
|
|||||||
|
Fusion Protein:KCNB1-ATP6V1E1 |
Fusion Gene and Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: KCNB1-ATP6V1E1 | FusionPDB ID: 41364 | FusionGDB2.0 ID: 41364 | Hgene | Tgene | Gene symbol | KCNB1 | ATP6V1E1 | Gene ID | 3745 | 529 |
| Gene name | potassium voltage-gated channel subfamily B member 1 | ATPase H+ transporting V1 subunit E1 | |
| Synonyms | DRK1|Kv2.1 | ARCL2C|ATP6E|ATP6E2|ATP6V1E|P31|Vma4 | |
| Cytomap | 20q13.13 | 22q11.21 | |
| Type of gene | protein-coding | protein-coding | |
| Description | potassium voltage-gated channel subfamily B member 1delayed rectifier potassium channel 1potassium voltage-gated channel, Shab-related subfamily, member 1voltage-gated potassium channel subunit Kv2.1 | V-type proton ATPase subunit E 1ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E1H(+)-transporting two-sector ATPase, 31kDa subunitH+-transporting ATP synthase chain E, vacuolarV-ATPase 31 kDa subunitV-ATPase subunit E 1V-ATPase, subunit Evac | |
| Modification date | 20200329 | 20200313 | |
| UniProtAcc | Q14721 Main function of 5'-partner protein: FUNCTION: Voltage-gated potassium channel that mediates transmembrane potassium transport in excitable membranes, primarily in the brain, but also in the pancreas and cardiovascular system. Contributes to the regulation of the action potential (AP) repolarization, duration and frequency of repetitive AP firing in neurons, muscle cells and endocrine cells and plays a role in homeostatic attenuation of electrical excitability throughout the brain (PubMed:23161216). Plays also a role in the regulation of exocytosis independently of its electrical function (By similarity). Forms tetrameric potassium-selective channels through which potassium ions pass in accordance with their electrochemical gradient. The channel alternates between opened and closed conformations in response to the voltage difference across the membrane. Homotetrameric channels mediate a delayed-rectifier voltage-dependent outward potassium current that display rapid activation and slow inactivation in response to membrane depolarization (PubMed:8081723, PubMed:1283219, PubMed:10484328, PubMed:12560340, PubMed:19074135, PubMed:19717558, PubMed:24901643). Can form functional homotetrameric and heterotetrameric channels that contain variable proportions of KCNB2; channel properties depend on the type of alpha subunits that are part of the channel (By similarity). Can also form functional heterotetrameric channels with other alpha subunits that are non-conducting when expressed alone, such as KCNF1, KCNG1, KCNG3, KCNG4, KCNH1, KCNH2, KCNS1, KCNS2, KCNS3 and KCNV1, creating a functionally diverse range of channel complexes (PubMed:10484328, PubMed:11852086, PubMed:12060745, PubMed:19074135, PubMed:19717558, PubMed:24901643). Heterotetrameric channel activity formed with KCNS3 show increased current