAAACAGCTGCTCCACAGCCCATTCTTTAAGGTACTGGTAGCTCTTGCTTGTGACCT GGAGCTGGACACTCTGCCTTGCTGTGCCGAGACGCACAAGTGGGCCTGGTTCCGGAGGTACTGCATGGCCTCCCGTGTTGCTGTGGCCCT TGACAAAAGAACACCGTTGCCCCGTCTGTTTCTTGATGAGGTGGCTAAGAAAATTCGTGAATTAATGGCAGACAGCGAAAACATGGATGT TCTGCATGAGAGCCATGACATTTTTAAAAGAGAGCAAGACGAACAACTTGTGCAGTGGATGAACAGGCGACCAGATGACTGGACTCTCTC TGCTGGTGGCAGTGGAACAATTTATGGATGGGGACATAATCACAGGGGCCAGCTCGGGGGCATTGAAGGCGCAAAAGTCAAAGTTCCCAC TCCCTGTGAAGCCCTTGCAACTCTCAGACCCGTGCAGTTAATCGGAGGGGAACAGACCCTCTTTGCTGTGACGGCTGATGGGAAGCTGTA TGCCACTGGGTATGGTGCAGGTGGCAGACTAGGCATTGGAGGGACAGAGTCGGTGTCCACCCCAACATTGCTTGAATCCATTCAGCATGT GTTTATTAAGAAAGTAGCTGTGAACTCTGGAGGAAAGCACTGCCTTGCCCTGTCTTCAGAAGGAGAAGTTTACTCTTGGGGTGAGGCAGA AGATGGGAAGTTGGGGCATGGCAACAGAAGTCCGTGTGACCGCCCTCGTGTCATCGAGTCTCTGAGAGGAATTGAAGTGGTCGATGTTGC TGCTGGCGGAGCCCACAGCGCCTGTGTCACAGCAGCCGGGGACCTCTACACATGGGGCAAAGGCCGCTACGGCCGGCTGGGGCACAGCGA CAGTGAGGACCAGCTGAAGCCGAAGCTGGTGGAGGCGCTGCAGGGCCACCGTGTGGTTGACATCGCCTGTGGCAGTGGAGATGCCCAGAC CCTCTGCCTCACAGATGACGACACTGTCTGGTCCTGGGGGGACGGGGACTACGGCAAGCTCGGCCGGGGAGGCAGCGATGGCTGTAAAGT GCCTATGAAGATTGATTCTCTTACTGGTCTTGGAGTAGTTAAAGTGGAATGCGGATCCCAGTTTTCTGTTGCCCTTACCAAATCTGGAGC TGTTTATACCTGGGGCAAAGGCGATTATCACAGGTTGGGCCATGGATCAGATGACCATGTTCGAAGGCCTCGGCAGGTCCAAGGGTTGCA GGGGAAGAAAGTCATCGCCATCGCCACTGGCTCCCTGCACTGTGTGTGCTGCACAGAGGATGGTGAGGTTTATACATGGGGCGACAATGA TGAGGGACAACTGGGAGACGGAACCACCAATGCCATCCAGAGGCCTCGGTTGGTAGCTGCCCTTCAGGGTAAGAAGGTCAACCGTGTGGC CTGTGGCTCAGCACATACCCTCGCCTGGTCGACCAGCAAGCCCGCCAGTGCTGGCAAACTCCCTGCACAGGTCCCCATGGAGTACAATCA CCTGCAGGAGATCCCCATCATTGCGCTGAGGAACCGTCTGCTGCTGCTGCACCACCTCTCCGAGCTCTTCTGCCCCTGCATCCCCATGTT CGACCTGGAAGGCTCGCTCGACGAAACTGGACTCGGGCCTTCTGTTGGGTTCGACACTCTCCGAGGAATTCTGATATCCCAGGGAAAGGA GGCGGCTTTCCGGAAAGTAGTACAAGCAACTATGGTACGCGATCGTCAGCATGGCCCCGTCGTGGAGCTGAACCGCATCCAGGTCAAACG ATCAAGGAGCAAAGGCGGGCTGGCCGGCCCCGACGGCACCAAGTCTGTCTTTGGGCAGATGTGTGCTAAGATGAGCTCGTTTGGTCCCGA