| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Kinase Fusion Gene:CHD1_MTOR |
Kinase Fusion Protein Summary |
 Kinase Fusion gene summary Kinase Fusion gene summary |
| Kinase Fusion partner gene information | Kinase Fusion gene name: CHD1_MTOR | KinaseFusionDB ID: KFG1259 | FusionGDB2.0 ID: KFG1259 | Hgene | Tgene | Gene symbol | CHD1 | MTOR | Gene ID | 1105 | 2475 | |
| Gene name | chromodomain helicase DNA binding protein 1 | mechanistic target of rapamycin kinase | ||||||||||
| Synonyms | CHD-1|PILBOS | FRAP|FRAP1|FRAP2|RAFT1|RAPT1|SKS | ||||||||||
| Cytomap | 5q15-q21.1 | 1p36.22 | ||||||||||
| Type of gene | protein-coding | protein-coding | ||||||||||
| Description | chromodomain-helicase-DNA-binding protein 1ATP-dependent helicase CHD1 | serine/threonine-protein kinase mTORFK506 binding protein 12-rapamycin associated protein 2FK506-binding protein 12-rapamycin complex-associated protein 1FKBP-rapamycin associated proteinFKBP12-rapamycin complex-associated protein 1mammalian target o | ||||||||||
| Modification date | 20240411 | 20240411 | ||||||||||
| UniProtAcc | Q86WJ1 | P42345 | ||||||||||
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000284049, ENST00000511067, | ENST00000376838, ENST00000495435, ENST00000361445, | |||||||||
| Context (manual curation of fusion genes in KinaseFusionDB) | PubMed: CHD1 [Title/Abstract] AND MTOR [Title/Abstract] AND fusion [Title/Abstract] | |||||||||||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | CHD1(98199112)-MTOR(11273623), # samples:4 | |||||||||||
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | MTOR | GO:0001558 | regulation of cell growth | 18762023 |
| Tgene | MTOR | GO:0006468 | protein phosphorylation | 15467718 |
| Tgene | MTOR | GO:0006468 | protein phosphorylation | 18925875 |
| Tgene | MTOR | GO:0007040 | lysosome organization | 22692423 |
| Tgene | MTOR | GO:0009267 | cellular response to starvation | 22343943|22576015|22692423|28223137 |
| Tgene | MTOR | GO:0010507 | negative regulation of autophagy | 22576015|30704899|32561715 |
| Tgene | MTOR | GO:0016242 | negative regulation of macroautophagy | 25327288 |
| Tgene | MTOR | GO:0016310 | phosphorylation | 25327288 |
| Tgene | MTOR | GO:0016310 | phosphorylation | 11853878 |
| Tgene | MTOR | GO:0018105 | peptidyl-serine phosphorylation | 22343943|22576015|22692423 |
| Tgene | MTOR | GO:0031648 | protein destabilization | 36608670 |
| Tgene | MTOR | GO:0031667 | response to nutrient levels | 29750193 |
| Tgene | MTOR | GO:0031669 | cellular response to nutrient levels | 29750193|32561715 |
| Tgene | MTOR | GO:0031670 | cellular response to nutrient | 22017875|22017876|22017877 |
| Tgene | MTOR | GO:0032869 | cellular response to insulin stimulus | 18372248 |
| Tgene | MTOR | GO:0034198 | cellular response to amino acid starvation | 22343943|22424946|22576015|22692423 |
| Tgene | MTOR | GO:0038202 | TORC1 signaling | 12087098|17517883|18372248|22017875|22017876|22017877|24403073|24448649|28223137|29236692|29750193|31112131|32612235|36608670 |
| Tgene | MTOR | GO:0043200 | response to amino acid | 18497260 |
| Tgene | MTOR | GO:0045727 | positive regulation of translation | 18762023 |
| Tgene | MTOR | GO:0045948 | positive regulation of translational initiation | 22578813|29750193 |
| Tgene | MTOR | GO:0046777 | protein autophosphorylation | 15467718 |
| Tgene | MTOR | GO:0051647 | nucleus localization | 22343943|22576015|22692423 |
| Tgene | MTOR | GO:0071230 | cellular response to amino acid stimulus | 22424946 |
| Tgene | MTOR | GO:0071233 | cellular response to L-leucine | 22424946 |
| Tgene | MTOR | GO:1900181 | negative regulation of protein localization to nucleus | 22692423|24448649|32612235|35662396 |
| Tgene | MTOR | GO:1905672 | negative regulation of lysosome organization | 24448649|32612235 |
| Tgene | MTOR | GO:1990253 | cellular response to leucine starvation | 22424946 |
| Tgene | MTOR | GO:2000785 | regulation of autophagosome assembly | 23524951 |
 Kinase Fusion gene breakpoints across CHD1 (5'-gene) Kinase Fusion gene breakpoints across CHD1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
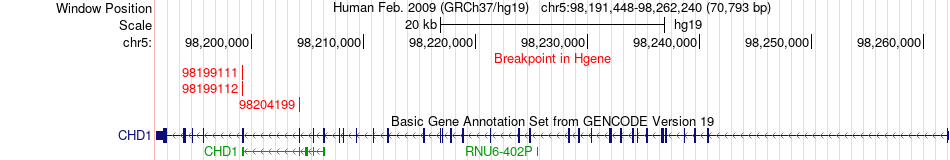 |
 Kinase Fusion gene breakpoints across MTOR (3'-gene) Kinase Fusion gene breakpoints across MTOR (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
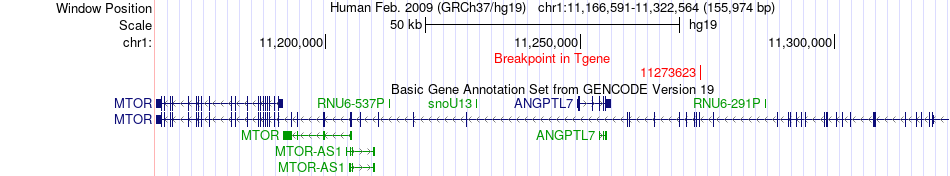 |
Top |
Kinase Fusion Gene Sample Information |
 Kinase Fusion gene information. Kinase Fusion gene information. |
 Kinase Fusion gene information from four resources (ChiTars 5.0, ChimerDB 4.0, COSMIC, and CCLE) Kinase Fusion gene information from four resources (ChiTars 5.0, ChimerDB 4.0, COSMIC, and CCLE)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Sample | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp |
| ChimerDB4 | TCGA-AB-2939-03A | CHD1 | chr5 | 98199112 | MTOR | chr1 | 11273623 |
| ChimerDB4 | TCGA-AB-2939_61FFWAAXX_7 | CHD1 | chr5 | 98204199 | MTOR | chr1 | 11273623 |
| ChimerDB4 | TCGA-AB-2939 | CHD1 | chr5 | 98199111 | MTOR | chr1 | 11273623 |
Top |
Kinase Fusion ORF Analysis |
 Kinase Fusion information from ORFfinder translation from full-length transcript sequence from KinaseFusionDB. Kinase Fusion information from ORFfinder translation from full-length transcript sequence from KinaseFusionDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp | Seq length (transcript) | Seq length (amino acids) |
