|
Home |
Download |
Statistics |
Examples |
Help |
Contact |
Terms of Use |
Navigation
0. Overview of FusionNeoAntigen pipeline.1. Fusion Gene Data Collection.
2. Open Reading Frame (ORF) Analysis
3. Creation of Fusion Transcript and Amino Acid Sequences.
4. Protean Features Retention Analysis.
5. Identification of the breakpoint in the fusion protein sequence.
6. Prediction of the fusion neoantigens.
7. Prediction of the 3D structures of fusion breakpoint 14AA peptides.
8. Prediction of the interaction between the fusion breakpoint 14AA peptides and HLAs.
9. Cell surface fusion proteins.
10. Manual curation of fusion neoantigen literature.
11. Understanding of FusionNeoAntigen's Annotation Category.
- Search Page, example: TMPRSS2 involved fusion genes (specifically, TMPRSS2-ERG).
- FusionGene Search Result Page.
- FusionGene Annotation Result Page.
-- 1) Fusion Gene and Fusion Protein Summary.
-- 2) Fusion Amino Acid Sequences (multiple BPs and multiple gene isoforms).
-- 3) Fusion Protein Breakpoint Sequences - (for the Screening of the FusionNeoAntigens).
-- 4) Potential FusionNeoAntigens in HLA I - (netMHCpan v4.1 + deepHLApan v1.1).
-- 5) Potential FusionNeoAntigens in HLA II - (netMHCIIpan v4.1).
-- 6) FusionNeoAntigen Structures - (RoseTTAFold).
-- 7) Filtering FusionNeoAntigens Through Checking the Interaction with HLAs in 3D - (Glide).
-- 8) Vaccine Design for the FusionNeoAntigens (RNA/protein sequences).
-- 9) Potential target of CAR-T therapy development.
-- 10) Information on the samples that have these potential fusion neoantigens.
-- 11) Fusion Protein Targeting Drugs - (Manual Curation).
-- 12) Fusion Protein Related diseases - (Manual Curation).
12. Download Data and Contact Us.
0. Overview of FusionNeoAntigen pipeline.
The neoantigen studies were reported increasingly since 2015. However, so far, there were only 23 studies reported on the neoantigens in 19 fusion genes out of 266 manually curated fusion genes. Compared to the studies on the neoantigens derived from SNVs or Indels, the studies on the fusion neoantigens are very small. This is because of the lack of fusion protein sequence resource and knowledge. From our previous study, FusionGDB2, we successfully made ~ 43K fusion protein sequences from ~ 82K full-length fusion transcripts among ~ 16K in-frame fusion genes. These fusion protein sequences are currently being imported into UniProt as the reference source of the human fusion protein sequences. Furthermore, in FusionPDB, we are sharing the 3D structures of more than 3K fusion proteins. In this study, based on the ~ 43K fusion protein sequences, we predicted the fusion gene-derived neoantigens using the HLA binding affinity-based prediction tools (i.e., NetMHCpan and deepHLApan). To provide the fusion-specific neoantigens, we filtered out the neoantigens that are not crossed over the fusion protein breakpoint position. After filtering out based on output values of those tools, we finally identified about 10K and 7K fusion breakpoints that have potential interaction with HLA-Is and HLA-IIs, respectively. Each of these fusion breakpoints in the fusion protein sequences has multiple neoantigens crossing the fusion protein breakpoints. Taking the fusion breakpoint 14 AA sequence (since the minimum protein sequence length for prediction of protein structure is 14 AA in the RoseTTAFold), we predicted the 3D structures of ~ 10K fusion breakpoint 14 AA peptides. For these predicted fusion breakpoint peptide structures, we performed the virtual screening against 25 different HLAs, which have known 3D structures from PDB. By doing this, we could identify about half of the fusion breakpoint 14 AA peptides that showed the interaction with these HLA-Is.
1. Fusion Gene Data Collection
We downloaded the fusion gene information from FusionGDB2.0. Detailed information is on the statistics page.2. Creation of Fusion Transcript and Amino Acid Sequences.
Two different genes can form fusion genes with multiple breakpoints based on multiple gene isoforms. Therefore, we considered all gene isoforms at each breakpoint. To help with the identification and validation of fusion genes, we focused on the in-frame fusion genes. For more reliable fusion genes, we checked the distance between the two breakpoints in case of intra-chromosomal rearrangements and created fusion sequences when those genes are apart more than 100kb. We also selected fusion genes when both of their breakpoints are aligned at the exon junction. To call each exon sequence of the given breakpoint, transcription start/end sites, and CDS start/end sites, we used the nibFrag utility from UCSC Genome Browser based on ENCODE hg19 genome structure. By adding these exon sequences, we made the full-length fusion transcript sequences of the in-frame fusion genes. For these fusion transcript sequences, we input to the ORFfinder and chose the longest ORF as the potential fusion protein sequence.3. Open Reading Frame (ORF) Analysis.
To check the coding potential, we analyzed the ORF of the fusion transcript sequences. First, we investigated the ORF whether in-frame or frame-shift if both breakpoints are located in the coding sequence (CDS) area. If not, we reported the location of individual breakpoint is in 5'-UTR, CDS, or 3'-UTR. Second, to have the potential amino acid sequence, we ran ORFfinder by NCBI. Third, we ran the in-house classifier (to be available soon) between the coding genes mapped by Ribo-seq reads with high reliability and non-coding genes not mapped by any Ribo-seq reads.4. Protein Features Retention Analysis.
