
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:BMPR2-ATG4B (FusionGDB2 ID:9922) |
Fusion Gene Summary for BMPR2-ATG4B |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: BMPR2-ATG4B | Fusion gene ID: 9922 | Hgene | Tgene | Gene symbol | BMPR2 | ATG4B | Gene ID | 659 | 23192 |
| Gene name | bone morphogenetic protein receptor type 2 | autophagy related 4B cysteine peptidase | |
| Synonyms | BMPR-II|BMPR3|BMR2|BRK-3|POVD1|PPH1|T-ALK | APG4B|AUTL1 | |
| Cytomap | 2q33.1-q33.2 | 2q37.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | bone morphogenetic protein receptor type-2BMP type II receptorBMP type-2 receptorbone morphogenetic protein receptor type IIbone morphogenetic protein receptor, type II (serine/threonine kinase)type II activin receptor-like kinasetype II receptor fo | cysteine protease ATG4BAPG4 autophagy 4 homolog BATG4 autophagy related 4 homolog BAUT-like 1 cysteine endopeptidaseautophagin-1autophagy-related cysteine endopeptidase 1autophagy-related protein 4 homolog BhAPG4B | |
| Modification date | 20200313 | 20200329 | |
| UniProtAcc | Q13873 | Q9Y4P1 | |
| Ensembl transtripts involved in fusion gene | ENST00000479069, ENST00000374574, ENST00000374580, | ENST00000405546, ENST00000491867, ENST00000396411, ENST00000402096, ENST00000404914, ENST00000474739, | |
| Fusion gene scores | * DoF score | 20 X 12 X 9=2160 | 4 X 4 X 3=48 |
| # samples | 21 | 5 | |
| ** MAII score | log2(21/2160*10)=-3.36257007938471 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(5/48*10)=0.0588936890535686 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: BMPR2 [Title/Abstract] AND ATG4B [Title/Abstract] AND fusion [Title/Abstract] | ||
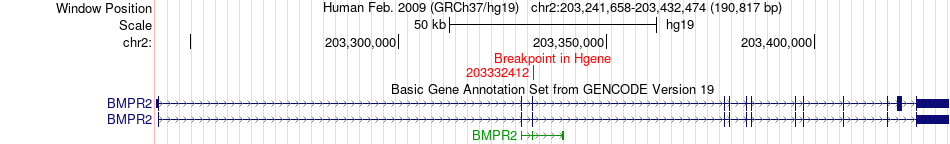
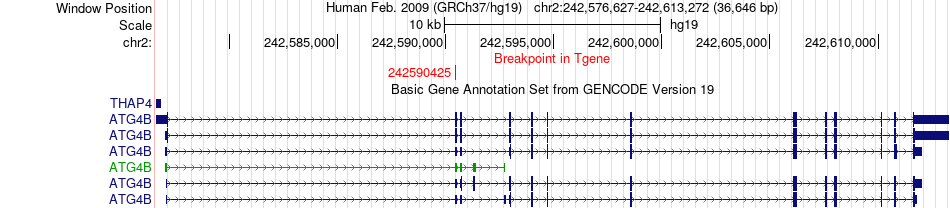
| Most frequent breakpoint | BMPR2(203332412)-ATG4B(242590425), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | BMPR2 | GO:0007178 | transmembrane receptor protein serine/threonine kinase signaling pathway | 12045205 |
| Hgene | BMPR2 | GO:0010634 | positive regulation of epithelial cell migration | 12819188 |
| Hgene | BMPR2 | GO:0030308 | negative regulation of cell growth | 12819188 |
| Hgene | BMPR2 | GO:0030509 | BMP signaling pathway | 18436533 |
| Tgene | ATG4B | GO:0006508 | proteolysis | 15169837|18387192 |
| Tgene | ATG4B | GO:0006914 | autophagy | 18387192 |
| Tgene | ATG4B | GO:0051697 | protein delipidation | 25327288 |
