
|
|||||||
|
Fusion Protein:BCR-ABL1 |
Fusion Gene and Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: BCR-ABL1 | FusionPDB ID: 9501 | FusionGDB2.0 ID: 9501 | Hgene | Tgene | Gene symbol | BCR | ABL1 | Gene ID | 613 | 25 |
| Gene name | BCR activator of RhoGEF and GTPase | ABL proto-oncogene 1, non-receptor tyrosine kinase | |
| Synonyms | ALL|BCR1|CML|D22S11|D22S662|PHL | ABL|BCR-ABL|CHDSKM|JTK7|bcr/abl|c-ABL|c-ABL1|p150|v-abl | |
| Cytomap | 22q11.23 | 9q34.12 | |
| Type of gene | protein-coding | protein-coding | |
| Description | breakpoint cluster region proteinBCR, RhoGEF and GTPase activating proteinBCR/FGFR1 chimera proteinFGFR1/BCR chimera proteinbreakpoint cluster regionrenal carcinoma antigen NY-REN-26 | tyrosine-protein kinase ABL1ABL protooncogene 1 nonreceptor tyrosine kinaseAbelson tyrosine-protein kinase 1bcr/c-abl oncogene proteinc-abl oncogene 1, receptor tyrosine kinaseproto-oncogene c-Ablproto-oncogene tyrosine-protein kinase ABL1truncated | |
| Modification date | 20200313 | 20200327 | |
| UniProtAcc | P11274 Main function of 5'-partner protein: FUNCTION: Protein with a unique structure having two opposing regulatory activities toward small GTP-binding proteins. The C-terminus is a GTPase-activating protein (GAP) domain which stimulates GTP hydrolysis by RAC1, RAC2 and CDC42. Accelerates the intrinsic rate of GTP hydrolysis of RAC1 or CDC42, leading to down-regulation of the active GTP-bound form (PubMed:7479768, PubMed:1903516, PubMed:17116687). The central Dbl homology (DH) domain functions as guanine nucleotide exchange factor (GEF) that modulates the GTPases CDC42, RHOA and RAC1. Promotes the conversion of CDC42, RHOA and RAC1 from the GDP-bound to the GTP-bound form (PubMed:7479768, PubMed:23940119). The amino terminus contains an intrinsic kinase activity (PubMed:1657398). Functions as an important negative regulator of neuronal RAC1 activity (By similarity). Regulates macrophage functions such as CSF1-directed motility and phagocytosis through the modulation of RAC1 activity (PubMed:17116687). Plays a major role as a RHOA GEF in keratinocytes being involved in focal adhesion formation and keratinocyte differentiation (PubMed:23940119). {ECO:0000250|UniProtKB:Q6PAJ1, ECO:0000269|PubMed:1657398, ECO:0000269|PubMed:17116687, ECO:0000269|PubMed:1903516, ECO:0000269|PubMed:23940119, ECO:0000269|PubMed:7479768}. | P00519 Main function of 5'-partner protein: FUNCTION: Non-receptor tyrosine-protein kinase that plays a role in many key processes linked to cell growth and survival such as cytoskeleton remodeling in response to extracellular stimuli, cell motility and adhesion, receptor endocytosis, autophagy, DNA damage response and apoptosis. Coordinates actin remodeling through tyrosine phosphorylation of proteins controlling cytoskeleton dynamics like WASF3 (involved in branch formation); ANXA1 (involved in membrane anchoring); DBN1, DBNL, CTTN, RAPH1 and ENAH (involved in signaling); or MAPT and PXN (microtubule-binding proteins). Phosphorylation of WASF3 is critical for the stimulation of lamellipodia formation and cell migration. Involved in the regulation of cell adhesion and motility through phosphorylation of key regulators of these processes such as BCAR1, CRK, CRKL, DOK1, EFS or NEDD9. Phosphorylates multiple receptor tyrosine kinases and more particularly promotes endocytosis of EGFR, facilitates the formation of neuromuscular synapses through MUSK, inhibits PDGFRB-mediated chemotaxis and modulates the endocytosis of activated B-cell receptor complexes. Other substrates which are involved in endocytosis regulation are the caveolin (CAV1) and RIN1. Moreover, ABL1 regulates the CBL family of ubiquitin ligases that drive receptor down-regulation and actin remodeling. Phosphorylation of CBL leads to increased EGFR stability. Involved in late-stage autophagy by regulating positively the trafficking and function of lysosomal components. ABL1 targets to mitochondria in response to oxidative stress and thereby mediates mitochondrial dysfunction and cell death. In response to oxidative stress, phosphorylates serine/threonine kinase PRKD2 at 'Tyr-717' (PubMed:28428613). ABL1 is also translocated in the nucleus where it has DNA-binding activity and is involved in DNA-damage response and apoptosis. Many substrates are known mediators of DNA repair: DDB1, DDB2, ERCC3, ERCC6, RAD9A, RAD51, RAD52 or WRN. Activates the proapoptotic pathway when the DNA damage is too severe to be repaired. Phosphorylates TP73, a primary regulator for this type of damage-induced apoptosis. Phosphorylates the caspase CASP9 on 'Tyr-153' and regulates its processing in the apoptotic response to DNA damage. Phosphorylates PSMA7 that leads to an inhibition of proteasomal activity and cell cycle transition blocks. ABL1 acts also as a regulator of multiple pathological signaling cascades during infection. Several known tyrosine-phosphorylated microbial proteins have been identified as ABL1 substrates. This is the case of A36R of Vaccinia virus, Tir (translocated intimin receptor) of pathogenic E.coli and possibly Citrobacter, CagA (cytotoxin-associated gene A) of H.pylori, or AnkA (ankyrin repeat-containing protein A) of A.phagocytophilum. Pathogens can highjack ABL1 kinase signaling to reorganize the host actin cytoskeleton for multiple purposes, like facilitating intracellular movement and host cell exit. Finally, functions as its own regulator through autocatalytic activity as well as through phosphorylation of its inhibitor, ABI1. Regulates T-cell differentiation in a TBX21-dependent manner. Phosphorylates TBX21 on tyrosine residues leading to an enhancement of its transcriptional activator activity (By similarity). {ECO:0000250|UniProtKB:P00520, ECO:0000269|PubMed:10391250, ECO:0000269|PubMed:11971963, ECO:0000269|PubMed:12379650, ECO:0000269|PubMed:12531427, ECO:0000269|PubMed:12672821, ECO:0000269|PubMed:15031292, ECO:0000269|PubMed:15556646, ECO:0000269|PubMed:15657060, ECO:0000269|PubMed:15886098, ECO:0000269|PubMed:16424036, ECO:0000269|PubMed:16678104, ECO:0000269|PubMed:16943190, ECO:0000269|PubMed:17306540, ECO:0000269|PubMed:17623672, ECO:0000269|PubMed:18328268, ECO:0000269|PubMed:18945674, ECO:0000269|PubMed:19891780, ECO:0000269|PubMed:20357770, ECO:0000269|PubMed:20417104, ECO:0000269|PubMed:28428613, ECO:0000269|PubMed:9037071, ECO:0000269|PubMed:9144171, ECO:0000269|PubMed:9461559}. | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000436990, ENST00000305877, ENST00000359540, ENST00000398512, | ENST00000318560, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 22 X 142 X 16=49984 | 21 X 149 X 8=25032 |
| # samples | 163 | 154 | |
| ** MAII score | log2(163/49984*10)=-4.93852248902354 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(154/25032*10)=-4.02277130765866 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Fusion gene context | PubMed: BCR [Title/Abstract] AND ABL1 [Title/Abstract] AND fusion [Title/Abstract] Expression of the ABL-BCR Fusion Gene in Philadelphia-Positive Acute Lymphoblastic Leukemia (pmid: 8490164)A Philadelphia-negative chronic myeloid leukemia with a BCR/ABL fusion gene on chromosome 9 (pmid: 9809034) | ||
| Fusion neoantigen context | PubMed: BCR [Title/Abstract] AND ABL1 [Title/Abstract] AND neoantigen [Title/Abstract] BCR-ABL-specific CD4+ T-helper cells promote the priming of antigen-specific cytotoxic T cells via dendritic cells', 'Mutated BCR-ABL generates immunogenic T-cell epitopes in CML patients.', 'Neoantigens in Hematologic Malignancies.', 'Fall of the mutants: T cells targeting BCR-ABL.', 'Applying Antibodies Inside Cells: Principles and Recent Advances in Neurobiology, Virology and Oncology.', 'BCR-ABL fusion peptides and cytotoxic T cells in chronic myeloid leukaemia.', 'Binding of BCR/ABL junctional peptides to major histocompatibility complex (MHC) class I molecules: studies in antigen-processing defective cell lines.' (pmid: 27181332, 22912393, 32117272, 28153834, 32301049, 11697642, 8289483) | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | BCR(23632600)-ABL1(133729451), # samples:12 ABL1(133589842)-BCR(23634728), # samples:6 | ||
| Anticipated loss of major functional domain due to fusion event. | BCR-ABL1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. BCR-ABL1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. BCR-ABL1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. BCR-ABL1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. BCR-ABL1 seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. BCR-ABL1 seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. BCR-ABL1 seems lost the major protein functional domain in Hgene partner, which is a kinase due to the frame-shifted ORF. BCR-ABL1 seems lost the major protein functional domain in Hgene partner, which is a tumor suppressor due to the frame-shifted ORF. BCR-ABL1 seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. BCR-ABL1 seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. BCR-ABL1 seems lost the major protein functional domain in Tgene partner, which is a kinase due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | BCR | GO:0090630 | activation of GTPase activity | 7479768 |
| Tgene | ABL1 | GO:0006974 | cellular response to DNA damage stimulus | 15657060 |
| Tgene | ABL1 | GO:0006975 | DNA damage induced protein phosphorylation | 18280240 |
| Tgene | ABL1 | GO:0018108 | peptidyl-tyrosine phosphorylation | 7590236|9144171|10713049|11121037 |
| Tgene | ABL1 | GO:0038083 | peptidyl-tyrosine autophosphorylation | 10518561 |
| Tgene | ABL1 | GO:0042770 | signal transduction in response to DNA damage | 9037071|15657060 |
| Tgene | ABL1 | GO:0043065 | positive regulation of apoptotic process | 9037071 |
| Tgene | ABL1 | GO:0046777 | protein autophosphorylation | 10713049 |
| Tgene | ABL1 | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | 15657060 |
| Tgene | ABL1 | GO:0051353 | positive regulation of oxidoreductase activity | 12893824 |
| Tgene | ABL1 | GO:0051444 | negative regulation of ubiquitin-protein transferase activity | 20823226 |
| Tgene | ABL1 | GO:0070301 | cellular response to hydrogen peroxide | 10713049 |
| Tgene | ABL1 | GO:0071103 | DNA conformation change | 9558345 |
| Tgene | ABL1 | GO:0071901 | negative regulation of protein serine/threonine kinase activity | 11121037 |
| Tgene | ABL1 | GO:1990051 | activation of protein kinase C activity | 10713049 |
| Tgene | ABL1 | GO:2001020 | regulation of response to DNA damage stimulus | 9461559 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr22:23632600/chr9:133729451) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
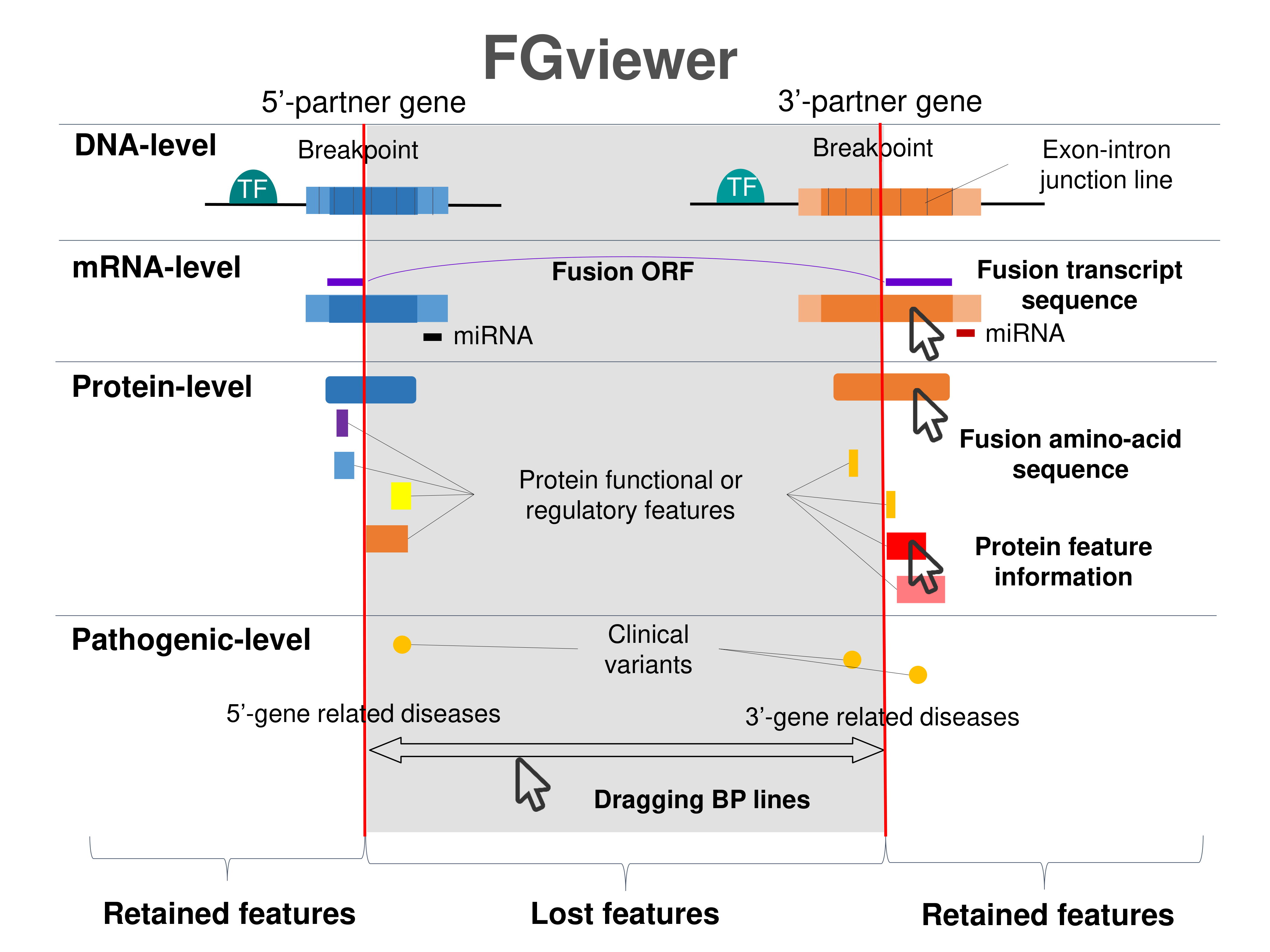 |
 Retention analysis results of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features, are available here. Retention analysis results of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features, are available here. |
 Fusion gene breakpoints across BCR (5'-gene) Fusion gene breakpoints across BCR (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
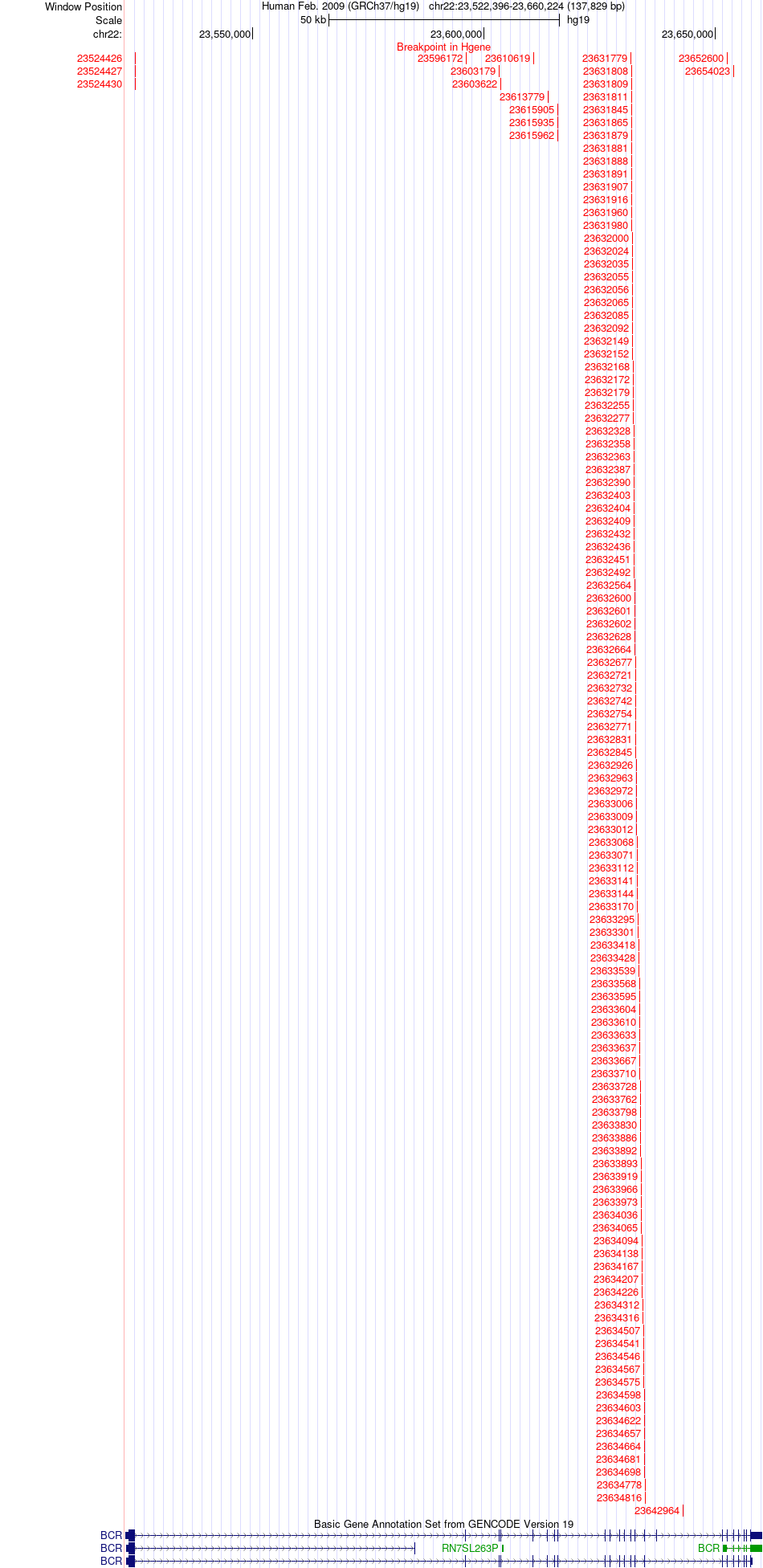 |
 Fusion gene breakpoints across ABL1 (3'-gene) Fusion gene breakpoints across ABL1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
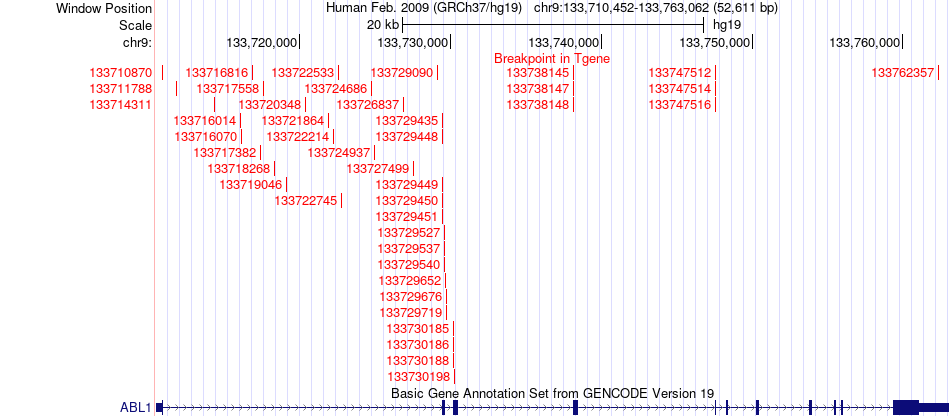 |
Top |
Fusion Amino Acid Sequences |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000305877 | BCR | chr22 | 23524426 | + | ENST00000318560 | ABL1 | chr9 | 133729448 | + | 7336 | 2030 | 751 | 5343 | 1530 |
| ENST00000359540 | BCR | chr22 | 23524426 | + | ENST00000318560 | ABL1 | chr9 | 133729448 | + | 7181 | 1875 | 596 | 5188 | 1530 |
| ENST00000398512 | BCR | chr22 | 23524426 | + | ENST00000318560 | ABL1 | chr9 | 133729448 | + | 7181 | 1875 | 596 | 5188 | 1530 |
| ENST00000305877 | BCR | chr22 | 23524426 | + | ENST00000318560 | ABL1 | chr9 | 133729450 | + | 7336 | 2030 | 751 | 5343 | 1530 |
| ENST00000359540 | BCR | chr22 | 23524426 | + | ENST00000318560 | ABL1 | chr9 | 133729450 | + | 7181 | 1875 | 596 | 5188 | 1530 |
| ENST00000398512 | BCR | chr22 | 23524426 | + | ENST00000318560 | ABL1 | chr9 | 133729450 | + | 7181 | 1875 | 596 | 5188 | 1530 |
| ENST00000305877 | BCR | chr22 | 23524430 | + | ENST00000318560 | ABL1 | chr9 | 133730185 | + | 7162 | 2030 | 751 | 5169 | 1472 |
| ENST00000359540 | BCR | chr22 | 23524430 | + | ENST00000318560 | ABL1 | chr9 | 133730185 | + | 7007 | 1875 | 596 | 5014 | 1472 |
| ENST00000398512 | BCR | chr22 | 23524430 | + | ENST00000318560 | ABL1 | chr9 | 133730185 | + | 7007 | 1875 | 596 | 5014 | 1472 |
| ENST00000305877 | BCR | chr22 | 23613779 | + | ENST00000318560 | ABL1 | chr9 | 133729451 | + | 7978 | 2672 | 751 | 5985 | 1744 |
| ENST00000359540 | BCR | chr22 | 23613779 | + | ENST00000318560 | ABL1 | chr9 | 133729451 | + | 7823 | 2517 | 596 | 5830 | 1744 |
| ENST00000305877 | BCR | chr22 | 23631779 | + | ENST00000318560 | ABL1 | chr9 | 133729451 | + | 8688 | 3382 | 3363 | 6695 | 1110 |
| ENST00000359540 | BCR | chr22 | 23631779 | + | ENST00000318560 | ABL1 | chr9 | 133729451 | + | 8533 | 3227 | 3208 | 6540 | 1110 |
| ENST00000305877 | BCR | chr22 | 23631808 | + | ENST00000318560 | ABL1 | chr9 | 133729449 | + | 8764 | 3458 | 751 | 6771 | 2006 |
| ENST00000359540 | BCR | chr22 | 23631808 | + | ENST00000318560 | ABL1 | chr9 | 133729449 | + | 8609 | 3303 | 596 | 6616 | 2006 |
| ENST00000305877 | BCR | chr22 | 23631811 | + | ENST00000318560 | ABL1 | chr9 | 133730186 | + | 8590 | 3458 | 751 | 6597 | 1948 |
| ENST00000359540 | BCR | chr22 | 23631811 | + | ENST00000318560 | ABL1 | chr9 | 133730186 | + | 8435 | 3303 | 596 | 6442 | 1948 |
| ENST00000305877 | BCR | chr22 | 23632600 | + | ENST00000318560 | ABL1 | chr9 | 133729450 | + | 8839 | 3533 | 751 | 6846 | 2031 |
| ENST00000359540 | BCR | chr22 | 23632600 | + | ENST00000318560 | ABL1 | chr9 | 133729450 | + | 8684 | 3378 | 596 | 6691 | 2031 |
| ENST00000305877 | BCR | chr22 | 23632600 | + | ENST00000318560 | ABL1 | chr9 | 133729451 | + | 8839 | 3533 | 751 | 6846 | 2031 |
| ENST00000359540 | BCR | chr22 | 23632600 | + | ENST00000318560 | ABL1 | chr9 | 133729451 | + | 8684 | 3378 | 596 | 6691 | 2031 |
| ENST00000305877 | BCR | chr22 | 23632602 | + | ENST00000318560 | ABL1 | chr9 | 133730188 | + | 8665 | 3533 | 751 | 6672 | 1973 |
| ENST00000359540 | BCR | chr22 | 23632602 | + | ENST00000318560 | ABL1 | chr9 | 133730188 | + | 8510 | 3378 | 596 | 6517 | 1973 |
| ENST00000305877 | BCR | chr22 | 23654023 | + | ENST00000318560 | ABL1 | chr9 | 133729451 | + | 9379 | 4073 | 751 | 7386 | 2211 |
| ENST00000359540 | BCR | chr22 | 23654023 | + | ENST00000318560 | ABL1 | chr9 | 133729451 | + | 9092 | 3786 | 596 | 7099 | 2167 |
| ENST00000305877 | BCR | chr22 | 23524426 | + | ENST00000318560 | ABL1 | chr9 | 133729451 | + | 7336 | 2030 | 751 | 5343 | 1530 |
| ENST00000359540 | BCR | chr22 | 23524426 | + | ENST00000318560 | ABL1 | chr9 | 133729451 | + | 7181 | 1875 | 596 | 5188 | 1530 |
| ENST00000398512 | BCR | chr22 | 23524426 | + | ENST00000318560 | ABL1 | chr9 | 133729451 | + | 7181 | 1875 | 596 | 5188 | 1530 |
| ENST00000305877 | BCR | chr22 | 23632600 | + | ENST00000318560 | ABL1 | chr9 | 133729451 | + | 8839 | 3533 | 751 | 6846 | 2031 |
| ENST00000359540 | BCR | chr22 | 23632600 | + | ENST00000318560 | ABL1 | chr9 | 133729451 | + | 8684 | 3378 | 596 | 6691 | 2031 |
| ENST00000305877 | BCR | chr22 | 23632600 | + | ENST00000318560 | ABL1 | chr9 | 133729450 | + | 8839 | 3533 | 751 | 6846 | 2031 |
| ENST00000359540 | BCR | chr22 | 23632600 | + | ENST00000318560 | ABL1 | chr9 | 133729450 | + | 8684 | 3378 | 596 | 6691 | 2031 |
| ENST00000305877 | BCR | chr22 | 23524426 | + | ENST00000318560 | ABL1 | chr9 | 133729450 | + | 7336 | 2030 | 751 | 5343 | 1530 |
| ENST00000359540 | BCR | chr22 | 23524426 | + | ENST00000318560 | ABL1 | chr9 | 133729450 | + | 7181 | 1875 | 596 | 5188 | 1530 |
| ENST00000398512 | BCR | chr22 | 23524426 | + | ENST00000318560 | ABL1 | chr9 | 133729450 | + | 7181 | 1875 | 596 | 5188 | 1530 |
| ENST00000305877 | BCR | chr22 | 23524426 | + | ENST00000318560 | ABL1 | chr9 | 133729448 | + | 7336 | 2030 | 751 | 5343 | 1530 |
| ENST00000359540 | BCR | chr22 | 23524426 | + | ENST00000318560 | ABL1 | chr9 | 133729448 | + | 7181 | 1875 | 596 | 5188 | 1530 |
| ENST00000398512 | BCR | chr22 | 23524426 | + | ENST00000318560 | ABL1 | chr9 | 133729448 | + | 7181 | 1875 | 596 | 5188 | 1530 |
| ENST00000305877 | BCR | chr22 | 23654023 | + | ENST00000318560 | ABL1 | chr9 | 133729451 | + | 9379 | 4073 | 751 | 7386 | 2211 |
| ENST00000359540 | BCR | chr22 | 23654023 | + | ENST00000318560 | ABL1 | chr9 | 133729451 | + | 9092 | 3786 | 596 | 7099 | 2167 |
| ENST00000305877 | BCR | chr22 | 23631808 | + | ENST00000318560 | ABL1 | chr9 | 133729449 | + | 8764 | 3458 | 751 | 6771 | 2006 |
| ENST00000359540 | BCR | chr22 | 23631808 | + | ENST00000318560 | ABL1 | chr9 | 133729449 | + | 8609 | 3303 | 596 | 6616 | 2006 |
| ENST00000305877 | BCR | chr22 | 23631811 | + | ENST00000318560 | ABL1 | chr9 | 133730186 | + | 8590 | 3458 | 751 | 6597 | 1948 |
| ENST00000359540 | BCR | chr22 | 23631811 | + | ENST00000318560 | ABL1 | chr9 | 133730186 | + | 8435 | 3303 | 596 | 6442 | 1948 |
| ENST00000305877 | BCR | chr22 | 23632602 | + | ENST00000318560 | ABL1 | chr9 | 133730188 | + | 8665 | 3533 | 751 | 6672 | 1973 |
| ENST00000359540 | BCR | chr22 | 23632602 | + | ENST00000318560 | ABL1 | chr9 | 133730188 | + | 8510 | 3378 | 596 | 6517 | 1973 |
| ENST00000305877 | BCR | chr22 | 23524430 | + | ENST00000318560 | ABL1 | chr9 | 133730185 | + | 7162 | 2030 | 751 | 5169 | 1472 |
| ENST00000359540 | BCR | chr22 | 23524430 | + | ENST00000318560 | ABL1 | chr9 | 133730185 | + | 7007 | 1875 | 596 | 5014 | 1472 |
| ENST00000398512 | BCR | chr22 | 23524430 | + | ENST00000318560 | ABL1 | chr9 | 133730185 | + | 7007 | 1875 | 596 | 5014 | 1472 |
| ENST00000305877 | BCR | chr22 | 23613779 | + | ENST00000318560 | ABL1 | chr9 | 133729451 | + | 7978 | 2672 | 751 | 5985 | 1744 |
| ENST00000359540 | BCR | chr22 | 23613779 | + | ENST00000318560 | ABL1 | chr9 | 133729451 | + | 7823 | 2517 | 596 | 5830 | 1744 |
| ENST00000305877 | BCR | chr22 | 23631779 | + | ENST00000318560 | ABL1 | chr9 | 133729451 | + | 8688 | 3382 | 3363 | 6695 | 1110 |
| ENST00000359540 | BCR | chr22 | 23631779 | + | ENST00000318560 | ABL1 | chr9 | 133729451 | + | 8533 | 3227 | 3208 | 6540 | 1110 |
| ENST00000305877 | BCR | chr22 | 23615905 | + | ENST00000318560 | ABL1 | chr9 | 133710870 | + | 8129 | 2781 | 2792 | 6136 | 1114 |
| ENST00000359540 | BCR | chr22 | 23615905 | + | ENST00000318560 | ABL1 | chr9 | 133710870 | + | 7974 | 2626 | 2637 | 5981 | 1114 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000305877 | ENST00000318560 | BCR | chr22 | 23524426 | + | ABL1 | chr9 | 133729451 | + | 0.004791084 | 0.9952089 |
| ENST00000359540 | ENST00000318560 | BCR | chr22 | 23524426 | + | ABL1 | chr9 | 133729451 | + | 0.004495037 | 0.99550503 |