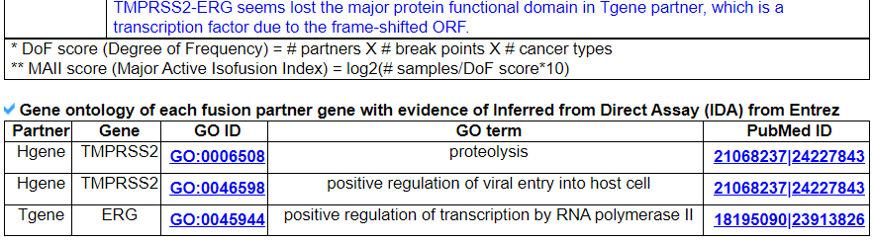
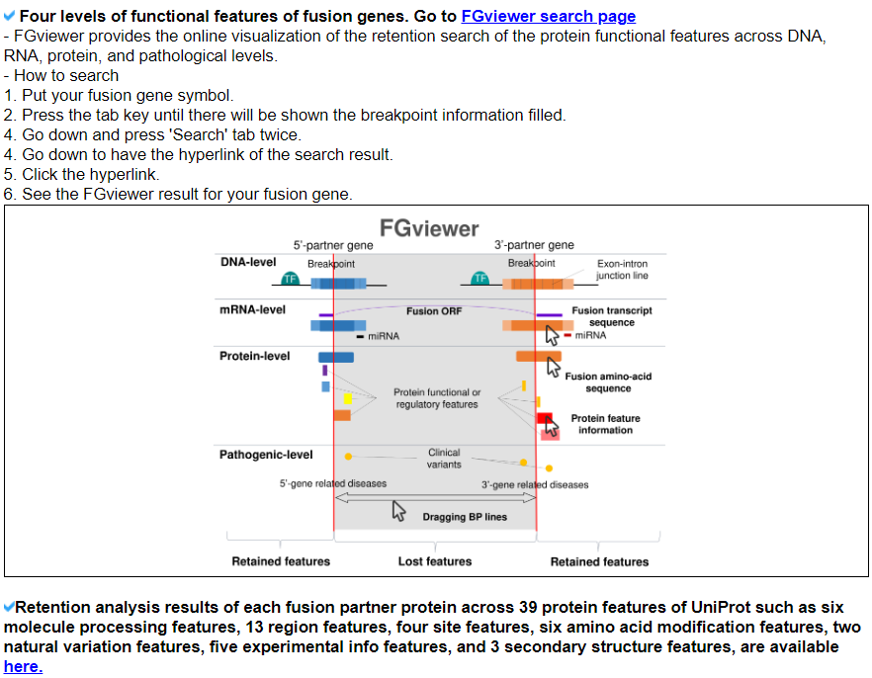
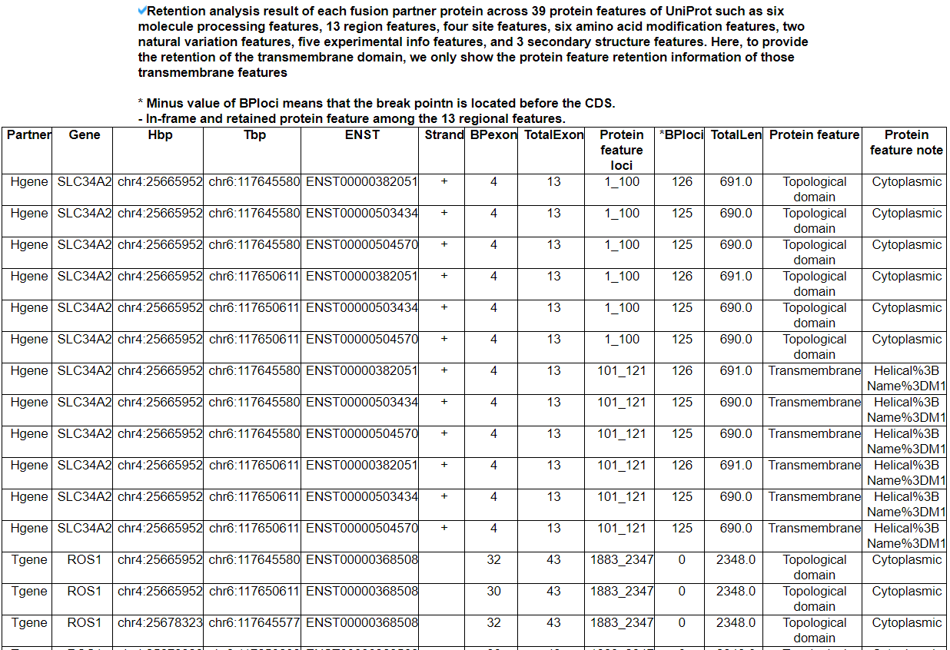
We searched the retention of 39 protein features of UniProt (six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features) at the fusion amino acid sequence level. Through this process, we also checked the retention of protein-protein interaction (PPI) at the fusion protein. Detailed information about all of the protein features is on the UniProt page. FGviewer provides functional feature annotations at four different levels: DNA-, RNA-, protein-, and pathogenic levels. The same breakpoint line across four tiers will classify between FG involving or non-involving zone with multiple types of functional features.5. Identification of the breakpoint in the fusion protein sequence.
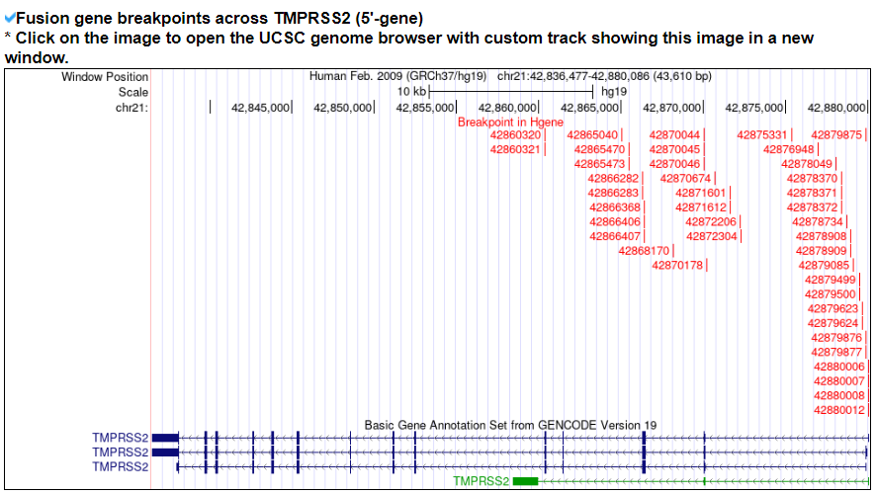
For individual fusion protein sequences, we predicted the breakpoints by running BLAT. Then, we matched the genomic position of the alignments with the exon junction breakpoint regions of the fusion genes. Then, to input to the binding affinity-based neoantigen prediction tools, we make +/-13AA sequences from the fusion protein breakpoints.6. Prediction of the fusion neoantigens.
To predict the fusion neoantigens that are interacting with HLA-Is, we used NetMHCpan v4.1 (%rank<0.5) and deepHLApan v1.1 (immunogenic score>0.5). To predict the fusion neoantigens that are interacting with HLA-IIs, We used NetMHCIIpan v4.1 (%rank<0.5).7. Prediction of the 3D structures of fusion breakpoint 14AA peptides.
