
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:YAP1-STIM1 (FusionGDB2 ID:99893) |
Fusion Gene Summary for YAP1-STIM1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: YAP1-STIM1 | Fusion gene ID: 99893 | Hgene | Tgene | Gene symbol | YAP1 | STIM1 | Gene ID | 10413 | 6786 |
| Gene name | Yes1 associated transcriptional regulator | stromal interaction molecule 1 | |
| Synonyms | COB1|YAP|YAP2|YAP65|YKI | D11S4896E|GOK|IMD10|STRMK|TAM|TAM1 | |
| Cytomap | 11q22.1 | 11p15.4 | |
| Type of gene | protein-coding | protein-coding | |
| Description | transcriptional coactivator YAP165 kDa Yes-associated proteinYes associated protein 1protein yorkie homologyes-associated protein 1yes-associated protein 2yes-associated protein YAP65 homologyorkie homolog | stromal interaction molecule 1 | |
| Modification date | 20200329 | 20200329 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000282441, ENST00000524575, ENST00000345877, ENST00000526343, ENST00000528834, ENST00000531439, ENST00000537274, | ENST00000527484, ENST00000533977, ENST00000300737, ENST00000527651, | |
| Fusion gene scores | * DoF score | 23 X 13 X 14=4186 | 11 X 11 X 4=484 |
| # samples | 25 | 12 | |
| ** MAII score | log2(25/4186*10)=-4.06557231159362 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(12/484*10)=-2.01197264166608 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: YAP1 [Title/Abstract] AND STIM1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | YAP1(102080295)-STIM1(4045103), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | YAP1 | GO:0006974 | cellular response to DNA damage stimulus | 18280240 |
| Hgene | YAP1 | GO:0008283 | cell proliferation | 17974916 |
| Hgene | YAP1 | GO:0032570 | response to progesterone | 16772533 |
| Hgene | YAP1 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway | 16772533 |
| Hgene | YAP1 | GO:0045893 | positive regulation of transcription, DNA-templated | 20368466 |
| Hgene | YAP1 | GO:0045944 | positive regulation of transcription by RNA polymerase II | 25796446 |
| Hgene | YAP1 | GO:0050767 | regulation of neurogenesis | 25433207 |
| Hgene | YAP1 | GO:0050847 | progesterone receptor signaling pathway | 16772533 |
| Hgene | YAP1 | GO:0060242 | contact inhibition | 17974916 |
| Hgene | YAP1 | GO:0065003 | protein-containing complex assembly | 20368466 |
| Hgene | YAP1 | GO:0071480 | cellular response to gamma radiation | 18280240 |
| Hgene | YAP1 | GO:0072091 | regulation of stem cell proliferation | 25433207 |
| Tgene | STIM1 | GO:0002115 | store-operated calcium entry | 28219928 |
| Tgene | STIM1 | GO:0005513 | detection of calcium ion | 16005298 |
| Tgene | STIM1 | GO:0032237 | activation of store-operated calcium channel activity | 16005298|26322679|28219928 |
| Tgene | STIM1 | GO:0045762 | positive regulation of adenylate cyclase activity | 19171672 |
| Tgene | STIM1 | GO:0051924 | regulation of calcium ion transport | 16005298 |
 Fusion gene breakpoints across YAP1 (5'-gene) Fusion gene breakpoints across YAP1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
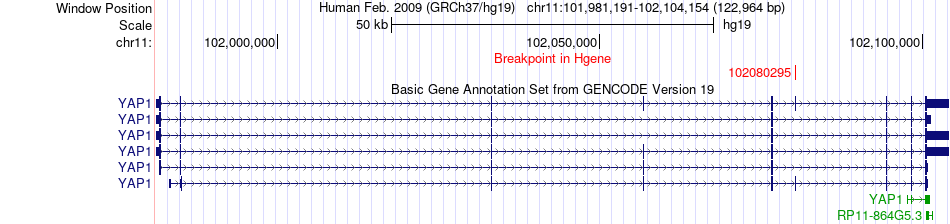 |
 Fusion gene breakpoints across STIM1 (3'-gene) Fusion gene breakpoints across STIM1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
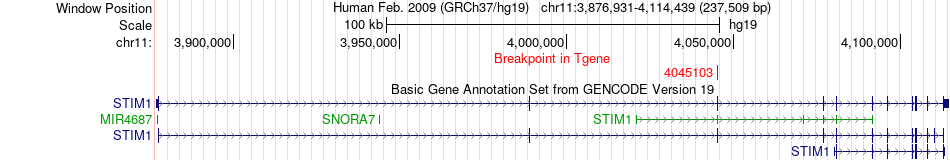 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-A8-A086-01A | YAP1 | chr11 | 102080295 | - | STIM1 | chr11 | 4045103 | + |
| ChimerDB4 | BRCA | TCGA-A8-A086-01A | YAP1 | chr11 | 102080295 | + | STIM1 | chr11 | 4045103 | + |
Top |
Fusion Gene ORF analysis for YAP1-STIM1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000282441 | ENST00000527484 | YAP1 | chr11 | 102080295 | + | STIM1 | chr11 | 4045103 | + |
| 5CDS-3UTR | ENST00000524575 | ENST00000527484 | YAP1 | chr11 | 102080295 | + | STIM1 | chr11 | 4045103 | + |
| 5CDS-intron | ENST00000282441 | ENST00000533977 | YAP1 | chr11 | 102080295 | + | STIM1 | chr11 | 4045103 | + |
| 5CDS-intron | ENST00000524575 | ENST00000533977 | YAP1 | chr11 | 102080295 | + | STIM1 | chr11 | 4045103 | + |
| In-frame | ENST00000282441 | ENST00000300737 | YAP1 | chr11 | 102080295 | + | STIM1 | chr11 | 4045103 | + |
| In-frame | ENST00000282441 | ENST00000527651 | YAP1 | chr11 | 102080295 | + | STIM1 | chr11 | 4045103 | + |