 Fusion gene breakpoints across BMPR2 (5'-gene) Fusion gene breakpoints across BMPR2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene breakpoints across ATG4B (3'-gene) Fusion gene breakpoints across ATG4B (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUSC | TCGA-63-6202-01A | BMPR2 | chr2 | 203332412 | - | ATG4B | chr2 | 242590425 | + |
Top |
Fusion Gene ORF analysis for BMPR2-ATG4B |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000479069 | ENST00000405546 | BMPR2 | chr2 | 203332412 | - | ATG4B | chr2 | 242590425 | + |
| 3UTR-3UTR | ENST00000479069 | ENST00000491867 | BMPR2 | chr2 | 203332412 | - | ATG4B | chr2 | 242590425 | + |
| 3UTR-5UTR | ENST00000479069 | ENST00000396411 | BMPR2 | chr2 | 203332412 | - | ATG4B | chr2 | 242590425 | + |
| 3UTR-5UTR | ENST00000479069 | ENST00000402096 | BMPR2 | chr2 | 203332412 | - | ATG4B | chr2 | 242590425 | + |
| 3UTR-5UTR | ENST00000479069 | ENST00000404914 | BMPR2 | chr2 | 203332412 | - | ATG4B | chr2 | 242590425 | + |
| 3UTR-5UTR | ENST00000479069 | ENST00000474739 | BMPR2 | chr2 | 203332412 | - | ATG4B | chr2 | 242590425 | + |
| 5CDS-3UTR | ENST00000374574 | ENST00000491867 | BMPR2 | chr2 | 203332412 | - | ATG4B | chr2 | 242590425 | + |
| 5CDS-3UTR | ENST00000374580 | ENST00000491867 | BMPR2 | chr2 | 203332412 | - | ATG4B | chr2 | 242590425 | + |
| 5CDS-5UTR | ENST00000374574 | ENST00000396411 | BMPR2 | chr2 | 203332412 | - | ATG4B | chr2 | 242590425 | + |
| 5CDS-5UTR | ENST00000374574 | ENST00000402096 | BMPR2 | chr2 | 203332412 | - | ATG4B | chr2 | 242590425 | + |
| 5CDS-5UTR | ENST00000374574 | ENST00000404914 | BMPR2 | chr2 | 203332412 | - | ATG4B | chr2 | 242590425 | + |
| 5CDS-5UTR | ENST00000374574 | ENST00000474739 | BMPR2 | chr2 | 203332412 | - | ATG4B | chr2 | 242590425 | + |
| 5CDS-5UTR | ENST00000374580 | ENST00000396411 | BMPR2 | chr2 | 203332412 | - | ATG4B | chr2 | 242590425 | + |
| 5CDS-5UTR | ENST00000374580 | ENST00000402096 | BMPR2 | chr2 | 203332412 | - | ATG4B | chr2 | 242590425 | + |
| 5CDS-5UTR | ENST00000374580 | ENST00000404914 | BMPR2 | chr2 | 203332412 | - | ATG4B | chr2 | 242590425 | + |
| 5CDS-5UTR | ENST00000374580 | ENST00000474739 | BMPR2 | chr2 | 203332412 | - | ATG4B | chr2 | 242590425 | + |
| In-frame | ENST00000374574 | ENST00000405546 | BMPR2 | chr2 | 203332412 | - | ATG4B | chr2 | 242590425 | + |
| In-frame | ENST00000374580 | ENST00000405546 | BMPR2 | chr2 | 203332412 | - | ATG4B | chr2 | 242590425 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000374580 | BMPR2 | chr2 | 203332412 | - | ENST00000405546 | ATG4B | chr2 | 242590425 | + | 3739 | 957 | 524 | 2089 | 521 |