To check the interaction between the fusion breakpoint peptides and HLAs, we predicted the 3D structures of individual fusion breakpoint peptides. Since the minimum length required in one of the accurate prediction of protein structure using AI, RoseTTAFold, was 14AA, we made the fusion breakpoint 14AA peptide sequences of ~ 10K fusion breakpoints that have the potential fusion neoantigens from running the binding affinity based prediction tools.8. Prediction of the interaction between the fusion breakpoint 14AA peptides and HLAs.
The grid size represents the volume of an active site where the ligand can search for binding while docking. The grids around the binding of 25 HLAs, which had the known 3D structures in PDB, were generated using a module named Receptor Grid Generation of the Schrödinger package. The dimensions of the grid were selected by considering the active site information available in the PDB database or predicted using the SiteMap module of the Schrödinger package. To perform the virtual screening, we constructed about 10K fusion neoantigen ligand libraries with the LigPrep of the Schrödinger package. Then, finally, we ran the GLIDE of the Schrödinger package against the known 25 HLA 3D structures.9. Cell surface fusion proteins.
We first downloaded the cell surface genes from The Cancer Surfaceome Atlas. There were 3,557 cell surface genes. Overlapping between these cell surface genes with the genes involved in 43K fusion proteins, we found ~ 14K fusion proteins that are potentially translated from 4,297 cell surface genes-involved fusion genes. Then, we investigated the retention of 'Transmembrane' in the fusion protein, we found 544 fusion genes that are keeping the transmembrane domain in their fusion protein structure.10. Manual curation of fusion neoantigen literature.
For 266 manually curated fusion genes, we searched PubMed literature that has previous results on neoantigen for individual fusion genes using this search terms, "BCR and ABL1 and neoantigen". We also considered gene synonyms.11. Understanding FusionNeoAntigen's Annotation Categories
Search page, example: TMPRSS2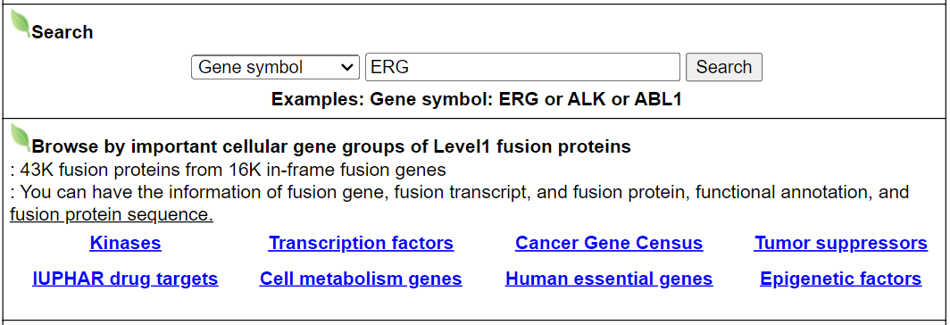
Input query: Official HUGO gene symbols.
FusionGene Search Result Page
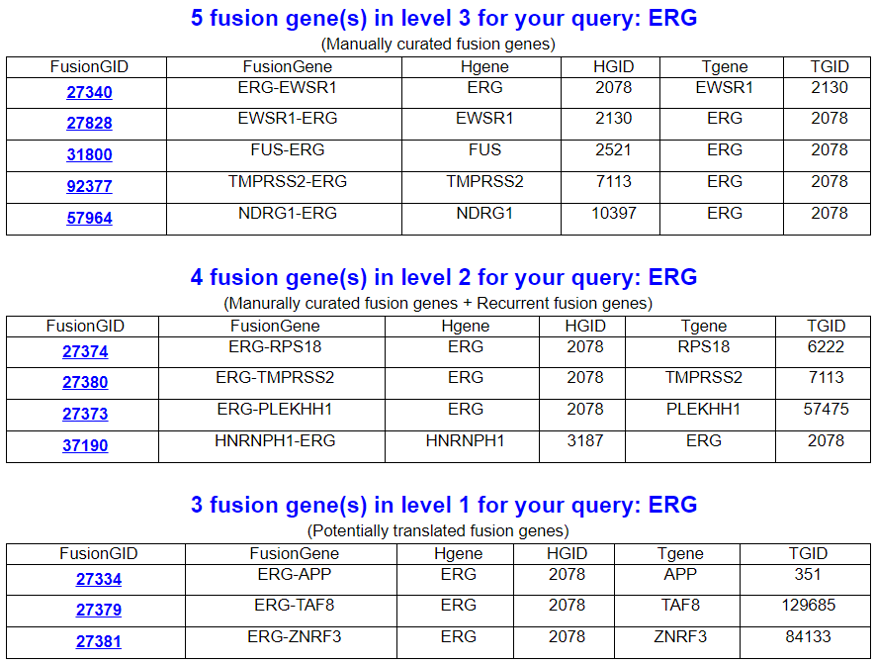
Select your fusion gene from the gene list. According to the fusion protein categories provided by the FusionPDB, we list the fusion protein names such as level 3, 2, and 1 for the manually curated fusions, manually curated + recurrent fusions, and the rest of these out of 43K fusion proteins, respectively.
FusionGene Annotation Result Page

These are FusionNeoAntigens's annotation categories for your query with links to their corresponding annotation parts.
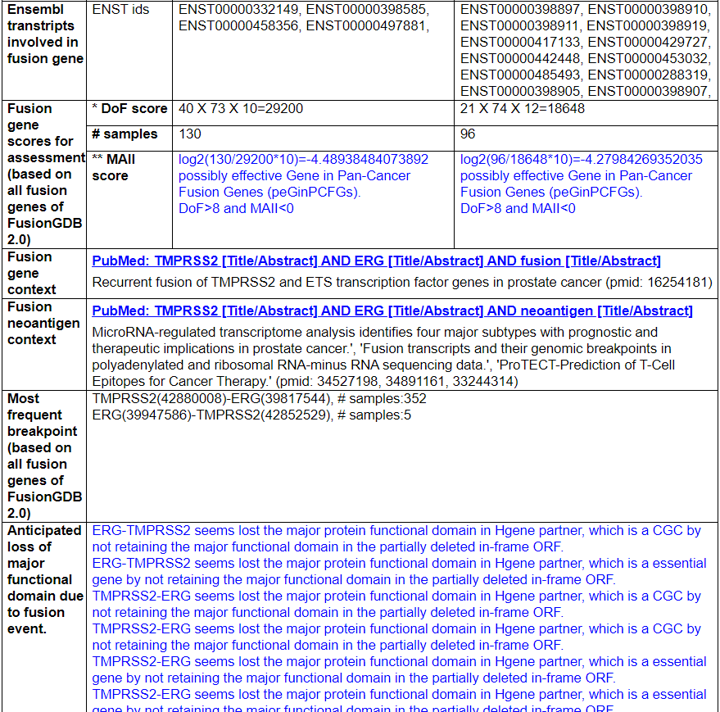
1) Fusion Gene and Fusion Protein Summary.