| In-frame | ENST00000524575 | ENST00000300737 | YAP1 | chr11 | 102080295 | + | STIM1 | chr11 | 4045103 | + |
| In-frame | ENST00000524575 | ENST00000527651 | YAP1 | chr11 | 102080295 | + | STIM1 | chr11 | 4045103 | + |
| intron-3CDS | ENST00000345877 | ENST00000300737 | YAP1 | chr11 | 102080295 | + | STIM1 | chr11 | 4045103 | + |
| intron-3CDS | ENST00000345877 | ENST00000527651 | YAP1 | chr11 | 102080295 | + | STIM1 | chr11 | 4045103 | + |
| intron-3CDS | ENST00000526343 | ENST00000300737 | YAP1 | chr11 | 102080295 | + | STIM1 | chr11 | 4045103 | + |
| intron-3CDS | ENST00000526343 | ENST00000527651 | YAP1 | chr11 | 102080295 | + | STIM1 | chr11 | 4045103 | + |
| intron-3CDS | ENST00000528834 | ENST00000300737 | YAP1 | chr11 | 102080295 | + | STIM1 | chr11 | 4045103 | + |
| intron-3CDS | ENST00000528834 | ENST00000527651 | YAP1 | chr11 | 102080295 | + | STIM1 | chr11 | 4045103 | + |
| intron-3CDS | ENST00000531439 | ENST00000300737 | YAP1 | chr11 | 102080295 | + | STIM1 | chr11 | 4045103 | + |
| intron-3CDS | ENST00000531439 | ENST00000527651 | YAP1 | chr11 | 102080295 | + | STIM1 | chr11 | 4045103 | + |
| intron-3CDS | ENST00000537274 | ENST00000300737 | YAP1 | chr11 | 102080295 | + | STIM1 | chr11 | 4045103 | + |
| intron-3CDS | ENST00000537274 | ENST00000527651 | YAP1 | chr11 | 102080295 | + | STIM1 | chr11 | 4045103 | + |
| intron-3UTR | ENST00000345877 | ENST00000527484 | YAP1 | chr11 | 102080295 | + | STIM1 | chr11 | 4045103 | + |
| intron-3UTR | ENST00000526343 | ENST00000527484 | YAP1 | chr11 | 102080295 | + | STIM1 | chr11 | 4045103 | + |
| intron-3UTR | ENST00000528834 | ENST00000527484 | YAP1 | chr11 | 102080295 | + | STIM1 | chr11 | 4045103 | + |
| intron-3UTR | ENST00000531439 | ENST00000527484 | YAP1 | chr11 | 102080295 | + | STIM1 | chr11 | 4045103 | + |
| intron-3UTR | ENST00000537274 | ENST00000527484 | YAP1 | chr11 | 102080295 | + | STIM1 | chr11 | 4045103 | + |
| intron-intron | ENST00000345877 | ENST00000533977 | YAP1 | chr11 | 102080295 | + | STIM1 | chr11 | 4045103 | + |
| intron-intron | ENST00000526343 | ENST00000533977 | YAP1 | chr11 | 102080295 | + | STIM1 | chr11 | 4045103 | + |
| intron-intron | ENST00000528834 | ENST00000533977 | YAP1 | chr11 | 102080295 | + | STIM1 | chr11 | 4045103 | + |
| intron-intron | ENST00000531439 | ENST00000533977 | YAP1 | chr11 | 102080295 | + | STIM1 | chr11 | 4045103 | + |
| intron-intron | ENST00000537274 | ENST00000533977 | YAP1 | chr11 | 102080295 | + | STIM1 | chr11 | 4045103 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000282441 | YAP1 | chr11 | 102080295 | + | ENST00000300737 | STIM1 | chr11 | 4045103 | + | 4619 | 1420 | 388 | 3207 | 939 |
| ENST00000282441 | YAP1 | chr11 | 102080295 | + | ENST00000527651 | STIM1 | chr11 | 4045103 | + | 3245 | 1420 | 388 | 2772 | 794 |
| ENST00000524575 | YAP1 | chr11 | 102080295 | + | ENST00000300737 | STIM1 | chr11 | 4045103 | + | 4142 | 943 | 262 | 2730 | 822 |
| ENST00000524575 | YAP1 | chr11 | 102080295 | + | ENST00000527651 | STIM1 | chr11 | 4045103 | + | 2768 | 943 | 262 | 2295 | 677 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000282441 | ENST00000300737 | YAP1 | chr11 | 102080295 | + | STIM1 | chr11 | 4045103 | + | 0.004838228 | 0.99516183 |
| ENST00000282441 | ENST00000527651 | YAP1 | chr11 | 102080295 | + | STIM1 | chr11 | 4045103 | + | 0.03085545 | 0.9691445 |
| ENST00000524575 | ENST00000300737 | YAP1 | chr11 | 102080295 | + | STIM1 | chr11 | 4045103 | + | 0.006155456 | 0.9938445 |
| ENST00000524575 | ENST00000527651 | YAP1 | chr11 | 102080295 | + | STIM1 | chr11 | 4045103 | + | 0.02557428 | 0.97442573 |
Top |
Fusion Genomic Features for YAP1-STIM1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| YAP1 | chr11 | 102080295 | + | STIM1 | chr11 | 4045102 | + | 4.27E-05 | 0.9999573 |
| YAP1 | chr11 | 102080295 | + | STIM1 | chr11 | 4045102 | + | 4.27E-05 | 0.9999573 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
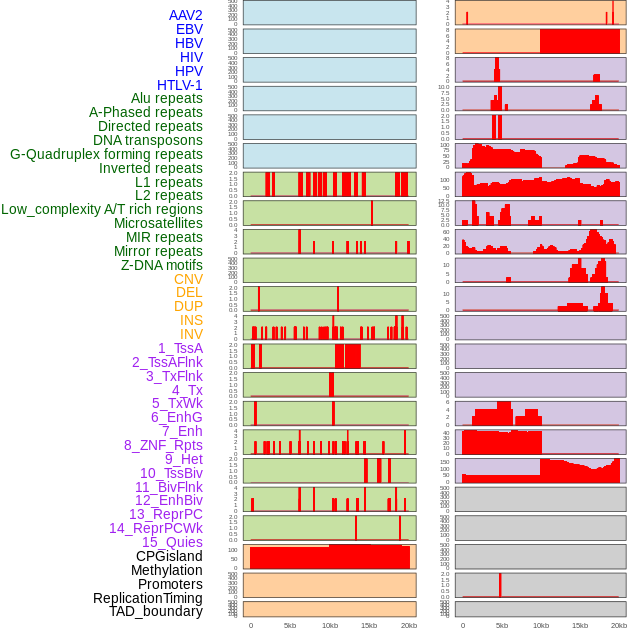 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
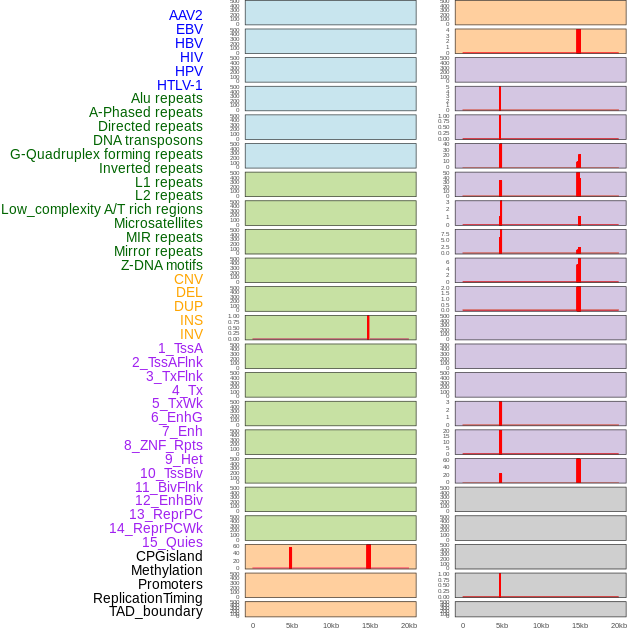 |
Top |