| ENST00000374574 | BMPR2 | chr2 | 203332412 | - | ENST00000405546 | ATG4B | chr2 | 242590425 | + | 3241 | 459 | 26 | 1591 | 521 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000374580 | ENST00000405546 | BMPR2 | chr2 | 203332412 | - | ATG4B | chr2 | 242590425 | + | 0.011874816 | 0.9881252 |
| ENST00000374574 | ENST00000405546 | BMPR2 | chr2 | 203332412 | - | ATG4B | chr2 | 242590425 | + | 0.010126697 | 0.9898733 |
Top |
Fusion Genomic Features for BMPR2-ATG4B |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for BMPR2-ATG4B |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr2:203332412/chr2:242590425) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| BMPR2 | ATG4B |
| FUNCTION: On ligand binding, forms a receptor complex consisting of two type II and two type I transmembrane serine/threonine kinases. Type II receptors phosphorylate and activate type I receptors which autophosphorylate, then bind and activate SMAD transcriptional regulators. Binds to BMP7, BMP2 and, less efficiently, BMP4. Binding is weak but enhanced by the presence of type I receptors for BMPs. Mediates induction of adipogenesis by GDF6. {ECO:0000250|UniProtKB:O35607}. | FUNCTION: Cysteine protease required for the cytoplasm to vacuole transport (Cvt) and autophagy. Cleaves the C-terminal amino acid of ATG8 family proteins MAP1LC3, GABARAPL1, GABARAPL2 and GABARAP, to reveal a C-terminal glycine. Exposure of the glycine at the C-terminus is essential for ATG8 proteins conjugation to phosphatidylethanolamine (PE) and insertion to membranes, which is necessary for autophagy. Has also an activity of delipidating enzyme for the PE-conjugated forms. {ECO:0000269|PubMed:15169837, ECO:0000269|PubMed:15187094, ECO:0000269|PubMed:17347651, ECO:0000269|PubMed:19322194, ECO:0000269|PubMed:21177865, ECO:0000269|PubMed:22302004}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | BMPR2 | chr2:203332412 | chr2:242590425 | ENST00000374580 | - | 3 | 13 | 547_550 | 139 | 1039.0 | Compositional bias | Note=Poly-Ser |
| Hgene | BMPR2 | chr2:203332412 | chr2:242590425 | ENST00000374580 | - | 3 | 13 | 610_618 | 139 | 1039.0 | Compositional bias | Note=Poly-Thr |
| Hgene | BMPR2 | chr2:203332412 | chr2:242590425 | ENST00000374580 | - | 3 | 13 | 901_908 | 139 | 1039.0 | Compositional bias | Note=Poly-Asn |