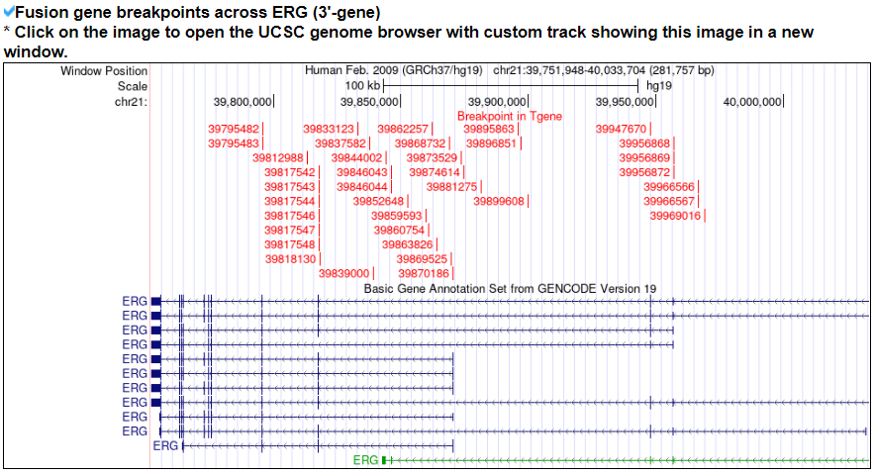
This category shows the information of the fusion gene/protein.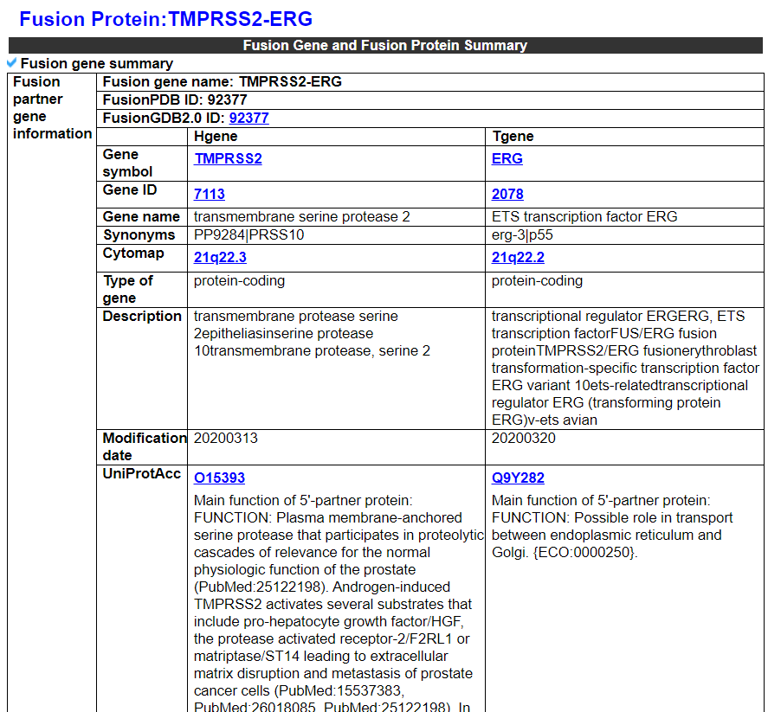
2) Fusion Amino Acid Sequences (multiple BPs and multiple gene isoforms).
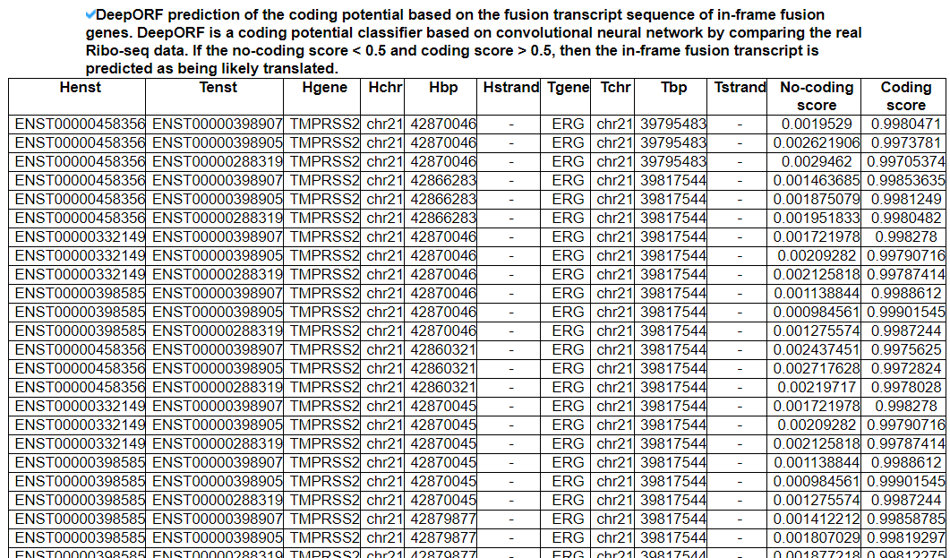
This category shows the coding potential study results from three approaches. First, we investigated the ORF whether in-frame or frame-shift if both breakpoints are located in the coding sequence (CDS) area. If not, we reported the location of individual breakpoints is in 5'-UTR, CDS, or 3'-UTR. Second, to have the potential amino acid sequence, we ran ORFfinder by