Fusion Protein Features for YAP1-STIM1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr11:102080295/chr11:4045103) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000282441 | + | 6 | 9 | 86_100 | 344 | 505.0 | Coiled coil | Ontology_term=ECO:0000269 |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000524575 | + | 6 | 9 | 86_100 | 166 | 327.0 | Coiled coil | Ontology_term=ECO:0000269 |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000282441 | + | 6 | 9 | 3_49 | 344 | 505.0 | Compositional bias | Note=Pro-rich |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000524575 | + | 6 | 9 | 3_49 | 166 | 327.0 | Compositional bias | Note=Pro-rich |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000282441 | + | 6 | 9 | 171_204 | 344 | 505.0 | Domain | WW 1 |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000282441 | + | 6 | 9 | 230_263 | 344 | 505.0 | Domain | WW 2 |
| Tgene | STIM1 | chr11:102080295 | chr11:4045103 | ENST00000300737 | 1 | 12 | 248_442 | 90 | 686.0 | Coiled coil | Ontology_term=ECO:0000269,ECO:0000269,ECO:0000269 | |
| Tgene | STIM1 | chr11:102080295 | chr11:4045103 | ENST00000300737 | 1 | 12 | 270_336 | 90 | 686.0 | Compositional bias | Note=Glu-rich | |
| Tgene | STIM1 | chr11:102080295 | chr11:4045103 | ENST00000300737 | 1 | 12 | 600_629 | 90 | 686.0 | Compositional bias | Note=Pro/Ser-rich | |
| Tgene | STIM1 | chr11:102080295 | chr11:4045103 | ENST00000300737 | 1 | 12 | 672_685 | 90 | 686.0 | Compositional bias | Note=Lys-rich | |
| Tgene | STIM1 | chr11:102080295 | chr11:4045103 | ENST00000300737 | 1 | 12 | 132_200 | 90 | 686.0 | Domain | SAM | |
| Tgene | STIM1 | chr11:102080295 | chr11:4045103 | ENST00000300737 | 1 | 12 | 642_645 | 90 | 686.0 | Motif | Note=Microtubule tip localization signal | |
| Tgene | STIM1 | chr11:102080295 | chr11:4045103 | ENST00000300737 | 1 | 12 | 334_444 | 90 | 686.0 | Region | Note=SOAR/CAD | |
| Tgene | STIM1 | chr11:102080295 | chr11:4045103 | ENST00000300737 | 1 | 12 | 235_685 | 90 | 686.0 | Topological domain | Cytoplasmic | |
| Tgene | STIM1 | chr11:102080295 | chr11:4045103 | ENST00000300737 | 1 | 12 | 214_234 | 90 | 686.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000282441 | + | 6 | 9 | 298_359 | 344 | 505.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000345877 | + | 1 | 7 | 298_359 | 0 | 455.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000345877 | + | 1 | 7 | 86_100 | 0 | 455.0 | Coiled coil | Ontology_term=ECO:0000269 |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000524575 | + | 6 | 9 | 298_359 | 166 | 327.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000526343 | + | 1 | 7 | 298_359 | 0 | 451.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000526343 | + | 1 | 7 | 86_100 | 0 | 451.0 | Coiled coil | Ontology_term=ECO:0000269 |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000531439 | + | 1 | 8 | 298_359 | 0 | 489.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000531439 | + | 1 | 8 | 86_100 | 0 | 489.0 | Coiled coil | Ontology_term=ECO:0000269 |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000537274 | + | 1 | 8 | 298_359 | 0 | 493.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000537274 | + | 1 | 8 | 86_100 | 0 | 493.0 | Coiled coil | Ontology_term=ECO:0000269 |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000345877 | + | 1 | 7 | 3_49 | 0 | 455.0 | Compositional bias | Note=Pro-rich |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000526343 | + | 1 | 7 | 3_49 | 0 | 451.0 | Compositional bias | Note=Pro-rich |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000531439 | + | 1 | 8 | 3_49 | 0 | 489.0 | Compositional bias | Note=Pro-rich |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000537274 | + | 1 | 8 | 3_49 | 0 | 493.0 | Compositional bias | Note=Pro-rich |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000345877 | + | 1 | 7 | 171_204 | 0 | 455.0 | Domain | WW 1 |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000345877 | + | 1 | 7 | 230_263 | 0 | 455.0 | Domain | WW 2 |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000524575 | + | 6 | 9 | 171_204 | 166 | 327.0 | Domain | WW 1 |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000524575 | + | 6 | 9 | 230_263 | 166 | 327.0 | Domain | WW 2 |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000526343 | + | 1 | 7 | 171_204 | 0 | 451.0 | Domain | WW 1 |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000526343 | + | 1 | 7 | 230_263 | 0 | 451.0 | Domain | WW 2 |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000531439 | + | 1 | 8 | 171_204 | 0 | 489.0 | Domain | WW 1 |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000531439 | + | 1 | 8 | 230_263 | 0 | 489.0 | Domain | WW 2 |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000537274 | + | 1 | 8 | 171_204 | 0 | 493.0 | Domain | WW 1 |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000537274 | + | 1 | 8 | 230_263 | 0 | 493.0 | Domain | WW 2 |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000282441 | + | 6 | 9 | 291_504 | 344 | 505.0 | Region | Note=Transactivation domain |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000345877 | + | 1 | 7 | 291_504 | 0 | 455.0 | Region | Note=Transactivation domain |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000524575 | + | 6 | 9 | 291_504 | 166 | 327.0 | Region | Note=Transactivation domain |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000526343 | + | 1 | 7 | 291_504 | 0 | 451.0 | Region | Note=Transactivation domain |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000531439 | + | 1 | 8 | 291_504 | 0 | 489.0 | Region | Note=Transactivation domain |
| Hgene | YAP1 | chr11:102080295 | chr11:4045103 | ENST00000537274 | + | 1 | 8 | 291_504 | 0 | 493.0 | Region | Note=Transactivation domain |
| Tgene | STIM1 | chr11:102080295 | chr11:4045103 | ENST00000300737 | 1 | 12 | 76_87 | 90 | 686.0 | Calcium binding | . | |
| Tgene | STIM1 | chr11:102080295 | chr11:4045103 | ENST00000300737 | 1 | 12 | 63_98 | 90 | 686.0 | Domain | Note=EF-hand | |
| Tgene | STIM1 | chr11:102080295 | chr11:4045103 | ENST00000300737 | 1 | 12 | 23_213 | 90 | 686.0 | Topological domain | Extracellular |
Top |
Fusion Gene Sequence for YAP1-STIM1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >99893_99893_1_YAP1-STIM1_YAP1_chr11_102080295_ENST00000282441_STIM1_chr11_4045103_ENST00000300737_length(transcript)=4619nt_BP=1420nt GCCGCCGCCAGGGAAAAGAAAGGGAGGAAGGAAGGAACAAGAAAAGGAAATAAAGAGAAAGGGGAGGCGGGGAAAGGCAACGAGCTGTCC GGCCTCCGTCAAGGGAGTTGGAGGGAAAAAGTTCTCAGGCGCCGCAGGTCCGAGTGCCTCGCAGCCCCTCCCGAGGCGCAGCCGCCAGAC CAGTGGAGCCGGGGCGCAGGGCGGGGGCGGAGGCGCCGGGGCGGGGGATGCGGGGCCGCGGCGCAGCCCCCCGGCCCTGAGAGCGAGGAC AGCGCCGCCCGGCCCGCAGCCGTCGCCGCTTCTCCACCTCGGCCCGTGGAGCCGGGGCGTCCGGGCGTAGCCCTCGCTCGCCTGGGTCAG GGGGTGCGCGTCGGGGGAGGCAGAAGCCATGGATCCCGGGCAGCAGCCGCCGCCTCAACCGGCCCCCCAGGGCCAAGGGCAGCCGCCTTC GCAGCCCCCGCAGGGGCAGGGCCCGCCGTCCGGACCCGGGCAACCGGCACCCGCGGCGACCCAGGCGGCGCCGCAGGCACCCCCCGCCGG GCATCAGATCGTGCACGTCCGCGGGGACTCGGAGACCGACCTGGAGGCGCTCTTCAACGCCGTCATGAACCCCAAGACGGCCAACGTGCC CCAGACCGTGCCCATGAGGCTCCGGAAGCTGCCCGACTCCTTCTTCAAGCCGCCGGAGCCCAAATCCCACTCCCGACAGGCCAGTACTGA TGCAGGCACTGCAGGAGCCCTGACTCCACAGCATGTTCGAGCTCATTCCTCTCCAGCTTCTCTGCAGTTGGGAGCTGTTTCTCCTGGGAC ACTGACCCCCACTGGAGTAGTCTCTGGCCCAGCAGCTACACCCACAGCTCAGCATCTTCGACAGTCTTCTTTTGAGATACCTGATGATGT ACCTCTGCCAGCAGGTTGGGAGATGGCAAAGACATCTTCTGGTCAGAGATACTTCTTAAATCACATCGATCAGACAACAACATGGCAGGA CCCCAGGAAGGCCATGCTGTCCCAGATGAACGTCACAGCCCCCACCAGTCCACCAGTGCAGCAGAATATGATGAACTCGGCTTCAGGTCC TCTTCCTGATGGATGGGAACAAGCCATGACTCAGGATGGAGAAATTTACTATATAAACCATAAGAACAAGACCACCTCTTGGCTAGACCC AAGGCTTGACCCTCGTTTTGCCATGAACCAGAGAATCAGTCAGAGTGCTCCAGTGAAACAGCCACCACCCCTGGCTCCCCAGAGCCCACA GGGAGGCGTCATGGGTGGCAGCAACTCCAACCAGCAGCAACAGATGCGACTGCAGCAACTGCAGATGGAGAAGGAGAGGCTGCGGCTGAA ACAGCAAGAACTGCTTCGGCAGGCAATGCGGAATATCAATCCCAGCACAGCAAATTCTCCAAAATGTCAGTTCCTGAGGGAAGACCTCAA TTACCATGACCCAACAGTGAAACACAGCACCTTCCATGGTGAGGATAAGCTCATCAGCGTGGAGGACCTGTGGAAGGCATGGAAGTCATC AGAAGTATACAATTGGACCGTGGATGAGGTGGTACAGTGGCTGATCACATATGTGGAGCTGCCTCAGTATGAGGAGACCTTCCGGAAGCT GCAGCTCAGTGGCCATGCCATGCCAAGGCTGGCTGTCACCAACACCACCATGACAGGGACTGTGCTGAAGATGACAGACCGGAGTCATCG GCAGAAGCTGCAGCTGAAGGCTCTGGATACAGTGCTCTTTGGGCCTCCTCTCTTGACTCGCCATAATCACCTCAAGGACTTCATGCTGGT GGTGTCTATCGTTATTGGTGTGGGCGGCTGCTGGTTTGCCTATATCCAGAACCGTTACTCCAAGGAGCACATGAAGAAGATGATGAAGGA CTTGGAGGGGTTACACCGAGCTGAGCAGAGTCTGCATGACCTTCAGGAAAGGCTGCACAAGGCCCAGGAGGAGCACCGCACAGTGGAGGT GGAGAAGGTCCATCTGGAAAAGAAGCTGCGCGATGAGATCAACCTTGCTAAGCAGGAAGCCCAGCGGCTGAAGGAGCTGCGGGAGGGTAC TGAGAATGAGCGGAGCCGCCAAAAATATGCTGAGGAGGAGTTGGAGCAGGTTCGGGAGGCCTTGAGGAAAGCAGAGAAGGAGCTAGAATC TCACAGCTCATGGTATGCTCCAGAGGCCCTTCAGAAGTGGCTGCAGCTGACACATGAGGTGGAGGTGCAATATTACAACATCAAGAAGCA