CAGCCTCCTCCTTCCTCACCGTGTCTGGAAAGTCAAGTTTGTGGGTGAATCTGTGGATGACTGTGGGGGCGGCTACAGCGAGTCCATAGC TGAGATCTGTGAGGAGCTGCAGAACGGACTCACGCCCCTGCTGATCGTGACACCCAACGGGAGGGATGAGTCTGGGGCCAACCGAGACTG CTACCTGCTCAGCCCGGCCGCCAGAGCACCCGTGCACAGCAGCATGTTCCGCTTCCTGGGTGTGTTGCTGGGCATTGCCATCCGAACCGG GAGTCCCCTGAGCCTCAACCTTGCCGAGCCTGTCTGGAAGCAGCTGGCTGGGATGAGCCTCACCATCGCGGACCTCAGTGAGGTTGATAA GGATTTTATTCCTGGACTCATGTACATCCGAGACAATGAAGCCACCTCAGAGGAGTTTGAAGCCATGAGCCTGCCCTTCACAGTGCCAAG TGCCAGTGGCCAGGACATTCAGTTGAGCTCCAAGCACACACACATCACCCTGGACAACCGCGCGGAGTACGTGCGGCTGGCGATAAACTA TAGACTCCATGAATTTGATGAGCAGGTGGCTGCTGTTCGGGAAGGAATGGCCCGCGTTGTGCCTGTTCCCCTCCTCTCTCTGTTCACCGG CTACGAACTGGAGACGATGGTGTGTGGCAGCCCTGACATCCCGCTGCACCTTCTCAAGTCGGTGGCCACCTATAAAGGCATCGAGCCTTC CGCATCGCTGATCCAGTGGTTCTGGGAGGTGATGGAGTCCTTCTCCAACACAGAGCGCTCTCTTTTCCTTCGCTTCGTCTGGGGCCGGAC GAGGCTGCCCAGGACCATCGCCGACTTCCGGGGCCGAGACTTCGTCATCCAGGTGTTGGATAAATACAACCCTCCAGACCACTTCCTCCC TGAGTCCTACACCTGTTTCTTCTTGCTGAAGCTGCCCAGGTATTCCTGCAAGCAGGTGCTGGAGGAGAAGCTCAAGTACGCCATCCACTT CTGCAAGTCCATAGACACAGATGACTACGCTCGCATCGCACTTACAGGAGAGCCAGCCGCCGACGACAGCAGCGACGATTCAGATAACGA GGATGTCGACTCCTTTGCTTCGGACTCTACACAAGATTATTTAACAGGACACTAAGATGGGGAAACGTCCTCGTGAGATGAGAGCCTGAG CCAGGCAGCAGAGCGCTCGCTGCTGTGTAGACTGTAGGCTGCCTGGTGTGTCTGATGAGAAGCGTCCGTCCTCGAGCCAGGCGGGAGGAG GGAGTGGAGAGACTGACTGGCCGTGATGGGAATGACAGTGAGAAGGTCCGCCTGTGCGCGTGGAACACTGTGGACGCTCGACTTCCAAGG GTCTTCTCACCCGTAATGCTGCATTACATGTAGGACTGTGTTTACTAAAGTGTGTAAATGTTTATATAAATACCAAATTGCAGCATCCCC AAAATGAATAAAGCCTTTTTACTTGTGGGTGCAATCGATTTTTTTTCTTTCTCCTTTCTTTCAAGTGTCGTGAGTCGTCTTGATTGTATA TTGGAAATAACTGTGTAACAAATCGTATTATAAATATTTCAATTAATTTTACTCTGAATTTGTTTATTAAAAGACTTTTGAACATGAAAT GATTAGTATTACTTGAATGCATCCAGAGGATATTTAAACCAAAATGAAAAACCAGAAGGCCATTTGGTGTCCCCCCTCCCAGGTGTCCCC TTGTAGCATATGCATTATGTCATCTGAATTGAGGCCTTTCTGTGAACAGCATCATAACTTCTATCATGGAAAGTGTACTATATATAATGT >99012_99012_1_WDR48-HERC2_WDR48_chr3_39093564_ENST00000302313_HERC2_chr15_28397976_ENST00000261609_length(amino acids)=1268AA_BP=599 