NCBI. Third, we ran the in-house classifier (to be available soon) between the coding genes mapped by Ribo-seq reads with high reliability and non-coding genes not mapped by any Ribo-seq reads.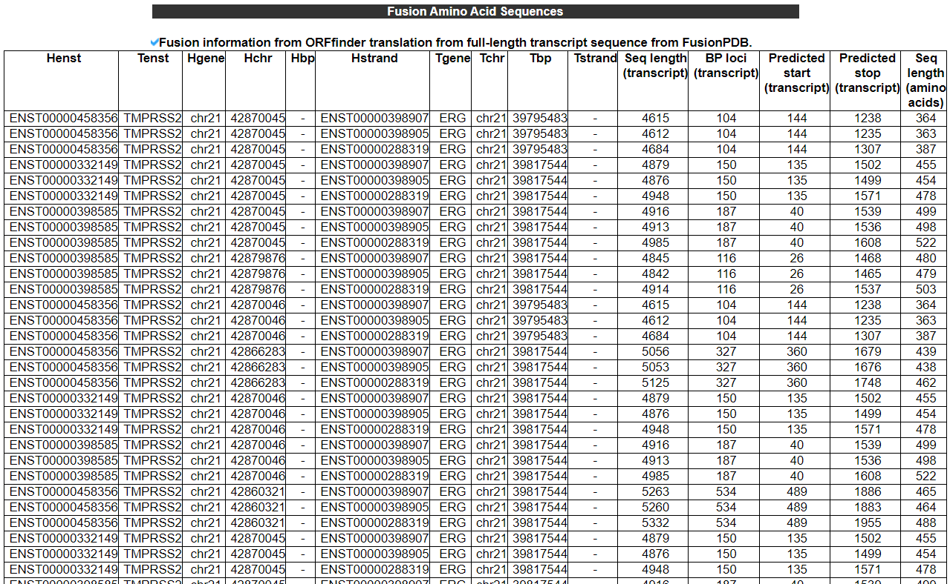
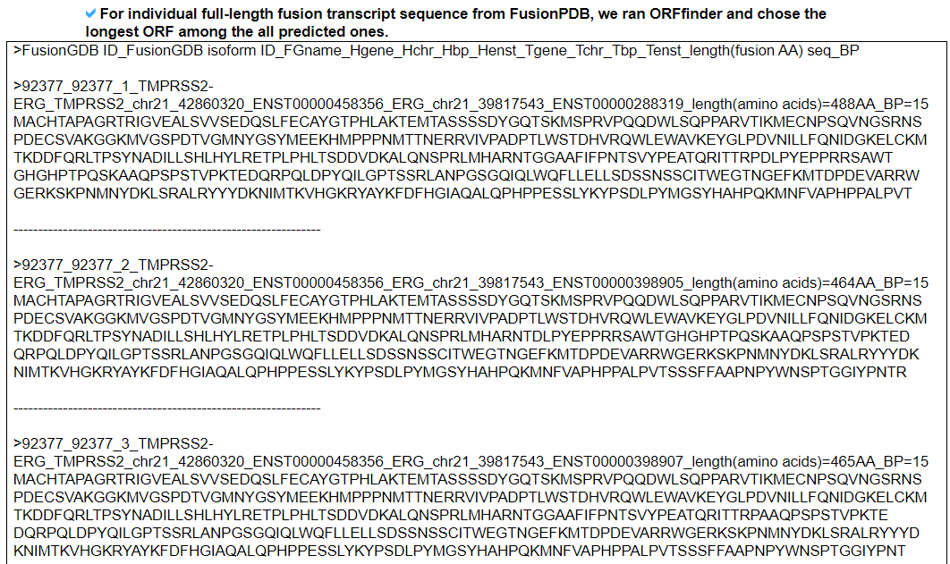
3) Fusion Protein Breakpoint Sequences - (for the Screening of the FusionNeoAntigens).
This category provides the fusion protein breakpoint sequences, the input material for the binding-affinity-based neoantigen prediction tools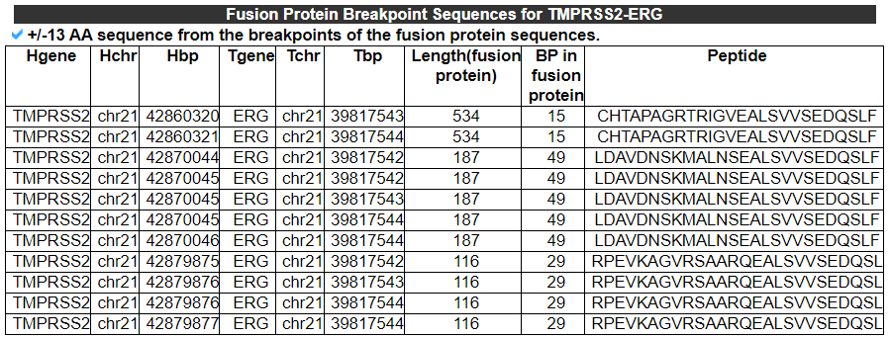
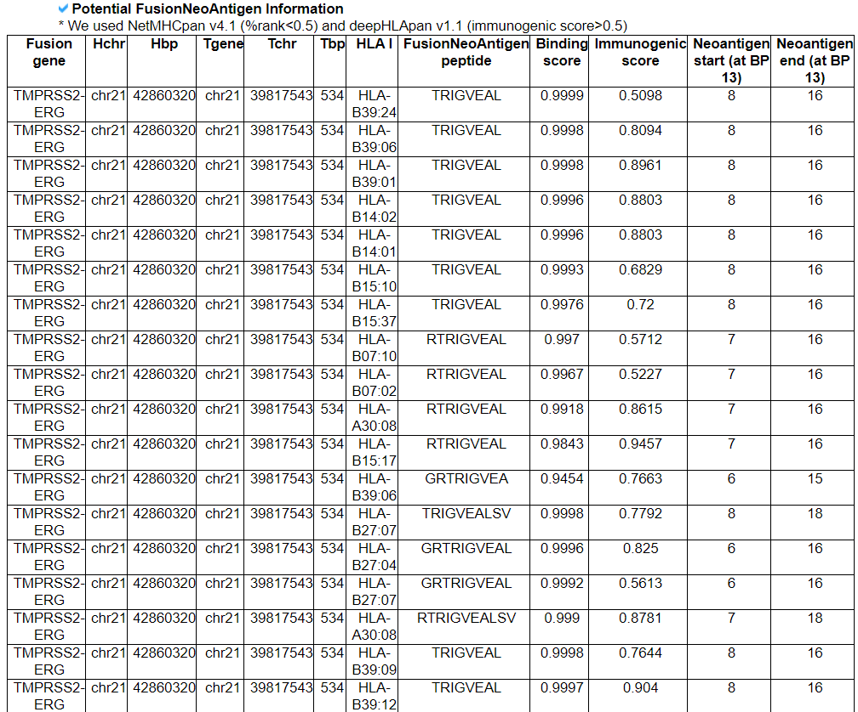
4) Potential FusionNeoAntigens in HLA-I.