AAATGCTGAGAAGCAGCTGCTGGTGGCCAAGGAGGGGGCTGAGAAGATAAAAAAGAAGAGAAACACACTCTTTGGCACCTTCCACGTGGC CCACAGCTCTTCCCTGGATGATGTAGATCATAAAATTCTAACAGCTAAGCAAGCACTGAGCGAGGTGACAGCAGCATTGCGGGAGCGCCT GCACCGCTGGCAACAGATCGAGATCCTCTGTGGCTTCCAGATTGTCAACAACCCTGGCATCCACTCACTGGTGGCTGCCCTCAACATAGA CCCCAGCTGGATGGGCAGTACACGCCCCAACCCTGCTCACTTCATCATGACTGACGACGTGGATGACATGGATGAGGAGATTGTGTCTCC CTTGTCCATGCAGTCCCCTAGCCTGCAGAGCAGTGTTCGGCAGCGCCTGACGGAGCCACAGCATGGCCTGGGATCTCAGAGGGATTTGAC CCATTCCGATTCGGAGTCCTCCCTCCACATGAGTGACCGCCAGCGTGTGGCCCCCAAACCTCCTCAGATGAGCCGTGCTGCAGACGAGGC TCTCAATGCCATGACTTCCAATGGCAGCCACCGGCTGATCGAGGGGGTCCACCCAGGGTCTCTGGTGGAGAAACTGCCTGACAGCCCTGC CCTGGCCAAGAAGGCATTACTGGCGCTGAACCATGGGCTGGACAAGGCCCACAGCCTGATGGAGCTGAGCCCCTCAGCCCCACCTGGTGG CTCTCCACATTTGGATTCTTCCCGTTCTCACAGCCCCAGCTCCCCAGACCCAGACACACCATCTCCAGTTGGGGACAGCCGAGCCCTGCA AGCCAGCCGAAACACACGCATTCCCCACCTGGCTGGCAAGAAGGCTGTGGCTGAGGAGGATAATGGCTCTATTGGCGAGGAAACAGACTC CAGCCCAGGCCGGAAGAAGTTTCCCCTCAAAATCTTTAAGAAGCCTCTTAAGAAGTAGGCAGGATGGGGTGGCAGTAAAGGGACAGCTTG TCCTTCCCTGGGTGTTCTGTCTCTCCTTCCCTCCCTTCCTTCAAGATAACTGGCCCCAAGAGTGGGGCATGGGAAGGGCTGGTCCAGGGG TCTGGGCACTGTACATACCTGCCCCCTCATCCTTGGGTCCTTCATTATTATTTATTAACTGACCACCATGGCCTGCCTGCCCTGCCTCCG TCCCAACCATGGGCTGCTGCTGTCACTCCCTCTCCACTTCAGTGCATGTCTTAGTTGCTGTTCCCTCAGCTCCCAGCTCCACCTCTGGGG TTCAGCTTCTGTCTCTGCTGTCCCAGTTTTGAGGTTTGGTTTCTTGTTTCTGTCTCTTGCTTTCGGGCTCCTCCCTCCCACCACTCCCCA ACTTCCCCTAGCAGTTGCAGGGAAGATAGGACGAGTAGCTTCTGACATGTGTGCCTCAGATCTGTTCCACCCCACTCACAGTGGTTCTGT TTGCTCCAGACTGGGGCTAGGGCCTAATCTTTGAAGTTTGTTCTTTGGTATTGATGTGGGTCAGAAGGAGCCTCATCCTAATCTCACTCA GGCCTCCAGGGATCCATGGGGGAGTGAAACCAATTCTCAGAGAACAACCCACCAGAGACTTTTAAAGAGAGGCCAGGCTTGGGAATGGGT TGGGAGAGGCATCTGTTCATTGGAGCATGAGTGGATGCCAGAACTGTAGGTTATAAGGCAGTCACTTTTTCTCTCTACTCCCACCCACAC CTGCCTCCCTCTTACCCCTGCTCCCCCACACTGCAGGAGGATTTGTCTCTAAGAGGTGCTGCCCCAAAGCTCCCCAAGCATCAATACTCC TAGGGCTCAGGACAAGTGGCTCCCCTGGCCAGGAGAGCCACAGCCATGATACAGGGCTCTTATGGAGCCCTGGAGTTGTTGGGCAAGGAT GCTGTCATTTTTTGAACCAAAAGACAAACAGGTTAAAAGGAAAAAAAGTAATCTGAATTTCCCAAGTGCCTACGCTGCATATTCCCCTTG TTAGATCCCATTTTCATGTTACTTTGTAGCCTTGGCCAGAGGCTCAAAAAGGACACAACCAGTTTGGGGAAGGGGTGGCTAAGGAAGATG GTATAGGTGAAGGCGGCTGTGTGACCACTTTCCCCCACCCTTCCCACCCTCTAGACAACTCTCTCCCTTACCTGTTTTTGCTATGGCTGT AAAGGTATTTTTCCTCTGCCCCACTCCCTGCCATACCTTTATCCTGGGATCCTATTTTGGGCCTGGGGTGGGTATACCTGGGGCTGGTCT TAGGAGGGTGCTAGGCTGCAGACTGCCTTGTACTCCCTGGACACCCTCAAATGGGGTTTTCTGTGTTATTTCATAAAATTCTTTGAAGTC >99893_99893_1_YAP1-STIM1_YAP1_chr11_102080295_ENST00000282441_STIM1_chr11_4045103_ENST00000300737_length(amino acids)=939AA_BP=343 MDPGQQPPPQPAPQGQGQPPSQPPQGQGPPSGPGQPAPAATQAAPQAPPAGHQIVHVRGDSETDLEALFNAVMNPKTANVPQTVPMRLRK LPDSFFKPPEPKSHSRQASTDAGTAGALTPQHVRAHSSPASLQLGAVSPGTLTPTGVVSGPAATPTAQHLRQSSFEIPDDVPLPAGWEMA KTSSGQRYFLNHIDQTTTWQDPRKAMLSQMNVTAPTSPPVQQNMMNSASGPLPDGWEQAMTQDGEIYYINHKNKTTSWLDPRLDPRFAMN QRISQSAPVKQPPPLAPQSPQGGVMGGSNSNQQQQMRLQQLQMEKERLRLKQQELLRQAMRNINPSTANSPKCQFLREDLNYHDPTVKHS TFHGEDKLISVEDLWKAWKSSEVYNWTVDEVVQWLITYVELPQYEETFRKLQLSGHAMPRLAVTNTTMTGTVLKMTDRSHRQKLQLKALD TVLFGPPLLTRHNHLKDFMLVVSIVIGVGGCWFAYIQNRYSKEHMKKMMKDLEGLHRAEQSLHDLQERLHKAQEEHRTVEVEKVHLEKKL RDEINLAKQEAQRLKELREGTENERSRQKYAEEELEQVREALRKAEKELESHSSWYAPEALQKWLQLTHEVEVQYYNIKKQNAEKQLLVA KEGAEKIKKKRNTLFGTFHVAHSSSLDDVDHKILTAKQALSEVTAALRERLHRWQQIEILCGFQIVNNPGIHSLVAALNIDPSWMGSTRP NPAHFIMTDDVDDMDEEIVSPLSMQSPSLQSSVRQRLTEPQHGLGSQRDLTHSDSESSLHMSDRQRVAPKPPQMSRAADEALNAMTSNGS HRLIEGVHPGSLVEKLPDSPALAKKALLALNHGLDKAHSLMELSPSAPPGGSPHLDSSRSHSPSSPDPDTPSPVGDSRALQASRNTRIPH -------------------------------------------------------------- >99893_99893_2_YAP1-STIM1_YAP1_chr11_102080295_ENST00000282441_STIM1_chr11_4045103_ENST00000527651_length(transcript)=3245nt_BP=1420nt GCCGCCGCCAGGGAAAAGAAAGGGAGGAAGGAAGGAACAAGAAAAGGAAATAAAGAGAAAGGGGAGGCGGGGAAAGGCAACGAGCTGTCC GGCCTCCGTCAAGGGAGTTGGAGGGAAAAAGTTCTCAGGCGCCGCAGGTCCGAGTGCCTCGCAGCCCCTCCCGAGGCGCAGCCGCCAGAC CAGTGGAGCCGGGGCGCAGGGCGGGGGCGGAGGCGCCGGGGCGGGGGATGCGGGGCCGCGGCGCAGCCCCCCGGCCCTGAGAGCGAGGAC AGCGCCGCCCGGCCCGCAGCCGTCGCCGCTTCTCCACCTCGGCCCGTGGAGCCGGGGCGTCCGGGCGTAGCCCTCGCTCGCCTGGGTCAG GGGGTGCGCGTCGGGGGAGGCAGAAGCCATGGATCCCGGGCAGCAGCCGCCGCCTCAACCGGCCCCCCAGGGCCAAGGGCAGCCGCCTTC GCAGCCCCCGCAGGGGCAGGGCCCGCCGTCCGGACCCGGGCAACCGGCACCCGCGGCGACCCAGGCGGCGCCGCAGGCACCCCCCGCCGG GCATCAGATCGTGCACGTCCGCGGGGACTCGGAGACCGACCTGGAGGCGCTCTTCAACGCCGTCATGAACCCCAAGACGGCCAACGTGCC