
|
|||||||
|
Fusion Protein:STK3-MTDH |
Fusion Gene and Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: STK3-MTDH | FusionPDB ID: 87574 | FusionGDB2.0 ID: 87574 | Hgene | Tgene | Gene symbol | STK3 | MTDH | Gene ID | 8428 | 92140 |
| Gene name | serine/threonine kinase 24 | metadherin | |
| Synonyms | HEL-S-95|MST3|MST3B|STE20|STK3 | 3D3|AEG-1|AEG1|LYRIC|LYRIC/3D3 | |
| Cytomap | 13q32.2 | 8q22.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | serine/threonine-protein kinase 24STE20-like kinase 3STE20-like kinase MST3epididymis secretory protein Li 95mammalian STE20-like protein kinase 3serine/threonine kinase 24 (STE20 homolog, yeast)sterile 20-like kinase 3 | protein LYRIC3D3/LYRICastrocyte elevated gene 1astrocyte elevated gene-1 proteinlysine-rich CEACAM1 co-isolated proteinmetastasis adhesion protein | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | Q86UE4 Main function of 5'-partner protein: FUNCTION: Downregulates SLC1A2/EAAT2 promoter activity when expressed ectopically. Activates the nuclear factor kappa-B (NF-kappa-B) transcription factor. Promotes anchorage-independent growth of immortalized melanocytes and astrocytes which is a key component in tumor cell expansion. Promotes lung metastasis and also has an effect on bone and brain metastasis, possibly by enhancing the seeding of tumor cells to the target organ endothelium. Induces chemoresistance. {ECO:0000269|PubMed:15927426, ECO:0000269|PubMed:16452207, ECO:0000269|PubMed:18316612, ECO:0000269|PubMed:19111877}. | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000523601, ENST00000419617, ENST00000521768, | ENST00000336273, ENST00000519934, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 24 X 16 X 11=4224 | 9 X 15 X 7=945 |
| # samples | 30 | 16 | |
| ** MAII score | log2(30/4224*10)=-3.81557542886257 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(16/945*10)=-2.56224242422107 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Fusion gene context | PubMed: STK3 [Title/Abstract] AND MTDH [Title/Abstract] AND fusion [Title/Abstract] | ||
| Fusion neoantigen context | PubMed: STK3 [Title/Abstract] AND MTDH [Title/Abstract] AND neoantigen [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | STK3(99718694)-MTDH(98673299), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | STK3-MTDH seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. STK3-MTDH seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. STK3-MTDH seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. STK3-MTDH seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. STK3-MTDH seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. STK3-MTDH seems lost the major protein functional domain in Hgene partner, which is a kinase due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | STK3 | GO:0006468 | protein phosphorylation | 19604147 |
| Hgene | STK3 | GO:0042542 | response to hydrogen peroxide | 22291017 |
| Hgene | STK3 | GO:0046777 | protein autophosphorylation | 17046825|17657516 |
| Tgene | MTDH | GO:0000122 | negative regulation of transcription by RNA polymerase II | 15927426 |
| Tgene | MTDH | GO:0010508 | positive regulation of autophagy | 21127263 |
| Tgene | MTDH | GO:0043066 | negative regulation of apoptotic process | 17704808 |
| Tgene | MTDH | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling | 16452207|18316612 |