This category provides the information of the predicted fusion neoantigens that have binding with HLA-Is.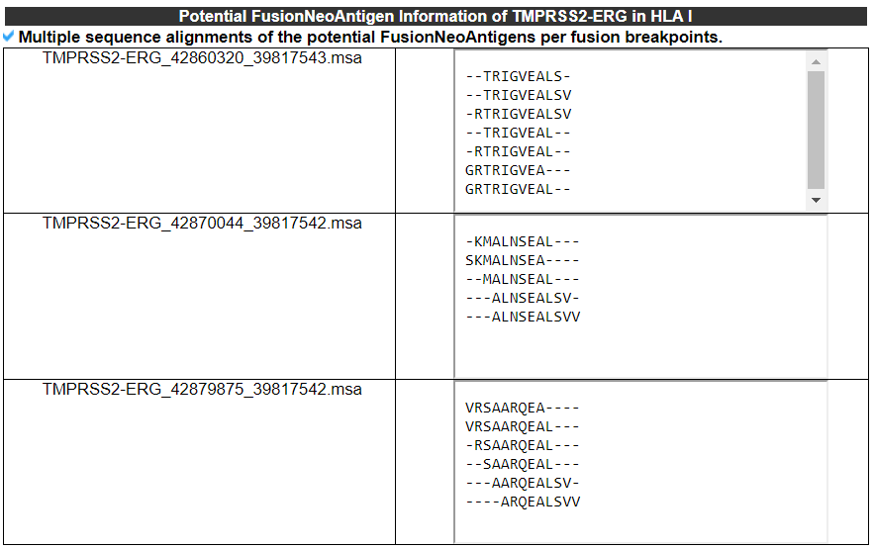
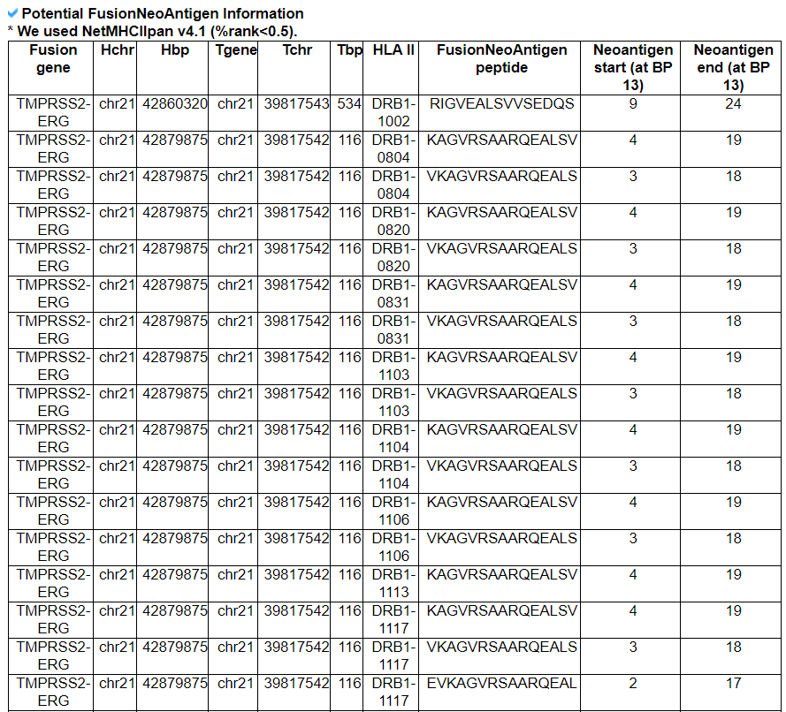
5) Potential FusionNeoAntigens in HLA-IIs.
This category provides the information of the predicted fusion neoantigens that have bind ing with HLA-IIs.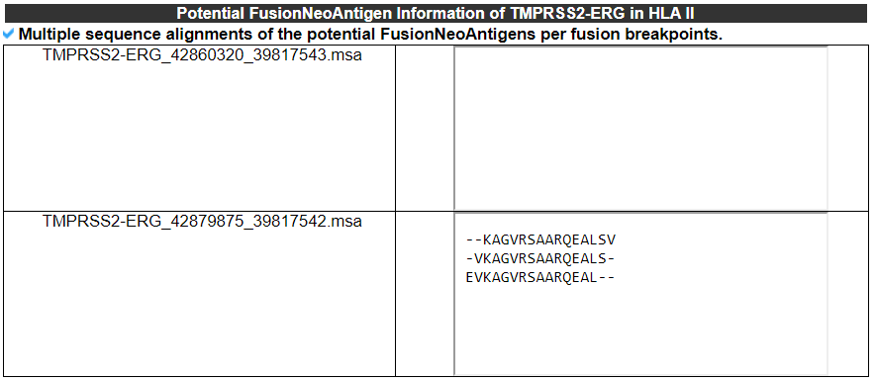
6) FusionNeoAntigen Structures - (RoseTTAFold).
This category shows the predicted 3D strutures of individul fusion breakpoint 14AA peptide sequences that have the potential fusion neoantigens. We used RoseTTAFold.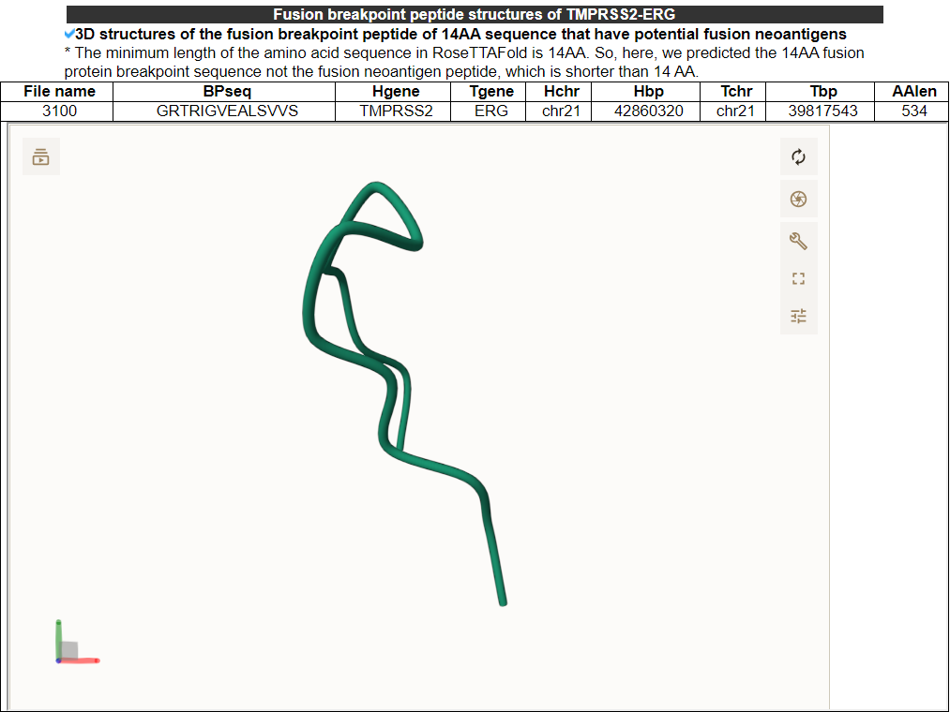
7) Filtering FusionNeoAntigens Through Checking the Interaction with HLAs in 3D - (Glide).