CCAGACCGTGCCCATGAGGCTCCGGAAGCTGCCCGACTCCTTCTTCAAGCCGCCGGAGCCCAAATCCCACTCCCGACAGGCCAGTACTGA TGCAGGCACTGCAGGAGCCCTGACTCCACAGCATGTTCGAGCTCATTCCTCTCCAGCTTCTCTGCAGTTGGGAGCTGTTTCTCCTGGGAC ACTGACCCCCACTGGAGTAGTCTCTGGCCCAGCAGCTACACCCACAGCTCAGCATCTTCGACAGTCTTCTTTTGAGATACCTGATGATGT ACCTCTGCCAGCAGGTTGGGAGATGGCAAAGACATCTTCTGGTCAGAGATACTTCTTAAATCACATCGATCAGACAACAACATGGCAGGA CCCCAGGAAGGCCATGCTGTCCCAGATGAACGTCACAGCCCCCACCAGTCCACCAGTGCAGCAGAATATGATGAACTCGGCTTCAGGTCC TCTTCCTGATGGATGGGAACAAGCCATGACTCAGGATGGAGAAATTTACTATATAAACCATAAGAACAAGACCACCTCTTGGCTAGACCC AAGGCTTGACCCTCGTTTTGCCATGAACCAGAGAATCAGTCAGAGTGCTCCAGTGAAACAGCCACCACCCCTGGCTCCCCAGAGCCCACA GGGAGGCGTCATGGGTGGCAGCAACTCCAACCAGCAGCAACAGATGCGACTGCAGCAACTGCAGATGGAGAAGGAGAGGCTGCGGCTGAA ACAGCAAGAACTGCTTCGGCAGGCAATGCGGAATATCAATCCCAGCACAGCAAATTCTCCAAAATGTCAGTTCCTGAGGGAAGACCTCAA TTACCATGACCCAACAGTGAAACACAGCACCTTCCATGGTGAGGATAAGCTCATCAGCGTGGAGGACCTGTGGAAGGCATGGAAGTCATC AGAAGTATACAATTGGACCGTGGATGAGGTGGTACAGTGGCTGATCACATATGTGGAGCTGCCTCAGTATGAGGAGACCTTCCGGAAGCT GCAGCTCAGTGGCCATGCCATGCCAAGGCTGGCTGTCACCAACACCACCATGACAGGGACTGTGCTGAAGATGACAGACCGGAGTCATCG GCAGAAGCTGCAGCTGAAGGCTCTGGATACAGTGCTCTTTGGGCCTCCTCTCTTGACTCGCCATAATCACCTCAAGGACTTCATGCTGGT GGTGTCTATCGTTATTGGTGTGGGCGGCTGCTGGTTTGCCTATATCCAGAACCGTTACTCCAAGGAGCACATGAAGAAGATGATGAAGGA CTTGGAGGGGTTACACCGAGCTGAGCAGAGTCTGCATGACCTTCAGGAAAGGCTGCACAAGGCCCAGGAGGAGCACCGCACAGTGGAGGT GGAGAAGGTCCATCTGGAAAAGAAGCTGCGCGATGAGATCAACCTTGCTAAGCAGGAAGCCCAGCGGCTGAAGGAGCTGCGGGAGGGTAC TGAGAATGAGCGGAGCCGCCAAAAATATGCTGAGGAGGAGTTGGAGCAGGTTCGGGAGGCCTTGAGGAAAGCAGAGAAGGAGCTAGAATC TCACAGCTCATGGTATGCTCCAGAGGCCCTTCAGAAGTGGCTGCAGCTGACACATGAGGTGGAGGTGCAATATTACAACATCAAGAAGCA AAATGCTGAGAAGCAGCTGCTGGTGGCCAAGGAGGGGGCTGAGAAGATAAAAAAGAAGAGAAACACACTCTTTGGCACCTTCCACGTGGC CCACAGCTCTTCCCTGGATGATGTAGATCATAAAATTCTAACAGCTAAGCAAGCACTGAGCGAGGTGACAGCAGCATTGCGGGAGCGCCT GCACCGCTGGCAACAGATCGAGATCCTCTGTGGCTTCCAGATTGTCAACAACCCTGGCATCCACTCACTGGTGGCTGCCCTCAACATAGA CCCCAGCTGGATGGGCAGTACACGCCCCAACCCTGCTCACTTCATCATGACTGACGACGTGGATGACATGGATGAGGAGATTGTGTCTCC CTTGTCCATGCAGTCCCCTAGCCTGCAGAGCAGTGTTCGGCAGCGCCTGACGGAGCCACAGCATGGCCTGGGATCTCAGAGAGGATCATC TCTAAAGGCAAACAGGCTCTCTAGTAAGGGATTTGACCCATTCCGATTCGGAGTCCTCCCTCCACATGAGTGACCGCCAGCGTGTGGCCC CCAAACCTCCTCAGATGAGCCGTGCTGCAGACGAGGCTCTCAATGCCATGACTTCCAATGGCAGCCACCGGCTGATCGAGGGGGTCCACC CAGGGTCTCTGGTGGAGAAACTGCCTGACAGCCCTGCCCTGGCCAAGAAGGCATTACTGGCGCTGAACCATGGGCTGGACAAGGCCCACA GCCTGATGGAGCTGAGCCCCTCAGCCCCACCTGGTGGCTCTCCACATTTGGATTCTTCCCGTTCTCACAGCCCCAGCTCCCCAGACCCAG ACACACCATCTCCAGTTGGGGACAGCCGAGCCCTGCAAGCCAGCCGAAACACACGCATTCCCCACCTGGCTGGCAAGAAGGCTGTGGCTG AGGAGGATAATGGCTCTATTGGCGAGGAAACAGACTCCAGCCCAGGCCGGAAGAAGTTTCCCCTCAAAATCTTTAAGAAGCCTCTTAAGA >99893_99893_2_YAP1-STIM1_YAP1_chr11_102080295_ENST00000282441_STIM1_chr11_4045103_ENST00000527651_length(amino acids)=794AA_BP=343 MDPGQQPPPQPAPQGQGQPPSQPPQGQGPPSGPGQPAPAATQAAPQAPPAGHQIVHVRGDSETDLEALFNAVMNPKTANVPQTVPMRLRK LPDSFFKPPEPKSHSRQASTDAGTAGALTPQHVRAHSSPASLQLGAVSPGTLTPTGVVSGPAATPTAQHLRQSSFEIPDDVPLPAGWEMA KTSSGQRYFLNHIDQTTTWQDPRKAMLSQMNVTAPTSPPVQQNMMNSASGPLPDGWEQAMTQDGEIYYINHKNKTTSWLDPRLDPRFAMN QRISQSAPVKQPPPLAPQSPQGGVMGGSNSNQQQQMRLQQLQMEKERLRLKQQELLRQAMRNINPSTANSPKCQFLREDLNYHDPTVKHS TFHGEDKLISVEDLWKAWKSSEVYNWTVDEVVQWLITYVELPQYEETFRKLQLSGHAMPRLAVTNTTMTGTVLKMTDRSHRQKLQLKALD TVLFGPPLLTRHNHLKDFMLVVSIVIGVGGCWFAYIQNRYSKEHMKKMMKDLEGLHRAEQSLHDLQERLHKAQEEHRTVEVEKVHLEKKL RDEINLAKQEAQRLKELREGTENERSRQKYAEEELEQVREALRKAEKELESHSSWYAPEALQKWLQLTHEVEVQYYNIKKQNAEKQLLVA KEGAEKIKKKRNTLFGTFHVAHSSSLDDVDHKILTAKQALSEVTAALRERLHRWQQIEILCGFQIVNNPGIHSLVAALNIDPSWMGSTRP -------------------------------------------------------------- >99893_99893_3_YAP1-STIM1_YAP1_chr11_102080295_ENST00000524575_STIM1_chr11_4045103_ENST00000300737_length(transcript)=4142nt_BP=943nt CTGCAATGTAGTTAGCCCACTCGGGATGTAACTTGAGTGGAAAGAAGAGTTACCGCCCCCACTCCCATTCCCTCTCCTATTTGCAGGCCT GTTGTATAGTCTCCTGTCGGAGACCAAAGGGTTTTGGAACTCAGAAAAAATCACTTAACCTGATGACAAAAACCCGGGTTAAGGAGGAGG AAAGAAGGAAAAACAATTTAGCTGCTTGCCTAAACACTTCAATTTGTCTTAGGCCAGTACTGATGCAGGCACTGCAGGAGCCCTGACTCC ACAGCATGTTCGAGCTCATTCCTCTCCAGCTTCTCTGCAGTTGGGAGCTGTTTCTCCTGGGACACTGACCCCCACTGGAGTAGTCTCTGG CCCAGCAGCTACACCCACAGCTCAGCATCTTCGACAGTCTTCTTTTGAGATACCTGATGATGTACCTCTGCCAGCAGGTTGGGAGATGGC AAAGACATCTTCTGGTCAGAGATACTTCTTAAATCACATCGATCAGACAACAACATGGCAGGACCCCAGGAAGGCCATGCTGTCCCAGAT