| ENST00000398512 | ENST00000318560 | BCR | chr22 | 23524426 | + | ABL1 | chr9 | 133729451 | + | 0.004495037 | 0.99550503 |
| ENST00000305877 | ENST00000318560 | BCR | chr22 | 23632600 | + | ABL1 | chr9 | 133729451 | + | 0.007060856 | 0.9929391 |
| ENST00000359540 | ENST00000318560 | BCR | chr22 | 23632600 | + | ABL1 | chr9 | 133729451 | + | 0.006598799 | 0.9934012 |
| ENST00000305877 | ENST00000318560 | BCR | chr22 | 23632600 | + | ABL1 | chr9 | 133729450 | + | 0.007060856 | 0.9929391 |
| ENST00000359540 | ENST00000318560 | BCR | chr22 | 23632600 | + | ABL1 | chr9 | 133729450 | + | 0.006598799 | 0.9934012 |
| ENST00000305877 | ENST00000318560 | BCR | chr22 | 23524426 | + | ABL1 | chr9 | 133729450 | + | 0.004791084 | 0.9952089 |
| ENST00000359540 | ENST00000318560 | BCR | chr22 | 23524426 | + | ABL1 | chr9 | 133729450 | + | 0.004495037 | 0.99550503 |
| ENST00000398512 | ENST00000318560 | BCR | chr22 | 23524426 | + | ABL1 | chr9 | 133729450 | + | 0.004495037 | 0.99550503 |
| ENST00000305877 | ENST00000318560 | BCR | chr22 | 23524426 | + | ABL1 | chr9 | 133729448 | + | 0.004791084 | 0.9952089 |
| ENST00000359540 | ENST00000318560 | BCR | chr22 | 23524426 | + | ABL1 | chr9 | 133729448 | + | 0.004495037 | 0.99550503 |
| ENST00000398512 | ENST00000318560 | BCR | chr22 | 23524426 | + | ABL1 | chr9 | 133729448 | + | 0.004495037 | 0.99550503 |
| ENST00000305877 | ENST00000318560 | BCR | chr22 | 23654023 | + | ABL1 | chr9 | 133729451 | + | 0.006147568 | 0.9938524 |
| ENST00000359540 | ENST00000318560 | BCR | chr22 | 23654023 | + | ABL1 | chr9 | 133729451 | + | 0.005948206 | 0.99405175 |
| ENST00000305877 | ENST00000318560 | BCR | chr22 | 23631808 | + | ABL1 | chr9 | 133729449 | + | 0.007167646 | 0.9928323 |
| ENST00000359540 | ENST00000318560 | BCR | chr22 | 23631808 | + | ABL1 | chr9 | 133729449 | + | 0.006694621 | 0.9933054 |
| ENST00000305877 | ENST00000318560 | BCR | chr22 | 23631811 | + | ABL1 | chr9 | 133730186 | + | 0.0073375 | 0.99266255 |
| ENST00000359540 | ENST00000318560 | BCR | chr22 | 23631811 | + | ABL1 | chr9 | 133730186 | + | 0.006870943 | 0.99312913 |
| ENST00000305877 | ENST00000318560 | BCR | chr22 | 23632602 | + | ABL1 | chr9 | 133730188 | + | 0.007320939 | 0.9926791 |
| ENST00000359540 | ENST00000318560 | BCR | chr22 | 23632602 | + | ABL1 | chr9 | 133730188 | + | 0.006857449 | 0.9931425 |
| ENST00000305877 | ENST00000318560 | BCR | chr22 | 23524430 | + | ABL1 | chr9 | 133730185 | + | 0.004913177 | 0.99508685 |
| ENST00000359540 | ENST00000318560 | BCR | chr22 | 23524430 | + | ABL1 | chr9 | 133730185 | + | 0.004611767 | 0.99538815 |
| ENST00000398512 | ENST00000318560 | BCR | chr22 | 23524430 | + | ABL1 | chr9 | 133730185 | + | 0.004611767 | 0.99538815 |
| ENST00000305877 | ENST00000318560 | BCR | chr22 | 23613779 | + | ABL1 | chr9 | 133729451 | + | 0.005766662 | 0.9942333 |
| ENST00000359540 | ENST00000318560 | BCR | chr22 | 23613779 | + | ABL1 | chr9 | 133729451 | + | 0.005396213 | 0.9946038 |
| ENST00000305877 | ENST00000318560 | BCR | chr22 | 23631779 | + | ABL1 | chr9 | 133729451 | + | 0.05435984 | 0.94564015 |
| ENST00000359540 | ENST00000318560 | BCR | chr22 | 23631779 | + | ABL1 | chr9 | 133729451 | + | 0.052943673 | 0.9470563 |
| ENST00000305877 | ENST00000318560 | BCR | chr22 | 23615905 | + | ABL1 | chr9 | 133710870 | + | 0.04901826 | 0.95098174 |
| ENST00000359540 | ENST00000318560 | BCR | chr22 | 23615905 | + | ABL1 | chr9 | 133710870 | + | 0.047086153 | 0.9529138 |
 Predicted full-length fusion amino acid sequences. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among all the predicted ones. Predicted full-length fusion amino acid sequences. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among all the predicted ones. |
Get the fusion protein sequences from here. |
| Fusion protein sequence information is available in the fasta format. >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP |
Top |
Fusion Protein Breakpoint Sequences for BCR-ABL1 |
 +/-13 AA sequence from the breakpoints of the fusion protein sequences. +/-13 AA sequence from the breakpoints of the fusion protein sequences. |
| Hgene | Hchr | Hbp | Tgene | Tchr | Tbp | Length(fusion protein) | BP in fusion protein | Peptide |
| BCR | chr22 | 23524426 | ABL1 | chr9 | 133729448 | 1875 | 426 | WPNDGEGAFHGDAEALQRPVASDFEP |
| BCR | chr22 | 23524426 | ABL1 | chr9 | 133729448 | 2030 | 426 | WPNDGEGAFHGDAEALQRPVASDFEP |
| BCR | chr22 | 23524426 | ABL1 | chr9 | 133729450 | 1875 | 426 | WPNDGEGAFHGDAEALQRPVASDFEP |
| BCR | chr22 | 23524426 | ABL1 | chr9 | 133729450 | 2030 | 426 | WPNDGEGAFHGDAEALQRPVASDFEP |
| BCR | chr22 | 23524426 | ABL1 | chr9 | 133729451 | 1875 | 426 | WPNDGEGAFHGDAEALQRPVASDFEP |
| BCR | chr22 | 23524426 | ABL1 | chr9 | 133729451 | 2030 | 426 | WPNDGEGAFHGDAEALQRPVASDFEP |
| BCR | chr22 | 23524430 | ABL1 | chr9 | 133730185 | 1875 | 426 | WPNDGEGAFHGDAGEKLRVLGYNHNG |
| BCR | chr22 | 23524430 | ABL1 | chr9 | 133730185 | 2030 | 426 | WPNDGEGAFHGDAGEKLRVLGYNHNG |
| BCR | chr22 | 23613779 | ABL1 | chr9 | 133729451 | 2517 | 637 | RSNKDAKDPTTKNSLEKALQRPVASD |
| BCR | chr22 | 23613779 | ABL1 | chr9 | 133729451 | 2672 | 637 | RSNKDAKDPTTKNSLEKALQRPVASD |
| BCR | chr22 | 23631808 | ABL1 | chr9 | 133729449 | 3303 | 902 | QTVHSIPLTINKEEALQRPVASDFEP |
| BCR | chr22 | 23631808 | ABL1 | chr9 | 133729449 | 3458 | 902 | QTVHSIPLTINKEEALQRPVASDFEP |
| BCR | chr22 | 23631811 | ABL1 | chr9 | 133730186 | 3303 | 902 | QTVHSIPLTINKEGEKLRVLGYNHNG |
| BCR | chr22 | 23631811 | ABL1 | chr9 | 133730186 | 3458 | 902 | QTVHSIPLTINKEGEKLRVLGYNHNG |
| BCR | chr22 | 23632600 | ABL1 | chr9 | 133729450 | 3378 | 928 | IVHSATGFKQSSKALQRPVASDFEPQ |
| BCR | chr22 | 23632600 | ABL1 | chr9 | 133729450 | 3533 | 928 | IVHSATGFKQSSKALQRPVASDFEPQ |
| BCR | chr22 | 23632600 | ABL1 | chr9 | 133729451 | 3378 | 928 | IVHSATGFKQSSKALQRPVASDFEPQ |
| BCR | chr22 | 23632600 | ABL1 | chr9 | 133729451 | 3533 | 928 | IVHSATGFKQSSKALQRPVASDFEPQ |
| BCR | chr22 | 23632602 | ABL1 | chr9 | 133730188 | 3378 | 926 | NVIVHSATGFKQSSSEKLRVLGYNHN |
| BCR | chr22 | 23632602 | ABL1 | chr9 | 133730188 | 3533 | 926 | NVIVHSATGFKQSSSEKLRVLGYNHN |
| BCR | chr22 | 23654023 | ABL1 | chr9 | 133729451 | 3786 | 1064 | TDIQALKAAFDVKALQRPVASDFEPQ |
| BCR | chr22 | 23654023 | ABL1 | chr9 | 133729451 | 4073 | 1108 | TDIQALKAAFDVKALQRPVASDFEPQ |
Top |
Potential FusionNeoAntigen Information of BCR-ABL1 in HLA I |
 Multiple sequence alignments of the potential FusionNeoAntigens per fusion breakpoints. If the MSA is empty, then it means that there were predicted fusion neoantigens in this fusion breakpoint, but those predicted fusion neoantigens were not across the breakpoint, which is not fusion-specific. Multiple sequence alignments of the potential FusionNeoAntigens per fusion breakpoints. If the MSA is empty, then it means that there were predicted fusion neoantigens in this fusion breakpoint, but those predicted fusion neoantigens were not across the breakpoint, which is not fusion-specific. |
| BCR-ABL1_23524426_133729448.msa | |
| BCR-ABL1_23524430_133730185.msa | |
| BCR-ABL1_23613779_133729451.msa | |
| BCR-ABL1_23631808_133729449.msa | |
| BCR-ABL1_23631811_133730186.msa | |
| BCR-ABL1_23632600_133729450.msa | |
| BCR-ABL1_23632602_133730188.msa | |
| BCR-ABL1_23654023_133729451.msa |
 Potential FusionNeoAntigen Information Potential FusionNeoAntigen Information* We used NetMHCpan v4.1 (%rank<0.5) and deepHLApan v1.1 (immunogenic score>0.5) |
| Fusion gene | Hchr | Hbp | Tgene | Tchr | Tbp | HLA I | FusionNeoAntigen peptide | Binding score | Immunogenic score | Neoantigen start (at BP 13) | Neoantigen end (at BP 13) |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | HLA-B39:24 | FHGDAEAL | 0.9992 | 0.652 | 8 | 16 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | HLA-B39:01 | FHGDAEAL | 0.999 | 0.9581 | 8 | 16 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | HLA-B38:02 | FHGDAEAL | 0.9976 | 0.9715 | 8 | 16 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | HLA-B38:01 | FHGDAEAL | 0.9972 | 0.9695 | 8 | 16 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | HLA-B15:10 | FHGDAEAL | 0.9968 | 0.6966 | 8 | 16 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | HLA-B15:37 | FHGDAEAL | 0.97 | 0.6914 | 8 | 16 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | HLA-B45:01 | AEALQRPVA | 0.9995 | 0.9947 | 12 | 21 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | HLA-B40:02 | AEALQRPVA | 0.9994 | 0.6377 | 12 | 21 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | HLA-B50:02 | AEALQRPVA | 0.9987 | 0.8589 | 12 | 21 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | HLA-B51:01 | DAEALQRPV | 0.9953 | 0.923 | 11 | 20 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | HLA-B41:01 | AEALQRPVA | 0.861 | 0.9818 | 12 | 21 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | HLA-B40:05 | AEALQRPVA | 0.6926 | 0.7268 | 12 | 21 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | HLA-B50:01 | AEALQRPVA | 0.5919 | 0.9632 | 12 | 21 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | HLA-B50:01 | GEGAFHGDAEA | 0.9988 | 0.9164 | 4 | 15 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | HLA-B39:09 | FHGDAEAL | 0.9993 | 0.8265 | 8 | 16 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | HLA-B39:05 | FHGDAEAL | 0.997 | 0.9514 | 8 | 16 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | HLA-B40:06 | AEALQRPVA | 0.9994 | 0.9767 | 12 | 21 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | HLA-B51:08 | DAEALQRPV | 0.9863 | 0.8577 | 11 | 20 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | HLA-B44:09 | AEALQRPVA | 0.947 | 0.9244 | 12 | 21 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | HLA-B40:03 | AEALQRPVA | 0.9017 | 0.663 | 12 | 21 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | HLA-B39:31 | FHGDAEAL | 0.9989 | 0.9578 | 8 | 16 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | HLA-B38:05 | FHGDAEAL | 0.9972 | 0.9695 | 8 | 16 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | HLA-B15:09 | FHGDAEAL | 0.9918 | 0.867 | 8 | 16 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | HLA-B39:11 | FHGDAEAL | 0.9716 | 0.8698 | 8 | 16 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | HLA-B51:13 | DAEALQRPV | 0.9946 | 0.8909 | 11 | 20 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | HLA-B40:04 | AEALQRPVA | 0.9906 | 0.9574 | 12 | 21 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | HLA-B51:29 | DAEALQRPV | 0.9874 | 0.8444 | 11 | 20 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | HLA-B41:03 | AEALQRPVA | 0.8783 | 0.8099 | 12 | 21 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | HLA-B50:04 | AEALQRPVA | 0.5919 | 0.9632 | 12 | 21 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | HLA-B50:05 | AEALQRPVA | 0.5919 | 0.9632 | 12 | 21 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | HLA-C14:02 | AFHGDAEAL | 0.0949 | 0.9728 | 7 | 16 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | HLA-C14:03 | AFHGDAEAL | 0.0949 | 0.9728 | 7 | 16 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | HLA-B50:05 | GEGAFHGDAEA | 0.9988 | 0.9164 | 4 | 15 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | HLA-B50:04 | GEGAFHGDAEA | 0.9988 | 0.9164 | 4 | 15 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-B51:01 | DAGEKLRV | 0.9996 | 0.781 | 11 | 19 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-B39:01 | FHGDAGEKL | 0.9966 | 0.975 | 8 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-B14:02 | DAGEKLRVL | 0.9956 | 0.724 | 11 | 20 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-B14:01 | DAGEKLRVL | 0.9956 | 0.724 | 11 | 20 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-B38:02 | FHGDAGEKL | 0.9918 | 0.9884 | 8 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-B38:01 | FHGDAGEKL | 0.9912 | 0.9895 | 8 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-B39:24 | FHGDAGEKL | 0.9911 | 0.8124 | 8 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-B39:06 | FHGDAGEKL | 0.9894 | 0.9597 | 8 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-B15:10 | FHGDAGEKL | 0.9363 | 0.7036 | 8 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-B08:09 | DAGEKLRVL | 0.9118 | 0.6573 | 11 | 20 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-B15:18 | FHGDAGEKL | 0.4336 | 0.8923 | 8 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-B15:37 | FHGDAGEKL | 0.2312 | 0.7531 | 8 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-B07:10 | FHGDAGEKL | 0.0005 | 0.5372 | 8 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-B38:01 | AFHGDAGEKL | 0.4792 | 0.9942 | 7 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-B38:02 | AFHGDAGEKL | 0.4774 | 0.9929 | 7 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-B38:01 | GAFHGDAGEKL | 0.9511 | 0.9926 | 6 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-C08:15 | HGDAGEKL | 1 | 0.9864 | 9 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-C05:09 | HGDAGEKL | 1 | 0.9812 | 9 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-B39:09 | FHGDAGEKL | 0.9977 | 0.8794 | 8 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-B39:12 | FHGDAGEKL | 0.9953 | 0.9759 | 8 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-B39:05 | FHGDAGEKL | 0.9926 | 0.9708 | 8 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-B14:03 | DAGEKLRVL | 0.9763 | 0.7916 | 11 | 20 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-B51:07 | DAGEKLRVL | 0.8622 | 0.8491 | 11 | 20 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-B78:01 | DAGEKLRVL | 0.8026 | 0.6366 | 11 | 20 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-C07:13 | FHGDAGEKL | 0.4521 | 0.9726 | 8 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-C07:29 | FHGDAGEKL | 0.3666 | 0.9516 | 8 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-B14:03 | FHGDAGEKL | 0.1732 | 0.963 | 8 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-B39:05 | AFHGDAGEKL | 0.4186 | 0.9702 | 7 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-C08:02 | HGDAGEKL | 1 | 0.9864 | 9 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-C05:01 | HGDAGEKL | 1 | 0.9812 | 9 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-C04:03 | HGDAGEKL | 1 | 0.9446 | 9 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-B51:21 | DAGEKLRV | 0.9993 | 0.7756 | 11 | 19 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-B39:02 | FHGDAGEKL | 0.9966 | 0.9809 | 8 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-B39:31 | FHGDAGEKL | 0.9959 | 0.9752 | 8 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-B38:05 | FHGDAGEKL | 0.9912 | 0.9895 | 8 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-B15:09 | FHGDAGEKL | 0.9362 | 0.8801 | 8 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-B39:11 | FHGDAGEKL | 0.7394 | 0.9575 | 8 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-C07:04 | FHGDAGEKL | 0.1859 | 0.9757 | 8 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-B39:11 | AFHGDAGEKL | 0.5065 | 0.9578 | 7 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-B38:05 | AFHGDAGEKL | 0.4792 | 0.9942 | 7 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | HLA-B38:05 | GAFHGDAGEKL | 0.9511 | 0.9926 | 6 | 17 |
| BCR-ABL1 | chr22 | 23613779 | chr9 | 133729451 | 2672 | HLA-B39:01 | TKNSLEKAL | 0.9936 | 0.9307 | 10 | 19 |
| BCR-ABL1 | chr22 | 23613779 | chr9 | 133729451 | 2672 | HLA-B14:02 | TKNSLEKAL | 0.9386 | 0.6188 | 10 | 19 |
| BCR-ABL1 | chr22 | 23613779 | chr9 | 133729451 | 2672 | HLA-B14:01 | TKNSLEKAL | 0.9386 | 0.6188 | 10 | 19 |
| BCR-ABL1 | chr22 | 23613779 | chr9 | 133729451 | 2672 | HLA-B15:10 | TKNSLEKAL | 0.7677 | 0.6746 | 10 | 19 |
| BCR-ABL1 | chr22 | 23613779 | chr9 | 133729451 | 2672 | HLA-B15:37 | TKNSLEKAL | 0.256 | 0.5558 | 10 | 19 |
| BCR-ABL1 | chr22 | 23613779 | chr9 | 133729451 | 2672 | HLA-B39:01 | AKDPTTKNSL | 0.9792 | 0.7519 | 5 | 15 |
| BCR-ABL1 | chr22 | 23613779 | chr9 | 133729451 | 2672 | HLA-B39:13 | AKDPTTKNSL | 0.9297 | 0.7968 | 5 | 15 |
| BCR-ABL1 | chr22 | 23613779 | chr9 | 133729451 | 2672 | HLA-B39:10 | DPTTKNSL | 0.5312 | 0.6233 | 7 | 15 |
| BCR-ABL1 | chr22 | 23613779 | chr9 | 133729451 | 2672 | HLA-B39:12 | TKNSLEKAL | 0.9931 | 0.9359 | 10 | 19 |
| BCR-ABL1 | chr22 | 23613779 | chr9 | 133729451 | 2672 | HLA-B39:05 | TKNSLEKAL | 0.9844 | 0.9198 | 10 | 19 |
| BCR-ABL1 | chr22 | 23613779 | chr9 | 133729451 | 2672 | HLA-B14:03 | TKNSLEKAL | 0.2392 | 0.7428 | 10 | 19 |
| BCR-ABL1 | chr22 | 23613779 | chr9 | 133729451 | 2672 | HLA-B39:08 | AKDPTTKNSL | 0.9299 | 0.576 | 5 | 15 |
| BCR-ABL1 | chr22 | 23613779 | chr9 | 133729451 | 2672 | HLA-B39:05 | AKDPTTKNSL | 0.9216 | 0.7259 | 5 | 15 |
| BCR-ABL1 | chr22 | 23613779 | chr9 | 133729451 | 2672 | HLA-B39:02 | TKNSLEKAL | 0.994 | 0.9057 | 10 | 19 |
| BCR-ABL1 | chr22 | 23613779 | chr9 | 133729451 | 2672 | HLA-B39:31 | TKNSLEKAL | 0.9939 | 0.9319 | 10 | 19 |
| BCR-ABL1 | chr22 | 23613779 | chr9 | 133729451 | 2672 | HLA-B15:09 | TKNSLEKAL | 0.8194 | 0.7278 | 10 | 19 |