This table provides the virtual screening results between 25 HLAs that have the known 3D structures and ~ 10K of our predicted fusion breakpoint 14AA peptide structures that have the potential fusion neoantigens.
8) Vaccine Design for the FusionNeoAntigens (RNA/protein sequences).
This table provides the corresponding RNA and protein sequences of individual fusion neoantigens that have potential binding to HLA-Is.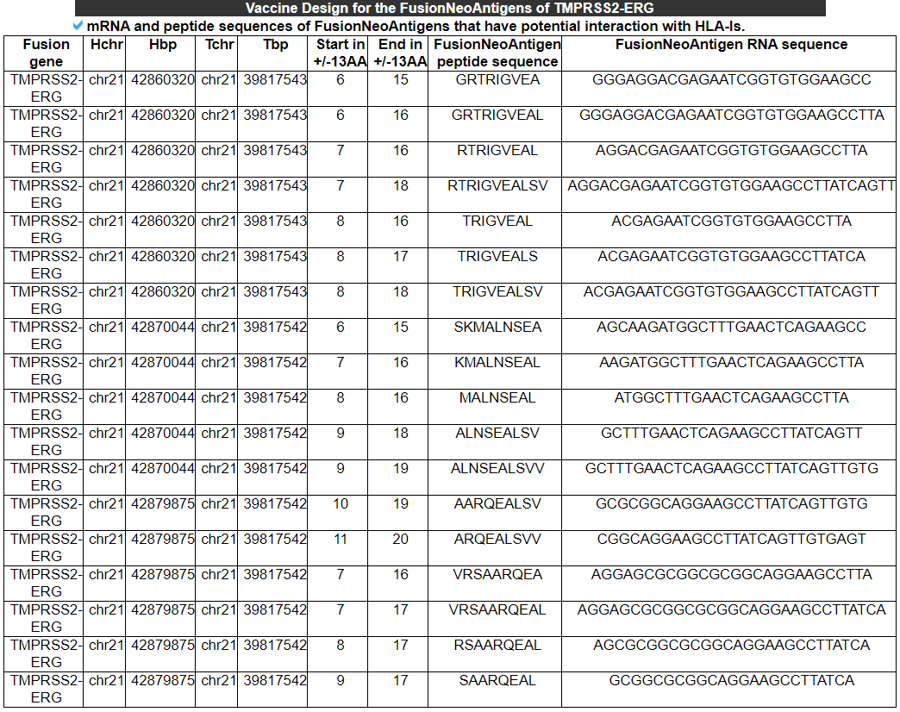
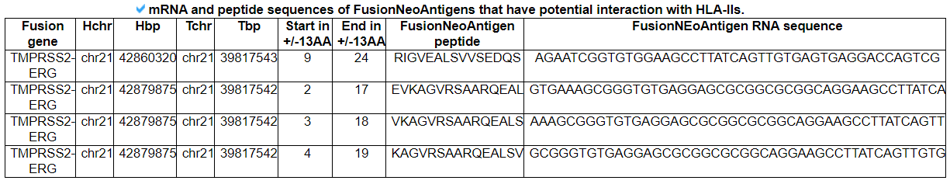
This table provides the corresponding RNA and protein sequences of individual fusion neoantigens that have potential binding to HLA-IIs.
9) 3D Structure of Fusion Protein - (in Case of Cell Surface Fusion Proteins as the Potential Target of the CAR-T Therapy) - (RoseTTAFold).
This part shows the predicted 3D structure of the cell surface fusion protein that retains the transmembrane domain in the fusion protein structure.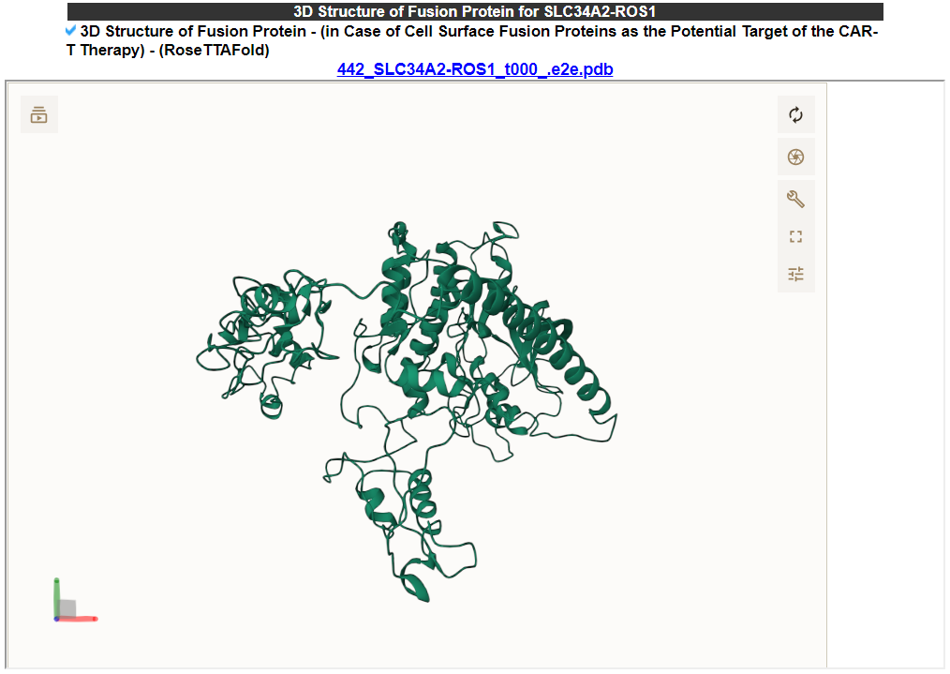
This table shows the transmembrane/topological domain retention information in the fusion proteins.