| ENST00000284049 | ENST00000361445 | CHD1 | chr5 | 98204199 | MTOR | chr1 | 11273623 | 9881 | 2926 |
Top |
Kinase Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from KinaseFusionDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from KinaseFusionDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >Henst_Tenst_Hgene_Hchr_Hbp_Tgene_Tchr_Tbp_length(fusion AA)_AAseq >ENST00000284049_ENST00000361445_CHD1_chr5_98204199_MTOR_chr1_11273623_length(amino acids)=2926 MNGHSDEESVRNSSGESSQSDDDSGSASGSGSGSSSGSSSDGSSSQSGSSDSDSGSESGSQSESESDTSRENKVQAKPPKVDGAEFWKSS PSILAVQRSAILKKQQQQQQQQQHQASSNSGSEEDSSSSEDSDDSSSEVKRKKHKDEDWQMSGSGSPSQSGSDSESEEEREKSSCDETES DYEPKNKVKSRKPQNRSKSKNGKKILGQKKRQIDSSEEDDDEEDYDNDKRSSRRQATVNVSYKEDEEMKTDSDDLLEVCGEDVPQPEEEE FETIERFMDCRIGRKGATGATTTIYAVEADGDPNAGFEKNKEPGEIQYLIKWKGWSHIHNTWETEETLKQQNVRGMKKLDNYKKKDQETK RWLKNASPEDVEYYNCQQELTDDLHKQYQIVERIIAHSNQKSAAGYPDYYCKWQGLPYSECSWEDGALISKKFQACIDEYFSRNQSKTTP FKDCKVLKQRPRFVALKKQPSYIGGHEGLELRDYQLNGLNWLAHSWCKGNSCILADEMGLGKTIQTISFLNYLFHEHQLYGPFLLVVPLS TLTSWQREIQTWASQMNAVVYLGDINSRNMIRTHEWTHHQTKRLKFNILLTTYEILLKDKAFLGGLNWAFIGVDEAHRLKNDDSLLYKTL IDFKSNHRLLITGTPLQNSLKELWSLLHFIMPEKFSSWEDFEEEHGKGREYGYASLHKELEPFLLRRVKKDVEKSLPAKVEQILRMEMSA LQKQYYKWILTRNYKALSKGSKGSTSGFLNIMMELKKCCNHCYLIKPPDNNEFYNKQEALQHLIRSSGKLILLDKLLIRLRERGNRVLIF SQMVRMLDILAEYLKYRQFPFQRLDGSIKGELRKQALDHFNAEGSEDFCFLLSTRAGGLGINLASADTVVIFDSDWNPQNDLQAQARAHR IGQKKQVNIYRLVTKGSVEEDILERAKKKMVLDHLVIQRMDTTGKTVLHTGSAPSSSTPFNKEELSAILKFGAEELFKEPEGEEQEPQEM DIDEILKRAETHENEPGPLTVGDELLSQFKVANFSNMDEDDIELEPERNSKNWEEIIPEDQRRRLEEEERQKELEEIYMLPRMRNCAKQI SFNGSEGRRSRSRRYSGSDSDSISEGKRPKKRGRPRTIPRENIKGFSDAEIRRFIKSYKKFGGPLERLDAIARDAELVDKSETDLRRLGE LVHNGCIKALKDSSSGTERTGGRLGKVKGPTFRISGVQVNAKLVISHEEELIPLHKSIPSDPEERKQYTIPCHTKAAHFDIDWGKEDDSN LLIGIYEYGYGSWEMIKMDPDLSLTHKILPDDPDKKPQAKQLQTRADYLIKLLSRDLAKKEALSGAGSSKRRKARAKKNKAMKSIKVKEE IKSDSSPLPSEKSDEDDDKLSESKSDGRERSKKSSVSDAPVHITASGEPVPISEESEELDQKTFSIEFWVMNTSIQSTIILLIEQIVVAL GGEFKLYLPQLIPHMLRVFMHDNSPGRIVSIKLLAAIQLFGANLDDYLHLLLPPIVKLFDAPEAPLPSRKAALETVDRLTESLDFTDYAS RIIHPIVRTLDQSPELRSTAMDTLSSLVFQLGKKYQIFIPMVNKVLVRHRINHQRYDVLICRIVKGYTLADEEEDPLIYQHRMLRSGQGD ALASGPVETGPMKKLHVSTINLQKAWGAARRVSKDDWLEWLRRLSLELLKDSSSPSLRSCWALAQAYNPMARDLFNAAFVSCWSELNEDQ QDELIRSIELALTSQDIAEVTQTLLNLAEFMEHSDKGPLPLRDDNGIVLLGERAAKCRAYAKALHYKELEFQKGPTPAILESLISINNKL QQPEAAAGVLEYAMKHFGELEIQATWYEKLHEWEDALVAYDKKMDTNKDDPELMLGRMRCLEALGEWGQLHQQCCEKWTLVNDETQAKMA RMAAAAAWGLGQWDSMEEYTCMIPRDTHDGAFYRAVLALHQDLFSLAQQCIDKARDLLDAELTAMAGESYSRAYGAMVSCHMLSELEEVI QYKLVPERREIIRQIWWERLQGCQRIVEDWQKILMVRSLVVSPHEDMRTWLKYASLCGKSGRLALAHKTLVLLLGVDPSRQLDHPLPTVH PQVTYAYMKNMWKSARKIDAFQHMQHFVQTMQQQAQHAIATEDQQHKQELHKLMARCFLKLGEWQLNLQGINESTIPKVLQYYSAATEHD RSWYKAWHAWAVMNFEAVLHYKHQNQARDEKKKLRHASGANITNATTAATTAATATTTASTEGSNSESEAESTENSPTPSPLQKKVTEDL SKTLLMYTVPAVQGFFRSISLSRGNNLQDTLRVLTLWFDYGHWPDVNEALVEGVKAIQIDTWLQVIPQLIARIDTPRPLVGRLIHQLLTD IGRYHPQALIYPLTVASKSTTTARHNAANKILKNMCEHSNTLVQQAMMVSEELIRVAILWHEMWHEGLEEASRLYFGERNVKGMFEVLEP LHAMMERGPQTLKETSFNQAYGRDLMEAQEWCRKYMKSGNVKDLTQAWDLYYHVFRRISKQLPQLTSLELQYVSPKLLMCRDLELAVPGT YDPNQPIIRIQSIAPSLQVITSKQRPRKLTLMGSNGHEFVFLLKGHEDLRQDERVMQLFGLVNTLLANDPTSLRKNLSIQRYAVIPLSTN SGLIGWVPHCDTLHALIRDYREKKKILLNIEHRIMLRMAPDYDHLTLMQKVEVFEHAVNNTAGDDLAKLLWLKSPSSEVWFDRRTNYTRS LAVMSMVGYILGLGDRHPSNLMLDRLSGKILHIDFGDCFEVAMTREKFPEKIPFRLTRMLTNAMEVTGLDGNYRITCHTVMEVLREHKDS VMAVLEAFVYDPLLNWRLMDTNTKGNKRSRTRTDSYSAGQSVEILDGVELGEPAHKKTGTTVPESIHSFIGDGLVKPEALNKKAIQIINR -------------------------------------------------------------- |
Multiple Sequence Alignment of All Fusion Protein Isoforms |
Top |
Kinase Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr5:98199112/chr1:11273623) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CHD1 | MTOR |
| FUNCTION: ATP-dependent chromatin remodeler that mediates chromatin-remodeling following DNA damage (PubMed:19661379, PubMed:29220652, PubMed:29220653, PubMed:33357431, PubMed:34486521, PubMed:34874266, PubMed:34210977). Recruited to DNA damage sites through interaction with poly-ADP-ribose: specifically recognizes and binds histones that are poly-ADP-ribosylated on serine residues in response to DNA damage (PubMed:19661379, PubMed:29220652, PubMed:29220653, PubMed:34874266, PubMed:34486521). Poly-ADP-ribose-binding activates the ATP-dependent chromatin remodeler activity, thereby regulating chromatin during DNA repair (PubMed:19661379, PubMed:29220652, PubMed:29220653, PubMed:34874266, PubMed:34486521). Catalyzes nucleosome sliding away from DNA breaks in an ATP-dependent manner (PubMed:19661379, PubMed:29220652, PubMed:29220653). Chromatin remodeling activity promotes PARP2 removal from chromatin (PubMed:33275888). {ECO:0000269|PubMed:19661379, ECO:0000269|PubMed:29220652, ECO:0000269|PubMed:29220653, ECO:0000269|PubMed:33275888, ECO:0000269|PubMed:33357431, ECO:0000269|PubMed:34210977, ECO:0000269|PubMed:34486521, ECO:0000269|PubMed:34874266}. | FUNCTION: Serine/threonine protein kinase which is a central regulator of cellular metabolism, growth and survival in response to hormones, growth factors, nutrients, energy and stress signals (PubMed:12087098, PubMed:12150925, PubMed:12150926, PubMed:12231510, PubMed:12718876, PubMed:14651849, PubMed:15268862, PubMed:15467718, PubMed:15545625, PubMed:15718470, PubMed:18497260, PubMed:18762023, PubMed:18925875, PubMed:20516213, PubMed:20537536, PubMed:21659604, PubMed:23429703, PubMed:23429704, PubMed:25799227, PubMed:26018084, PubMed:29150432, PubMed:31112131, PubMed:31601708, PubMed:32561715, PubMed:34519269, PubMed:29236692, PubMed:37751742). MTOR directly or indirectly regulates the phosphorylation of at least 800 proteins (PubMed:15268862, PubMed:15467718, PubMed:17517883, PubMed:18925875, PubMed:18372248, PubMed:18497260, PubMed:20516213, PubMed:21576368, PubMed:21659604, PubMed:23429704, PubMed:29236692, PubMed:37751742). Functions as part of 2 structurally and functionally distinct signaling complexes mTORC1 and mTORC2 (mTOR complex 1 and 2) (PubMed:15268862, PubMed:15467718, PubMed:18925875, PubMed:18497260, PubMed:20516213, PubMed:21576368, PubMed:21659604, PubMed:23429704). In response to nutrients, growth factors or amino acids, mTORC1 is recruited to the lysosome membrane and promotes protein, lipid and nucleotide synthesis by phosphorylating key regulators of mRNA translation and ribosome synthesis (PubMed:12087098, PubMed:12150925, PubMed:12150926, PubMed:12231510, PubMed:12718876, PubMed:14651849, PubMed:15268862, PubMed:15467718, PubMed:15545625, PubMed:15718470, PubMed:18497260, PubMed:18762023, PubMed:18925875, PubMed:20516213, PubMed:20537536, PubMed:21659604, PubMed:23429703, PubMed:23429704, PubMed:25799227, PubMed:26018084, PubMed:29150432, PubMed:31112131, PubMed:34519269, PubMed:29236692). This includes phosphorylation of EIF4EBP1 and release of its inhibition toward the elongation initiation factor 4E (eiF4E) (PubMed:24403073, PubMed:29236692). Moreover, phosphorylates and activates RPS6KB1 and RPS6KB2 that promote protein synthesis by modulating the activity of their downstream targets including ribosomal protein S6, eukaryotic translation initiation factor EIF4B, and the inhibitor of translation initiation PDCD4 (PubMed:12150925, PubMed:12087098, PubMed:18925875, PubMed:29150432, PubMed:29236692). Stimulates the pyrimidine biosynthesis pathway, both by acute regulation through RPS6KB1-mediated phosphorylation of the biosynthetic enzyme CAD, and delayed regulation, through transcriptional enhancement of the pentose phosphate pathway which produces 5-phosphoribosyl-1-pyrophosphate (PRPP), an allosteric activator of CAD at a later step in synthesis, this function is dependent on the mTORC1 complex (PubMed:23429704, PubMed:23429703). Regulates ribosome synthesis by activating RNA polymerase III-dependent transcription through phosphorylation and inhibition of MAF1 an RNA polymerase III-repressor (PubMed:20516213). Activates dormant ribosomes by mediating phosphorylation of SERBP1, leading to SERBP1 inactivation and reactivation of translation (PubMed:36691768). In parallel to protein synthesis, also regulates lipid synthesis through SREBF1/SREBP1 and LPIN1 (By similarity). To maintain energy homeostasis mTORC1 may also regulate mitochondrial biogenesis through regulation of PPARGC1A (By similarity). In the same time, mTORC1 inhibits catabolic pathways: negatively regulates autophagy through phosphorylation of ULK1 (PubMed:32561715). Under nutrient sufficiency, phosphorylates ULK1 at 'Ser-758', disrupting the interaction with AMPK and preventing activation of ULK1 (PubMed:32561715). Also prevents autophagy through phosphorylation of the autophagy inhibitor DAP (PubMed:20537536). Also prevents autophagy by phosphorylating RUBCNL/Pacer under nutrient-rich conditions (PubMed:30704899). Prevents autophagy by mediating phosphorylation of AMBRA1, thereby inhibiting AMBRA1 ability to mediate ubiquitination of ULK1 and interaction between AMBRA1 and PPP2CA (PubMed:23524951, PubMed:25438055). mTORC1 exerts a feedback control on upstream growth factor signaling that includes phosphorylation and activation of GRB10 a INSR-dependent signaling suppressor (PubMed:21659604). Among other potential targets mTORC1 may phosphorylate CLIP1 and regulate microtubules (PubMed:12231510). The mTORC1 complex is inhibited in response to starvation and amino acid depletion (PubMed:12150925, PubMed:12150926, PubMed:24403073). The non-canonical mTORC1 complex, which acts independently of RHEB, specifically mediates phosphorylation of MiT/TFE factors MITF, TFEB and TFE3 in the presence of nutrients, promoting their cytosolic retention and inactivation (PubMed:22576015, PubMed:22343943, PubMed:22692423, PubMed:24448649, PubMed:32612235, PubMed:36608670, PubMed:36697823). Upon starvation or lysosomal stress, inhibition of mTORC1 induces dephosphorylation and nuclear translocation of TFEB and TFE3, promoting their transcription factor activity (PubMed:22576015, PubMed:22343943, PubMed:22692423, PubMed:24448649, PubMed:32612235, PubMed:36608670). The mTORC1 complex regulates pyroptosis in macrophages by promoting GSDMD oligomerization (PubMed:34289345). MTOR phosphorylates RPTOR which in turn inhibits mTORC1 (By similarity). As part of the mTORC2 complex MTOR may regulate other cellular processes including survival and organization of the cytoskeleton (PubMed:15268862, PubMed:15467718). mTORC2 plays a critical role in the phosphorylation at 'Ser-473' of AKT1, a pro-survival effector of