| Hgene | BMPR2 | chr2:203332412 | chr2:242590425 | ENST00000374580 | - | 3 | 13 | 203_504 | 139 | 1039.0 | Domain | Protein kinase |
| Hgene | BMPR2 | chr2:203332412 | chr2:242590425 | ENST00000374580 | - | 3 | 13 | 209_217 | 139 | 1039.0 | Nucleotide binding | ATP |
| Hgene | BMPR2 | chr2:203332412 | chr2:242590425 | ENST00000374580 | - | 3 | 13 | 280_282 | 139 | 1039.0 | Nucleotide binding | ATP |
| Hgene | BMPR2 | chr2:203332412 | chr2:242590425 | ENST00000374580 | - | 3 | 13 | 337_338 | 139 | 1039.0 | Nucleotide binding | ATP |
| Hgene | BMPR2 | chr2:203332412 | chr2:242590425 | ENST00000374580 | - | 3 | 13 | 172_1038 | 139 | 1039.0 | Topological domain | Cytoplasmic |
| Hgene | BMPR2 | chr2:203332412 | chr2:242590425 | ENST00000374580 | - | 3 | 13 | 27_150 | 139 | 1039.0 | Topological domain | Extracellular |
| Hgene | BMPR2 | chr2:203332412 | chr2:242590425 | ENST00000374580 | - | 3 | 13 | 151_171 | 139 | 1039.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for BMPR2-ATG4B |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >9922_9922_1_BMPR2-ATG4B_BMPR2_chr2_203332412_ENST00000374574_ATG4B_chr2_242590425_ENST00000405546_length(transcript)=3241nt_BP=459nt ATTTCTTTTCTTTGCCCTCCTGATTCTTGGCTGGCCCAGGGATGACTTCCTCGCTGCAGCGGCCCTGGCGGGTGCCCTGGCTACCATGGA CCATCCTGCTGGTCAGCACTGCGGCTGCTTCGCAGAATCAAGAACGGCTATGTGCGTTTAAAGATCCGTATCAGCAAGACCTTGGGATAG GTGAGAGTAGAATCTCTCATGAAAATGGGACAATATTATGCTCGAAAGGTAGCACCTGCTATGGCCTTTGGGAGAAATCAAAAGGGGACA TAAATCTTGTAAAACAAGGATGTTGGTCTCACATTGGAGATCCCCAAGAGTGTCACTATGAAGAATGTGTAGTAACTACCACTCCTCCCT CAATTCAGAATGGAACATACCGTTTCTGCTGTTGTAGCACAGATTTATGTAATGTCAACTTTACTGAGAATTTTCCACCTCCTGACACAA CACCACTCACTACTCTGACCTACGACACTCTCCGGTTTGCTGAGTTTGAAGATTTTCCTGAGACCTCAGAGCCCGTTTGGATACTGGGTA GAAAATACAGCATTTTCACAGAAAAGGACGAGATCTTGTCTGATGTGGCATCTAGACTTTGGTTTACATACAGGAAAAACTTTCCAGCCA TTGGGGGGACAGGCCCCACCTCGGACACAGGCTGGGGCTGCATGCTGCGGTGTGGACAGATGATCTTTGCCCAAGCCCTGGTGTGCCGGC ACCTAGGCCGAGATTGGAGGTGGACACAAAGGAAGAGGCAGCCAGACAGCTACTTCAGCGTCCTCAACGCATTCATCGACAGGAAGGACA GTTACTACTCCATTCACCAGATAGCGCAAATGGGAGTTGGCGAAGGCAAGTCCATAGGCCAGTGGTACGGGCCCAACACTGTCGCCCAGG TCCTGAAGAAGCTTGCTGTCTTCGATACGTGGAGCTCCTTGGCGGTCCACATTGCAATGGACAACACTGTTGTGATGGAGGAAATCAGAA GGTTGTGCAGGACCAGCGTTCCCTGTGCAGGCGCCACTGCGTTTCCTGCAGATTCCGACCGGCACTGCAACGGATTCCCTGCCGGAGCTG AGGTCACCAACAGGCCGTCGCCATGGAGACCCCTGGTACTTCTCATTCCCCTGCGCCTGGGGCTCACGGACATCAACGAGGCCTACGTGG AGACGCTGAAGCACTGCTTCATGATGCCCCAGTCCCTGGGCGTCATCGGAGGGAAGCCCAACAGCGCCCACTACTTCATCGGCTACGTTG