amplitude with the threshold for action potential activation shifted towards more negative values in hypoxic-treated pulmonary artery smooth muscle cells (By similarity). Channel properties are also modulated by cytoplasmic ancillary beta subunits such as AMIGO1, KCNE1, KCNE2 and KCNE3, slowing activation and inactivation rate of the delayed rectifier potassium channels (By similarity). In vivo, membranes probably contain a mixture of heteromeric potassium channel complexes, making it difficult to assign currents observed in intact tissues to any particular potassium channel family member. Major contributor to the slowly inactivating delayed-rectifier voltage-gated potassium current in neurons of the central nervous system, sympathetic ganglion neurons, neuroendocrine cells, pancreatic beta cells, cardiomyocytes and smooth muscle cells. Mediates the major part of the somatodendritic delayed-rectifier potassium current in hippocampal and cortical pyramidal neurons and sympathetic superior cervical ganglion (CGC) neurons that acts to slow down periods of firing, especially during high frequency stimulation. Plays a role in the induction of long-term potentiation (LTP) of neuron excitability in the CA3 layer of the hippocampus (By similarity). Contributes to the regulation of glucose-induced action potential amplitude and duration in pancreatic beta cells, hence limiting calcium influx and insulin secretion (PubMed:23161216). Plays a role in the regulation of resting membrane potential and contraction in hypoxia-treated pulmonary artery smooth muscle cells. May contribute to the regulation of the duration of both the action potential of cardiomyocytes and the heart ventricular repolarization QT interval. Contributes to the pronounced pro-apoptotic potassium current surge during neuronal apoptotic cell death in response to oxidative injury. May confer neuroprotection in response to hypoxia/ischemic insults by suppressing pyramidal neurons hyperexcitability in hippocampal and cortical regions (By similarity). Promotes trafficking of KCNG3, KCNH1 and KCNH2 to the cell surface membrane, presumably by forming heterotetrameric channels with these subunits (PubMed:12060745). Plays a role in the calcium-dependent recruitment and release of fusion-competent vesicles from the soma of neurons, neuroendocrine and glucose-induced pancreatic beta cells by binding key components of the fusion machinery in a pore-independent manner (By similarity). {ECO:0000250|UniProtKB:P15387, ECO:0000250|UniProtKB:Q03717, ECO:0000269|PubMed:10484328, ECO:0000269|PubMed:11852086, ECO:0000269|PubMed:12060745, ECO:0000269|PubMed:12560340, ECO:0000269|PubMed:1283219, ECO:0000269|PubMed:19074135, ECO:0000269|PubMed:19717558, ECO:0000269|PubMed:23161216, ECO:0000269|PubMed:24901643, ECO:0000269|PubMed:8081723}. | P36543 Main function of 5'-partner protein: FUNCTION: Subunit of the peripheral V1 complex of vacuolar ATPase essential for assembly or catalytic function. V-ATPase is responsible for acidifying a variety of intracellular compartments in eukaryotic cells. | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000371741, | ENST00000399796, ENST00000399798, ENST00000478963, ENST00000253413, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 6 X 5 X 4=120 | 7 X 8 X 4=224 |