phosphoinositide 3-kinase, facilitating its activation by PDK1 (PubMed:15718470). mTORC2 may regulate the actin cytoskeleton, through phosphorylation of PRKCA, PXN and activation of the Rho-type guanine nucleotide exchange factors RHOA and RAC1A or RAC1B (PubMed:15268862). mTORC2 also regulates the phosphorylation of SGK1 at 'Ser-422' (PubMed:18925875). Regulates osteoclastogenesis by adjusting the expression of CEBPB isoforms (By similarity). Plays an important regulatory role in the circadian clock function; regulates period length and rhythm amplitude of the suprachiasmatic nucleus (SCN) and liver clocks (By similarity). Phosphorylates SQSTM1, promoting interaction between SQSTM1 and KEAP1 and subsequent inactivation of the BCR(KEAP1) complex (By similarity). {ECO:0000250|UniProtKB:Q9JLN9, ECO:0000269|PubMed:12087098, ECO:0000269|PubMed:12150925, ECO:0000269|PubMed:12150926, ECO:0000269|PubMed:12231510, ECO:0000269|PubMed:12718876, ECO:0000269|PubMed:14651849, ECO:0000269|PubMed:15268862, ECO:0000269|PubMed:15467718, ECO:0000269|PubMed:15545625, ECO:0000269|PubMed:15718470, ECO:0000269|PubMed:17517883, ECO:0000269|PubMed:18372248, ECO:0000269|PubMed:18497260, ECO:0000269|PubMed:18762023, ECO:0000269|PubMed:18925875, ECO:0000269|PubMed:20516213, ECO:0000269|PubMed:20537536, ECO:0000269|PubMed:21576368, ECO:0000269|PubMed:21659604, ECO:0000269|PubMed:22343943, ECO:0000269|PubMed:22576015, ECO:0000269|PubMed:22692423, ECO:0000269|PubMed:23429703, ECO:0000269|PubMed:23429704, ECO:0000269|PubMed:23524951, ECO:0000269|PubMed:24403073, ECO:0000269|PubMed:24448649, ECO:0000269|PubMed:25438055, ECO:0000269|PubMed:25799227, ECO:0000269|PubMed:26018084, ECO:0000269|PubMed:29150432, ECO:0000269|PubMed:29236692, ECO:0000269|PubMed:30704899, ECO:0000269|PubMed:31112131, ECO:0000269|PubMed:31601708, ECO:0000269|PubMed:32561715, ECO:0000269|PubMed:32612235, ECO:0000269|PubMed:34289345, ECO:0000269|PubMed:34519269, ECO:0000269|PubMed:36608670, ECO:0000269|PubMed:36691768, ECO:0000269|PubMed:36697823, ECO:0000269|PubMed:37751742}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
 - Retained domain in the 5'-partner of fusion protein (protein functional feature from UniProt). - Retained domain in the 5'-partner of fusion protein (protein functional feature from UniProt). |
| Partner | Hgeneene | Hbp | Tgeneene | Tbp | ENST | BPexon | TotalExon | Protein feature loci | BPloci | TotalLen | Feature | Note |
 - Retained domain in the 3'-partner of fusion protein (protein functional feature from UniProt). - Retained domain in the 3'-partner of fusion protein (protein functional feature from UniProt). |
| Partner | Hgeneene | Hbp | Tgeneene | Tbp | ENST | BPexon | TotalExon | Protein feature loci | BPloci | TotalLen | Feature | Note |
| Tgene | CHD1 | 98204199 | MTOR | 11273623 | ENST00000284049 | 19 | 58 | 1382_1982 | 1039 | 2550 | Domain | Note=FAT;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00534 |
| Tgene | CHD1 | 98204199 | MTOR | 11273623 | ENST00000284049 | 19 | 58 | 2517_2549 | 1039 | 2550 | Domain | Note=FATC;Ontology_term=ECO:0000255,ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00534,ECO:0000255|PROSITE-ProRule:PRU00535 |
| Tgene | CHD1 | 98204199 | MTOR | 11273623 | ENST00000284049 | 19 | 58 | 2156_2469 | 1039 | 2550 | Domain | Note=PI3K/PI4K catalytic;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00269 |
Top |
Kinase Fusion Protein Structures |
 CIF files of the predicted kinase fusion proteins CIF files of the predicted kinase fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Kinase Fusion protein CIF link (fusion AA seq ID in KinaseFusionDB) | Henst | Tenst | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp | AA seq | Len(AA seq) |
| PDB file >>>101_CHD1_MTOR | ENST00000284049 | ENST00000361445 | CHD1 | chr5 | 98204199 | MTOR | chr1 | 11273623 | MNGHSDEESVRNSSGESSQSDDDSGSASGSGSGSSSGSSSDGSSSQSGSSDSDSGSESGSQSESESDTSRENKVQAKPPKVDGAEFWKSS PSILAVQRSAILKKQQQQQQQQQHQASSNSGSEEDSSSSEDSDDSSSEVKRKKHKDEDWQMSGSGSPSQSGSDSESEEEREKSSCDETES DYEPKNKVKSRKPQNRSKSKNGKKILGQKKRQIDSSEEDDDEEDYDNDKRSSRRQATVNVSYKEDEEMKTDSDDLLEVCGEDVPQPEEEE FETIERFMDCRIGRKGATGATTTIYAVEADGDPNAGFEKNKEPGEIQYLIKWKGWSHIHNTWETEETLKQQNVRGMKKLDNYKKKDQETK RWLKNASPEDVEYYNCQQELTDDLHKQYQIVERIIAHSNQKSAAGYPDYYCKWQGLPYSECSWEDGALISKKFQACIDEYFSRNQSKTTP FKDCKVLKQRPRFVALKKQPSYIGGHEGLELRDYQLNGLNWLAHSWCKGNSCILADEMGLGKTIQTISFLNYLFHEHQLYGPFLLVVPLS TLTSWQREIQTWASQMNAVVYLGDINSRNMIRTHEWTHHQTKRLKFNILLTTYEILLKDKAFLGGLNWAFIGVDEAHRLKNDDSLLYKTL IDFKSNHRLLITGTPLQNSLKELWSLLHFIMPEKFSSWEDFEEEHGKGREYGYASLHKELEPFLLRRVKKDVEKSLPAKVEQILRMEMSA LQKQYYKWILTRNYKALSKGSKGSTSGFLNIMMELKKCCNHCYLIKPPDNNEFYNKQEALQHLIRSSGKLILLDKLLIRLRERGNRVLIF SQMVRMLDILAEYLKYRQFPFQRLDGSIKGELRKQALDHFNAEGSEDFCFLLSTRAGGLGINLASADTVVIFDSDWNPQNDLQAQARAHR IGQKKQVNIYRLVTKGSVEEDILERAKKKMVLDHLVIQRMDTTGKTVLHTGSAPSSSTPFNKEELSAILKFGAEELFKEPEGEEQEPQEM DIDEILKRAETHENEPGPLTVGDELLSQFKVANFSNMDEDDIELEPERNSKNWEEIIPEDQRRRLEEEERQKELEEIYMLPRMRNCAKQI SFNGSEGRRSRSRRYSGSDSDSISEGKRPKKRGRPRTIPRENIKGFSDAEIRRFIKSYKKFGGPLERLDAIARDAELVDKSETDLRRLGE LVHNGCIKALKDSSSGTERTGGRLGKVKGPTFRISGVQVNAKLVISHEEELIPLHKSIPSDPEERKQYTIPCHTKAAHFDIDWGKEDDSN LLIGIYEYGYGSWEMIKMDPDLSLTHKILPDDPDKKPQAKQLQTRADYLIKLLSRDLAKKEALSGAGSSKRRKARAKKNKAMKSIKVKEE IKSDSSPLPSEKSDEDDDKLSESKSDGRERSKKSSVSDAPVHITASGEPVPISEESEELDQKTFSIEFWVMNTSIQSTIILLIEQIVVAL