| BCR-ABL1 | chr22 | 23613779 | chr9 | 133729451 | 2672 | HLA-B39:02 | AKDPTTKNSL | 0.9812 | 0.7953 | 5 | 15 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-B45:01 | EEALQRPVA | 0.9993 | 0.9882 | 12 | 21 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-B50:02 | EEALQRPVA | 0.998 | 0.8068 | 12 | 21 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-B45:01 | KEEALQRPV | 0.9976 | 0.9489 | 11 | 20 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-B50:02 | KEEALQRPV | 0.9965 | 0.7966 | 11 | 20 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-B41:01 | EEALQRPVA | 0.9687 | 0.9891 | 12 | 21 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-B18:01 | EEALQRPVA | 0.7678 | 0.9579 | 12 | 21 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-B41:01 | KEEALQRPV | 0.749 | 0.963 | 11 | 20 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-B50:01 | EEALQRPVA | 0.6828 | 0.9368 | 12 | 21 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-B50:01 | KEEALQRPV | 0.5422 | 0.8282 | 11 | 20 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-B45:01 | KEEALQRPVA | 0.9958 | 0.9597 | 11 | 21 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-B50:02 | KEEALQRPVA | 0.9814 | 0.8073 | 11 | 21 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-B41:01 | KEEALQRPVA | 0.9421 | 0.9848 | 11 | 21 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-B50:01 | KEEALQRPVA | 0.8952 | 0.9123 | 11 | 21 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-B35:03 | IPLTINKEEAL | 0.9973 | 0.8525 | 5 | 16 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-B40:06 | KEEALQRPV | 0.9996 | 0.8683 | 11 | 20 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-C03:07 | LTINKEEAL | 0.9995 | 0.9794 | 7 | 16 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-C03:08 | LTINKEEAL | 0.9994 | 0.9416 | 7 | 16 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-B40:06 | EEALQRPVA | 0.9971 | 0.9396 | 12 | 21 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-B40:06 | KEEALQRPVA | 0.9958 | 0.94 | 11 | 21 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-B42:01 | IPLTINKEEAL | 0.9969 | 0.7192 | 5 | 16 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-B39:10 | IPLTINKEEAL | 0.979 | 0.9066 | 5 | 16 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-C03:04 | LTINKEEAL | 0.9996 | 0.9876 | 7 | 16 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-C03:03 | LTINKEEAL | 0.9996 | 0.9876 | 7 | 16 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-C03:17 | LTINKEEAL | 0.9988 | 0.9581 | 7 | 16 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-C03:06 | LTINKEEAL | 0.9919 | 0.9896 | 7 | 16 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-B41:03 | KEEALQRPV | 0.8537 | 0.8639 | 11 | 20 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-B18:05 | EEALQRPVA | 0.7678 | 0.9579 | 12 | 21 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-B18:06 | EEALQRPVA | 0.7614 | 0.9658 | 12 | 21 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-B50:04 | EEALQRPVA | 0.6828 | 0.9368 | 12 | 21 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-B50:05 | EEALQRPVA | 0.6828 | 0.9368 | 12 | 21 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-B18:03 | EEALQRPVA | 0.682 | 0.9533 | 12 | 21 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-B35:13 | LTINKEEAL | 0.6498 | 0.9609 | 7 | 16 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-B50:05 | KEEALQRPV | 0.5422 | 0.8282 | 11 | 20 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-B50:04 | KEEALQRPV | 0.5422 | 0.8282 | 11 | 20 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-B50:05 | KEEALQRPVA | 0.8952 | 0.9123 | 11 | 21 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-B50:04 | KEEALQRPVA | 0.8952 | 0.9123 | 11 | 21 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | HLA-B67:01 | IPLTINKEEAL | 0.9732 | 0.6693 | 5 | 16 |
| BCR-ABL1 | chr22 | 23631811 | chr9 | 133730186 | 3458 | HLA-B39:13 | KEGEKLRVL | 0.5125 | 0.9 | 11 | 20 |
| BCR-ABL1 | chr22 | 23631811 | chr9 | 133730186 | 3458 | HLA-B41:01 | KEGEKLRVL | 0.4547 | 0.8085 | 11 | 20 |
| BCR-ABL1 | chr22 | 23631811 | chr9 | 133730186 | 3458 | HLA-B40:06 | KEGEKLRVL | 0.9994 | 0.5749 | 11 | 20 |
| BCR-ABL1 | chr22 | 23631811 | chr9 | 133730186 | 3458 | HLA-B39:08 | KEGEKLRVL | 0.5859 | 0.787 | 11 | 20 |
| BCR-ABL1 | chr22 | 23631811 | chr9 | 133730186 | 3458 | HLA-B40:04 | KEGEKLRVL | 0.9993 | 0.6821 | 11 | 20 |
| BCR-ABL1 | chr22 | 23631811 | chr9 | 133730186 | 3458 | HLA-B39:02 | KEGEKLRVL | 0.5527 | 0.9112 | 11 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | HLA-A30:08 | SSKALQRPV | 0.9933 | 0.6977 | 10 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | HLA-A30:01 | SSKALQRPV | 0.9933 | 0.7397 | 10 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | HLA-C14:02 | GFKQSSKAL | 0.1969 | 0.9757 | 6 | 15 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | HLA-C14:03 | GFKQSSKAL | 0.1969 | 0.9757 | 6 | 15 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-B39:13 | KQSSSEKL | 0.7644 | 0.8928 | 10 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-B39:01 | FKQSSSEKL | 0.9446 | 0.8375 | 9 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-B38:02 | FKQSSSEKL | 0.8979 | 0.9295 | 9 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-B38:01 | FKQSSSEKL | 0.8941 | 0.9191 | 9 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-B39:13 | FKQSSSEKL | 0.8816 | 0.8602 | 9 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-B14:02 | SSSEKLRVL | 0.8546 | 0.6569 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-B14:01 | SSSEKLRVL | 0.8546 | 0.6569 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-B15:37 | FKQSSSEKL | 0.1528 | 0.5118 | 9 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-B13:02 | KQSSSEKLRV | 0.744 | 0.7413 | 10 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-B39:08 | KQSSSEKL | 0.8586 | 0.7677 | 10 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C03:07 | SSSEKLRVL | 0.9997 | 0.9211 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C15:04 | SSSEKLRVL | 0.9997 | 0.7788 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C03:19 | SSSEKLRVL | 0.9996 | 0.9766 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C15:06 | SSSEKLRVL | 0.9995 | 0.7969 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C03:08 | SSSEKLRVL | 0.9995 | 0.8901 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C01:17 | SSSEKLRVL | 0.9984 | 0.9115 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C12:12 | SSSEKLRVL | 0.9972 | 0.8781 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C04:06 | SSSEKLRVL | 0.9966 | 0.8372 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C06:03 | SSSEKLRVL | 0.9953 | 0.9813 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C12:04 | SSSEKLRVL | 0.9946 | 0.9748 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C08:04 | SSSEKLRVL | 0.9926 | 0.8556 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C08:13 | SSSEKLRVL | 0.9926 | 0.8556 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C01:30 | SSSEKLRVL | 0.9777 | 0.9451 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-B39:09 | FKQSSSEKL | 0.958 | 0.5044 | 9 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C08:03 | SSSEKLRVL | 0.9493 | 0.9245 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-B39:12 | FKQSSSEKL | 0.9271 | 0.843 | 9 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C07:13 | SSSEKLRVL | 0.9224 | 0.8883 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C03:14 | SSSEKLRVL | 0.9119 | 0.9479 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C12:16 | SSSEKLRVL | 0.8879 | 0.91 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C07:29 | SSSEKLRVL | 0.8859 | 0.9381 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-B39:05 | FKQSSSEKL | 0.8774 | 0.8142 | 9 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C07:19 | SSSEKLRVL | 0.8663 | 0.7238 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C07:27 | SSSEKLRVL | 0.8427 | 0.9511 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C07:29 | FKQSSSEKL | 0.8398 | 0.912 | 9 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C07:95 | SSSEKLRVL | 0.8217 | 0.764 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C02:06 | SSSEKLRVL | 0.8148 | 0.8691 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C07:13 | FKQSSSEKL | 0.7726 | 0.9157 | 9 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-B14:03 | SSSEKLRVL | 0.7483 | 0.8257 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-B39:08 | FKQSSSEKL | 0.3587 | 0.7397 | 9 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C15:09 | SSSEKLRVL | 0.9997 | 0.7788 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C03:04 | SSSEKLRVL | 0.9996 | 0.9782 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C03:03 | SSSEKLRVL | 0.9996 | 0.9782 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C15:05 | SSSEKLRVL | 0.9995 | 0.7979 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C06:06 | SSSEKLRVL | 0.9995 | 0.969 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C15:02 | SSSEKLRVL | 0.9994 | 0.7537 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C03:17 | SSSEKLRVL | 0.9993 | 0.948 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C03:05 | SSSEKLRVL | 0.9991 | 0.8969 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C03:67 | SSSEKLRVL | 0.999 | 0.9677 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C15:02 | QSSSEKLRV | 0.9988 | 0.8212 | 11 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C01:03 | SSSEKLRVL | 0.9984 | 0.8946 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C15:05 | QSSSEKLRV | 0.9984 | 0.8633 | 11 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C01:02 | SSSEKLRVL | 0.9983 | 0.9062 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C03:02 | SSSEKLRVL | 0.9978 | 0.9379 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C16:04 | SSSEKLRVL | 0.9978 | 0.9537 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C12:03 | SSSEKLRVL | 0.995 | 0.9442 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C07:04 | SSSEKLRVL | 0.9937 | 0.8799 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C03:06 | SSSEKLRVL | 0.9887 | 0.9759 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C06:17 | SSSEKLRVL | 0.9876 | 0.975 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C06:02 | SSSEKLRVL | 0.9876 | 0.975 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C16:02 | SSSEKLRVL | 0.9808 | 0.9717 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C12:02 | SSSEKLRVL | 0.9614 | 0.9149 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C08:01 | SSSEKLRVL | 0.9493 | 0.9245 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-B58:06 | SSSEKLRVL | 0.9453 | 0.6993 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-B39:31 | FKQSSSEKL | 0.943 | 0.8388 | 9 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-B39:02 | FKQSSSEKL | 0.9369 | 0.8593 | 9 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-A30:01 | GFKQSSSEK | 0.9309 | 0.7809 | 8 | 17 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C07:22 | SSSEKLRVL | 0.9175 | 0.7022 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-B38:05 | FKQSSSEKL | 0.8941 | 0.9191 | 9 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C16:01 | SSSEKLRVL | 0.8577 | 0.9615 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C07:01 | SSSEKLRVL | 0.8114 | 0.7161 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C02:10 | SSSEKLRVL | 0.7134 | 0.9034 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C02:02 | SSSEKLRVL | 0.7134 | 0.9034 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C06:08 | SSSEKLRVL | 0.5933 | 0.9424 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-C17:01 | SSSEKLRVL | 0.4468 | 0.6648 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-B15:09 | FKQSSSEKL | 0.4418 | 0.5813 | 9 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-B39:11 | FKQSSSEKL | 0.3694 | 0.6916 | 9 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-B07:13 | SSSEKLRVL | 0.2969 | 0.6249 | 12 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | HLA-A30:01 | ATGFKQSSSEK | 0.9815 | 0.8513 | 6 | 17 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-A31:02 | AFDVKALQR | 0.9571 | 0.682 | 8 | 17 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-A31:06 | AFDVKALQR | 0.8876 | 0.5741 | 8 | 17 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-A74:09 | AAFDVKALQR | 0.9775 | 0.7419 | 7 | 17 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-A74:11 | AAFDVKALQR | 0.9775 | 0.7419 | 7 | 17 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-A74:03 | AAFDVKALQR | 0.9775 | 0.7419 | 7 | 17 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-A31:06 | AAFDVKALQR | 0.9761 | 0.5698 | 7 | 17 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-A31:02 | AAFDVKALQR | 0.9597 | 0.7226 | 7 | 17 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-C03:08 | AAFDVKAL | 0.9999 | 0.9542 | 7 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-C03:19 | AAFDVKAL | 0.9999 | 0.9908 | 7 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-C12:12 | AAFDVKAL | 0.9988 | 0.943 | 7 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-C03:07 | KAAFDVKAL | 0.9995 | 0.9536 | 6 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-C03:19 | KAAFDVKAL | 0.9993 | 0.9795 | 6 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-C03:08 | KAAFDVKAL | 0.999 | 0.8887 | 6 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-C15:06 | KAAFDVKAL | 0.9987 | 0.8668 | 6 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-C12:12 | KAAFDVKAL | 0.9666 | 0.9031 | 6 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-C12:04 | KAAFDVKAL | 0.9555 | 0.9871 | 6 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-C06:03 | KAAFDVKAL | 0.954 | 0.9879 | 6 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-C08:13 | KAAFDVKAL | 0.9426 | 0.9446 | 6 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-C08:04 | KAAFDVKAL | 0.9426 | 0.9446 | 6 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-C02:06 | KAAFDVKAL | 0.918 | 0.9297 | 6 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-C03:14 | KAAFDVKAL | 0.9136 | 0.9577 | 6 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-C08:03 | KAAFDVKAL | 0.7996 | 0.9757 | 6 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-A31:01 | AAFDVKALQR | 0.9796 | 0.6961 | 7 | 17 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-C03:17 | AAFDVKAL | 0.9999 | 0.981 | 7 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-C03:04 | AAFDVKAL | 0.9999 | 0.992 | 7 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-C03:03 | AAFDVKAL | 0.9999 | 0.992 | 7 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-C03:05 | AAFDVKAL | 0.9998 | 0.9496 | 7 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-C16:04 | AAFDVKAL | 0.9991 | 0.9796 | 7 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-C12:03 | AAFDVKAL | 0.9974 | 0.9785 | 7 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-C16:01 | AAFDVKAL | 0.9871 | 0.985 | 7 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-C03:03 | KAAFDVKAL | 0.9991 | 0.9813 | 6 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-C03:04 | KAAFDVKAL | 0.9991 | 0.9813 | 6 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-C03:05 | KAAFDVKAL | 0.9989 | 0.9428 | 6 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-C03:17 | KAAFDVKAL | 0.9989 | 0.9662 | 6 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-C03:02 | KAAFDVKAL | 0.9983 | 0.9621 | 6 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-C03:06 | KAAFDVKAL | 0.9664 | 0.9825 | 6 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-C16:04 | KAAFDVKAL | 0.959 | 0.9683 | 6 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-C12:03 | KAAFDVKAL | 0.9527 | 0.9687 | 6 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-C16:01 | KAAFDVKAL | 0.9495 | 0.9652 | 6 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-C16:02 | KAAFDVKAL | 0.9338 | 0.9813 | 6 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-B07:13 | KAAFDVKAL | 0.9093 | 0.8296 | 6 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-C12:02 | KAAFDVKAL | 0.868 | 0.95 | 6 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-C08:01 | KAAFDVKAL | 0.7996 | 0.9757 | 6 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-B35:13 | KAAFDVKAL | 0.6822 | 0.8552 | 6 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | HLA-A74:01 | AAFDVKALQR | 0.9775 | 0.7419 | 7 | 17 |
Top |
Potential FusionNeoAntigen Information of BCR-ABL1 in HLA II |
 Multiple sequence alignments of the potential FusionNeoAntigens per fusion breakpoints. If the MSA is empty, then it means that there were predicted fusion neoantigens in this fusion breakpoint, but those predicted fusion neoantigens were not across the breakpoint, which is not fusion-specific. Multiple sequence alignments of the potential FusionNeoAntigens per fusion breakpoints. If the MSA is empty, then it means that there were predicted fusion neoantigens in this fusion breakpoint, but those predicted fusion neoantigens were not across the breakpoint, which is not fusion-specific. |
| BCR-ABL1_23524426_133729448.msa | |
| BCR-ABL1_23524430_133730185.msa | |
| BCR-ABL1_23613779_133729451.msa | |
| BCR-ABL1_23631808_133729449.msa | |
| BCR-ABL1_23632600_133729450.msa | |
| BCR-ABL1_23632602_133730188.msa | |
| BCR-ABL1_23654023_133729451.msa |
 Potential FusionNeoAntigen Information Potential FusionNeoAntigen Information * We used NetMHCIIpan v4.1 (%rank<0.5). |
| Fusion gene | Hchr | Hbp | Tgene | Tchr | Tbp | HLA II | FusionNeoAntigen peptide | Neoantigen start (at BP 13) | Neoantigen end (at BP 13) |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | DRB1-0305 | EGAFHGDAEALQRPV | 5 | 20 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | DRB1-0434 | EGAFHGDAEALQRPV | 5 | 20 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | DRB1-0434 | GEGAFHGDAEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | DRB1-0435 | EGAFHGDAEALQRPV | 5 | 20 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | DRB1-0464 | EGAFHGDAEALQRPV | 5 | 20 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | DRB1-0464 | GEGAFHGDAEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | DRB1-0466 | EGAFHGDAEALQRPV | 5 | 20 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | DRB1-0466 | GEGAFHGDAEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | DRB3-0101 | EGAFHGDAEALQRPV | 5 | 20 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | DRB3-0101 | GEGAFHGDAEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | DRB3-0104 | EGAFHGDAEALQRPV | 5 | 20 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | DRB3-0104 | GEGAFHGDAEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | DRB3-0108 | EGAFHGDAEALQRPV | 5 | 20 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | DRB3-0108 | GEGAFHGDAEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | DRB3-0109 | EGAFHGDAEALQRPV | 5 | 20 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | DRB3-0111 | EGAFHGDAEALQRPV | 5 | 20 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | DRB3-0111 | GEGAFHGDAEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | DRB3-0112 | EGAFHGDAEALQRPV | 5 | 20 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | DRB3-0112 | GEGAFHGDAEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | DRB3-0113 | EGAFHGDAEALQRPV | 5 | 20 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | DRB3-0113 | GEGAFHGDAEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | DRB3-0114 | EGAFHGDAEALQRPV | 5 | 20 |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 | DRB3-0114 | GEGAFHGDAEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB1-0338 | GEGAFHGDAGEKLRV | 4 | 19 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB1-0338 | EGAFHGDAGEKLRVL | 5 | 20 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB1-0338 | DGEGAFHGDAGEKLR | 3 | 18 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0101 | GEGAFHGDAGEKLRV | 4 | 19 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0101 | EGAFHGDAGEKLRVL | 5 | 20 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0101 | DGEGAFHGDAGEKLR | 3 | 18 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0101 | GAFHGDAGEKLRVLG | 6 | 21 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0101 | NDGEGAFHGDAGEKL | 2 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0104 | GEGAFHGDAGEKLRV | 4 | 19 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0104 | EGAFHGDAGEKLRVL | 5 | 20 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0104 | DGEGAFHGDAGEKLR | 3 | 18 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0104 | GAFHGDAGEKLRVLG | 6 | 21 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0104 | NDGEGAFHGDAGEKL | 2 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0105 | GEGAFHGDAGEKLRV | 4 | 19 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0105 | EGAFHGDAGEKLRVL | 5 | 20 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0105 | DGEGAFHGDAGEKLR | 3 | 18 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0105 | GAFHGDAGEKLRVLG | 6 | 21 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0105 | NDGEGAFHGDAGEKL | 2 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0108 | GEGAFHGDAGEKLRV | 4 | 19 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0108 | EGAFHGDAGEKLRVL | 5 | 20 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0108 | DGEGAFHGDAGEKLR | 3 | 18 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0108 | GAFHGDAGEKLRVLG | 6 | 21 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0108 | NDGEGAFHGDAGEKL | 2 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0109 | GEGAFHGDAGEKLRV | 4 | 19 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0109 | EGAFHGDAGEKLRVL | 5 | 20 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0109 | DGEGAFHGDAGEKLR | 3 | 18 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0109 | GAFHGDAGEKLRVLG | 6 | 21 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0111 | GEGAFHGDAGEKLRV | 4 | 19 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0111 | EGAFHGDAGEKLRVL | 5 | 20 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0111 | DGEGAFHGDAGEKLR | 3 | 18 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0111 | GAFHGDAGEKLRVLG | 6 | 21 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0111 | NDGEGAFHGDAGEKL | 2 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0112 | GEGAFHGDAGEKLRV | 4 | 19 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0112 | EGAFHGDAGEKLRVL | 5 | 20 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0112 | DGEGAFHGDAGEKLR | 3 | 18 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0112 | GAFHGDAGEKLRVLG | 6 | 21 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0112 | NDGEGAFHGDAGEKL | 2 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0113 | GEGAFHGDAGEKLRV | 4 | 19 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0113 | EGAFHGDAGEKLRVL | 5 | 20 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0113 | DGEGAFHGDAGEKLR | 3 | 18 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0113 | GAFHGDAGEKLRVLG | 6 | 21 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0113 | NDGEGAFHGDAGEKL | 2 | 17 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0114 | GEGAFHGDAGEKLRV | 4 | 19 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0114 | EGAFHGDAGEKLRVL | 5 | 20 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0114 | DGEGAFHGDAGEKLR | 3 | 18 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0114 | GAFHGDAGEKLRVLG | 6 | 21 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0209 | GEGAFHGDAGEKLRV | 4 | 19 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0209 | EGAFHGDAGEKLRVL | 5 | 20 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0209 | DGEGAFHGDAGEKLR | 3 | 18 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0216 | GEGAFHGDAGEKLRV | 4 | 19 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0221 | GEGAFHGDAGEKLRV | 4 | 19 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0221 | EGAFHGDAGEKLRVL | 5 | 20 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0221 | DGEGAFHGDAGEKLR | 3 | 18 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0303 | GEGAFHGDAGEKLRV | 4 | 19 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0303 | EGAFHGDAGEKLRVL | 5 | 20 |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 | DRB3-0303 | DGEGAFHGDAGEKLR | 3 | 18 |
| BCR-ABL1 | chr22 | 23613779 | chr9 | 133729451 | 2672 | DRB1-0906 | KNSLEKALQRPVASD | 11 | 26 |
| BCR-ABL1 | chr22 | 23613779 | chr9 | 133729451 | 2672 | DRB1-0906 | TKNSLEKALQRPVAS | 10 | 25 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-0303 | SIPLTINKEEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-0303 | PLTINKEEALQRPVA | 6 | 21 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-0303 | IPLTINKEEALQRPV | 5 | 20 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-0307 | PLTINKEEALQRPVA | 6 | 21 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-0307 | IPLTINKEEALQRPV | 5 | 20 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-0307 | SIPLTINKEEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-0315 | PLTINKEEALQRPVA | 6 | 21 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-0324 | SIPLTINKEEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-0338 | SIPLTINKEEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-0338 | HSIPLTINKEEALQR | 3 | 18 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-0342 | PLTINKEEALQRPVA | 6 | 21 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-0342 | IPLTINKEEALQRPV | 5 | 20 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-0342 | SIPLTINKEEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-1107 | PLTINKEEALQRPVA | 6 | 21 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-1107 | IPLTINKEEALQRPV | 5 | 20 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-1107 | SIPLTINKEEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-1331 | SIPLTINKEEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-1438 | SIPLTINKEEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-1439 | SIPLTINKEEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-1439 | HSIPLTINKEEALQR | 3 | 18 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-1448 | SIPLTINKEEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-1450 | SIPLTINKEEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-1450 | HSIPLTINKEEALQR | 3 | 18 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-1452 | PLTINKEEALQRPVA | 6 | 21 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-1470 | SIPLTINKEEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-1470 | HSIPLTINKEEALQR | 3 | 18 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-1476 | SIPLTINKEEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-1479 | SIPLTINKEEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-1493 | SIPLTINKEEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-1503 | SIPLTINKEEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-1503 | HSIPLTINKEEALQR | 3 | 18 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-1523 | SIPLTINKEEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-1523 | HSIPLTINKEEALQR | 3 | 18 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-1525 | SIPLTINKEEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-1525 | IPLTINKEEALQRPV | 5 | 20 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB1-1525 | HSIPLTINKEEALQR | 3 | 18 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB3-0109 | SIPLTINKEEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB3-0201 | SIPLTINKEEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB3-0201 | HSIPLTINKEEALQR | 3 | 18 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB3-0204 | SIPLTINKEEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB3-0204 | HSIPLTINKEEALQR | 3 | 18 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB3-0204 | IPLTINKEEALQRPV | 5 | 20 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB3-0209 | SIPLTINKEEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB3-0214 | SIPLTINKEEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB3-0216 | SIPLTINKEEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB3-0221 | SIPLTINKEEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB3-0224 | SIPLTINKEEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB3-0224 | HSIPLTINKEEALQR | 3 | 18 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB3-0301 | SIPLTINKEEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB5-0101 | PLTINKEEALQRPVA | 6 | 21 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB5-0101 | IPLTINKEEALQRPV | 5 | 20 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB5-0105 | PLTINKEEALQRPVA | 6 | 21 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB5-0105 | IPLTINKEEALQRPV | 5 | 20 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB5-0106 | PLTINKEEALQRPVA | 6 | 21 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB5-0106 | IPLTINKEEALQRPV | 5 | 20 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB5-0106 | SIPLTINKEEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB5-0106 | LTINKEEALQRPVAS | 7 | 22 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB5-0111 | PLTINKEEALQRPVA | 6 | 21 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB5-0111 | IPLTINKEEALQRPV | 5 | 20 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB5-0113 | PLTINKEEALQRPVA | 6 | 21 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB5-0113 | IPLTINKEEALQRPV | 5 | 20 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB5-0114 | PLTINKEEALQRPVA | 6 | 21 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB5-0114 | IPLTINKEEALQRPV | 5 | 20 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB5-0202 | PLTINKEEALQRPVA | 6 | 21 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB5-0202 | IPLTINKEEALQRPV | 5 | 20 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB5-0203 | PLTINKEEALQRPVA | 6 | 21 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB5-0204 | PLTINKEEALQRPVA | 6 | 21 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB5-0204 | IPLTINKEEALQRPV | 5 | 20 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB5-0205 | PLTINKEEALQRPVA | 6 | 21 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB5-0205 | IPLTINKEEALQRPV | 5 | 20 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB5-0205 | SIPLTINKEEALQRP | 4 | 19 |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 | DRB5-0205 | LTINKEEALQRPVAS | 7 | 22 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0466 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0466 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0801 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0801 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0802 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0802 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0802 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0802 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0803 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0803 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0805 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0805 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0807 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0807 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0808 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0808 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0809 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0809 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0809 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0809 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0811 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0811 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0813 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0813 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0814 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0814 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0815 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0815 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0816 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0816 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0818 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0818 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0821 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0821 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0821 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0821 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0823 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0823 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0824 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0824 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0824 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0824 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0826 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0826 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0827 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0827 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0829 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0829 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0829 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0830 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0830 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0833 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0833 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0835 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0835 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0836 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0836 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0837 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0837 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0838 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0838 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0839 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0839 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0840 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-0840 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1101 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1101 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1101 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1101 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1105 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1105 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1105 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1105 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1108 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1108 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1108 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1108 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1109 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1109 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1109 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1109 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1110 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1110 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1110 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1110 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1111 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1111 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1112 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1112 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1112 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1112 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1114 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1114 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1115 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1115 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1115 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1115 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1119 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1119 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1119 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1119 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1120 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1120 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1124 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1124 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1124 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1124 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1127 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1127 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1127 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1127 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1128 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1128 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1128 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1128 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1129 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1129 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1129 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1129 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1130 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1130 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1131 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1131 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1131 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1131 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1132 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1132 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1132 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1132 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1133 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1133 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1133 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1133 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1137 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1137 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1137 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1137 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1139 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1139 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1139 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1139 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1145 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1145 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1149 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1149 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1149 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1149 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1151 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1151 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1151 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1151 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1153 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1153 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1153 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1153 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1154 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1154 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1161 