GTGAGGAGCTCATCTACCTGGACCCCCACACCACGCAGCCAGCCGTGGAGCCCACTGATGGCTGCTTCATCCCGGACGAGAGCTTCCACT GCCAGCACCCGCCGTGCCGCATGAGCATCGCGGAGCTTGACCCGTCCATCGCTGTGGGGTTTTTCTGTAAGACTGAAGATGACTTCAATG ATTGGTGCCAGCAAGTCAAAAAGCTGTCTCTGCTTGGAGGTGCCCTGCCCATGTTTGAGCTGGTGGAGCTGCAGCCTTCACATCTGGCCT GCCCCGACGTCCTGAACCTGTCCCTAGGTGAGAGCTGCCAAGTCCAGATTCTTCTGATGTAGAGCGACTGGAAAGATTCTTCGACTCAGA AGATGAAGACTTTGAAATCCTGTCCCTTTGAAAATCCTGGGGTCGGGGGTGGCACCTGTGAGAGCCTGGGGCTCCTGGTGCCGCTGCGTT TCATCCATCCCGCCCGCTCGCCTGCCGAGGGCTGCGCCCCGTGCTGCCTCCCCCCAGAGGGCCACCCGCTGTGCTCGTGGACTGAGGCTG CGCTGCCCGGGAGGCCTTACTGCTTGGTGTCAGACTGCCCAGCTCAGAGTGCCCGTCAGGGCCTGTGCATCCGCACGCGGAGCCGTCTGT TAGGAGCTTCCAGAGTGTTCTCTCGACACTGCCAGCCCCGTGTTAGCACCTGGGCCTCAGTCCCACTTGCTCCCAGGCGCCGGTTCTGTG GTTGGTTTGGAATTAAAGTCCTGTTTGAAGTTGTCAGACACAGACATGAATTTCTGGGCGCTCCCTGAGTCAGAGTCTCAGAAGACCTGT GCAGGCTGGCGTGAGAGGAGCGGCAGCCACACTGCGGCCCCACGCCCAAGGACTGGGCTGCTCTCGAGGGGGGCGCGCCCACCGCTGTGT CCTCTCTGCCCAGCCTGGCTTACCAAGGGCTACCTCAGTGGGAGATGAGGTTGGAGGAACGAAGGCGAGGTTCCTCCTTGCTTTGGGGAG AAAAGTATTCAGGAAGTGGGTGTGTGGGAAACCTGAAGATGGCGTGCACAGGACACAGCGTGGGCGGCCTGGGCAGAAGGGCGGCTGGCT GTCCTGGAGCTGCTGCTGGAGCCTGCCCTCAGAGTGTCCCTTTCCAGTGCTGTGGCATTCTGTGGCAGCTTCCCCAGGTGTGGTGACGGG GGGGGGGCGGGGCCTCCACCTGTGACAGCCAGGCTTGAGGGTGGACGGCGTGCCTCTCCCAGGAGCCTTCCCCATGTCCTTGCCTTGCTG AGAATTGCCCTCCCATGCCGCTGAGGTGTTAGGTGGTTTAGGGCCAAAAGGGGAAAACCACTTGAGTCTTGTGGTGTGTGGTGGGCAGAC ACCACAGGGTGGCATCACCTGGTGGCATTTCCAGAACCTCAGCCCCGATTCCAGCACCCACCACCGCCTGACCCTGTGTAACCTGCTGTC CCGGGTCCCAGAGTGCACTCTGCCCCGCTGCTCTGCTGCCTGTCCTGGGAAAGTATCTTTGCCCCACTAGGAAATGTAAACAGGAGGGCT TGGGGAGCGTGGGCACTTTTCTCATGAGCAGCTACTGCGGCGTTGGCAGGACTCGCTGCTGCTGCTGCTGCTTGTGTAGGTCGGGGAGCC AGAGATCCCCGAGGACGCGCGCCGGACAGTCGGCACTGACCGGCCCACCTGGTAGCAGAGGACACCCCCAGCCCCCCAAGCATTGAAGAC ATAGTGTATTTCCTCGTATCCTTTCTCCCTTGGGTGTAGTTGGGGTGGGGAAGCAGGGAAGGCTGGTGCGATCTCCATTCCTTGGGCTCC ACGTCCGAGTTCATGGTGCGCCGCTGTGCTGGGAGCTGCAGTGGTAATGTGTGGGACACCTTGACCAAAGGGGAGCTTTGTCTCGTGTGT TTTGAAAAAGGCTTAATGAAGAGAATGTTGTTCATTCTTAGTAGTATAGTTTGCAATTCTTAATGGCAAATAATAAGTTTCAGTAGAAAA >9922_9922_1_BMPR2-ATG4B_BMPR2_chr2_203332412_ENST00000374574_ATG4B_chr2_242590425_ENST00000405546_length(amino acids)=521AA_BP=24 MAGPGMTSSLQRPWRVPWLPWTILLVSTAAASQNQERLCAFKDPYQQDLGIGESRISHENGTILCSKGSTCYGLWEKSKGDINLVKQGCW SHIGDPQECHYEECVVTTTPPSIQNGTYRFCCCSTDLCNVNFTENFPPPDTTPLTTLTYDTLRFAEFEDFPETSEPVWILGRKYSIFTEK DEILSDVASRLWFTYRKNFPAIGGTGPTSDTGWGCMLRCGQMIFAQALVCRHLGRDWRWTQRKRQPDSYFSVLNAFIDRKDSYYSIHQIA QMGVGEGKSIGQWYGPNTVAQVLKKLAVFDTWSSLAVHIAMDNTVVMEEIRRLCRTSVPCAGATAFPADSDRHCNGFPAGAEVTNRPSPW