GGEFKLYLPQLIPHMLRVFMHDNSPGRIVSIKLLAAIQLFGANLDDYLHLLLPPIVKLFDAPEAPLPSRKAALETVDRLTESLDFTDYAS RIIHPIVRTLDQSPELRSTAMDTLSSLVFQLGKKYQIFIPMVNKVLVRHRINHQRYDVLICRIVKGYTLADEEEDPLIYQHRMLRSGQGD ALASGPVETGPMKKLHVSTINLQKAWGAARRVSKDDWLEWLRRLSLELLKDSSSPSLRSCWALAQAYNPMARDLFNAAFVSCWSELNEDQ QDELIRSIELALTSQDIAEVTQTLLNLAEFMEHSDKGPLPLRDDNGIVLLGERAAKCRAYAKALHYKELEFQKGPTPAILESLISINNKL QQPEAAAGVLEYAMKHFGELEIQATWYEKLHEWEDALVAYDKKMDTNKDDPELMLGRMRCLEALGEWGQLHQQCCEKWTLVNDETQAKMA RMAAAAAWGLGQWDSMEEYTCMIPRDTHDGAFYRAVLALHQDLFSLAQQCIDKARDLLDAELTAMAGESYSRAYGAMVSCHMLSELEEVI QYKLVPERREIIRQIWWERLQGCQRIVEDWQKILMVRSLVVSPHEDMRTWLKYASLCGKSGRLALAHKTLVLLLGVDPSRQLDHPLPTVH PQVTYAYMKNMWKSARKIDAFQHMQHFVQTMQQQAQHAIATEDQQHKQELHKLMARCFLKLGEWQLNLQGINESTIPKVLQYYSAATEHD RSWYKAWHAWAVMNFEAVLHYKHQNQARDEKKKLRHASGANITNATTAATTAATATTTASTEGSNSESEAESTENSPTPSPLQKKVTEDL SKTLLMYTVPAVQGFFRSISLSRGNNLQDTLRVLTLWFDYGHWPDVNEALVEGVKAIQIDTWLQVIPQLIARIDTPRPLVGRLIHQLLTD IGRYHPQALIYPLTVASKSTTTARHNAANKILKNMCEHSNTLVQQAMMVSEELIRVAILWHEMWHEGLEEASRLYFGERNVKGMFEVLEP LHAMMERGPQTLKETSFNQAYGRDLMEAQEWCRKYMKSGNVKDLTQAWDLYYHVFRRISKQLPQLTSLELQYVSPKLLMCRDLELAVPGT YDPNQPIIRIQSIAPSLQVITSKQRPRKLTLMGSNGHEFVFLLKGHEDLRQDERVMQLFGLVNTLLANDPTSLRKNLSIQRYAVIPLSTN SGLIGWVPHCDTLHALIRDYREKKKILLNIEHRIMLRMAPDYDHLTLMQKVEVFEHAVNNTAGDDLAKLLWLKSPSSEVWFDRRTNYTRS LAVMSMVGYILGLGDRHPSNLMLDRLSGKILHIDFGDCFEVAMTREKFPEKIPFRLTRMLTNAMEVTGLDGNYRITCHTVMEVLREHKDS VMAVLEAFVYDPLLNWRLMDTNTKGNKRSRTRTDSYSAGQSVEILDGVELGEPAHKKTGTTVPESIHSFIGDGLVKPEALNKKAIQIINR | 2926 |
| 3D view using mol* of 101_CHD1_MTOR | ||||||||||
| PDB file >>>TKFP_173_CHD1_MTOR | ENST00000284049 | ENST00000361445 | CHD1 | chr5 | 98204199 | MTOR | chr1 | 11273623 | MNGHSDEESVRNSSGESSQSDDDSGSASGSGSGSSSGSSSDGSSSQSGSSDSDSGSESGSQSESESDTSRENKVQAKPPKVDGAEFWKSS PSILAVQRSAILKKQQQQQQQQQHQASSNSGSEEDSSSSEDSDDSSSEVKRKKHKDEDWQMSGSGSPSQSGSDSESEEEREKSSCDETES DYEPKNKVKSRKPQNRSKSKNGKKILGQKKRQIDSSEEDDDEEDYDNDKRSSRRQATVNVSYKEDEEMKTDSDDLLEVCGEDVPQPEEEE FETIERFMDCRIGRKGATGATTTIYAVEADGDPNAGFEKNKEPGEIQYLIKWKGWSHIHNTWETEETLKQQNVRGMKKLDNYKKKDQETK RWLKNASPEDVEYYNCQQELTDDLHKQYQIVERIIAHSNQKSAAGYPDYYCKWQGLPYSECSWEDGALISKKFQACIDEYFSRNQSKTTP FKDCKVLKQRPRFVALKKQPSYIGGHEGLELRDYQLNGLNWLAHSWCKGNSCILADEMGLGKTIQTISFLNYLFHEHQLYGPFLLVVPLS TLTSWQREIQTWASQMNAVVYLGDINSRNMIRTHEWTHHQTKRLKFNILLTTYEILLKDKAFLGGLNWAFIGVDEAHRLKNDDSLLYKTL IDFKSNHRLLITGTPLQNSLKELWSLLHFIMPEKFSSWEDFEEEHGKGREYGYASLHKELEPFLLRRVKKDVEKSLPAKVEQILRMEMSA LQKQYYKWILTRNYKALSKGSKGSTSGFLNIMMELKKCCNHCYLIKPPDNNEFYNKQEALQHLIRSSGKLILLDKLLIRLRERGNRVLIF SQMVRMLDILAEYLKYRQFPFQRLDGSIKGELRKQALDHFNAEGSEDFCFLLSTRAGGLGINLASADTVVIFDSDWNPQNDLQAQARAHR IGQKKQVNIYRLVTKGSVEEDILERAKKKMVLDHLVIQRMDTTGKTVLHTGSAPSSSTPFNKEELSAILKFGAEELFKEPEGEEQEPQEM DIDEILKRAETHENEPGPLTVGDELLSQFKVANFSNMDEDDIELEPERNSKNWEEIIPEDQRRRLEEEERQKELEEIYMLPRMRNCAKQI SFNGSEGRRSRSRRYSGSDSDSISEGKRPKKRGRPRTIPRENIKGFSDAEIRRFIKSYKKFGGPLERLDAIARDAELVDKSETDLRRLGE LVHNGCIKALKDSSSGTERTGGRLGKVKGPTFRISGVQVNAKLVISHEEELIPLHKSIPSDPEERKQYTIPCHTKAAHFDIDWGKEDDSN LLIGIYEYGYGSWEMIKMDPDLSLTHKILPDDPDKKPQAKQLQTRADYLIKLLSRDLAKKEALSGAGSSKRRKARAKKNKAMKSIKVKEE IKSDSSPLPSEKSDEDDDKLSESKSDGRERSKKSSVSDAPVHITASGEPVPISEESEELDQKTFSIEFWVMNTSIQSTIILLIEQIVVAL GGEFKLYLPQLIPHMLRVFMHDNSPGRIVSIKLLAAIQLFGANLDDYLHLLLPPIVKLFDAPEAPLPSRKAALETVDRLTESLDFTDYAS RIIHPIVRTLDQSPELRSTAMDTLSSLVFQLGKKYQIFIPMVNKVLVRHRINHQRYDVLICRIVKGYTLADEEEDPLIYQHRMLRSGQGD ALASGPVETGPMKKLHVSTINLQKAWGAARRVSKDDWLEWLRRLSLELLKDSSSPSLRSCWALAQAYNPMARDLFNAAFVSCWSELNEDQ QDELIRSIELALTSQDIAEVTQTLLNLAEFMEHSDKGPLPLRDDNGIVLLGERAAKCRAYAKALHYKELEFQKGPTPAILESLISINNKL QQPEAAAGVLEYAMKHFGELEIQATWYEKLHEWEDALVAYDKKMDTNKDDPELMLGRMRCLEALGEWGQLHQQCCEKWTLVNDETQAKMA RMAAAAAWGLGQWDSMEEYTCMIPRDTHDGAFYRAVLALHQDLFSLAQQCIDKARDLLDAELTAMAGESYSRAYGAMVSCHMLSELEEVI QYKLVPERREIIRQIWWERLQGCQRIVEDWQKILMVRSLVVSPHEDMRTWLKYASLCGKSGRLALAHKTLVLLLGVDPSRQLDHPLPTVH PQVTYAYMKNMWKSARKIDAFQHMQHFVQTMQQQAQHAIATEDQQHKQELHKLMARCFLKLGEWQLNLQGINESTIPKVLQYYSAATEHD RSWYKAWHAWAVMNFEAVLHYKHQNQARDEKKKLRHASGANITNATTAATTAATATTTASTEGSNSESEAESTENSPTPSPLQKKVTEDL SKTLLMYTVPAVQGFFRSISLSRGNNLQDTLRVLTLWFDYGHWPDVNEALVEGVKAIQIDTWLQVIPQLIARIDTPRPLVGRLIHQLLTD IGRYHPQALIYPLTVASKSTTTARHNAANKILKNMCEHSNTLVQQAMMVSEELIRVAILWHEMWHEGLEEASRLYFGERNVKGMFEVLEP LHAMMERGPQTLKETSFNQAYGRDLMEAQEWCRKYMKSGNVKDLTQAWDLYYHVFRRISKQLPQLTSLELQYVSPKLLMCRDLELAVPGT YDPNQPIIRIQSIAPSLQVITSKQRPRKLTLMGSNGHEFVFLLKGHEDLRQDERVMQLFGLVNTLLANDPTSLRKNLSIQRYAVIPLSTN SGLIGWVPHCDTLHALIRDYREKKKILLNIEHRIMLRMAPDYDHLTLMQKVEVFEHAVNNTAGDDLAKLLWLKSPSSEVWFDRRTNYTRS LAVMSMVGYILGLGDRHPSNLMLDRLSGKILHIDFGDCFEVAMTREKFPEKIPFRLTRMLTNAMEVTGLDGNYRITCHTVMEVLREHKDS VMAVLEAFVYDPLLNWRLMDTNTKGNKRSRTRTDSYSAGQSVEILDGVELGEPAHKKTGTTVPESIHSFIGDGLVKPEALNKKAIQIINR | 2926_CHD1_MTOR |
Top |
Comparison of Fusion Protein Isoforms |
 Superimpose the 3D Structures Among All Fusion Protein Isoforms Superimpose the 3D Structures Among All Fusion Protein Isoforms * Download the pdb file and open it from the molstar online viewer. |
 Comparison of the Secondary Structures of Fusion Protein Isoforms Comparison of the Secondary Structures of Fusion Protein Isoforms |
Top |
Comparison of Fusion Protein Sequences/Structures with Known Sequences/Structures from PDB |
Top |
pLDDT score distribution |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. * The blue color at the bottom marks the best active site residues. |
| 101_CHD1_MTOR.png |
 |
| 101_CHD1_MTOR.png |
 |