MAAHHRQNTAGRRKVQVADMLLELCVTELEDVATDSQSGRLSSQPVVVESSHPYTDDTSTSGTVKIPGAEGLRVEFDRQCSTERRHDPLT VMDGVNRIVSVRSGREWSDWSSELRIPGDELKWKFISDGSVNGWGWRFTVYPIMPAAGPKELLSDRCVLSCPSMDLVTCLLDFRLNLASN RSIVPRLAASLAACAQLSALAASHRMWALQRLRKLLTTEFGQSININRLLGENDGETRALSFTGSALAALVKGLPEALQRQFEYEDPIVR GGKQLLHSPFFKVLVALACDLELDTLPCCAETHKWAWFRRYCMASRVAVALDKRTPLPRLFLDEVAKKIRELMADSENMDVLHESHDIFK REQDEQLVQWMNRRPDDWTLSAGGSGTIYGWGHNHRGQLGGIEGAKVKVPTPCEALATLRPVQLIGGEQTLFAVTADGKLYATGYGAGGR LGIGGTESVSTPTLLESIQHVFIKKVAVNSGGKHCLALSSEGEVYSWGEAEDGKLGHGNRSPCDRPRVIESLRGIEVVDVAAGGAHSACV TAAGDLYTWGKGRYGRLGHSDSEDQLKPKLVEALQGHRVVDIACGSGDAQTLCLTDDDTVWSWGDGDYGKLGRGGSDGCKVPMKIDSLTG LGVVKVECGSQFSVALTKSGAVYTWGKGDYHRLGHGSDDHVRRPRQVQGLQGKKVIAIATGSLHCVCCTEDGEVYTWGDNDEGQLGDGTT NAIQRPRLVAALQGKKVNRVACGSAHTLAWSTSKPASAGKLPAQVPMEYNHLQEIPIIALRNRLLLLHHLSELFCPCIPMFDLEGSLDET GLGPSVGFDTLRGILISQGKEAAFRKVVQATMVRDRQHGPVVELNRIQVKRSRSKGGLAGPDGTKSVFGQMCAKMSSFGPDSLLLPHRVW KVKFVGESVDDCGGGYSESIAEICEELQNGLTPLLIVTPNGRDESGANRDCYLLSPAARAPVHSSMFRFLGVLLGIAIRTGSPLSLNLAE PVWKQLAGMSLTIADLSEVDKDFIPGLMYIRDNEATSEEFEAMSLPFTVPSASGQDIQLSSKHTHITLDNRAEYVRLAINYRLHEFDEQV AAVREGMARVVPVPLLSLFTGYELETMVCGSPDIPLHLLKSVATYKGIEPSASLIQWFWEVMESFSNTERSLFLRFVWGRTRLPRTIADF RGRDFVIQVLDKYNPPDHFLPESYTCFFLLKLPRYSCKQVLEEKLKYAIHFCKSIDTDDYARIALTGEPAADDSSDDSDNEDVDSFASDS -------------------------------------------------------------- >99012_99012_2_WDR48-HERC2_WDR48_chr3_39093564_ENST00000544962_HERC2_chr15_28397976_ENST00000261609_length(transcript)=4540nt_BP=58nt AACATGCAAGATGGCGGCCCATCACCGGCAGAACACAGCAGGGCGGAGGAAAGTGCAGGTGGCAGATATGCTGTTGGAGCTCTGTGTCAC CGAGTTGGAGGATGTGGCCACAGACTCGCAGAGCGGCCGCCTCTCTTCTCAGCCTGTGGTGGTGGAGAGTAGCCACCCTTACACCGACGA CACCTCCACCAGTGGCACAGTGAAGATACCAGGTGCAGAAGGACTCAGGGTAGAATTTGACCGGCAGTGCTCCACAGAGAGGCGCCACGA CCCTCTCACAGTCATGGACGGCGTCAACAGGATCGTCTCCGTGCGGTCAGGCCGAGAGTGGTCCGACTGGTCCAGCGAGCTGCGCATCCC AGGGGATGAGTTAAAGTGGAAGTTCATCAGCGATGGGTCTGTGAATGGCTGGGGCTGGCGCTTCACCGTCTATCCCATCATGCCAGCTGC