| # samples | 6 | 10 | |
| ** MAII score | log2(6/120*10)=-1 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(10/224*10)=-1.16349873228288 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Fusion gene context | PubMed: KCNB1 [Title/Abstract] AND ATP6V1E1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Fusion neoantigen context | PubMed: KCNB1 [Title/Abstract] AND ATP6V1E1 [Title/Abstract] AND neoantigen [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | KCNB1(48098450)-ATP6V1E1(18083947), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | KCNB1-ATP6V1E1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. KCNB1-ATP6V1E1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. KCNB1-ATP6V1E1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. KCNB1-ATP6V1E1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | KCNB1 | GO:0001508 | action potential | 19223394 |
| Hgene | KCNB1 | GO:0071805 | potassium ion transmembrane transport | 19074135|19223394 |
| Hgene | KCNB1 | GO:0090314 | positive regulation of protein targeting to membrane | 19074135 |
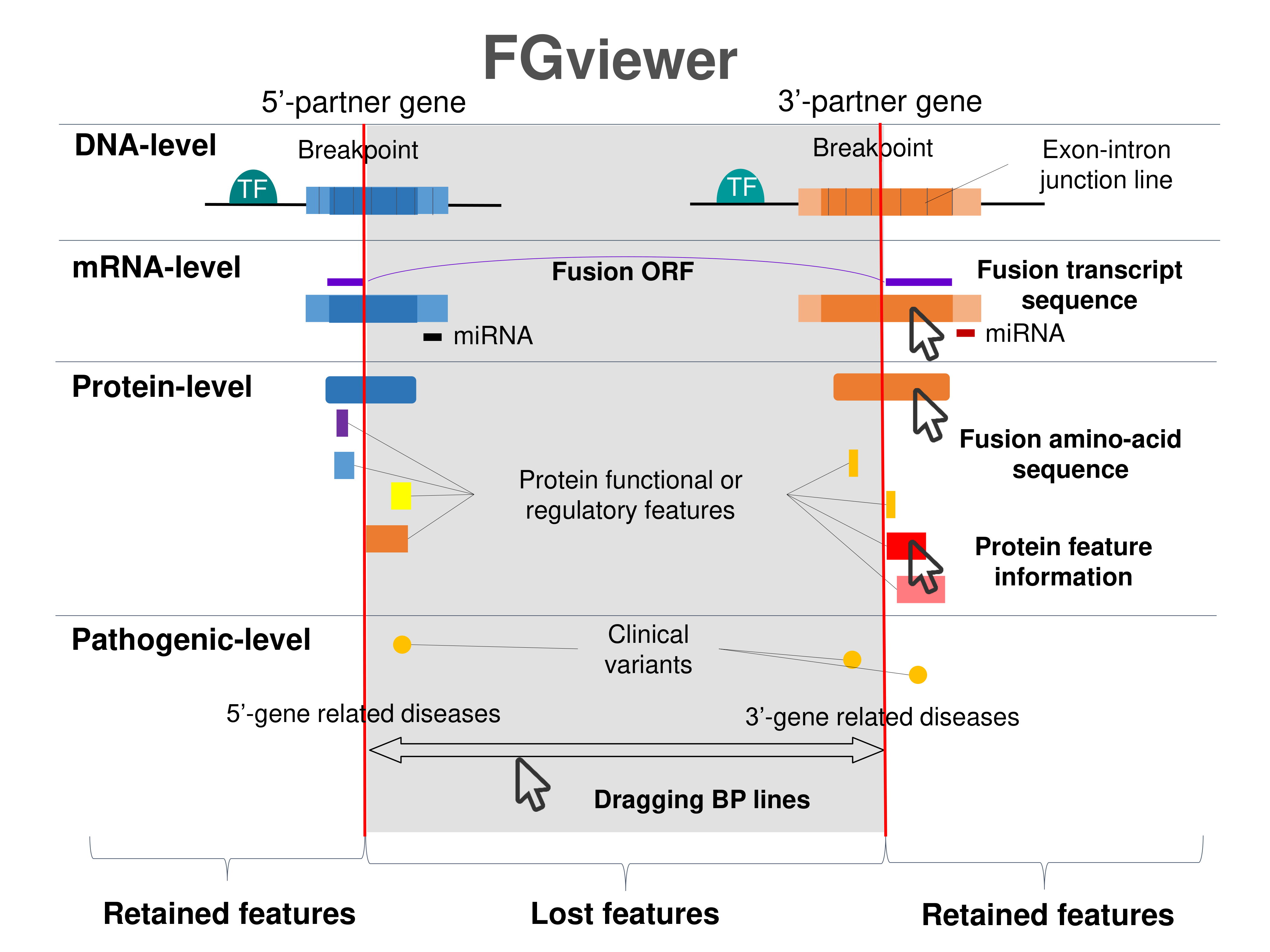
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr20:48098450/chr22:18083947) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Retention analysis results of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features, are available here. Retention analysis results of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features, are available here. |
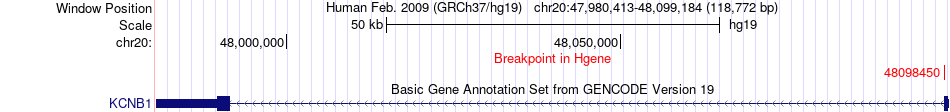
 Fusion gene breakpoints across KCNB1 (5'-gene) Fusion gene breakpoints across KCNB1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
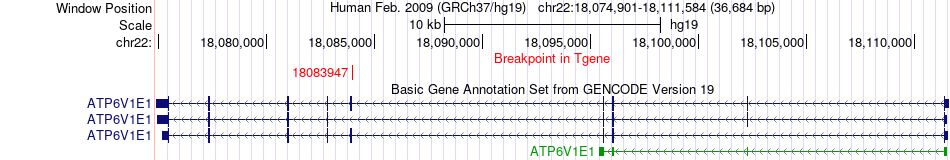
 Fusion gene breakpoints across ATP6V1E1 (3'-gene) Fusion gene breakpoints across ATP6V1E1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
Top |
Fusion Amino Acid Sequences |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000371741 | KCNB1 | chr20 | 48098450 | - | ENST00000253413 | ATP6V1E1 | chr22 | 18083947 | - | 1677 | 734 | 167 | 1138 | 323 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000371741 | ENST00000253413 | KCNB1 | chr20 | 48098450 | - | ATP6V1E1 | chr22 | 18083947 | - | 0.003351424 | 0.9966486 |
 Predicted full-length fusion amino acid sequences. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among all the predicted ones. Predicted full-length fusion amino acid sequences. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among all the predicted ones. |