Top |
Potential Active Site Information |
 The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. |
| Kinase Fusion AA seq ID in KinaseFusionDB | Site score | Size | Dscore | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
Top |
Ramachandran Plot of Kinase Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| 101_CHD1_MTOR_ramachandran.png |
 |
Top |
Virtual Screening Results |
 Distribution of the average docking score across all approved kinase inhibitors. Distribution of the average docking score across all approved kinase inhibitors.Distribution of the number of occurrence across all approved kinase inhibitors. |
| 5'-kinase fusion protein case |
| 3'-kinase fusion protein case |
 |
Top |
 Drug information from DrugBank of the top 20 interacting small molecules. Drug information from DrugBank of the top 20 interacting small molecules.* The detailed information of individual kinase inhibitors are available in the download page. |
| Fusion gene name info | Drug | Docking score | Glide g score | Glide energy |
| 101_CHD1_MTOR-DOCK_HTVS_1-001 | Crizotinib | -6.38428 | -6.88018 | -42.9757 |
| 101_CHD1_MTOR-DOCK_HTVS_1-001 | Crizotinib | -6.38428 | -6.88018 | -42.9757 |
| 101_CHD1_MTOR-DOCK_HTVS_1-001 | Vandetanib | -6.37434 | -6.37434 | -39.9729 |
| 101_CHD1_MTOR-DOCK_HTVS_1-001 | Vandetanib | -6.3529599999999995 | -6.3529599999999995 | -36.7263 |
| 101_CHD1_MTOR-DOCK_HTVS_1-001 | Pralsetinib | -6.19346 | -6.28496 | -42.1743 |
| 101_CHD1_MTOR-DOCK_HTVS_1-001 | Nilotinib | -6.13823 | -6.277830000000001 | -51.2568 |
| 101_CHD1_MTOR-DOCK_HTVS_1-001 | Nilotinib | -6.13823 | -6.277830000000001 | -51.2568 |
| 101_CHD1_MTOR-DOCK_HTVS_1-001 | Lapatinib | -5.93938 | -6.02818 | -54.2932 |
| 101_CHD1_MTOR-DOCK_HTVS_1-001 | Lapatinib | -5.73787 | -5.82667 | -49.4771 |
| 101_CHD1_MTOR-DOCK_HTVS_1-001 | Pralsetinib | -5.67788 | -5.76938 | -33.5879 |
| 101_CHD1_MTOR-DOCK_HTVS_1-001 | Belumosudil | -5.51393 | -5.52163 | -27.1343 |
| 101_CHD1_MTOR-DOCK_HTVS_1-001 | Pralsetinib | -5.45437 | -5.54587 | -39.1125 |
| 101_CHD1_MTOR-DOCK_HTVS_1-001 | Zanubrutinib | -5.26627 | -5.26627 | -35.7842 |
| 101_CHD1_MTOR-DOCK_HTVS_1-001 | Upadacitinib | -5.25355 | -5.25455 | -28.1047 |
| 101_CHD1_MTOR-DOCK_HTVS_1-001 | Neratinib | -5.2247900000000005 | -5.40709 | -43.6152 |
| 101_CHD1_MTOR-DOCK_HTVS_1-001 | Neratinib | -5.2247900000000005 | -5.40709 | -43.6152 |
| 101_CHD1_MTOR-DOCK_HTVS_1-001 | Neratinib | -5.22371 | -5.40961 | -42.145 |
| 101_CHD1_MTOR-DOCK_HTVS_1-001 | Pralsetinib | -4.87351 | -4.9650099999999995 | -40.4922 |
| 101_CHD1_MTOR-DOCK_HTVS_1-001 | Pexidartinib | -4.791919999999999 | -5.008719999999999 | -31.5873 |
| 101_CHD1_MTOR-DOCK_HTVS_1-001 | Pexidartinib | -4.791919999999999 | -5.008719999999999 | -31.5873 |
Top |
Kinase-Substrate Information of CHD1_MTOR |
 Phosphorylation target of the kinase Phosphorylation target of the kinase(phosphosite, 03-17-2024) |
| Kinase | Kinase UniProt Acc | Kinase species | Substrate | Substrate UniProt Acc | Substrate phosphorylated residues | Substrate phosphorylated sites (+/-7AA) | Domain |
| MTOR | P42345 | human | ANKRD17 | O75179 | S2047 | VssPssPsPPAQPGG | |
| MTOR | P42345 | human | IRS1 | P35568 | S636 | sGDyMPMsPKsVSAP | |
| MTOR | P42345 | human | RRAGC | Q9HB90 | S21 | GsYGAADsFPKDFGY | |
| MTOR | P42345 | human | MKNK2 | Q9HBH9 | S74 | KRGRAtDsFsGRFED | |
| MTOR | P42345 | human | ULK1 | O75385 | S758 | PVVFtVGsPPsGStP | |
| MTOR | P42345 | human | BRD9 | Q9H8M2 | S588 | DPYEFLQsPEPAAsA | |
| MTOR | P42345 | human | WASHC2A | Q641Q2 | S700 | DDVDSGGsLFGsPPT | |
| MTOR | P42345 | human | ESR1 | P03372 | S104 | FPPLNsVsPsPLMLL | Oest_recep |
| MTOR | P42345 | human | UVRAG | Q9P2Y5 | S549 | RKItsLSssLDTsLD | |
| MTOR | P42345 | human | TFEB | P19484 | S122 | PkPPPAAsPGVRAGH | MITF_TFEB_C_3_N |
| MTOR | P42345 | human | SRRM1 | Q8IYB3 | T574 | PRRRRtPtPPPRRRt | |
| MTOR | P42345 | human | SENP3 | Q9H4L4 | S143 | LLYsKstsLtFHWKL | |
| MTOR | P42345 | human | SENP3 | Q9H4L4 | T145 | YsKstsLtFHWKLWG | |
| MTOR | P42345 | human | MTOR | P42345 | S2448 | RsRtRtDsysAGQsV | |
| MTOR | P42345 | human | DAP | P51397 | S51 | DQEWEsPsPPkPtVF | DAP |
| MTOR | P42345 | human | SENP3 | Q9H4L4 | S25 | PGIPPAyssPRRERL | |
| MTOR | P42345 | human | PKN2 | Q16513 | T958 | TsEAPILtPPREPRI | Pkinase_C |
| MTOR | P42345 | human | RPS6KB1 | P23443 | T412 | NQVFLGFtyVAPsVL | Pkinase_C |
| MTOR | P42345 | human | MCL1 | Q07820 | S64 | IGGSAGAsPPStLtP | |
| MTOR | P42345 | human | AKT1 | P31749 | S473 | RPHFPQFsysAsGtA | Pkinase_C |
| MTOR | P42345 | human | NRBF2 | Q96F24 | S120 | sPLsQkYsPSTEKCL | NRBF2 |
| MTOR | P42345 | human | SRRM1 | Q8IYB3 | T572 | PPPRRRRtPtPPPRR | |
| MTOR | P42345 | human | ELP1 | O95163 | S1174 | SETSsVVsGSEMSGK | |
| MTOR | P42345 | human | NAA10 | P41227 | S228 | StDVKDssEAsDSAS | |
| MTOR | P42345 | human | TFEB | P19484 | S211 | LVGVTSSsCPADLTQ | |
| MTOR | P42345 | human | PRKN | O60260 | S127 | AVILHTDsRkDsPPA | |
| MTOR | P42345 | human | AR | P10275 | S96 | QQQGEDGsPQAHRRG | Androgen_recep |
| MTOR | P42345 | human | MTOR | P42345 | S2478 | tGttVPEsIHsFIGD | |
| MTOR | P42345 | human | PASK | Q96RG2 | T640 | MAGLSFGtPtLDEPW | |
| MTOR | P42345 | human | AKT1S1 | Q96B36 | S183 | PTQQYAKsLPVSVPV | PRAS |