| Tgene | MTDH | GO:0045766 | positive regulation of angiogenesis | 19940250 |
| Tgene | MTDH | GO:0051092 | positive regulation of NF-kappaB transcription factor activity | 16452207|18316612 |
| Tgene | MTDH | GO:0051897 | positive regulation of protein kinase B signaling | 17704808 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr8:99718694/chr8:98673299) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
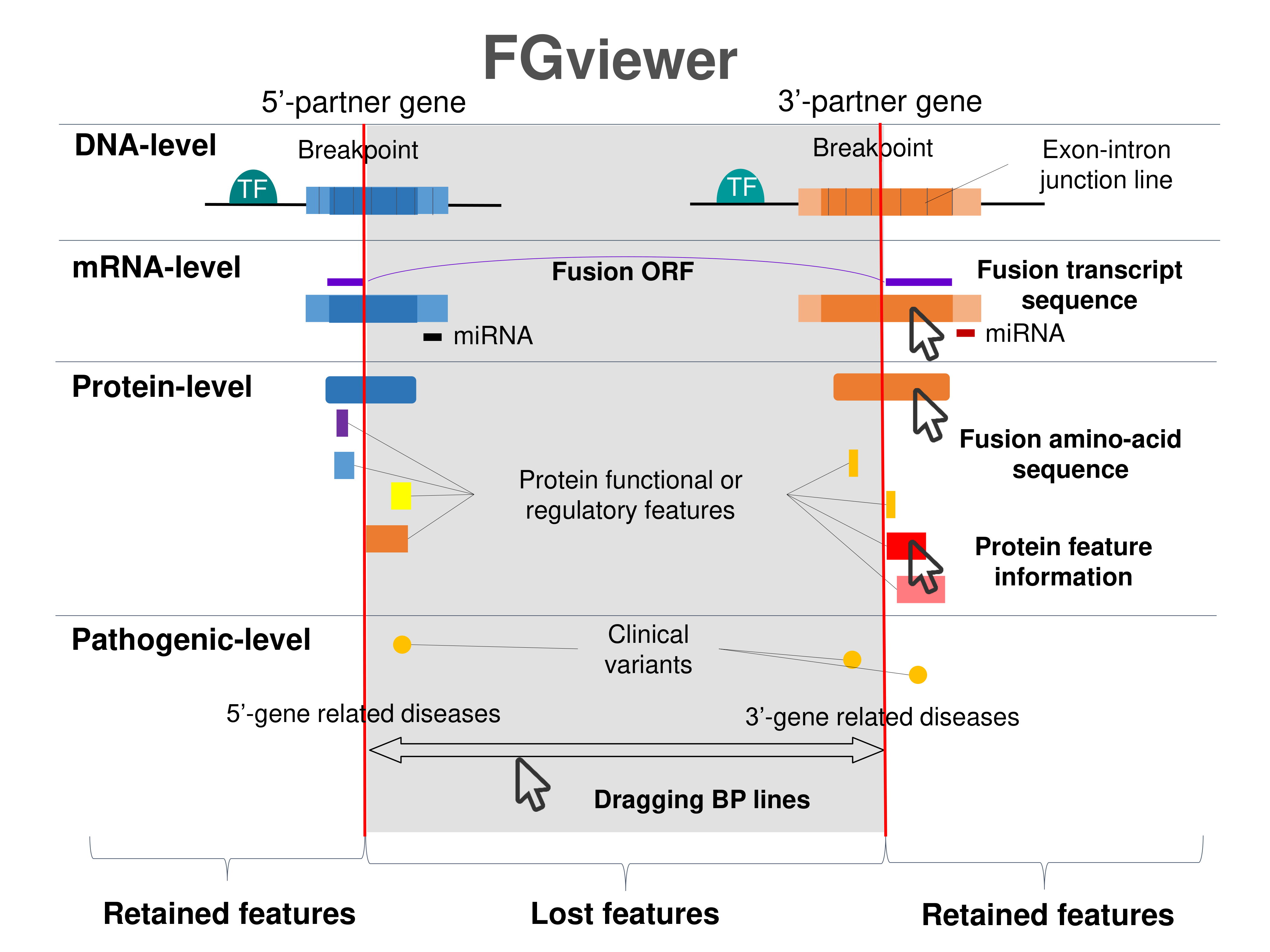 |
 Retention analysis results of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features, are available here. Retention analysis results of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features, are available here. |
 Fusion gene breakpoints across STK3 (5'-gene) Fusion gene breakpoints across STK3 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
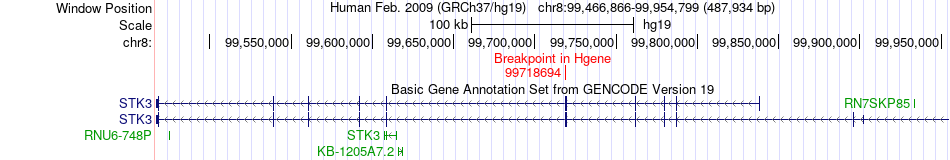 |
 Fusion gene breakpoints across MTDH (3'-gene) Fusion gene breakpoints across MTDH (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
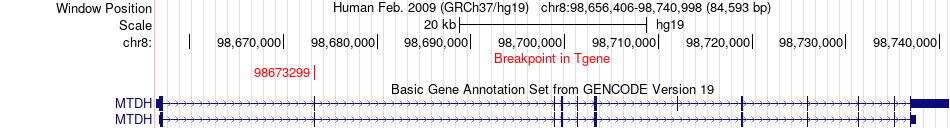 |
Top |
Fusion Amino Acid Sequences |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000419617 | STK3 | chr8 | 99718694 | - | ENST00000336273 | MTDH | chr8 | 98673299 | + | 6293 | 825 | 45 | 2192 | 715 |
| ENST00000419617 | STK3 | chr8 | 99718694 | - | ENST00000519934 | MTDH | chr8 | 98673299 | + | 2667 | 825 | 45 | 2093 | 682 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000419617 | ENST00000336273 | STK3 | chr8 | 99718694 | - | MTDH | chr8 | 98673299 | + | 0.000384093 | 0.9996159 |
| ENST00000419617 | ENST00000519934 | STK3 | chr8 | 99718694 | - | MTDH | chr8 | 98673299 | + | 0.000822269 | 0.99917775 |
 Predicted full-length fusion amino acid sequences. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among all the predicted ones. Predicted full-length fusion amino acid sequences. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among all the predicted ones. |
Get the fusion protein sequences from here. |
| Fusion protein sequence information is available in the fasta format. >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP |
Top |
Fusion Protein Breakpoint Sequences for STK3-MTDH |
 +/-13 AA sequence from the breakpoints of the fusion protein sequences. +/-13 AA sequence from the breakpoints of the fusion protein sequences. |
| Hgene | Hchr | Hbp | Tgene | Tchr | Tbp | Length(fusion protein) | BP in fusion protein | Peptide |
| STK3 | chr8 | 99718694 | MTDH | chr8 | 98673299 | 825 | 260 | EGKPPYADIHPMRPNGRTVEVAEGEA |
Top |
Potential FusionNeoAntigen Information of STK3-MTDH in HLA I |
 Multiple sequence alignments of the potential FusionNeoAntigens per fusion breakpoints. If the MSA is empty, then it means that there were predicted fusion neoantigens in this fusion breakpoint, but those predicted fusion neoantigens were not across the breakpoint, which is not fusion-specific. Multiple sequence alignments of the potential FusionNeoAntigens per fusion breakpoints. If the MSA is empty, then it means that there were predicted fusion neoantigens in this fusion breakpoint, but those predicted fusion neoantigens were not across the breakpoint, which is not fusion-specific. |
| STK3-MTDH_99718694_98673299.msa |