Get the fusion protein sequences from here. |
| Fusion protein sequence information is available in the fasta format. >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP |
Top |
Fusion Protein Breakpoint Sequences for KCNB1-ATP6V1E1 |
 +/-13 AA sequence from the breakpoints of the fusion protein sequences. +/-13 AA sequence from the breakpoints of the fusion protein sequences. |
| Hgene | Hchr | Hbp | Tgene | Tchr | Tbp | Length(fusion protein) | BP in fusion protein | Peptide |
| KCNB1 | chr20 | 48098450 | ATP6V1E1 | chr22 | 18083947 | 734 | 189 | DLLEKPNSSVAAKDLLNEAKQRLSKV |
Top |
Potential FusionNeoAntigen Information of KCNB1-ATP6V1E1 in HLA I |
 Multiple sequence alignments of the potential FusionNeoAntigens per fusion breakpoints. If the MSA is empty, then it means that there were predicted fusion neoantigens in this fusion breakpoint, but those predicted fusion neoantigens were not across the breakpoint, which is not fusion-specific. Multiple sequence alignments of the potential FusionNeoAntigens per fusion breakpoints. If the MSA is empty, then it means that there were predicted fusion neoantigens in this fusion breakpoint, but those predicted fusion neoantigens were not across the breakpoint, which is not fusion-specific. |
| KCNB1-ATP6V1E1_48098450_18083947.msa |
 Potential FusionNeoAntigen Information Potential FusionNeoAntigen Information* We used NetMHCpan v4.1 (%rank<0.5) and deepHLApan v1.1 (immunogenic score>0.5) |
| Fusion gene | Hchr | Hbp | Tgene | Tchr | Tbp | HLA I | FusionNeoAntigen peptide | Binding score | Immunogenic score | Neoantigen start (at BP 13) | Neoantigen end (at BP 13) |
| KCNB1-ATP6V1E1 | chr20 | 48098450 | chr22 | 18083947 | 734 | HLA-B67:01 | KPNSSVAAKDL | 0.9266 | 0.5802 | 4 | 15 |
Top |
Potential FusionNeoAntigen Information of KCNB1-ATP6V1E1 in HLA II |
 Multiple sequence alignments of the potential FusionNeoAntigens per fusion breakpoints. If the MSA is empty, then it means that there were predicted fusion neoantigens in this fusion breakpoint, but those predicted fusion neoantigens were not across the breakpoint, which is not fusion-specific. Multiple sequence alignments of the potential FusionNeoAntigens per fusion breakpoints. If the MSA is empty, then it means that there were predicted fusion neoantigens in this fusion breakpoint, but those predicted fusion neoantigens were not across the breakpoint, which is not fusion-specific. |
| KCNB1-ATP6V1E1_48098450_18083947.msa |
 Potential FusionNeoAntigen Information Potential FusionNeoAntigen Information * We used NetMHCIIpan v4.1 (%rank<0.5). |
| Fusion gene | Hchr | Hbp | Tgene | Tchr | Tbp | HLA II | FusionNeoAntigen peptide | Neoantigen start (at BP 13) | Neoantigen end (at BP 13) |
| KCNB1-ATP6V1E1 | chr20 | 48098450 | chr22 | 18083947 | 734 | DRB1-0303 | AKDLLNEAKQRLSKV | 11 | 26 |