| MTOR | P42345 | human | LARP6 | Q9BRS8 | S409 | GRLNCStsPEIFRKC | |
| MTOR | P42345 | human | SRRM2 | Q9UQ35 | S1318 | sPEHKELsNsPLREN | |
| MTOR | P42345 | human | UNK | Q9C0B0 | S611 | GtsAsHGsLGLNGMN | |
| MTOR | P42345 | human | UVRAG | Q9P2Y5 | S498 | GFsGGIPsPDKGHRK | |
| MTOR | P42345 | human | DNMT1 | P26358 | S714 | DNIPEMPsPkKMHQG | |
| MTOR | P42345 | human | ULK1 | O75385 | S638 | FDFPKtPssQNLLAL | |
| MTOR | P42345 | human | MAPKAP1 | Q9BPZ7 | S260 | PIHKFGFsTLALVEK | CRIM |
| MTOR | P42345 | human | UNK | Q9C0B0 | S606 | ENTFLGtsAsHGsLG | |
| MTOR | P42345 | human | HOXB13 | Q92826 | S31 | GRNLVAHsPLTSHPA | |
| MTOR | P42345 | human | AKT1 | P31749 | T450 | tAQMItItPPDQDDs | Pkinase_C |
| MTOR | P42345 | human | RRM1 | P23921 | S631 | IYTRRVLsGEFQIVN | Ribonuc_red_lgC |
| MTOR | P42345 | human | GRB10 | Q13322 | S476 | MNILGsQsPLHPSTL | |
| MTOR | P42345 | human | DUSP10 | Q9Y6W6 | S224 | SCREGKDsFKRIFsK | Rhodanese |
| MTOR | P42345 | human | WASHC2A | Q641Q2 | S704 | SGGsLFGsPPTSVPP | |
| MTOR | P42345 | human | PRKCE | Q02156 | T710 | TREEPVLtLVDEAIV | Pkinase_C |
| MTOR | P42345 | human | STK11IP | Q8N1F8 | S404 | EPRTLNPsPAGWFVQ | |
| MTOR | P42345 | human | LARP1 | Q6PKG0 | S774 | LPttVPEsPNyRNtR | |
| MTOR | P42345 | human | UNK | Q9C0B0 | S336 | QPSsAVssPtQPGPV | |
| MTOR | P42345 | human | UNK | Q9C0B0 | S608 | TFLGtsAsHGsLGLN | |
| MTOR | P42345 | human | MAF1 | Q9H063 | S60 | PHVLEALsPPQtsGL | Maf1 |
| MTOR | P42345 | human | AMOTL2 | Q9Y2J4 | S759 | SsSQRAAsLDsVATS | |
| MTOR | P42345 | human | EIF4EBP1 | Q13541 | S44 | tPGGtLFsttPGGtR | eIF_4EBP |
| MTOR | P42345 | human | PASK | Q96RG2 | S953 | FLAsLPGstHsTAAE | |
| MTOR | P42345 | human | UVRAG | Q9P2Y5 | S582 | GHANVHPsQEQGEAL | |
| MTOR | P42345 | human | SENP3 | Q9H4L4 | S141 | RMLLYsKstsLtFHW | |
| MTOR | P42345 | human | MTOR | P42345 | T2473 | PAHKKtGttVPEsIH | |
| MTOR | P42345 | human | MYCN | P04198 | S62 | LLPtPPLsPsrGFAE | Myc_N |
| MTOR | P42345 | human | RPTOR | Q8N122 | S859 | DtssLtQsAPAsPtN | |
| MTOR | P42345 | human | EIF4EBP1 | Q13541 | S65 | FLMECrNsPVtktPP | eIF_4EBP |
| MTOR | P42345 | human | NDRG1 | Q92597 | T346 | GtRsRsHtsEGtRsR | |
| MTOR | P42345 | human | SOD1 | P00441 | T40 | WGsIkGLtEGLHGFH | Sod_Cu |
| MTOR | P42345 | human | PRKCB | P05771 | T642 | TRQPVELtPtDKLFI | Pkinase_C |
| MTOR | P42345 | human | EEF2K | O00418 | S72 | ksERysssGsPANsF | |
| MTOR | P42345 | human | SRRM2 | Q9UQ35 | S1329 | LRENsFGsPLEFRNs | |
| MTOR | P42345 | human | MTOR | P42345 | S2454 | DsysAGQsVEILDGV | |
| MTOR | P42345 | human | MTOR | P42345 | S2481 | tVPEsIHsFIGDGLV | |
| MTOR | P42345 | human | ELL | P55199 | S309 | LGDPAAssPPGERGR | |
| MTOR | P42345 | human | EEF2K | O00418 | S74 | ERysssGsPANsFHF | |
| MTOR | P42345 | human | ESR1 | P03372 | S118 | LHPPPQLsPFLQPHG | Oest_recep |
| MTOR | P42345 | human | DEPTOR | Q8TB45 | T295 | GyFsssPtLsssPPV | |
| MTOR | P42345 | human | RPS6KB1 | P23443 | S394 | TRQtPVDsPDDStLS | Pkinase_C |
| MTOR | P42345 | human | PASK | Q96RG2 | S956 | sLPGstHsTAAELTG | |
| MTOR | P42345 | human | ESR1 | P03372 | S106 | PLNsVsPsPLMLLHP | Oest_recep |
| MTOR | P42345 | human | SENP3 | Q9H4L4 | S26 | GIPPAyssPRRERLR | |
| MTOR | P42345 | human | DEPTOR | Q8TB45 | S293 | ssGyFsssPtLsssP | |
| MTOR | P42345 | human | SENP3 | Q9H4L4 | S139 | AFRMLLYsKstsLtF | |
| MTOR | P42345 | human | HOXB13 | Q92826 | T41 | TSHPAAPtLMPAVNY | |
| MTOR | P42345 | human | BAG3 | O95817 | T285 | GsPARsstPLHsPsP | |
| MTOR | P42345 | human | RPRD1B | Q9NQG5 | S166 | DDyPGsysPQDPsAG | |
| MTOR | P42345 | human | SLC7A11 | Q9UPY5 | S26 | NVNGRLPsLGNkEPP | |
| MTOR | P42345 | human | PASK | Q96RG2 | T642 | GLSFGtPtLDEPWLG | |
| MTOR | P42345 | human | EIF4EBP1 | Q13541 | T70 | rNsPVtktPPRDLPt | eIF_4EBP |
| MTOR | P42345 | human | PRKCE | Q02156 | S729 | QEEFKGFsYFGEDLM | Pkinase_C |
| MTOR | P42345 | human | SRRM2 | Q9UQ35 | S1326 | NsPLRENsFGsPLEF | |
| MTOR | P42345 | human | UNK | Q9C0B0 | S255 | RKHKYRSsPCPNVKH | |
| MTOR | P42345 | human | PATL1 | Q86TB9 | S179 | ALPRRstsPIIGsPP | |
| MTOR | P42345 | human | RRAGC | Q9HB90 | T394 | kALTHNGtPRNAI__ | |
| MTOR | P42345 | human | GRB10 | Q13322 | S428 | stPVRsVsENsLVAM | BPS |
| MTOR | P42345 | human | UVRAG | Q9P2Y5 | S571 | KGEDLVGsLNGGHAN | |
| MTOR | P42345 | human | SGK1 | O00141 | S422 | AEAFLGFsYAPPTDS | Pkinase_C |
| MTOR | P42345 | human | APBA3 | O96018 | S7 | _MDFPtIsRsPsGPP | |
| MTOR | P42345 | human | YAP1 | P46937 | S436 | INQSTLPsQQNRFPD | |
| MTOR | P42345 | human | ZNRF2 | Q8NHG8 | S145 | GPRLVIGsLPAHLsP | |
| MTOR | P42345 | human | RPS6 | P62753 | S236 | AKRRRLssLRAstsK | |
| MTOR | P42345 | human | RPTOR | Q8N122 | S863 | LtQsAPAsPtNkGVH | |
| MTOR | P42345 | human | OXSR1 | O95747 | S339 | EDGGWEWsDDEFDEE | |
| MTOR | P42345 | human | ISCU | Q9H1K1 | S14 | FRLRRAAsALLLRsP | |
| MTOR | P42345 | human | PATL1 | Q86TB9 | S184 | stsPIIGsPPVRAVP | |
| MTOR | P42345 | human | UVRAG | Q9P2Y5 | S550 | KItsLSssLDTsLDF | |
| MTOR | P42345 | human | BAG3 | O95817 | S289 | RsstPLHsPsPIRVH | |
| MTOR | P42345 | human | DEPTOR | Q8TB45 | S286 | ssMssCGssGyFsss | |
| MTOR | P42345 | human | IRS1 | P35568 | S639 | yMPMsPKsVSAPQQI | |
| MTOR | P42345 | human | EIF4EBP1 | Q13541 | T37 | PPGDysttPGGtLFs | eIF_4EBP |