 Potential FusionNeoAntigen Information Potential FusionNeoAntigen Information* We used NetMHCpan v4.1 (%rank<0.5) and deepHLApan v1.1 (immunogenic score>0.5) |
| Fusion gene | Hchr | Hbp | Tgene | Tchr | Tbp | HLA I | FusionNeoAntigen peptide | Binding score | Immunogenic score | Neoantigen start (at BP 13) | Neoantigen end (at BP 13) |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B07:02 | RPNGRTVEV | 0.9996 | 0.7097 | 12 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B07:05 | RPNGRTVEV | 0.9996 | 0.611 | 12 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B07:10 | RPNGRTVEV | 0.9996 | 0.7298 | 12 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B08:09 | RPNGRTVEV | 0.9902 | 0.6592 | 12 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B14:01 | RPNGRTVEV | 0.8938 | 0.8535 | 12 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B14:02 | RPNGRTVEV | 0.8938 | 0.8535 | 12 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B35:04 | RPNGRTVEV | 0.643 | 0.9013 | 12 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B35:02 | RPNGRTVEV | 0.643 | 0.9013 | 12 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B82:01 | RPNGRTVEV | 0.356 | 0.5006 | 12 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B81:01 | RPNGRTVEV | 0.3422 | 0.5126 | 12 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B07:02 | RPNGRTVEVA | 0.9975 | 0.7298 | 12 | 22 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B07:05 | RPNGRTVEVA | 0.9974 | 0.6242 | 12 | 22 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B07:02 | HPMRPNGRTV | 0.996 | 0.6343 | 9 | 19 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B07:05 | HPMRPNGRTV | 0.9959 | 0.5567 | 9 | 19 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-A33:01 | DIHPMRPNGR | 0.9906 | 0.8538 | 7 | 17 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-A33:05 | DIHPMRPNGR | 0.9906 | 0.8538 | 7 | 17 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-A68:03 | DIHPMRPNGR | 0.9773 | 0.7045 | 7 | 17 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B08:09 | HPMRPNGRTV | 0.9697 | 0.7456 | 9 | 19 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B08:09 | MRPNGRTVEV | 0.9056 | 0.8071 | 11 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B35:03 | HPMRPNGRTV | 0.778 | 0.7655 | 9 | 19 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B82:01 | MRPNGRTVEV | 0.7132 | 0.6631 | 11 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B81:01 | HPMRPNGRTV | 0.6582 | 0.5792 | 9 | 19 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B82:01 | HPMRPNGRTV | 0.5966 | 0.6035 | 9 | 19 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B07:12 | RPNGRTVEV | 0.9983 | 0.7468 | 12 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-C08:15 | YADIHPMRP | 0.9965 | 0.9376 | 5 | 14 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B07:04 | RPNGRTVEV | 0.9892 | 0.642 | 12 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B14:03 | RPNGRTVEV | 0.9282 | 0.8271 | 12 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B54:01 | RPNGRTVEV | 0.9153 | 0.5266 | 12 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B35:12 | RPNGRTVEV | 0.643 | 0.9013 | 12 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B39:10 | RPNGRTVEV | 0.5553 | 0.7626 | 12 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B78:01 | RPNGRTVEV | 0.5498 | 0.5141 | 12 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-C04:14 | RPNGRTVEV | 0.2563 | 0.9637 | 12 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-C04:10 | RPNGRTVEV | 0.0064 | 0.9386 | 12 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-C04:07 | RPNGRTVEV | 0.0063 | 0.9367 | 12 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B54:01 | HPMRPNGRTV | 0.9885 | 0.6517 | 9 | 19 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B07:12 | HPMRPNGRTV | 0.9882 | 0.7302 | 9 | 19 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B07:12 | RPNGRTVEVA | 0.985 | 0.7413 | 12 | 22 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B42:02 | HPMRPNGRTV | 0.9819 | 0.6444 | 9 | 19 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B42:01 | HPMRPNGRTV | 0.9764 | 0.6403 | 9 | 19 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B07:04 | RPNGRTVEVA | 0.9752 | 0.6834 | 12 | 22 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B54:01 | MRPNGRTVEV | 0.9732 | 0.7008 | 11 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B07:04 | HPMRPNGRTV | 0.9697 | 0.5801 | 9 | 19 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B42:02 | MRPNGRTVEV | 0.9329 | 0.7984 | 11 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B56:04 | HPMRPNGRTV | 0.9242 | 0.5407 | 9 | 19 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B42:01 | MRPNGRTVEV | 0.9202 | 0.7868 | 11 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-A33:03 | DIHPMRPNGR | 0.8932 | 0.5482 | 7 | 17 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B54:01 | RPNGRTVEVA | 0.884 | 0.6499 | 12 | 22 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B39:10 | HPMRPNGRTV | 0.6915 | 0.911 | 9 | 19 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B78:01 | MRPNGRTVEV | 0.3424 | 0.7937 | 11 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-C06:02 | MRPNGRTV | 0.909 | 0.9917 | 11 | 19 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-C06:17 | MRPNGRTV | 0.909 | 0.9917 | 11 | 19 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B07:22 | RPNGRTVEV | 0.9996 | 0.7097 | 12 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B07:09 | RPNGRTVEV | 0.9996 | 0.6848 | 12 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B07:13 | RPNGRTVEV | 0.999 | 0.912 | 12 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-C08:02 | YADIHPMRP | 0.9965 | 0.9376 | 5 | 14 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B08:12 | RPNGRTVEV | 0.8962 | 0.5134 | 12 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B35:09 | RPNGRTVEV | 0.643 | 0.9013 | 12 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B67:01 | RPNGRTVEV | 0.6388 | 0.7525 | 12 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B35:13 | RPNGRTVEV | 0.6178 | 0.6975 | 12 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-C04:04 | RPNGRTVEV | 0.5474 | 0.9179 | 12 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B78:02 | RPNGRTVEV | 0.5267 | 0.621 | 12 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-C07:04 | RPNGRTVEV | 0.4027 | 0.9575 | 12 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B82:02 | RPNGRTVEV | 0.356 | 0.5006 | 12 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-C18:01 | RPNGRTVEV | 0.01 | 0.9493 | 12 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-C04:01 | RPNGRTVEV | 0.0063 | 0.9367 | 12 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B07:22 | RPNGRTVEVA | 0.9975 | 0.7298 | 12 | 22 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B07:09 | RPNGRTVEVA | 0.9963 | 0.6929 | 12 | 22 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B07:22 | HPMRPNGRTV | 0.996 | 0.6343 | 9 | 19 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B07:09 | HPMRPNGRTV | 0.9954 | 0.6125 | 9 | 19 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B55:04 | MRPNGRTVEV | 0.9902 | 0.6013 | 11 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B55:02 | HPMRPNGRTV | 0.973 | 0.5234 | 9 | 19 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B59:01 | HPMRPNGRTV | 0.9452 | 0.5556 | 9 | 19 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B55:04 | HPMRPNGRTV | 0.9435 | 0.5281 | 9 | 19 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B56:02 | HPMRPNGRTV | 0.9242 | 0.5407 | 9 | 19 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B78:02 | HPMRPNGRTV | 0.8254 | 0.5734 | 9 | 19 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B67:01 | HPMRPNGRTV | 0.7612 | 0.8162 | 9 | 19 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B82:02 | MRPNGRTVEV | 0.7132 | 0.6631 | 11 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | HLA-B82:02 | HPMRPNGRTV | 0.5966 | 0.6035 | 9 | 19 |
Top |
Potential FusionNeoAntigen Information of STK3-MTDH in HLA II |
 Multiple sequence alignments of the potential FusionNeoAntigens per fusion breakpoints. If the MSA is empty, then it means that there were predicted fusion neoantigens in this fusion breakpoint, but those predicted fusion neoantigens were not across the breakpoint, which is not fusion-specific. Multiple sequence alignments of the potential FusionNeoAntigens per fusion breakpoints. If the MSA is empty, then it means that there were predicted fusion neoantigens in this fusion breakpoint, but those predicted fusion neoantigens were not across the breakpoint, which is not fusion-specific. |
| STK3-MTDH_99718694_98673299.msa |
 Potential FusionNeoAntigen Information Potential FusionNeoAntigen Information * We used NetMHCIIpan v4.1 (%rank<0.5). |
| Fusion gene | Hchr | Hbp | Tgene | Tchr | Tbp | HLA II | FusionNeoAntigen peptide | Neoantigen start (at BP 13) | Neoantigen end (at BP 13) |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | DRB1-1452 | YADIHPMRPNGRTVE | 5 | 20 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | DRB5-0101 | YADIHPMRPNGRTVE | 5 | 20 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | DRB5-0105 | YADIHPMRPNGRTVE | 5 | 20 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | DRB5-0106 | YADIHPMRPNGRTVE | 5 | 20 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | DRB5-0106 | PYADIHPMRPNGRTV | 4 | 19 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | DRB5-0106 | ADIHPMRPNGRTVEV | 6 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | DRB5-0106 | PPYADIHPMRPNGRT | 3 | 18 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | DRB5-0111 | YADIHPMRPNGRTVE | 5 | 20 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | DRB5-0113 | YADIHPMRPNGRTVE | 5 | 20 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | DRB5-0114 | YADIHPMRPNGRTVE | 5 | 20 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | DRB5-0202 | YADIHPMRPNGRTVE | 5 | 20 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | DRB5-0202 | PYADIHPMRPNGRTV | 4 | 19 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | DRB5-0204 | YADIHPMRPNGRTVE | 5 | 20 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | DRB5-0204 | PYADIHPMRPNGRTV | 4 | 19 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | DRB5-0204 | ADIHPMRPNGRTVEV | 6 | 21 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | DRB5-0205 | YADIHPMRPNGRTVE | 5 | 20 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | DRB5-0205 | PYADIHPMRPNGRTV | 4 | 19 |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 | DRB5-0205 | ADIHPMRPNGRTVEV | 6 | 21 |
Top |
Fusion breakpoint peptide structures of STK3-MTDH |
 3D structures of the fusion breakpoint peptide of 14AA sequence that have potential fusion neoantigens 3D structures of the fusion breakpoint peptide of 14AA sequence that have potential fusion neoantigens* The minimum length of the amino acid sequence in RoseTTAFold is 14AA. Here, we predicted the 14AA fusion protein breakpoint sequence not the fusion neoantigen peptide, which is shorter than 14 AA. |
| File name | BPseq | Hgene | Tgene | Hchr | Hbp | Tchr | Tbp | AAlen |
| 117 | ADIHPMRPNGRTVE | STK3 | MTDH | chr8 | 99718694 | chr8 | 98673299 | 825 |
Top |
Filtering FusionNeoAntigens Through Checking the Interaction with HLAs in 3D of STK3-MTDH |
 Virtual screening between 25 HLAs (from PDB) and FusionNeoAntigens Virtual screening between 25 HLAs (from PDB) and FusionNeoAntigens* We used Glide to predict the interaction between HLAs and neoantigens. |
| HLA allele | PDB ID | File name | BPseq | Docking score | Glide score |
| HLA-B14:02 | 3BVN | 117 | ADIHPMRPNGRTVE | -8.60826 | -8.72166 |
| HLA-B14:02 | 3BVN | 117 | ADIHPMRPNGRTVE | -2.66483 | -3.70013 |
| HLA-B52:01 | 3W39 | 117 | ADIHPMRPNGRTVE | -6.90747 | -7.02087 |
| HLA-B52:01 | 3W39 | 117 | ADIHPMRPNGRTVE | -1.88432 | -2.91962 |
| HLA-A11:01 | 4UQ2 | 117 | ADIHPMRPNGRTVE | -11.2418 | -11.3552 |
| HLA-A24:02 | 5HGA | 117 | ADIHPMRPNGRTVE | -9.24546 | -9.35886 |
| HLA-A24:02 | 5HGA | 117 | ADIHPMRPNGRTVE | -4.08507 | -5.12037 |
| HLA-B44:05 | 3DX8 | 117 | ADIHPMRPNGRTVE | -7.05529 | -7.16869 |
| HLA-B44:05 | 3DX8 | 117 | ADIHPMRPNGRTVE | -4.33606 | -5.37136 |
| HLA-A02:01 | 6TDR | 117 | ADIHPMRPNGRTVE | -6.57504 | -6.68844 |
Top |
Vaccine Design for the FusionNeoAntigens of STK3-MTDH |
 mRNA and peptide sequences of FusionNeoAntigens that have potential interaction with HLA-Is. mRNA and peptide sequences of FusionNeoAntigens that have potential interaction with HLA-Is. |
| Fusion gene | Hchr | Hbp | Tchr | Tbp | Start in +/-13AA | End in +/-13AA | FusionNeoAntigen peptide sequence | FusionNeoAntigen RNA sequence |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 11 | 19 | MRPNGRTV | ATGAGGCCAAATGGGCGGACTGTT |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 11 | 21 | MRPNGRTVEV | ATGAGGCCAAATGGGCGGACTGTTGAAGTG |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 12 | 21 | RPNGRTVEV | AGGCCAAATGGGCGGACTGTTGAAGTG |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 12 | 22 | RPNGRTVEVA | AGGCCAAATGGGCGGACTGTTGAAGTGGCT |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 5 | 14 | YADIHPMRP | TATGCTGATATACATCCAATGAGGCCA |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 7 | 17 | DIHPMRPNGR | GATATACATCCAATGAGGCCAAATGGGCGG |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 9 | 19 | HPMRPNGRTV | CATCCAATGAGGCCAAATGGGCGGACTGTT |