TGGCCCTAAAGAACTCCTCTCTGACCGCTGCGTCCTCTCCTGTCCATCCATGGACTTGGTGACGTGTCTGTTAGACTTCCGACTCAACCT TGCCTCTAACAGAAGCATCGTCCCTCGCCTTGCGGCCTCGCTGGCAGCTTGTGCACAGCTGAGTGCCCTAGCTGCCAGTCACAGAATGTG GGCCCTTCAGAGACTGAGGAAGCTGCTTACAACTGAATTTGGGCAGTCAATTAACATAAATAGGCTGCTTGGAGAAAATGATGGGGAAAC AAGAGCTTTGAGTTTTACAGGTAGTGCTCTTGCTGCTTTGGTGAAAGGTCTTCCAGAAGCTTTGCAAAGGCAGTTTGAATATGAAGATCC TATTGTGAGGGGTGGCAAACAGCTGCTCCACAGCCCATTCTTTAAGGTACTGGTAGCTCTTGCTTGTGACCTGGAGCTGGACACTCTGCC TTGCTGTGCCGAGACGCACAAGTGGGCCTGGTTCCGGAGGTACTGCATGGCCTCCCGTGTTGCTGTGGCCCTTGACAAAAGAACACCGTT GCCCCGTCTGTTTCTTGATGAGGTGGCTAAGAAAATTCGTGAATTAATGGCAGACAGCGAAAACATGGATGTTCTGCATGAGAGCCATGA CATTTTTAAAAGAGAGCAAGACGAACAACTTGTGCAGTGGATGAACAGGCGACCAGATGACTGGACTCTCTCTGCTGGTGGCAGTGGAAC AATTTATGGATGGGGACATAATCACAGGGGCCAGCTCGGGGGCATTGAAGGCGCAAAAGTCAAAGTTCCCACTCCCTGTGAAGCCCTTGC AACTCTCAGACCCGTGCAGTTAATCGGAGGGGAACAGACCCTCTTTGCTGTGACGGCTGATGGGAAGCTGTATGCCACTGGGTATGGTGC AGGTGGCAGACTAGGCATTGGAGGGACAGAGTCGGTGTCCACCCCAACATTGCTTGAATCCATTCAGCATGTGTTTATTAAGAAAGTAGC TGTGAACTCTGGAGGAAAGCACTGCCTTGCCCTGTCTTCAGAAGGAGAAGTTTACTCTTGGGGTGAGGCAGAAGATGGGAAGTTGGGGCA TGGCAACAGAAGTCCGTGTGACCGCCCTCGTGTCATCGAGTCTCTGAGAGGAATTGAAGTGGTCGATGTTGCTGCTGGCGGAGCCCACAG CGCCTGTGTCACAGCAGCCGGGGACCTCTACACATGGGGCAAAGGCCGCTACGGCCGGCTGGGGCACAGCGACAGTGAGGACCAGCTGAA GCCGAAGCTGGTGGAGGCGCTGCAGGGCCACCGTGTGGTTGACATCGCCTGTGGCAGTGGAGATGCCCAGACCCTCTGCCTCACAGATGA CGACACTGTCTGGTCCTGGGGGGACGGGGACTACGGCAAGCTCGGCCGGGGAGGCAGCGATGGCTGTAAAGTGCCTATGAAGATTGATTC TCTTACTGGTCTTGGAGTAGTTAAAGTGGAATGCGGATCCCAGTTTTCTGTTGCCCTTACCAAATCTGGAGCTGTTTATACCTGGGGCAA AGGCGATTATCACAGGTTGGGCCATGGATCAGATGACCATGTTCGAAGGCCTCGGCAGGTCCAAGGGTTGCAGGGGAAGAAAGTCATCGC CATCGCCACTGGCTCCCTGCACTGTGTGTGCTGCACAGAGGATGGTGAGGTTTATACATGGGGCGACAATGATGAGGGACAACTGGGAGA CGGAACCACCAATGCCATCCAGAGGCCTCGGTTGGTAGCTGCCCTTCAGGGTAAGAAGGTCAACCGTGTGGCCTGTGGCTCAGCACATAC CCTCGCCTGGTCGACCAGCAAGCCCGCCAGTGCTGGCAAACTCCCTGCACAGGTCCCCATGGAGTACAATCACCTGCAGGAGATCCCCAT