RPLVLLIPLRLGLTDINEAYVETLKHCFMMPQSLGVIGGKPNSAHYFIGYVGEELIYLDPHTTQPAVEPTDGCFIPDESFHCQHPPCRMS -------------------------------------------------------------- >9922_9922_2_BMPR2-ATG4B_BMPR2_chr2_203332412_ENST00000374580_ATG4B_chr2_242590425_ENST00000405546_length(transcript)=3739nt_BP=957nt GGAAGCACCGAAGCGAAACTTAAGGAATCCTGCCTTCCCGGAGCCGCGGGCGATGCGACTAGGGCTGCCGGGCGCCGCCGCCGCCCGTCC GGCTTCGTCCTTCCCGGCAGTCGGGAACTAGTTCTGACCCTCGCCCCCCGACCCCGGATCGAATCCCCGCCCTCCGCACCCTGGATATGT TTTCTCCCAGACCTGGATATTTTTTTGATATCGTGAAACTACGAGGGAAATAATTTGGGGGATTTCTTCTTGGCTCCCTGCTTTCCCCAC AGACATGCCTTCCGTTTGGAGGGCCGCGGCACCCCGTCCGAGGCGAAGGAACCCCCCCAGCCGCGAGGGAGAGAAATGAAGGGAATTTCT GCAGCGGCATGAAAGCTCTGCAGCTAGGTCCTCTCATCAGCCATTTGTCCTTTCAAACTGTATTGTGATACGGGCAGGATCAGTCCACGG GAGAGAAGACGAGCCTCCCGGCTGTTTCTCCGCCGGTCTACTTCCCATATTTCTTTTCTTTGCCCTCCTGATTCTTGGCTGGCCCAGGGA TGACTTCCTCGCTGCAGCGGCCCTGGCGGGTGCCCTGGCTACCATGGACCATCCTGCTGGTCAGCACTGCGGCTGCTTCGCAGAATCAAG AACGGCTATGTGCGTTTAAAGATCCGTATCAGCAAGACCTTGGGATAGGTGAGAGTAGAATCTCTCATGAAAATGGGACAATATTATGCT CGAAAGGTAGCACCTGCTATGGCCTTTGGGAGAAATCAAAAGGGGACATAAATCTTGTAAAACAAGGATGTTGGTCTCACATTGGAGATC CCCAAGAGTGTCACTATGAAGAATGTGTAGTAACTACCACTCCTCCCTCAATTCAGAATGGAACATACCGTTTCTGCTGTTGTAGCACAG ATTTATGTAATGTCAACTTTACTGAGAATTTTCCACCTCCTGACACAACACCACTCACTACTCTGACCTACGACACTCTCCGGTTTGCTG AGTTTGAAGATTTTCCTGAGACCTCAGAGCCCGTTTGGATACTGGGTAGAAAATACAGCATTTTCACAGAAAAGGACGAGATCTTGTCTG ATGTGGCATCTAGACTTTGGTTTACATACAGGAAAAACTTTCCAGCCATTGGGGGGACAGGCCCCACCTCGGACACAGGCTGGGGCTGCA TGCTGCGGTGTGGACAGATGATCTTTGCCCAAGCCCTGGTGTGCCGGCACCTAGGCCGAGATTGGAGGTGGACACAAAGGAAGAGGCAGC CAGACAGCTACTTCAGCGTCCTCAACGCATTCATCGACAGGAAGGACAGTTACTACTCCATTCACCAGATAGCGCAAATGGGAGTTGGCG AAGGCAAGTCCATAGGCCAGTGGTACGGGCCCAACACTGTCGCCCAGGTCCTGAAGAAGCTTGCTGTCTTCGATACGTGGAGCTCCTTGG CGGTCCACATTGCAATGGACAACACTGTTGTGATGGAGGAAATCAGAAGGTTGTGCAGGACCAGCGTTCCCTGTGCAGGCGCCACTGCGT TTCCTGCAGATTCCGACCGGCACTGCAACGGATTCCCTGCCGGAGCTGAGGTCACCAACAGGCCGTCGCCATGGAGACCCCTGGTACTTC TCATTCCCCTGCGCCTGGGGCTCACGGACATCAACGAGGCCTACGTGGAGACGCTGAAGCACTGCTTCATGATGCCCCAGTCCCTGGGCG TCATCGGAGGGAAGCCCAACAGCGCCCACTACTTCATCGGCTACGTTGGTGAGGAGCTCATCTACCTGGACCCCCACACCACGCAGCCAG CCGTGGAGCCCACTGATGGCTGCTTCATCCCGGACGAGAGCTTCCACTGCCAGCACCCGCCGTGCCGCATGAGCATCGCGGAGCTTGACC CGTCCATCGCTGTGGGGTTTTTCTGTAAGACTGAAGATGACTTCAATGATTGGTGCCAGCAAGTCAAAAAGCTGTCTCTGCTTGGAGGTG CCCTGCCCATGTTTGAGCTGGTGGAGCTGCAGCCTTCACATCTGGCCTGCCCCGACGTCCTGAACCTGTCCCTAGGTGAGAGCTGCCAAG