GAACGTCACAGCCCCCACCAGTCCACCAGTGCAGCAGAATATGATGAACTCGGCTTCAGGTCCTCTTCCTGATGGATGGGAACAAGCCAT GACTCAGGATGGAGAAATTTACTATATAAACCATAAGAACAAGACCACCTCTTGGCTAGACCCAAGGCTTGACCCTCGTTTTGCCATGAA CCAGAGAATCAGTCAGAGTGCTCCAGTGAAACAGCCACCACCCCTGGCTCCCCAGAGCCCACAGGGAGGCGTCATGGGTGGCAGCAACTC CAACCAGCAGCAACAGATGCGACTGCAGCAACTGCAGATGGAGAAGGAGAGGCTGCGGCTGAAACAGCAAGAACTGCTTCGGCAGGCAAT GCGGAATATCAATCCCAGCACAGCAAATTCTCCAAAATGTCAGTTCCTGAGGGAAGACCTCAATTACCATGACCCAACAGTGAAACACAG CACCTTCCATGGTGAGGATAAGCTCATCAGCGTGGAGGACCTGTGGAAGGCATGGAAGTCATCAGAAGTATACAATTGGACCGTGGATGA GGTGGTACAGTGGCTGATCACATATGTGGAGCTGCCTCAGTATGAGGAGACCTTCCGGAAGCTGCAGCTCAGTGGCCATGCCATGCCAAG GCTGGCTGTCACCAACACCACCATGACAGGGACTGTGCTGAAGATGACAGACCGGAGTCATCGGCAGAAGCTGCAGCTGAAGGCTCTGGA TACAGTGCTCTTTGGGCCTCCTCTCTTGACTCGCCATAATCACCTCAAGGACTTCATGCTGGTGGTGTCTATCGTTATTGGTGTGGGCGG CTGCTGGTTTGCCTATATCCAGAACCGTTACTCCAAGGAGCACATGAAGAAGATGATGAAGGACTTGGAGGGGTTACACCGAGCTGAGCA GAGTCTGCATGACCTTCAGGAAAGGCTGCACAAGGCCCAGGAGGAGCACCGCACAGTGGAGGTGGAGAAGGTCCATCTGGAAAAGAAGCT GCGCGATGAGATCAACCTTGCTAAGCAGGAAGCCCAGCGGCTGAAGGAGCTGCGGGAGGGTACTGAGAATGAGCGGAGCCGCCAAAAATA TGCTGAGGAGGAGTTGGAGCAGGTTCGGGAGGCCTTGAGGAAAGCAGAGAAGGAGCTAGAATCTCACAGCTCATGGTATGCTCCAGAGGC CCTTCAGAAGTGGCTGCAGCTGACACATGAGGTGGAGGTGCAATATTACAACATCAAGAAGCAAAATGCTGAGAAGCAGCTGCTGGTGGC CAAGGAGGGGGCTGAGAAGATAAAAAAGAAGAGAAACACACTCTTTGGCACCTTCCACGTGGCCCACAGCTCTTCCCTGGATGATGTAGA TCATAAAATTCTAACAGCTAAGCAAGCACTGAGCGAGGTGACAGCAGCATTGCGGGAGCGCCTGCACCGCTGGCAACAGATCGAGATCCT CTGTGGCTTCCAGATTGTCAACAACCCTGGCATCCACTCACTGGTGGCTGCCCTCAACATAGACCCCAGCTGGATGGGCAGTACACGCCC CAACCCTGCTCACTTCATCATGACTGACGACGTGGATGACATGGATGAGGAGATTGTGTCTCCCTTGTCCATGCAGTCCCCTAGCCTGCA GAGCAGTGTTCGGCAGCGCCTGACGGAGCCACAGCATGGCCTGGGATCTCAGAGGGATTTGACCCATTCCGATTCGGAGTCCTCCCTCCA CATGAGTGACCGCCAGCGTGTGGCCCCCAAACCTCCTCAGATGAGCCGTGCTGCAGACGAGGCTCTCAATGCCATGACTTCCAATGGCAG CCACCGGCTGATCGAGGGGGTCCACCCAGGGTCTCTGGTGGAGAAACTGCCTGACAGCCCTGCCCTGGCCAAGAAGGCATTACTGGCGCT GAACCATGGGCTGGACAAGGCCCACAGCCTGATGGAGCTGAGCCCCTCAGCCCCACCTGGTGGCTCTCCACATTTGGATTCTTCCCGTTC TCACAGCCCCAGCTCCCCAGACCCAGACACACCATCTCCAGTTGGGGACAGCCGAGCCCTGCAAGCCAGCCGAAACACACGCATTCCCCA CCTGGCTGGCAAGAAGGCTGTGGCTGAGGAGGATAATGGCTCTATTGGCGAGGAAACAGACTCCAGCCCAGGCCGGAAGAAGTTTCCCCT CAAAATCTTTAAGAAGCCTCTTAAGAAGTAGGCAGGATGGGGTGGCAGTAAAGGGACAGCTTGTCCTTCCCTGGGTGTTCTGTCTCTCCT TCCCTCCCTTCCTTCAAGATAACTGGCCCCAAGAGTGGGGCATGGGAAGGGCTGGTCCAGGGGTCTGGGCACTGTACATACCTGCCCCCT CATCCTTGGGTCCTTCATTATTATTTATTAACTGACCACCATGGCCTGCCTGCCCTGCCTCCGTCCCAACCATGGGCTGCTGCTGTCACT CCCTCTCCACTTCAGTGCATGTCTTAGTTGCTGTTCCCTCAGCTCCCAGCTCCACCTCTGGGGTTCAGCTTCTGTCTCTGCTGTCCCAGT TTTGAGGTTTGGTTTCTTGTTTCTGTCTCTTGCTTTCGGGCTCCTCCCTCCCACCACTCCCCAACTTCCCCTAGCAGTTGCAGGGAAGAT AGGACGAGTAGCTTCTGACATGTGTGCCTCAGATCTGTTCCACCCCACTCACAGTGGTTCTGTTTGCTCCAGACTGGGGCTAGGGCCTAA TCTTTGAAGTTTGTTCTTTGGTATTGATGTGGGTCAGAAGGAGCCTCATCCTAATCTCACTCAGGCCTCCAGGGATCCATGGGGGAGTGA AACCAATTCTCAGAGAACAACCCACCAGAGACTTTTAAAGAGAGGCCAGGCTTGGGAATGGGTTGGGAGAGGCATCTGTTCATTGGAGCA TGAGTGGATGCCAGAACTGTAGGTTATAAGGCAGTCACTTTTTCTCTCTACTCCCACCCACACCTGCCTCCCTCTTACCCCTGCTCCCCC ACACTGCAGGAGGATTTGTCTCTAAGAGGTGCTGCCCCAAAGCTCCCCAAGCATCAATACTCCTAGGGCTCAGGACAAGTGGCTCCCCTG GCCAGGAGAGCCACAGCCATGATACAGGGCTCTTATGGAGCCCTGGAGTTGTTGGGCAAGGATGCTGTCATTTTTTGAACCAAAAGACAA ACAGGTTAAAAGGAAAAAAAGTAATCTGAATTTCCCAAGTGCCTACGCTGCATATTCCCCTTGTTAGATCCCATTTTCATGTTACTTTGT AGCCTTGGCCAGAGGCTCAAAAAGGACACAACCAGTTTGGGGAAGGGGTGGCTAAGGAAGATGGTATAGGTGAAGGCGGCTGTGTGACCA CTTTCCCCCACCCTTCCCACCCTCTAGACAACTCTCTCCCTTACCTGTTTTTGCTATGGCTGTAAAGGTATTTTTCCTCTGCCCCACTCC CTGCCATACCTTTATCCTGGGATCCTATTTTGGGCCTGGGGTGGGTATACCTGGGGCTGGTCTTAGGAGGGTGCTAGGCTGCAGACTGCC TTGTACTCCCTGGACACCCTCAAATGGGGTTTTCTGTGTTATTTCATAAAATTCTTTGAAGTCCAATAAAGCATGTAGGAGATTTTAACC >99893_99893_3_YAP1-STIM1_YAP1_chr11_102080295_ENST00000524575_STIM1_chr11_4045103_ENST00000300737_length(amino acids)=822AA_BP=226 MTPQHVRAHSSPASLQLGAVSPGTLTPTGVVSGPAATPTAQHLRQSSFEIPDDVPLPAGWEMAKTSSGQRYFLNHIDQTTTWQDPRKAML SQMNVTAPTSPPVQQNMMNSASGPLPDGWEQAMTQDGEIYYINHKNKTTSWLDPRLDPRFAMNQRISQSAPVKQPPPLAPQSPQGGVMGG SNSNQQQQMRLQQLQMEKERLRLKQQELLRQAMRNINPSTANSPKCQFLREDLNYHDPTVKHSTFHGEDKLISVEDLWKAWKSSEVYNWT VDEVVQWLITYVELPQYEETFRKLQLSGHAMPRLAVTNTTMTGTVLKMTDRSHRQKLQLKALDTVLFGPPLLTRHNHLKDFMLVVSIVIG VGGCWFAYIQNRYSKEHMKKMMKDLEGLHRAEQSLHDLQERLHKAQEEHRTVEVEKVHLEKKLRDEINLAKQEAQRLKELREGTENERSR QKYAEEELEQVREALRKAEKELESHSSWYAPEALQKWLQLTHEVEVQYYNIKKQNAEKQLLVAKEGAEKIKKKRNTLFGTFHVAHSSSLD