CATTGCGCTGAGGAACCGTCTGCTGCTGCTGCACCACCTCTCCGAGCTCTTCTGCCCCTGCATCCCCATGTTCGACCTGGAAGGCTCGCT CGACGAAACTGGACTCGGGCCTTCTGTTGGGTTCGACACTCTCCGAGGAATTCTGATATCCCAGGGAAAGGAGGCGGCTTTCCGGAAAGT AGTACAAGCAACTATGGTACGCGATCGTCAGCATGGCCCCGTCGTGGAGCTGAACCGCATCCAGGTCAAACGATCAAGGAGCAAAGGCGG GCTGGCCGGCCCCGACGGCACCAAGTCTGTCTTTGGGCAGATGTGTGCTAAGATGAGCTCGTTTGGTCCCGACAGCCTCCTCCTTCCTCA CCGTGTCTGGAAAGTCAAGTTTGTGGGTGAATCTGTGGATGACTGTGGGGGCGGCTACAGCGAGTCCATAGCTGAGATCTGTGAGGAGCT GCAGAACGGACTCACGCCCCTGCTGATCGTGACACCCAACGGGAGGGATGAGTCTGGGGCCAACCGAGACTGCTACCTGCTCAGCCCGGC CGCCAGAGCACCCGTGCACAGCAGCATGTTCCGCTTCCTGGGTGTGTTGCTGGGCATTGCCATCCGAACCGGGAGTCCCCTGAGCCTCAA CCTTGCCGAGCCTGTCTGGAAGCAGCTGGCTGGGATGAGCCTCACCATCGCGGACCTCAGTGAGGTTGATAAGGATTTTATTCCTGGACT CATGTACATCCGAGACAATGAAGCCACCTCAGAGGAGTTTGAAGCCATGAGCCTGCCCTTCACAGTGCCAAGTGCCAGTGGCCAGGACAT TCAGTTGAGCTCCAAGCACACACACATCACCCTGGACAACCGCGCGGAGTACGTGCGGCTGGCGATAAACTATAGACTCCATGAATTTGA TGAGCAGGTGGCTGCTGTTCGGGAAGGAATGGCCCGCGTTGTGCCTGTTCCCCTCCTCTCTCTGTTCACCGGCTACGAACTGGAGACGAT GGTGTGTGGCAGCCCTGACATCCCGCTGCACCTTCTCAAGTCGGTGGCCACCTATAAAGGCATCGAGCCTTCCGCATCGCTGATCCAGTG GTTCTGGGAGGTGATGGAGTCCTTCTCCAACACAGAGCGCTCTCTTTTCCTTCGCTTCGTCTGGGGCCGGACGAGGCTGCCCAGGACCAT CGCCGACTTCCGGGGCCGAGACTTCGTCATCCAGGTGTTGGATAAATACAACCCTCCAGACCACTTCCTCCCTGAGTCCTACACCTGTTT CTTCTTGCTGAAGCTGCCCAGGTATTCCTGCAAGCAGGTGCTGGAGGAGAAGCTCAAGTACGCCATCCACTTCTGCAAGTCCATAGACAC AGATGACTACGCTCGCATCGCACTTACAGGAGAGCCAGCCGCCGACGACAGCAGCGACGATTCAGATAACGAGGATGTCGACTCCTTTGC TTCGGACTCTACACAAGATTATTTAACAGGACACTAAGATGGGGAAACGTCCTCGTGAGATGAGAGCCTGAGCCAGGCAGCAGAGCGCTC GCTGCTGTGTAGACTGTAGGCTGCCTGGTGTGTCTGATGAGAAGCGTCCGTCCTCGAGCCAGGCGGGAGGAGGGAGTGGAGAGACTGACT GGCCGTGATGGGAATGACAGTGAGAAGGTCCGCCTGTGCGCGTGGAACACTGTGGACGCTCGACTTCCAAGGGTCTTCTCACCCGTAATG CTGCATTACATGTAGGACTGTGTTTACTAAAGTGTGTAAATGTTTATATAAATACCAAATTGCAGCATCCCCAAAATGAATAAAGCCTTT TTACTTGTGGGTGCAATCGATTTTTTTTCTTTCTCCTTTCTTTCAAGTGTCGTGAGTCGTCTTGATTGTATATTGGAAATAACTGTGTAA