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1161 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1161 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1161 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1162 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1162 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1162 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1162 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1164 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1164 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1166 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1166 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1166 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1166 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1168 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1168 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1169 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1169 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1169 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1169 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1172 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1172 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1173 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1173 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1173 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1173 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1174 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1174 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1174 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1174 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1175 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1175 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1175 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1175 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1179 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1179 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1179 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1180 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1180 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1180 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1180 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1181 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1181 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1181 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1181 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1182 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1182 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1186 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1187 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1187 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1187 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1187 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1190 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1190 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1190 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1190 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1191 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1191 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1191 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1191 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1193 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1193 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1193 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1193 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1194 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1194 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1194 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1194 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1195 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1195 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1195 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1195 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1196 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1196 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1196 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1196 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1302 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1302 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1303 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1303 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1305 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1305 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1305 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1305 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1307 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1307 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1307 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1307 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-13100 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-13100 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-13101 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-13101 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-13101 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1310 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1310 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1310 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1312 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1312 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1313 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1313 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1313 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1314 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1314 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1314 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1314 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1316 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1321 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1321 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1321 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1323 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1323 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1326 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1326 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1326 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1326 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1329 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1329 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1330 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1330 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1333 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1333 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1334 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1334 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1336 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1336 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1337 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1337 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1337 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1337 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1338 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1338 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1339 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1339 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1341 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1341 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1346 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1346 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1347 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1347 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1347 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1347 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1349 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1349 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1349 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1349 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1350 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1350 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1350 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1350 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1355 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1355 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1355 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1355 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1356 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1356 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1356 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1356 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1360 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1360 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1360 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1360 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1362 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1362 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1362 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1362 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1363 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1363 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1365 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1365 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1366 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1366 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1366 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1367 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1373 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1373 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1374 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1374 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1382 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1382 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1382 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1382 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1385 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1385 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1385 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1385 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1386 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1386 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1388 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1388 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1388 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1390 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1390 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1395 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1395 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1397 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1397 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1399 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1399 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1402 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1402 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1403 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1403 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1403 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1403 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1407 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1407 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1409 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1409 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1409 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1413 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1413 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1414 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1414 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1414 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1414 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1419 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1419 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1422 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1422 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1422 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1424 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1424 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1425 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1425 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1425 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1427 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1427 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1427 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1427 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1430 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1430 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1430 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1436 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1436 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1436 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1436 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1440 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1440 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1440 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1440 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1441 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1441 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1442 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1442 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1442 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1442 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1444 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1444 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1444 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1444 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1447 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1449 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1451 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1451 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1451 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1451 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1453 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1453 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1453 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1463 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1463 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1467 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1467 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1468 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1468 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1469 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1469 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1469 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1477 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1477 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1477 | TGFKQSSKALQRPVA | 5 | 20 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1477 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1485 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1485 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1485 | HSATGFKQSSKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1489 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1489 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1493 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1493 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1494 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1494 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1498 | ATGFKQSSKALQRPV | 4 | 19 |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 | DRB1-1498 | SATGFKQSSKALQRP | 3 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0113 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0113 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0117 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0117 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0121 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0121 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0469 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0469 | HSATGFKQSSSEKLR | 4 | 19 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0469 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0701 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0701 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0701 | HSATGFKQSSSEKLR | 4 | 19 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0701 | TGFKQSSSEKLRVLG | 7 | 22 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0701 | VHSATGFKQSSSEKL | 3 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0703 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0703 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0703 | HSATGFKQSSSEKLR | 4 | 19 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0703 | TGFKQSSSEKLRVLG | 7 | 22 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0703 | VHSATGFKQSSSEKL | 3 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0704 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0704 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0704 | HSATGFKQSSSEKLR | 4 | 19 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0704 | TGFKQSSSEKLRVLG | 7 | 22 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0704 | VHSATGFKQSSSEKL | 3 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0705 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0705 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0705 | HSATGFKQSSSEKLR | 4 | 19 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0705 | TGFKQSSSEKLRVLG | 7 | 22 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0705 | VHSATGFKQSSSEKL | 3 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0706 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0706 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0706 | HSATGFKQSSSEKLR | 4 | 19 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0706 | TGFKQSSSEKLRVLG | 7 | 22 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0706 | VHSATGFKQSSSEKL | 3 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0707 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0707 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0707 | HSATGFKQSSSEKLR | 4 | 19 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0707 | TGFKQSSSEKLRVLG | 7 | 22 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0707 | VHSATGFKQSSSEKL | 3 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0708 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0708 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0708 | HSATGFKQSSSEKLR | 4 | 19 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0708 | TGFKQSSSEKLRVLG | 7 | 22 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0708 | VHSATGFKQSSSEKL | 3 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0709 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0709 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0709 | HSATGFKQSSSEKLR | 4 | 19 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0709 | TGFKQSSSEKLRVLG | 7 | 22 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0709 | VHSATGFKQSSSEKL | 3 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0711 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0711 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0711 | HSATGFKQSSSEKLR | 4 | 19 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0711 | TGFKQSSSEKLRVLG | 7 | 22 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0711 | VHSATGFKQSSSEKL | 3 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0712 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0712 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0712 | HSATGFKQSSSEKLR | 4 | 19 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0712 | TGFKQSSSEKLRVLG | 7 | 22 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0712 | VHSATGFKQSSSEKL | 3 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0713 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0713 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0713 | HSATGFKQSSSEKLR | 4 | 19 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0713 | TGFKQSSSEKLRVLG | 7 | 22 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0713 | VHSATGFKQSSSEKL | 3 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0714 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0714 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0714 | HSATGFKQSSSEKLR | 4 | 19 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0714 | TGFKQSSSEKLRVLG | 7 | 22 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0714 | VHSATGFKQSSSEKL | 3 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0715 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0715 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0715 | HSATGFKQSSSEKLR | 4 | 19 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0715 | TGFKQSSSEKLRVLG | 7 | 22 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0715 | VHSATGFKQSSSEKL | 3 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0716 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0716 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0716 | HSATGFKQSSSEKLR | 4 | 19 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0716 | TGFKQSSSEKLRVLG | 7 | 22 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0716 | VHSATGFKQSSSEKL | 3 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0717 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0717 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0717 | HSATGFKQSSSEKLR | 4 | 19 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0717 | TGFKQSSSEKLRVLG | 7 | 22 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0717 | VHSATGFKQSSSEKL | 3 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0719 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0719 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0719 | HSATGFKQSSSEKLR | 4 | 19 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0719 | TGFKQSSSEKLRVLG | 7 | 22 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0719 | VHSATGFKQSSSEKL | 3 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0901 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0901 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0901 | HSATGFKQSSSEKLR | 4 | 19 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0901 | TGFKQSSSEKLRVLG | 7 | 22 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0901 | VHSATGFKQSSSEKL | 3 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0902 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0902 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0902 | HSATGFKQSSSEKLR | 4 | 19 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0903 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0903 | HSATGFKQSSSEKLR | 4 | 19 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0903 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0904 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0904 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0904 | HSATGFKQSSSEKLR | 4 | 19 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0904 | TGFKQSSSEKLRVLG | 7 | 22 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0904 | VHSATGFKQSSSEKL | 3 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0905 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0905 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0905 | HSATGFKQSSSEKLR | 4 | 19 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0905 | TGFKQSSSEKLRVLG | 7 | 22 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0907 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0907 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0907 | HSATGFKQSSSEKLR | 4 | 19 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0907 | TGFKQSSSEKLRVLG | 7 | 22 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0907 | VHSATGFKQSSSEKL | 3 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0908 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0908 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0908 | HSATGFKQSSSEKLR | 4 | 19 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0909 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0909 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0909 | HSATGFKQSSSEKLR | 4 | 19 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0909 | TGFKQSSSEKLRVLG | 7 | 22 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-0909 | VHSATGFKQSSSEKL | 3 | 18 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-1407 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-1468 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-1468 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-1468 | HSATGFKQSSSEKLR | 4 | 19 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-1493 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-1493 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-1493 | HSATGFKQSSSEKLR | 4 | 19 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-1527 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-1527 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-1534 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-1534 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-1534 | HSATGFKQSSSEKLR | 4 | 19 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-1601 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-1601 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-1602 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-1602 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-1603 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-1603 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-1605 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-1605 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-1607 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-1607 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-1608 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-1608 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-1611 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-1611 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-1612 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-1612 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-1614 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-1614 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-1616 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB1-1616 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB3-0109 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB3-0209 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB3-0209 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB3-0221 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB3-0221 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB3-0303 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB3-0303 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB5-0112 | SATGFKQSSSEKLRV | 5 | 20 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB5-0112 | ATGFKQSSSEKLRVL | 6 | 21 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB5-0112 | HSATGFKQSSSEKLR | 4 | 19 |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 | DRB5-0112 | TGFKQSSSEKLRVLG | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0478 | IQALKAAFDVKALQR | 2 | 17 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0802 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0802 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0804 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0804 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0804 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0804 | FDVKALQRPVASDFE | 9 | 24 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0806 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0806 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0808 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0809 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0809 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0810 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0810 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0811 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0812 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0812 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0813 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0813 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0815 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0815 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0820 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0820 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0820 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0820 | FDVKALQRPVASDFE | 9 | 24 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0821 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0821 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0822 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0822 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0822 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0824 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0824 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0828 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0828 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0828 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0830 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0830 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0831 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0831 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0831 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-0831 | FDVKALQRPVASDFE | 9 | 24 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1002 | TDIQALKAAFDVKAL | 0 | 15 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1101 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1101 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1103 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1103 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1104 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1104 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1104 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1104 | FDVKALQRPVASDFE | 9 | 24 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1105 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1105 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1106 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1106 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1106 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1106 | FDVKALQRPVASDFE | 9 | 24 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1108 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1108 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1109 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1109 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1110 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1110 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1112 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1112 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1113 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1113 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1115 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1115 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1117 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1117 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1118 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1118 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1118 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1119 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1119 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1124 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1124 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1125 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1125 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1125 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1125 | FDVKALQRPVASDFE | 9 | 24 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1127 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1127 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1128 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1128 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1129 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1129 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1131 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1131 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1132 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1132 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1133 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1133 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1134 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1134 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1135 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1135 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1135 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1135 | FDVKALQRPVASDFE | 9 | 24 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1137 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1137 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1138 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1138 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1138 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1138 | FDVKALQRPVASDFE | 9 | 24 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1139 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1139 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1141 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1141 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1142 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1142 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1142 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1143 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1143 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1143 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1143 | FDVKALQRPVASDFE | 9 | 24 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1144 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1144 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1144 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1144 | FDVKALQRPVASDFE | 9 | 24 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1146 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1146 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1146 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1146 | FDVKALQRPVASDFE | 9 | 24 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1147 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1147 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1147 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1147 | FDVKALQRPVASDFE | 9 | 24 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1149 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1149 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1150 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1150 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1150 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1150 | FDVKALQRPVASDFE | 9 | 24 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1151 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1151 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1152 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1152 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1154 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1154 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1154 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1156 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1156 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1156 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1156 | FDVKALQRPVASDFE | 9 | 24 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1157 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1157 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1157 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1157 | FDVKALQRPVASDFE | 9 | 24 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1158 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1158 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1158 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1158 | FDVKALQRPVASDFE | 9 | 24 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1159 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1159 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1160 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1160 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1160 