| KCNB1-ATP6V1E1 | chr20 | 48098450 | chr22 | 18083947 | 734 | DRB1-0303 | AAKDLLNEAKQRLSK | 10 | 25 |
| KCNB1-ATP6V1E1 | chr20 | 48098450 | chr22 | 18083947 | 734 | DRB1-0307 | AKDLLNEAKQRLSKV | 11 | 26 |
| KCNB1-ATP6V1E1 | chr20 | 48098450 | chr22 | 18083947 | 734 | DRB1-1107 | AKDLLNEAKQRLSKV | 11 | 26 |
| KCNB1-ATP6V1E1 | chr20 | 48098450 | chr22 | 18083947 | 734 | DRB1-1421 | AKDLLNEAKQRLSKV | 11 | 26 |
Top |
Fusion breakpoint peptide structures of KCNB1-ATP6V1E1 |
 3D structures of the fusion breakpoint peptide of 14AA sequence that have potential fusion neoantigens 3D structures of the fusion breakpoint peptide of 14AA sequence that have potential fusion neoantigens* The minimum length of the amino acid sequence in RoseTTAFold is 14AA. Here, we predicted the 14AA fusion protein breakpoint sequence not the fusion neoantigen peptide, which is shorter than 14 AA. |
| File name | BPseq | Hgene | Tgene | Hchr | Hbp | Tchr | Tbp | AAlen |
| 6400 | NSSVAAKDLLNEAK | KCNB1 | ATP6V1E1 | chr20 | 48098450 | chr22 | 18083947 | 734 |
Top |
Filtering FusionNeoAntigens Through Checking the Interaction with HLAs in 3D of KCNB1-ATP6V1E1 |
 Virtual screening between 25 HLAs (from PDB) and FusionNeoAntigens Virtual screening between 25 HLAs (from PDB) and FusionNeoAntigens* We used Glide to predict the interaction between HLAs and neoantigens. |
| HLA allele | PDB ID | File name | BPseq | Docking score | Glide score |
| HLA-B14:02 | 3BVN | 6400 | NSSVAAKDLLNEAK | -7.32466 | -7.43806 |
| HLA-B14:02 | 3BVN | 6400 | NSSVAAKDLLNEAK | -5.39793 | -6.43323 |
| HLA-B52:01 | 3W39 | 6400 | NSSVAAKDLLNEAK | -6.70926 | -6.82266 |
| HLA-B52:01 | 3W39 | 6400 | NSSVAAKDLLNEAK | -5.9249 | -6.9602 |
| HLA-A11:01 | 4UQ2 | 6400 | NSSVAAKDLLNEAK | -5.79744 | -6.83274 |
| HLA-A24:02 | 5HGA | 6400 | NSSVAAKDLLNEAK | -8.07755 | -8.19095 |
| HLA-A24:02 | 5HGA | 6400 | NSSVAAKDLLNEAK | -6.21161 | -7.24691 |
| HLA-B27:05 | 6PYJ | 6400 | NSSVAAKDLLNEAK | -5.0741 | -6.1094 |
| HLA-B27:05 | 6PYJ | 6400 | NSSVAAKDLLNEAK | -4.35973 | -4.47313 |
| HLA-B44:05 | 3DX8 | 6400 | NSSVAAKDLLNEAK | -8.39671 | -8.51011 |
| HLA-B44:05 | 3DX8 | 6400 | NSSVAAKDLLNEAK | -3.34895 | -4.38425 |
Top |
Vaccine Design for the FusionNeoAntigens of KCNB1-ATP6V1E1 |
 mRNA and peptide sequences of FusionNeoAntigens that have potential interaction with HLA-Is. mRNA and peptide sequences of FusionNeoAntigens that have potential interaction with HLA-Is. |
| Fusion gene | Hchr | Hbp | Tchr | Tbp | Start in +/-13AA | End in +/-13AA | FusionNeoAntigen peptide sequence | FusionNeoAntigen RNA sequence |
| KCNB1-ATP6V1E1 | chr20 | 48098450 | chr22 | 18083947 | 4 | 15 | KPNSSVAAKDL | AAGCCCAATTCCTCTGTGGCTGCCAAGGACCTA |
 mRNA and peptide sequences of FusionNeoAntigens that have potential interaction with HLA-IIs. mRNA and peptide sequences of FusionNeoAntigens that have potential interaction with HLA-IIs. |