DVDHKILTAKQALSEVTAALRERLHRWQQIEILCGFQIVNNPGIHSLVAALNIDPSWMGSTRPNPAHFIMTDDVDDMDEEIVSPLSMQSP SLQSSVRQRLTEPQHGLGSQRDLTHSDSESSLHMSDRQRVAPKPPQMSRAADEALNAMTSNGSHRLIEGVHPGSLVEKLPDSPALAKKAL LALNHGLDKAHSLMELSPSAPPGGSPHLDSSRSHSPSSPDPDTPSPVGDSRALQASRNTRIPHLAGKKAVAEEDNGSIGEETDSSPGRKK -------------------------------------------------------------- >99893_99893_4_YAP1-STIM1_YAP1_chr11_102080295_ENST00000524575_STIM1_chr11_4045103_ENST00000527651_length(transcript)=2768nt_BP=943nt CTGCAATGTAGTTAGCCCACTCGGGATGTAACTTGAGTGGAAAGAAGAGTTACCGCCCCCACTCCCATTCCCTCTCCTATTTGCAGGCCT GTTGTATAGTCTCCTGTCGGAGACCAAAGGGTTTTGGAACTCAGAAAAAATCACTTAACCTGATGACAAAAACCCGGGTTAAGGAGGAGG AAAGAAGGAAAAACAATTTAGCTGCTTGCCTAAACACTTCAATTTGTCTTAGGCCAGTACTGATGCAGGCACTGCAGGAGCCCTGACTCC ACAGCATGTTCGAGCTCATTCCTCTCCAGCTTCTCTGCAGTTGGGAGCTGTTTCTCCTGGGACACTGACCCCCACTGGAGTAGTCTCTGG CCCAGCAGCTACACCCACAGCTCAGCATCTTCGACAGTCTTCTTTTGAGATACCTGATGATGTACCTCTGCCAGCAGGTTGGGAGATGGC AAAGACATCTTCTGGTCAGAGATACTTCTTAAATCACATCGATCAGACAACAACATGGCAGGACCCCAGGAAGGCCATGCTGTCCCAGAT GAACGTCACAGCCCCCACCAGTCCACCAGTGCAGCAGAATATGATGAACTCGGCTTCAGGTCCTCTTCCTGATGGATGGGAACAAGCCAT GACTCAGGATGGAGAAATTTACTATATAAACCATAAGAACAAGACCACCTCTTGGCTAGACCCAAGGCTTGACCCTCGTTTTGCCATGAA CCAGAGAATCAGTCAGAGTGCTCCAGTGAAACAGCCACCACCCCTGGCTCCCCAGAGCCCACAGGGAGGCGTCATGGGTGGCAGCAACTC CAACCAGCAGCAACAGATGCGACTGCAGCAACTGCAGATGGAGAAGGAGAGGCTGCGGCTGAAACAGCAAGAACTGCTTCGGCAGGCAAT GCGGAATATCAATCCCAGCACAGCAAATTCTCCAAAATGTCAGTTCCTGAGGGAAGACCTCAATTACCATGACCCAACAGTGAAACACAG CACCTTCCATGGTGAGGATAAGCTCATCAGCGTGGAGGACCTGTGGAAGGCATGGAAGTCATCAGAAGTATACAATTGGACCGTGGATGA GGTGGTACAGTGGCTGATCACATATGTGGAGCTGCCTCAGTATGAGGAGACCTTCCGGAAGCTGCAGCTCAGTGGCCATGCCATGCCAAG GCTGGCTGTCACCAACACCACCATGACAGGGACTGTGCTGAAGATGACAGACCGGAGTCATCGGCAGAAGCTGCAGCTGAAGGCTCTGGA TACAGTGCTCTTTGGGCCTCCTCTCTTGACTCGCCATAATCACCTCAAGGACTTCATGCTGGTGGTGTCTATCGTTATTGGTGTGGGCGG CTGCTGGTTTGCCTATATCCAGAACCGTTACTCCAAGGAGCACATGAAGAAGATGATGAAGGACTTGGAGGGGTTACACCGAGCTGAGCA GAGTCTGCATGACCTTCAGGAAAGGCTGCACAAGGCCCAGGAGGAGCACCGCACAGTGGAGGTGGAGAAGGTCCATCTGGAAAAGAAGCT GCGCGATGAGATCAACCTTGCTAAGCAGGAAGCCCAGCGGCTGAAGGAGCTGCGGGAGGGTACTGAGAATGAGCGGAGCCGCCAAAAATA TGCTGAGGAGGAGTTGGAGCAGGTTCGGGAGGCCTTGAGGAAAGCAGAGAAGGAGCTAGAATCTCACAGCTCATGGTATGCTCCAGAGGC CCTTCAGAAGTGGCTGCAGCTGACACATGAGGTGGAGGTGCAATATTACAACATCAAGAAGCAAAATGCTGAGAAGCAGCTGCTGGTGGC CAAGGAGGGGGCTGAGAAGATAAAAAAGAAGAGAAACACACTCTTTGGCACCTTCCACGTGGCCCACAGCTCTTCCCTGGATGATGTAGA TCATAAAATTCTAACAGCTAAGCAAGCACTGAGCGAGGTGACAGCAGCATTGCGGGAGCGCCTGCACCGCTGGCAACAGATCGAGATCCT CTGTGGCTTCCAGATTGTCAACAACCCTGGCATCCACTCACTGGTGGCTGCCCTCAACATAGACCCCAGCTGGATGGGCAGTACACGCCC CAACCCTGCTCACTTCATCATGACTGACGACGTGGATGACATGGATGAGGAGATTGTGTCTCCCTTGTCCATGCAGTCCCCTAGCCTGCA GAGCAGTGTTCGGCAGCGCCTGACGGAGCCACAGCATGGCCTGGGATCTCAGAGAGGATCATCTCTAAAGGCAAACAGGCTCTCTAGTAA GGGATTTGACCCATTCCGATTCGGAGTCCTCCCTCCACATGAGTGACCGCCAGCGTGTGGCCCCCAAACCTCCTCAGATGAGCCGTGCTG CAGACGAGGCTCTCAATGCCATGACTTCCAATGGCAGCCACCGGCTGATCGAGGGGGTCCACCCAGGGTCTCTGGTGGAGAAACTGCCTG ACAGCCCTGCCCTGGCCAAGAAGGCATTACTGGCGCTGAACCATGGGCTGGACAAGGCCCACAGCCTGATGGAGCTGAGCCCCTCAGCCC CACCTGGTGGCTCTCCACATTTGGATTCTTCCCGTTCTCACAGCCCCAGCTCCCCAGACCCAGACACACCATCTCCAGTTGGGGACAGCC GAGCCCTGCAAGCCAGCCGAAACACACGCATTCCCCACCTGGCTGGCAAGAAGGCTGTGGCTGAGGAGGATAATGGCTCTATTGGCGAGG >99893_99893_4_YAP1-STIM1_YAP1_chr11_102080295_ENST00000524575_STIM1_chr11_4045103_ENST00000527651_length(amino acids)=677AA_BP=226 MTPQHVRAHSSPASLQLGAVSPGTLTPTGVVSGPAATPTAQHLRQSSFEIPDDVPLPAGWEMAKTSSGQRYFLNHIDQTTTWQDPRKAML SQMNVTAPTSPPVQQNMMNSASGPLPDGWEQAMTQDGEIYYINHKNKTTSWLDPRLDPRFAMNQRISQSAPVKQPPPLAPQSPQGGVMGG SNSNQQQQMRLQQLQMEKERLRLKQQELLRQAMRNINPSTANSPKCQFLREDLNYHDPTVKHSTFHGEDKLISVEDLWKAWKSSEVYNWT VDEVVQWLITYVELPQYEETFRKLQLSGHAMPRLAVTNTTMTGTVLKMTDRSHRQKLQLKALDTVLFGPPLLTRHNHLKDFMLVVSIVIG VGGCWFAYIQNRYSKEHMKKMMKDLEGLHRAEQSLHDLQERLHKAQEEHRTVEVEKVHLEKKLRDEINLAKQEAQRLKELREGTENERSR QKYAEEELEQVREALRKAEKELESHSSWYAPEALQKWLQLTHEVEVQYYNIKKQNAEKQLLVAKEGAEKIKKKRNTLFGTFHVAHSSSLD DVDHKILTAKQALSEVTAALRERLHRWQQIEILCGFQIVNNPGIHSLVAALNIDPSWMGSTRPNPAHFIMTDDVDDMDEEIVSPLSMQSP -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for YAP1-STIM1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for YAP1-STIM1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for YAP1-STIM1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