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1160 | FDVKALQRPVASDFE | 9 | 24 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1161 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1161 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1162 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1162 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1163 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1163 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1164 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1164 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1166 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1166 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1167 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1167 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1167 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1169 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1169 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1172 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1172 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1174 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1174 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1175 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1175 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1176 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1176 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1177 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1177 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1177 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1177 | FDVKALQRPVASDFE | 9 | 24 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1178 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1178 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1178 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1178 | FDVKALQRPVASDFE | 9 | 24 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1180 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1180 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1181 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1181 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1183 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1183 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1183 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1183 | FDVKALQRPVASDFE | 9 | 24 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1184 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1184 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1184 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1184 | FDVKALQRPVASDFE | 9 | 24 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1185 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1185 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1187 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1187 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1188 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1188 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1188 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1188 | FDVKALQRPVASDFE | 9 | 24 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1189 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1189 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1190 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1190 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1191 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1191 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1192 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1192 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1192 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1192 | FDVKALQRPVASDFE | 9 | 24 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1193 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1193 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1194 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1194 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1195 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1195 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1196 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1196 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1204 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1204 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1204 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1209 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1209 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1209 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1220 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1220 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1305 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1305 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1306 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1306 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1306 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1307 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1307 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1309 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1309 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-13100 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1311 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1311 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1311 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1311 | FDVKALQRPVASDFE | 9 | 24 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1314 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1314 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1318 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1318 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1318 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1318 | FDVKALQRPVASDFE | 9 | 24 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1324 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1324 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1326 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1326 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1342 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1342 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1342 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1342 | FDVKALQRPVASDFE | 9 | 24 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1344 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1344 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1346 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1347 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1347 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1350 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1350 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1356 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1356 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1358 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1358 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1360 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1360 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1362 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1362 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1377 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1377 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1381 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1382 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1382 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1402 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1402 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1403 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1403 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1405 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1405 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1406 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1406 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1406 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1408 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1408 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1409 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1409 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1411 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1411 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1412 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1412 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1412 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1415 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1415 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1415 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1415 | FDVKALQRPVASDFE | 9 | 24 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1417 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1417 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1418 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1418 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1420 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1420 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1420 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1421 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1421 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1422 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1422 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1423 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1423 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1425 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1425 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1427 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1427 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1429 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1429 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1429 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1430 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1431 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1431 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1432 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1432 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1433 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1433 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1434 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1434 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1434 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1437 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1437 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1440 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1440 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1441 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1441 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1443 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1443 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1445 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1445 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1452 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1452 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1453 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1456 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1456 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1459 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1459 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1464 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1465 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1465 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1467 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1467 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1472 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1473 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1473 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1473 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1474 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1474 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1477 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1477 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1478 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1478 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1480 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1480 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1480 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1481 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1481 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1481 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1483 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1483 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1483 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1484 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1484 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1484 | KAAFDVKALQRPVAS | 6 | 21 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1484 | FDVKALQRPVASDFE | 9 | 24 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1491 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1491 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1494 | AFDVKALQRPVASDF | 8 | 23 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1494 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1496 | AAFDVKALQRPVASD | 7 | 22 |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 | DRB1-1496 | AFDVKALQRPVASDF | 8 | 23 |
Top |
Fusion breakpoint peptide structures of BCR-ABL1 |
 3D structures of the fusion breakpoint peptide of 14AA sequence that have potential fusion neoantigens 3D structures of the fusion breakpoint peptide of 14AA sequence that have potential fusion neoantigens* The minimum length of the amino acid sequence in RoseTTAFold is 14AA. Here, we predicted the 14AA fusion protein breakpoint sequence not the fusion neoantigen peptide, which is shorter than 14 AA. |
| File name | BPseq | Hgene | Tgene | Hchr | Hbp | Tchr | Tbp | AAlen |
| 650 | ATGFKQSSSEKLRV | BCR | ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3533 |
| File name | BPseq | Hgene | Tgene | Hchr | Hbp | Tchr | Tbp | AAlen |
| 2644 | GAFHGDAEALQRPV | BCR | ABL1 | chr22 | 23524426 | chr9 | 133729448 | 2030 |
| File name | BPseq | Hgene | Tgene | Hchr | Hbp | Tchr | Tbp | AAlen |
| 2646 | GAFHGDAGEKLRVL | BCR | ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2030 |
| File name | BPseq | Hgene | Tgene | Hchr | Hbp | Tchr | Tbp | AAlen |
| 2776 | GFKQSSKALQRPVA | BCR | ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3533 |
| File name | BPseq | Hgene | Tgene | Hchr | Hbp | Tchr | Tbp | AAlen |
| 4091 | KAAFDVKALQRPVA | BCR | ABL1 | chr22 | 23654023 | chr9 | 133729451 | 4073 |
| File name | BPseq | Hgene | Tgene | Hchr | Hbp | Tchr | Tbp | AAlen |
| 4161 | KDPTTKNSLEKALQ | BCR | ABL1 | chr22 | 23613779 | chr9 | 133729451 | 2672 |
| File name | BPseq | Hgene | Tgene | Hchr | Hbp | Tchr | Tbp | AAlen |
| 6801 | PLTINKEEALQRPV | BCR | ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3458 |
| File name | BPseq | Hgene | Tgene | Hchr | Hbp | Tchr | Tbp | AAlen |
| 6802 | PLTINKEGEKLRVL | BCR | ABL1 | chr22 | 23631811 | chr9 | 133730186 | 3458 |
Top |
Filtering FusionNeoAntigens Through Checking the Interaction with HLAs in 3D of BCR-ABL1 |
 Virtual screening between 25 HLAs (from PDB) and FusionNeoAntigens Virtual screening between 25 HLAs (from PDB) and FusionNeoAntigens* We used Glide to predict the interaction between HLAs and neoantigens. |
| HLA allele | PDB ID | File name | BPseq | Docking score | Glide score |
| HLA-B14:02 | 3BVN | 650 | ATGFKQSSSEKLRV | -5.57779 | -5.57779 |
| HLA-A11:01 | 4UQ2 | 650 | ATGFKQSSSEKLRV | -6.58038 | -6.58038 |
| HLA-A24:02 | 5HGA | 650 | ATGFKQSSSEKLRV | -6.97152 | -6.97152 |
| HLA-B53:01 | 1A1O | 2644 | GAFHGDAEALQRPV | -4.17128 | -4.28468 |
| HLA-B53:01 | 1A1O | 2644 | GAFHGDAEALQRPV | -3.4824 | -4.5177 |
| HLA-B51:01 | 1E28 | 2644 | GAFHGDAEALQRPV | -3.8039 | -3.9173 |
| HLA-B51:01 | 1E28 | 2644 | GAFHGDAEALQRPV | -3.0982 | -4.1335 |
| HLA-B57:03 | 2BVO | 2644 | GAFHGDAEALQRPV | -2.25252 | -3.28782 |
| HLA-B57:03 | 2BVO | 2644 | GAFHGDAEALQRPV | -1.1301 | -1.2435 |
| HLA-A03:01 | 2XPG | 2644 | GAFHGDAEALQRPV | -4.44786 | -4.56126 |
| HLA-A03:01 | 2XPG | 2644 | GAFHGDAEALQRPV | -4.14835 | -5.18365 |
| HLA-B14:02 | 3BVN | 2644 | GAFHGDAEALQRPV | -5.80238 | -5.91578 |
| HLA-B14:02 | 3BVN | 2644 | GAFHGDAEALQRPV | -3.55223 | -4.58753 |
| HLA-B52:01 | 3W39 | 2644 | GAFHGDAEALQRPV | -5.11856 | -5.23196 |
| HLA-B52:01 | 3W39 | 2644 | GAFHGDAEALQRPV | -3.25922 | -4.29452 |
| HLA-B18:01 | 4JQV | 2644 | GAFHGDAEALQRPV | -4.41315 | -4.52655 |
| HLA-B18:01 | 4JQV | 2644 | GAFHGDAEALQRPV | -3.36095 | -4.39625 |
| HLA-A11:01 | 4UQ2 | 2644 | GAFHGDAEALQRPV | -8.56989 | -8.68329 |
| HLA-A11:01 | 4UQ2 | 2644 | GAFHGDAEALQRPV | -6.72556 | -7.76086 |
| HLA-A24:02 | 5HGA | 2644 | GAFHGDAEALQRPV | -6.2734 | -6.3868 |
| HLA-A24:02 | 5HGA | 2644 | GAFHGDAEALQRPV | -4.24193 | -5.27723 |
| HLA-B57:01 | 5VUD | 2644 | GAFHGDAEALQRPV | -3.95679 | -4.99209 |
| HLA-B57:01 | 5VUD | 2644 | GAFHGDAEALQRPV | -2.97944 | -3.09284 |
| HLA-C08:02 | 6JTP | 2644 | GAFHGDAEALQRPV | -4.74128 | -4.85468 |
| HLA-C08:02 | 6JTP | 2644 | GAFHGDAEALQRPV | -2.44773 | -3.48303 |
| HLA-B37:01 | 6MT4 | 2644 | GAFHGDAEALQRPV | -3.77829 | -4.81359 |
| HLA-B27:05 | 6PYJ | 2644 | GAFHGDAEALQRPV | -3.40019 | -3.51359 |
| HLA-B27:05 | 6PYJ | 2644 | GAFHGDAEALQRPV | -1.99024 | -3.02554 |
| HLA-B27:03 | 6PZ5 | 2644 | GAFHGDAEALQRPV | -3.1249 | -3.2383 |
| HLA-B27:03 | 6PZ5 | 2644 | GAFHGDAEALQRPV | -0.148491 | -1.18379 |
| HLA-B44:05 | 3DX8 | 2644 | GAFHGDAEALQRPV | -5.76333 | -5.87673 |
| HLA-B44:05 | 3DX8 | 2644 | GAFHGDAEALQRPV | -3.98513 | -5.02043 |
| HLA-B44:02 | 1M6O | 2644 | GAFHGDAEALQRPV | -1.76802 | -2.80332 |
| HLA-B07:02 | 5EO0 | 2644 | GAFHGDAEALQRPV | -3.47003 | -3.58343 |
| HLA-B07:02 | 5EO0 | 2644 | GAFHGDAEALQRPV | 0.983323 | -0.0519774 |
| HLA-A02:01 | 6TDR | 2644 | GAFHGDAEALQRPV | -2.71064 | -3.74594 |
| HLA-A02:01 | 6TDR | 2644 | GAFHGDAEALQRPV | -2.80613 | -2.91953 |
| HLA-B14:02 | 3BVN | 2646 | GAFHGDAGEKLRVL | -6.35143 | -7.38673 |
| HLA-B14:02 | 3BVN | 2646 | GAFHGDAGEKLRVL | -5.20162 | -5.31502 |
| HLA-B52:01 | 3W39 | 2646 | GAFHGDAGEKLRVL | -6.03049 | -6.14389 |
| HLA-B52:01 | 3W39 | 2646 | GAFHGDAGEKLRVL | -5.12323 | -6.15853 |
| HLA-A24:02 | 5HGA | 2646 | GAFHGDAGEKLRVL | -9.81198 | -9.92538 |
| HLA-A24:02 | 5HGA | 2646 | GAFHGDAGEKLRVL | -7.16433 | -8.19963 |
| HLA-B44:05 | 3DX8 | 2646 | GAFHGDAGEKLRVL | -6.46131 | -6.57471 |