| Fusion gene | Hchr | Hbp | Tchr | Tbp | Start in +/-13AA | End in +/-13AA | FusionNeoAntigen peptide | FusionNEoAntigen RNA sequence |
| KCNB1-ATP6V1E1 | chr20 | 48098450 | chr22 | 18083947 | 10 | 25 | AAKDLLNEAKQRLSK | GCTGCCAAGGACCTACTAAATGAAGCAAAACAGAGACTCAGCAAG |
| KCNB1-ATP6V1E1 | chr20 | 48098450 | chr22 | 18083947 | 11 | 26 | AKDLLNEAKQRLSKV | GCCAAGGACCTACTAAATGAAGCAAAACAGAGACTCAGCAAGGTG |
Top |
Information of the samples that have these potential fusion neoantigens of KCNB1-ATP6V1E1 |
 These samples were reported as having these fusion breakpoints. For individual breakpoints, we checked the open reading frames considering multiple gene isoforms and chose the in-frame fusion genes only. Then, we made fusion protein sequences and predicted the fusion neoantigens. These fusion-positive samples may have these potential fusion neoantigens. These samples were reported as having these fusion breakpoints. For individual breakpoints, we checked the open reading frames considering multiple gene isoforms and chose the in-frame fusion genes only. Then, we made fusion protein sequences and predicted the fusion neoantigens. These fusion-positive samples may have these potential fusion neoantigens. |
| Cancer type | Fusion gene | Hchr | Hbp | Henst | Tchr | Tbp | Tenst | Sample |
| LGG | KCNB1-ATP6V1E1 | chr20 | 48098450 | ENST00000371741 | chr22 | 18083947 | ENST00000253413 | TCGA-HT-8113 |
Top |
Potential target of CAR-T therapy development for KCNB1-ATP6V1E1 |
 Predicted 3D structure. We used RoseTTAFold. Predicted 3D structure. We used RoseTTAFold. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, to provide the retention of the transmembrane domain, we only show the protein feature retention information of those transmembrane features Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, to provide the retention of the transmembrane domain, we only show the protein feature retention information of those transmembrane features* Minus value of BPloci means that the break point is located before the CDS. |
| - In-frame and retained 'Transmembrane'. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
 Subcellular localization prediction of the transmembrane domain retained fusion proteins Subcellular localization prediction of the transmembrane domain retained fusion proteins* We used DeepLoc 1.0. The order of the X-axis of the barplot is as follows: Entry_ID, Localization, Type, Nucleus, Cytoplasm, Extracellular, Mitochondrion, Cell_membrane, Endoplasmic_reticulum, Plastid, Golgi.apparatus, Lysosome.Vacuole, Peroxisome. Y-axis is the output score of DeepLoc. Clicking the image will open a new tab with a large image. |
| Hgene | Hchr | Hbp | Henst | Tgene | Tchr | Tbp | Tenst | DeepLoc result |
Top |
Related Drugs to KCNB1-ATP6V1E1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to KCNB1-ATP6V1E1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