 mRNA and peptide sequences of FusionNeoAntigens that have potential interaction with HLA-IIs. mRNA and peptide sequences of FusionNeoAntigens that have potential interaction with HLA-IIs. |
| Fusion gene | Hchr | Hbp | Tchr | Tbp | Start in +/-13AA | End in +/-13AA | FusionNeoAntigen peptide | FusionNEoAntigen RNA sequence |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 3 | 18 | PPYADIHPMRPNGRT | CCTCCTTATGCTGATATACATCCAATGAGGCCAAATGGGCGGACT |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 4 | 19 | PYADIHPMRPNGRTV | CCTTATGCTGATATACATCCAATGAGGCCAAATGGGCGGACTGTT |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 5 | 20 | YADIHPMRPNGRTVE | TATGCTGATATACATCCAATGAGGCCAAATGGGCGGACTGTTGAA |
| STK3-MTDH | chr8 | 99718694 | chr8 | 98673299 | 6 | 21 | ADIHPMRPNGRTVEV | GCTGATATACATCCAATGAGGCCAAATGGGCGGACTGTTGAAGTG |
Top |
Information of the samples that have these potential fusion neoantigens of STK3-MTDH |
 These samples were reported as having these fusion breakpoints. For individual breakpoints, we checked the open reading frames considering multiple gene isoforms and chose the in-frame fusion genes only. Then, we made fusion protein sequences and predicted the fusion neoantigens. These fusion-positive samples may have these potential fusion neoantigens. These samples were reported as having these fusion breakpoints. For individual breakpoints, we checked the open reading frames considering multiple gene isoforms and chose the in-frame fusion genes only. Then, we made fusion protein sequences and predicted the fusion neoantigens. These fusion-positive samples may have these potential fusion neoantigens. |
| Cancer type | Fusion gene | Hchr | Hbp | Henst | Tchr | Tbp | Tenst | Sample |
| Non-Cancer | STK3-MTDH | chr8 | 99718694 | ENST00000419617 | chr8 | 98673299 | ENST00000336273 | 135N |
Top |
Potential target of CAR-T therapy development for STK3-MTDH |
 Predicted 3D structure. We used RoseTTAFold. Predicted 3D structure. We used RoseTTAFold. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, to provide the retention of the transmembrane domain, we only show the protein feature retention information of those transmembrane features Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, to provide the retention of the transmembrane domain, we only show the protein feature retention information of those transmembrane features* Minus value of BPloci means that the break point is located before the CDS. |
| - In-frame and retained 'Transmembrane'. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
 Subcellular localization prediction of the transmembrane domain retained fusion proteins Subcellular localization prediction of the transmembrane domain retained fusion proteins* We used DeepLoc 1.0. The order of the X-axis of the barplot is as follows: Entry_ID, Localization, Type, Nucleus, Cytoplasm, Extracellular, Mitochondrion, Cell_membrane, Endoplasmic_reticulum, Plastid, Golgi.apparatus, Lysosome.Vacuole, Peroxisome. Y-axis is the output score of DeepLoc. Clicking the image will open a new tab with a large image. |
| Hgene | Hchr | Hbp | Henst | Tgene | Tchr | Tbp | Tenst | DeepLoc result |
Top |
Related Drugs to STK3-MTDH |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to STK3-MTDH |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