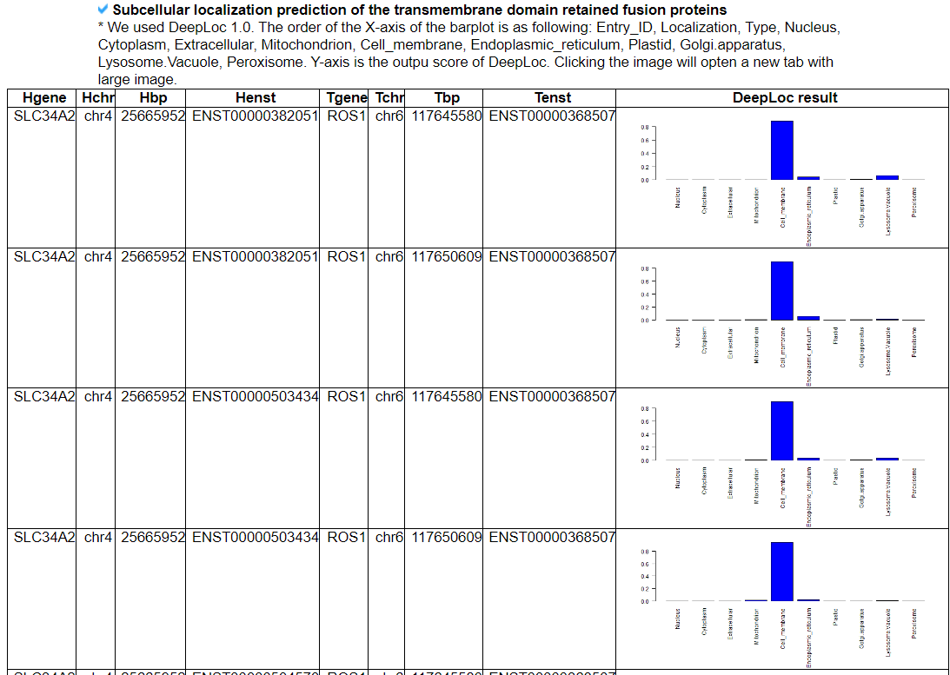
This part shows the potential location of the fusion protein in the cell.
10) Information on the samples that have these potential fusion neoantigens
This table provides the information of the samples that have potential fusion neoantigens from our analyses.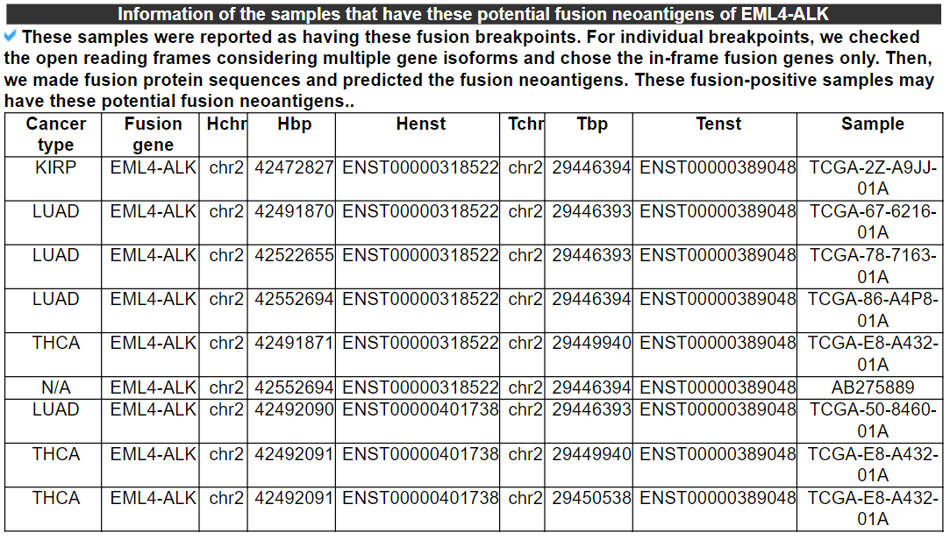
11) Fusion Protein Targeting Drugs - (Manual Curation)
This table provides information of drugs that were used to treat fusion gene patient from manual curation and multiple resources.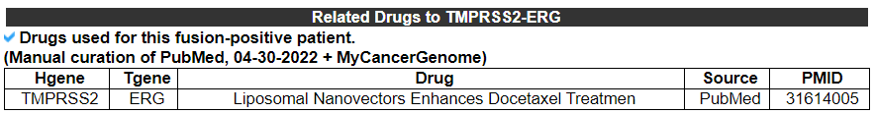
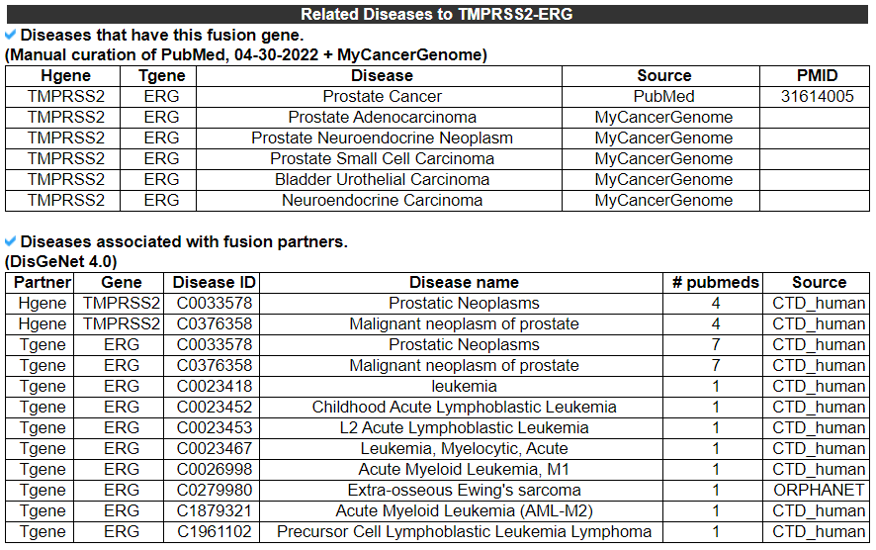
12) Fusion Protein Related diseases - (Manual Curation)
This table provides information of diseases that were expressed fusion gene from manual curation and multiple resources.