CAAATCGTATTATAAATATTTCAATTAATTTTACTCTGAATTTGTTTATTAAAAGACTTTTGAACATGAAATGATTAGTATTACTTGAAT GCATCCAGAGGATATTTAAACCAAAATGAAAAACCAGAAGGCCATTTGGTGTCCCCCCTCCCAGGTGTCCCCTTGTAGCATATGCATTAT GTCATCTGAATTGAGGCCTTTCTGTGAACAGCATCATAACTTCTATCATGGAAAGTGTACTATATATAATGTTTGTGTCATGTATATGCC >99012_99012_2_WDR48-HERC2_WDR48_chr3_39093564_ENST00000544962_HERC2_chr15_28397976_ENST00000261609_length(amino acids)=1268AA_BP=599 MAAHHRQNTAGRRKVQVADMLLELCVTELEDVATDSQSGRLSSQPVVVESSHPYTDDTSTSGTVKIPGAEGLRVEFDRQCSTERRHDPLT VMDGVNRIVSVRSGREWSDWSSELRIPGDELKWKFISDGSVNGWGWRFTVYPIMPAAGPKELLSDRCVLSCPSMDLVTCLLDFRLNLASN RSIVPRLAASLAACAQLSALAASHRMWALQRLRKLLTTEFGQSININRLLGENDGETRALSFTGSALAALVKGLPEALQRQFEYEDPIVR GGKQLLHSPFFKVLVALACDLELDTLPCCAETHKWAWFRRYCMASRVAVALDKRTPLPRLFLDEVAKKIRELMADSENMDVLHESHDIFK REQDEQLVQWMNRRPDDWTLSAGGSGTIYGWGHNHRGQLGGIEGAKVKVPTPCEALATLRPVQLIGGEQTLFAVTADGKLYATGYGAGGR LGIGGTESVSTPTLLESIQHVFIKKVAVNSGGKHCLALSSEGEVYSWGEAEDGKLGHGNRSPCDRPRVIESLRGIEVVDVAAGGAHSACV TAAGDLYTWGKGRYGRLGHSDSEDQLKPKLVEALQGHRVVDIACGSGDAQTLCLTDDDTVWSWGDGDYGKLGRGGSDGCKVPMKIDSLTG LGVVKVECGSQFSVALTKSGAVYTWGKGDYHRLGHGSDDHVRRPRQVQGLQGKKVIAIATGSLHCVCCTEDGEVYTWGDNDEGQLGDGTT NAIQRPRLVAALQGKKVNRVACGSAHTLAWSTSKPASAGKLPAQVPMEYNHLQEIPIIALRNRLLLLHHLSELFCPCIPMFDLEGSLDET GLGPSVGFDTLRGILISQGKEAAFRKVVQATMVRDRQHGPVVELNRIQVKRSRSKGGLAGPDGTKSVFGQMCAKMSSFGPDSLLLPHRVW KVKFVGESVDDCGGGYSESIAEICEELQNGLTPLLIVTPNGRDESGANRDCYLLSPAARAPVHSSMFRFLGVLLGIAIRTGSPLSLNLAE PVWKQLAGMSLTIADLSEVDKDFIPGLMYIRDNEATSEEFEAMSLPFTVPSASGQDIQLSSKHTHITLDNRAEYVRLAINYRLHEFDEQV AAVREGMARVVPVPLLSLFTGYELETMVCGSPDIPLHLLKSVATYKGIEPSASLIQWFWEVMESFSNTERSLFLRFVWGRTRLPRTIADF RGRDFVIQVLDKYNPPDHFLPESYTCFFLLKLPRYSCKQVLEEKLKYAIHFCKSIDTDDYARIALTGEPAADDSSDDSDNEDVDSFASDS -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for WDR48-HERC2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for WDR48-HERC2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for WDR48-HERC2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