| HLA-B44:05 | 3DX8 | 2646 | GAFHGDAGEKLRVL | -3.08576 | -4.12106 |
| HLA-A02:01 | 6TDR | 2646 | GAFHGDAGEKLRVL | -5.03933 | -5.15273 |
| HLA-B14:02 | 3BVN | 2776 | GFKQSSKALQRPVA | -7.9962 | -8.1096 |
| HLA-B14:02 | 3BVN | 2776 | GFKQSSKALQRPVA | -5.70842 | -6.74372 |
| HLA-B52:01 | 3W39 | 2776 | GFKQSSKALQRPVA | -6.83737 | -6.95077 |
| HLA-B52:01 | 3W39 | 2776 | GFKQSSKALQRPVA | -4.4836 | -5.5189 |
| HLA-A11:01 | 4UQ2 | 2776 | GFKQSSKALQRPVA | -10.0067 | -10.1201 |
| HLA-A11:01 | 4UQ2 | 2776 | GFKQSSKALQRPVA | -9.03915 | -10.0745 |
| HLA-A24:02 | 5HGA | 2776 | GFKQSSKALQRPVA | -6.56204 | -6.67544 |
| HLA-A24:02 | 5HGA | 2776 | GFKQSSKALQRPVA | -5.42271 | -6.45801 |
| HLA-B44:05 | 3DX8 | 2776 | GFKQSSKALQRPVA | -7.85648 | -8.89178 |
| HLA-B44:05 | 3DX8 | 2776 | GFKQSSKALQRPVA | -5.3978 | -5.5112 |
| HLA-A02:01 | 6TDR | 2776 | GFKQSSKALQRPVA | -3.37154 | -4.40684 |
| HLA-B14:02 | 3BVN | 4091 | KAAFDVKALQRPVA | -9.38616 | -9.49956 |
| HLA-B14:02 | 3BVN | 4091 | KAAFDVKALQRPVA | -2.96308 | -3.99838 |
| HLA-B52:01 | 3W39 | 4091 | KAAFDVKALQRPVA | -6.86378 | -6.97718 |
| HLA-B52:01 | 3W39 | 4091 | KAAFDVKALQRPVA | -2.9876 | -4.0229 |
| HLA-A11:01 | 4UQ2 | 4091 | KAAFDVKALQRPVA | -7.95435 | -8.06775 |
| HLA-A24:02 | 5HGA | 4091 | KAAFDVKALQRPVA | -7.59672 | -7.71012 |
| HLA-A24:02 | 5HGA | 4091 | KAAFDVKALQRPVA | -6.33737 | -7.37267 |
| HLA-B44:05 | 3DX8 | 4091 | KAAFDVKALQRPVA | -7.16165 | -7.27505 |
| HLA-B44:05 | 3DX8 | 4091 | KAAFDVKALQRPVA | -2.8843 | -3.9196 |
| HLA-A02:01 | 6TDR | 4091 | KAAFDVKALQRPVA | -5.10343 | -5.21683 |
| HLA-B14:02 | 3BVN | 4161 | KDPTTKNSLEKALQ | -7.15543 | -7.26883 |
| HLA-B14:02 | 3BVN | 4161 | KDPTTKNSLEKALQ | -4.77435 | -5.80965 |
| HLA-B52:01 | 3W39 | 4161 | KDPTTKNSLEKALQ | -6.80875 | -6.92215 |
| HLA-B52:01 | 3W39 | 4161 | KDPTTKNSLEKALQ | -4.20386 | -5.23916 |
| HLA-A11:01 | 4UQ2 | 4161 | KDPTTKNSLEKALQ | -7.5194 | -8.5547 |
| HLA-A11:01 | 4UQ2 | 4161 | KDPTTKNSLEKALQ | -6.9601 | -7.0735 |
| HLA-A24:02 | 5HGA | 4161 | KDPTTKNSLEKALQ | -7.52403 | -7.63743 |
| HLA-A24:02 | 5HGA | 4161 | KDPTTKNSLEKALQ | -5.82433 | -6.85963 |
| HLA-B27:05 | 6PYJ | 4161 | KDPTTKNSLEKALQ | -3.28285 | -4.31815 |
| HLA-B44:05 | 3DX8 | 4161 | KDPTTKNSLEKALQ | -5.91172 | -6.94702 |
| HLA-B44:05 | 3DX8 | 4161 | KDPTTKNSLEKALQ | -4.24346 | -4.35686 |
| HLA-B14:02 | 3BVN | 6801 | PLTINKEEALQRPV | -7.9962 | -8.1096 |
| HLA-B14:02 | 3BVN | 6801 | PLTINKEEALQRPV | -5.70842 | -6.74372 |
| HLA-B52:01 | 3W39 | 6801 | PLTINKEEALQRPV | -6.83737 | -6.95077 |
| HLA-B52:01 | 3W39 | 6801 | PLTINKEEALQRPV | -4.4836 | -5.5189 |
| HLA-A11:01 | 4UQ2 | 6801 | PLTINKEEALQRPV | -10.0067 | -10.1201 |
| HLA-A11:01 | 4UQ2 | 6801 | PLTINKEEALQRPV | -9.03915 | -10.0745 |
| HLA-A24:02 | 5HGA | 6801 | PLTINKEEALQRPV | -6.56204 | -6.67544 |
| HLA-A24:02 | 5HGA | 6801 | PLTINKEEALQRPV | -5.42271 | -6.45801 |
| HLA-B44:05 | 3DX8 | 6801 | PLTINKEEALQRPV | -7.85648 | -8.89178 |
| HLA-B44:05 | 3DX8 | 6801 | PLTINKEEALQRPV | -5.3978 | -5.5112 |
| HLA-A02:01 | 6TDR | 6801 | PLTINKEEALQRPV | -3.37154 | -4.40684 |
| HLA-B14:02 | 3BVN | 6802 | PLTINKEGEKLRVL | -6.01316 | -6.12656 |
| HLA-B14:02 | 3BVN | 6802 | PLTINKEGEKLRVL | -3.92858 | -4.96388 |
| HLA-B52:01 | 3W39 | 6802 | PLTINKEGEKLRVL | -5.92102 | -6.95632 |
| HLA-B52:01 | 3W39 | 6802 | PLTINKEGEKLRVL | -4.84472 | -4.95812 |
| HLA-A24:02 | 5HGA | 6802 | PLTINKEGEKLRVL | -8.26357 | -9.29887 |
| HLA-A24:02 | 5HGA | 6802 | PLTINKEGEKLRVL | -7.03366 | -7.14706 |
| HLA-B44:05 | 3DX8 | 6802 | PLTINKEGEKLRVL | -6.13971 | -6.25311 |
| HLA-B44:05 | 3DX8 | 6802 | PLTINKEGEKLRVL | -5.20728 | -6.24258 |
| HLA-A02:01 | 6TDR | 6802 | PLTINKEGEKLRVL | -5.28864 | -5.40204 |
Top |
Vaccine Design for the FusionNeoAntigens of BCR-ABL1 |
 mRNA and peptide sequences of FusionNeoAntigens that have potential interaction with HLA-Is. mRNA and peptide sequences of FusionNeoAntigens that have potential interaction with HLA-Is. |
| Fusion gene | Hchr | Hbp | Tchr | Tbp | Start in +/-13AA | End in +/-13AA | FusionNeoAntigen peptide sequence | FusionNeoAntigen RNA sequence |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 11 | 20 | DAEALQRPV | ACGCAGAAGCCCTTCAGCGGCCAGTAG |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 12 | 21 | AEALQRPVA | CAGAAGCCCTTCAGCGGCCAGTAGCAT |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 4 | 15 | GEGAFHGDAEA | GCGAGGGCGCCTTCCATGGAGACGCAGAAGCCC |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 7 | 16 | AFHGDAEAL | CCTTCCATGGAGACGCAGAAGCCCTTC |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 8 | 16 | FHGDAEAL | TCCATGGAGACGCAGAAGCCCTTC |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 11 | 19 | DAGEKLRV | ACGCAGGTGAAAAGCTCCGGGTCT |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 11 | 20 | DAGEKLRVL | ACGCAGGTGAAAAGCTCCGGGTCTTAG |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 6 | 17 | GAFHGDAGEKL | GCGCCTTCCATGGAGACGCAGGTGAAAAGCTCC |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 7 | 17 | AFHGDAGEKL | CCTTCCATGGAGACGCAGGTGAAAAGCTCC |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 8 | 17 | FHGDAGEKL | TCCATGGAGACGCAGGTGAAAAGCTCC |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 9 | 17 | HGDAGEKL | ATGGAGACGCAGGTGAAAAGCTCC |
| BCR-ABL1 | chr22 | 23613779 | chr9 | 133729451 | 10 | 19 | TKNSLEKAL | CTCTGGAAAAAGCCCTTCAGCGGCCAG |
| BCR-ABL1 | chr22 | 23613779 | chr9 | 133729451 | 5 | 15 | AKDPTTKNSL | CAACGACCAAGAACTCTCTGGAAAAAGCCC |
| BCR-ABL1 | chr22 | 23613779 | chr9 | 133729451 | 7 | 15 | DPTTKNSL | CCAAGAACTCTCTGGAAAAAGCCC |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 11 | 20 | KEEALQRPV | AGGAAGAAGCCCTTCAGCGGCCAGTAG |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 11 | 21 | KEEALQRPVA | AGGAAGAAGCCCTTCAGCGGCCAGTAGCAT |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 12 | 21 | EEALQRPVA | AAGAAGCCCTTCAGCGGCCAGTAGCAT |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 5 | 16 | IPLTINKEEAL | TTCCGCTGACCATCAATAAGGAAGAAGCCCTTC |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 7 | 16 | LTINKEEAL | TGACCATCAATAAGGAAGAAGCCCTTC |
| BCR-ABL1 | chr22 | 23631811 | chr9 | 133730186 | 11 | 20 | KEGEKLRVL | AGGAAGGTGAAAAGCTCCGGGTCTTAG |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 10 | 19 | SSKALQRPV | AGAGTTCAAAAGCCCTTCAGCGGCCAG |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 6 | 15 | GFKQSSKAL | CTGGATTTAAGCAGAGTTCAAAAGCCC |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 10 | 18 | KQSSSEKL | AGAGTTCAAGTGAAAAGCTCCGGG |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 10 | 20 | KQSSSEKLRV | AGAGTTCAAGTGAAAAGCTCCGGGTCTTAG |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 11 | 20 | QSSSEKLRV | GTTCAAGTGAAAAGCTCCGGGTCTTAG |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 12 | 21 | SSSEKLRVL | CAAGTGAAAAGCTCCGGGTCTTAGGCT |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 6 | 17 | ATGFKQSSSEK | CTGGATTTAAGCAGAGTTCAAGTGAAAAGCTCC |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 8 | 17 | GFKQSSSEK | TTAAGCAGAGTTCAAGTGAAAAGCTCC |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 9 | 18 | FKQSSSEKL | AGCAGAGTTCAAGTGAAAAGCTCCGGG |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 6 | 15 | KAAFDVKAL | TGAAGGCAGCCTTCGACGTCAAAGCCC |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 7 | 15 | AAFDVKAL | AGGCAGCCTTCGACGTCAAAGCCC |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 7 | 17 | AAFDVKALQR | AGGCAGCCTTCGACGTCAAAGCCCTTCAGC |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 8 | 17 | AFDVKALQR | CAGCCTTCGACGTCAAAGCCCTTCAGC |
 mRNA and peptide sequences of FusionNeoAntigens that have potential interaction with HLA-IIs. mRNA and peptide sequences of FusionNeoAntigens that have potential interaction with HLA-IIs. |
| Fusion gene | Hchr | Hbp | Tchr | Tbp | Start in +/-13AA | End in +/-13AA | FusionNeoAntigen peptide | FusionNEoAntigen RNA sequence |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 4 | 19 | GEGAFHGDAEALQRP | GCGAGGGCGCCTTCCATGGAGACGCAGAAGCCCTTCAGCGGCCAG |
| BCR-ABL1 | chr22 | 23524426 | chr9 | 133729448 | 5 | 20 | EGAFHGDAEALQRPV | AGGGCGCCTTCCATGGAGACGCAGAAGCCCTTCAGCGGCCAGTAG |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 2 | 17 | NDGEGAFHGDAGEKL | ACGATGGCGAGGGCGCCTTCCATGGAGACGCAGGTGAAAAGCTCC |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 3 | 18 | DGEGAFHGDAGEKLR | ATGGCGAGGGCGCCTTCCATGGAGACGCAGGTGAAAAGCTCCGGG |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 4 | 19 | GEGAFHGDAGEKLRV | GCGAGGGCGCCTTCCATGGAGACGCAGGTGAAAAGCTCCGGGTCT |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 5 | 20 | EGAFHGDAGEKLRVL | AGGGCGCCTTCCATGGAGACGCAGGTGAAAAGCTCCGGGTCTTAG |
| BCR-ABL1 | chr22 | 23524430 | chr9 | 133730185 | 6 | 21 | GAFHGDAGEKLRVLG | GCGCCTTCCATGGAGACGCAGGTGAAAAGCTCCGGGTCTTAGGCT |
| BCR-ABL1 | chr22 | 23613779 | chr9 | 133729451 | 10 | 25 | TKNSLEKALQRPVAS | CTCTGGAAAAAGCCCTTCAGCGGCCAGTAGCATCTGACTTTGAGC |
| BCR-ABL1 | chr22 | 23613779 | chr9 | 133729451 | 11 | 26 | KNSLEKALQRPVASD | TGGAAAAAGCCCTTCAGCGGCCAGTAGCATCTGACTTTGAGCCTC |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 3 | 18 | HSIPLTINKEEALQR | ACAGCATTCCGCTGACCATCAATAAGGAAGAAGCCCTTCAGCGGC |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 4 | 19 | SIPLTINKEEALQRP | GCATTCCGCTGACCATCAATAAGGAAGAAGCCCTTCAGCGGCCAG |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 5 | 20 | IPLTINKEEALQRPV | TTCCGCTGACCATCAATAAGGAAGAAGCCCTTCAGCGGCCAGTAG |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 6 | 21 | PLTINKEEALQRPVA | CGCTGACCATCAATAAGGAAGAAGCCCTTCAGCGGCCAGTAGCAT |
| BCR-ABL1 | chr22 | 23631808 | chr9 | 133729449 | 7 | 22 | LTINKEEALQRPVAS | TGACCATCAATAAGGAAGAAGCCCTTCAGCGGCCAGTAGCATCTG |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 2 | 17 | HSATGFKQSSKALQR | TCCACTCAGCCACTGGATTTAAGCAGAGTTCAAAAGCCCTTCAGC |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 3 | 18 | SATGFKQSSKALQRP | ACTCAGCCACTGGATTTAAGCAGAGTTCAAAAGCCCTTCAGCGGC |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 4 | 19 | ATGFKQSSKALQRPV | CAGCCACTGGATTTAAGCAGAGTTCAAAAGCCCTTCAGCGGCCAG |
| BCR-ABL1 | chr22 | 23632600 | chr9 | 133729450 | 5 | 20 | TGFKQSSKALQRPVA | CCACTGGATTTAAGCAGAGTTCAAAAGCCCTTCAGCGGCCAGTAG |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 3 | 18 | VHSATGFKQSSSEKL | ACTCAGCCACTGGATTTAAGCAGAGTTCAAGTGAAAAGCTCCGGG |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 4 | 19 | HSATGFKQSSSEKLR | CAGCCACTGGATTTAAGCAGAGTTCAAGTGAAAAGCTCCGGGTCT |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 5 | 20 | SATGFKQSSSEKLRV | CCACTGGATTTAAGCAGAGTTCAAGTGAAAAGCTCCGGGTCTTAG |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 6 | 21 | ATGFKQSSSEKLRVL | CTGGATTTAAGCAGAGTTCAAGTGAAAAGCTCCGGGTCTTAGGCT |
| BCR-ABL1 | chr22 | 23632602 | chr9 | 133730188 | 7 | 22 | TGFKQSSSEKLRVLG | GATTTAAGCAGAGTTCAAGTGAAAAGCTCCGGGTCTTAGGCTATA |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 0 | 15 | TDIQALKAAFDVKAL | CCACGGACATCCAGGCACTGAAGGCAGCCTTCGACGTCAAAGCCC |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 2 | 17 | IQALKAAFDVKALQR | ACATCCAGGCACTGAAGGCAGCCTTCGACGTCAAAGCCCTTCAGC |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 6 | 21 | KAAFDVKALQRPVAS | TGAAGGCAGCCTTCGACGTCAAAGCCCTTCAGCGGCCAGTAGCAT |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 7 | 22 | AAFDVKALQRPVASD | AGGCAGCCTTCGACGTCAAAGCCCTTCAGCGGCCAGTAGCATCTG |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 8 | 23 | AFDVKALQRPVASDF | CAGCCTTCGACGTCAAAGCCCTTCAGCGGCCAGTAGCATCTGACT |
| BCR-ABL1 | chr22 | 23654023 | chr9 | 133729451 | 9 | 24 | FDVKALQRPVASDFE | CCTTCGACGTCAAAGCCCTTCAGCGGCCAGTAGCATCTGACTTTG |
Top |
Information of the samples that have these potential fusion neoantigens of BCR-ABL1 |
 These samples were reported as having these fusion breakpoints. For individual breakpoints, we checked the open reading frames considering multiple gene isoforms and chose the in-frame fusion genes only. Then, we made fusion protein sequences and predicted the fusion neoantigens. These fusion-positive samples may have these potential fusion neoantigens. These samples were reported as having these fusion breakpoints. For individual breakpoints, we checked the open reading frames considering multiple gene isoforms and chose the in-frame fusion genes only. Then, we made fusion protein sequences and predicted the fusion neoantigens. These fusion-positive samples may have these potential fusion neoantigens. |
| Cancer type | Fusion gene | Hchr | Hbp | Henst | Tchr | Tbp | Tenst | Sample |
| LAML | BCR-ABL1 | chr22 | 23632600 | ENST00000305877 | chr9 | 133729450 | ENST00000318560 | TCGA-AB-2901 |
| N/A | BCR-ABL1 | chr22 | 23524426 | ENST00000305877 | chr9 | 133729448 | ENST00000318560 | AF113911 |
| N/A | BCR-ABL1 | chr22 | 23654023 | ENST00000305877 | chr9 | 133729451 | ENST00000318560 | AF487522 |
| N/A | BCR-ABL1 | chr22 | 23631808 | ENST00000305877 | chr9 | 133729449 | ENST00000318560 | AJ131467 |
| N/A | BCR-ABL1 | chr22 | 23631811 | ENST00000305877 | chr9 | 133730186 | ENST00000318560 | AM491359 |
| N/A | BCR-ABL1 | chr22 | 23632602 | ENST00000305877 | chr9 | 133730188 | ENST00000318560 | AM491360 |
| N/A | BCR-ABL1 | chr22 | 23524430 | ENST00000305877 | chr9 | 133730185 | ENST00000318560 | AM491361 |
| N/A | BCR-ABL1 | chr22 | 23613779 | ENST00000305877 | chr9 | 133729451 | ENST00000318560 | AM491362 |
Top |
Potential target of CAR-T therapy development for BCR-ABL1 |
 Predicted 3D structure. We used RoseTTAFold. Predicted 3D structure. We used RoseTTAFold. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, to provide the retention of the transmembrane domain, we only show the protein feature retention information of those transmembrane features Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, to provide the retention of the transmembrane domain, we only show the protein feature retention information of those transmembrane features* Minus value of BPloci means that the break point is located before the CDS. |
| - In-frame and retained 'Transmembrane'. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
 Subcellular localization prediction of the transmembrane domain retained fusion proteins Subcellular localization prediction of the transmembrane domain retained fusion proteins* We used DeepLoc 1.0. The order of the X-axis of the barplot is as follows: Entry_ID, Localization, Type, Nucleus, Cytoplasm, Extracellular, Mitochondrion, Cell_membrane, Endoplasmic_reticulum, Plastid, Golgi.apparatus, Lysosome.Vacuole, Peroxisome. Y-axis is the output score of DeepLoc. Clicking the image will open a new tab with a large image. |
| Hgene | Hchr | Hbp | Henst | Tgene | Tchr | Tbp | Tenst | DeepLoc result |
Top |
Related Drugs to BCR-ABL1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
| BCR | ABL1 | Imatinib | PubMed | 32000382 |
| BCR | ABL1 | Dasatinib | MyCancerGenome | |
| BCR | ABL1 | Bosutinib | MyCancerGenome | |
| BCR | ABL1 | Nilotinib | MyCancerGenome | |
| BCR | ABL1 | Imatinib | MyCancerGenome | |
| BCR | ABL1 | Ponatinib | MyCancerGenome |
Top |
Related Diseases to BCR-ABL1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
| BCR | ABL1 | Chronic Myelogenous Leukemi | PubMed | 32000382 |
| BCR | ABL1 | Chronic Myeloid Leukemia | MyCancerGenome | |
| BCR | ABL1 | Breast Invasive Ductal Carcinoma | MyCancerGenome | |
| BCR | ABL1 | Unknown | MyCancerGenome | |
| BCR | ABL1 | B-Cell Lymphoblastic Leukemia/Lymphoma | MyCancerGenome | |
| BCR | ABL1 | Acute Myeloid Leukemia | MyCancerGenome |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | BCR | C0005586 | Bipolar Disorder | 4 | PSYGENET |
| Hgene | BCR | C0023473 | Myeloid Leukemia, Chronic | 3 | CTD_human;ORPHANET |
| Hgene | BCR | C0005699 | Blast Phase | 1 | CTD_human |
| Hgene | BCR | C0006413 | Burkitt Lymphoma | 1 | ORPHANET |
| Hgene | BCR | C0023893 | Liver Cirrhosis, Experimental | 1 | CTD_human |
| Hgene | BCR | C0027022 | Myeloproliferative disease | 1 | CTD_human |
| Hgene | BCR | C0027540 | Necrosis | 1 | CTD_human |
| Hgene | BCR | C0027659 | Neoplasms, Experimental | 1 | CTD_human |
| Hgene | BCR | C0041696 | Unipolar Depression | 1 | PSYGENET |
| Hgene | BCR | C1269683 | Major Depressive Disorder | 1 | PSYGENET |
| Hgene | BCR | C1292769 | Precursor B-cell lymphoblastic leukemia | 1 | ORPHANET |
| Tgene | ABL1 | C0023473 | Myeloid Leukemia, Chronic | 3 | CGI;CTD_human;ORPHANET |
| Tgene | ABL1 | C0023452 | Childhood Acute Lymphoblastic Leukemia | 2 | CTD_human |
| Tgene | ABL1 | C0023453 | L2 Acute Lymphoblastic Leukemia | 2 | CTD_human |
| Tgene | ABL1 | C1961102 | Precursor Cell Lymphoblastic Leukemia Lymphoma | 2 | CGI;CTD_human |
| Tgene | ABL1 | C0001418 | Adenocarcinoma | 1 | CTD_human |
| Tgene | ABL1 | C0003706 | Arachnodactyly | 1 | GENOMICS_ENGLAND |
| Tgene | ABL1 | C0005941 | Bone Diseases, Developmental | 1 | CTD_human |
| Tgene | ABL1 | C0006142 | Malignant neoplasm of breast | 1 | CTD_human |
| Tgene | ABL1 | C0006413 | Burkitt Lymphoma | 1 | ORPHANET |
| Tgene | ABL1 | C0014859 | Esophageal Neoplasms | 1 | CTD_human |
| Tgene | ABL1 | C0015544 | Failure to Thrive | 1 | CTD_human |
| Tgene | ABL1 | C0018798 | Congenital Heart Defects | 1 | CTD_human |
| Tgene | ABL1 | C0023903 | Liver neoplasms | 1 | CTD_human |
| Tgene | ABL1 | C0027659 | Neoplasms, Experimental | 1 | CTD_human |
| Tgene | ABL1 | C0032927 | Precancerous Conditions | 1 | CTD_human |
| Tgene | ABL1 | C0039075 | Syndactyly | 1 | GENOMICS_ENGLAND |
| Tgene | ABL1 | C0151491 | Congenital musculoskeletal anomalies | 1 | CTD_human |
| Tgene | ABL1 | C0205641 | Adenocarcinoma, Basal Cell | 1 | CTD_human |
| Tgene | ABL1 | C0205642 | Adenocarcinoma, Oxyphilic | 1 | CTD_human |
| Tgene | ABL1 | C0205643 | Carcinoma, Cribriform | 1 | CTD_human |
| Tgene | ABL1 | C0205644 | Carcinoma, Granular Cell | 1 | CTD_human |
| Tgene | ABL1 | C0205645 | Adenocarcinoma, Tubular | 1 | CTD_human |
| Tgene | ABL1 | C0265610 | Clinodactyly of fingers | 1 | GENOMICS_ENGLAND |
| Tgene | ABL1 | C0282313 | Condition, Preneoplastic | 1 | CTD_human |
| Tgene | ABL1 | C0345904 | Malignant neoplasm of liver | 1 | CTD_human |
| Tgene | ABL1 | C0546837 | Malignant neoplasm of esophagus | 1 | CTD_human |
| Tgene | ABL1 | C0596263 | Carcinogenesis | 1 | CTD_human |
| Tgene | ABL1 | C0678222 | Breast Carcinoma | 1 | CTD_human |
| Tgene | ABL1 | C1257931 | Mammary Neoplasms, Human | 1 | CTD_human |
| Tgene | ABL1 | C1292769 | Precursor B-cell lymphoblastic leukemia | 1 | ORPHANET |
| Tgene | ABL1 | C1458155 | Mammary Neoplasms | 1 | CTD_human |
| Tgene | ABL1 | C1961099 | Precursor T-Cell Lymphoblastic Leukemia-Lymphoma | 1 | CGI;ORPHANET |
| Tgene | ABL1 | C4539857 | CONGENITAL HEART DEFECTS AND SKELETAL MALFORMATIONS SYNDROME | 1 | GENOMICS_ENGLAND;UNIPROT |
| Tgene | ABL1 | C4551485 | Clinodactyly | 1 | GENOMICS_ENGLAND |
| Tgene | ABL1 | C4704874 | Mammary Carcinoma, Human | 1 | CTD_human |