TCCAGATTCTTCTGATGTAGAGCGACTGGAAAGATTCTTCGACTCAGAAGATGAAGACTTTGAAATCCTGTCCCTTTGAAAATCCTGGGG TCGGGGGTGGCACCTGTGAGAGCCTGGGGCTCCTGGTGCCGCTGCGTTTCATCCATCCCGCCCGCTCGCCTGCCGAGGGCTGCGCCCCGT GCTGCCTCCCCCCAGAGGGCCACCCGCTGTGCTCGTGGACTGAGGCTGCGCTGCCCGGGAGGCCTTACTGCTTGGTGTCAGACTGCCCAG CTCAGAGTGCCCGTCAGGGCCTGTGCATCCGCACGCGGAGCCGTCTGTTAGGAGCTTCCAGAGTGTTCTCTCGACACTGCCAGCCCCGTG TTAGCACCTGGGCCTCAGTCCCACTTGCTCCCAGGCGCCGGTTCTGTGGTTGGTTTGGAATTAAAGTCCTGTTTGAAGTTGTCAGACACA GACATGAATTTCTGGGCGCTCCCTGAGTCAGAGTCTCAGAAGACCTGTGCAGGCTGGCGTGAGAGGAGCGGCAGCCACACTGCGGCCCCA CGCCCAAGGACTGGGCTGCTCTCGAGGGGGGCGCGCCCACCGCTGTGTCCTCTCTGCCCAGCCTGGCTTACCAAGGGCTACCTCAGTGGG AGATGAGGTTGGAGGAACGAAGGCGAGGTTCCTCCTTGCTTTGGGGAGAAAAGTATTCAGGAAGTGGGTGTGTGGGAAACCTGAAGATGG CGTGCACAGGACACAGCGTGGGCGGCCTGGGCAGAAGGGCGGCTGGCTGTCCTGGAGCTGCTGCTGGAGCCTGCCCTCAGAGTGTCCCTT TCCAGTGCTGTGGCATTCTGTGGCAGCTTCCCCAGGTGTGGTGACGGGGGGGGGGCGGGGCCTCCACCTGTGACAGCCAGGCTTGAGGGT GGACGGCGTGCCTCTCCCAGGAGCCTTCCCCATGTCCTTGCCTTGCTGAGAATTGCCCTCCCATGCCGCTGAGGTGTTAGGTGGTTTAGG GCCAAAAGGGGAAAACCACTTGAGTCTTGTGGTGTGTGGTGGGCAGACACCACAGGGTGGCATCACCTGGTGGCATTTCCAGAACCTCAG CCCCGATTCCAGCACCCACCACCGCCTGACCCTGTGTAACCTGCTGTCCCGGGTCCCAGAGTGCACTCTGCCCCGCTGCTCTGCTGCCTG TCCTGGGAAAGTATCTTTGCCCCACTAGGAAATGTAAACAGGAGGGCTTGGGGAGCGTGGGCACTTTTCTCATGAGCAGCTACTGCGGCG TTGGCAGGACTCGCTGCTGCTGCTGCTGCTTGTGTAGGTCGGGGAGCCAGAGATCCCCGAGGACGCGCGCCGGACAGTCGGCACTGACCG GCCCACCTGGTAGCAGAGGACACCCCCAGCCCCCCAAGCATTGAAGACATAGTGTATTTCCTCGTATCCTTTCTCCCTTGGGTGTAGTTG GGGTGGGGAAGCAGGGAAGGCTGGTGCGATCTCCATTCCTTGGGCTCCACGTCCGAGTTCATGGTGCGCCGCTGTGCTGGGAGCTGCAGT GGTAATGTGTGGGACACCTTGACCAAAGGGGAGCTTTGTCTCGTGTGTTTTGAAAAAGGCTTAATGAAGAGAATGTTGTTCATTCTTAGT >9922_9922_2_BMPR2-ATG4B_BMPR2_chr2_203332412_ENST00000374580_ATG4B_chr2_242590425_ENST00000405546_length(amino acids)=521AA_BP=24 MAGPGMTSSLQRPWRVPWLPWTILLVSTAAASQNQERLCAFKDPYQQDLGIGESRISHENGTILCSKGSTCYGLWEKSKGDINLVKQGCW SHIGDPQECHYEECVVTTTPPSIQNGTYRFCCCSTDLCNVNFTENFPPPDTTPLTTLTYDTLRFAEFEDFPETSEPVWILGRKYSIFTEK DEILSDVASRLWFTYRKNFPAIGGTGPTSDTGWGCMLRCGQMIFAQALVCRHLGRDWRWTQRKRQPDSYFSVLNAFIDRKDSYYSIHQIA QMGVGEGKSIGQWYGPNTVAQVLKKLAVFDTWSSLAVHIAMDNTVVMEEIRRLCRTSVPCAGATAFPADSDRHCNGFPAGAEVTNRPSPW RPLVLLIPLRLGLTDINEAYVETLKHCFMMPQSLGVIGGKPNSAHYFIGYVGEELIYLDPHTTQPAVEPTDGCFIPDESFHCQHPPCRMS -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for BMPR2-ATG4B |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for BMPR2-ATG4B |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for BMPR2-ATG4B |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