| MTOR | P42345 | human | HSF1 | Q00613 | S303 | RVkEEPPsPPQsPRV | Vert_HS_TF |
| MTOR | P42345 | human | RPS6KB1 | P23443 | S434 | sFEPKIRsPRRFIGs | |
| MTOR | P42345 | human | MYC | P01106 | S77 | LLPtPPLsPsRRsGL | Myc_N |
| MTOR | P42345 | human | EIF4EBP1 | Q13541 | T41 | ysttPGGtLFsttPG | eIF_4EBP |
| MTOR | P42345 | human | DUSP10 | Q9Y6W6 | S230 | DsFKRIFsKEIIVyD | Rhodanese |
| MTOR | P42345 | human | UVRAG | Q9P2Y5 | S522 | QyktPPPsyNSALAQ | |
| MTOR | P42345 | human | AKT1S1 | Q96B36 | S221 | DLDRIAAsMRALVLR | PRAS |
| MTOR | P42345 | human | UVRAG | Q9P2Y5 | S508 | KGHRKRAssENERLQ | |
| MTOR | P42345 | human | UNK | Q9C0B0 | S359 | DSVPVsPssPHAPDL | |
| MTOR | P42345 | human | DEPTOR | Q8TB45 | S265 | stsFMsVsPsKEIKI | |
| MTOR | P42345 | human | PIP4K2C | Q8TBX8 | S328 | LVGsYGTsPEGIGGY | PIP5K |
| MTOR | P42345 | human | AMBRA1 | Q9C0C7 | S52 | KRVELPDsPRSTFLL | |
| MTOR | P42345 | human | ANKRD17 | O75179 | S2045 | yPVssPssPsPPAQP | |
| MTOR | P42345 | human | HSF1 | Q00613 | S326 | ssVDtLLsPTALIDs | Vert_HS_TF |
| MTOR | P42345 | human | UBR4 | Q5T4S7 | S2932 | GHPAGPGsVsSStGA | |
| MTOR | P42345 | human | RUBCNL | Q9H714 | S157 | sPGILATsPYPETDS | |
| MTOR | P42345 | human | SCYL1 | Q96KG9 | S754 | sTQPRPDsWGEDNWE | |
| MTOR | P42345 | human | RPS6KB1 | P23443-2 | T389 | NQVFLGFtYVAPSVL | Pkinase_C |
| MTOR | P42345 | human | EIF4EBP1 | Q13541 | T46 | GGtLFsttPGGtRII | eIF_4EBP |
| MTOR | P42345 | human | ZNF768 | Q9H5H4 | S139 | sPGyEPRsPGyEsES | RNA_pol_Rpb1_R |
| MTOR | P42345 | human | SENP3 | Q9H4L4 | T142 | MLLYsKstsLtFHWK | |
| MTOR | P42345 | human | LARP6 | Q9BRS8 | S348 | DPESNPTsPMAGRRH | |
| MTOR | P42345 | human | RPS6 | P62753 | S235 | IAKRRRLssLRAsts | |
| MTOR | P42345 | human | NRBF2 | Q96F24 | S113 | AEDAEGQsPLsQkYs | NRBF2 |
| MTOR | P42345 | human | MAF1 | Q9H063 | S75 | sPsRLsksQGGEEEG | Maf1 |
| MTOR | P42345 | human | MAF1 | Q9H063 | S68 | PPQtsGLsPsRLsks | Maf1 |
| MTOR | P42345 | human | APBA3 | O96018 | T5 | ___MDFPtIsRsPsG | |
| MTOR | P42345 | human | UVRAG | Q9P2Y5 | S493 | GGADVGFsGGIPsPD | |
| MTOR | P42345 | human | PASK | Q96RG2 | S949 | RTRLFLAsLPGstHs | |
| MTOR | P42345 | human | PIP4K2C | Q8TBX8 | S324 | GPPALVGsYGTsPEG | PIP5K |
| MTOR | P42345 | human | GRB10 | Q13322 | T155 | PGsPPVLtPGsLPPS | |
| MTOR | P42345 | human | DEPTOR | Q8TB45 | S299 | ssPtLsssPPVLCNP | |
| MTOR | P42345 | human | HOXB13 | Q92826 | T8 | MEPGNYAtLDGAKDI | |
| MTOR | P42345 | human | MTOR | P42345 | T2474 | AHKKtGttVPEsIHs | |
| MTOR | P42345 | human | WNK1 | Q9H4A3 | S2032 | DGsGsPHsPHQLssK | |
| MTOR | P42345 | human | LARP1 | Q6PKG0 | S766 | EPStIARsLPttVPE | |
| MTOR | P42345 | human | PRKCE | Q02156 | T566 | LNGVTTTtFCGTPDy | Pkinase |
 Biological Network Integration of This Kinase and Substrates Biological Network Integration of This Kinase and Substrates (GeneMANIA website) |
 Enriched GO biological processes of the phosphorylation target genes of the kinase Enriched GO biological processes of the phosphorylation target genes of the kinase |
| Kinase | GOID | GO term | P.adjust |
| MTOR | ID | Description | 0.00e+00 |
| MTOR | GO:0010506 | regulation of autophagy | 2.32e-10 |
| MTOR | GO:0031929 | TOR signaling | 2.02e-08 |
| MTOR | GO:0031331 | positive regulation of cellular catabolic process | 8.18e-08 |
| MTOR | GO:0018105 | peptidyl-serine phosphorylation | 3.10e-07 |
| MTOR | GO:0018209 | peptidyl-serine modification | 4.05e-07 |
| MTOR | GO:0038202 | TORC1 signaling | 8.92e-07 |
| MTOR | GO:0010508 | positive regulation of autophagy | 1.02e-06 |
| MTOR | GO:0045931 | positive regulation of mitotic cell cycle | 4.85e-06 |
| MTOR | GO:0031667 | response to nutrient levels | 4.85e-06 |
| MTOR | GO:0010507 | negative regulation of autophagy | 7.39e-06 |
| MTOR | GO:1901654 | response to ketone | 1.12e-05 |
| MTOR | GO:0006359 | regulation of transcription by RNA polymerase III | 1.26e-05 |
| MTOR | GO:0016241 | regulation of macroautophagy | 1.72e-05 |
| MTOR | GO:0062197 | cellular response to chemical stress | 2.97e-05 |
| MTOR | GO:0031669 | cellular response to nutrient levels | 3.52e-05 |
| MTOR | GO:0016236 | macroautophagy | 3.52e-05 |
| MTOR | GO:0016239 | positive regulation of macroautophagy | 3.52e-05 |
| MTOR | GO:0000082 | G1/S transition of mitotic cell cycle | 3.52e-05 |
| MTOR | GO:0045787 | positive regulation of cell cycle | 3.54e-05 |
| MTOR | GO:0046627 | negative regulation of insulin receptor signaling pathway | 3.85e-05 |
| MTOR | GO:0006109 | regulation of carbohydrate metabolic process | 3.85e-05 |
| MTOR | GO:1900077 | negative regulation of cellular response to insulin stimulus | 3.98e-05 |
| MTOR | GO:0071496 | cellular response to external stimulus | 4.05e-05 |
| MTOR | GO:0045945 | positive regulation of transcription by RNA polymerase III | 4.13e-05 |
| MTOR | GO:0043467 | regulation of generation of precursor metabolites and energy | 5.72e-05 |
| MTOR | GO:0044843 | cell cycle G1/S phase transition | 5.72e-05 |
| MTOR | GO:0031668 | cellular response to extracellular stimulus | 5.78e-05 |
| MTOR | GO:0032869 | cellular response to insulin stimulus | 5.78e-05 |
| MTOR | GO:0031330 | negative regulation of cellular catabolic process | 6.00e-05 |
| MTOR | GO:0042594 | response to starvation | 8.27e-05 |
| MTOR | GO:1990928 | response to amino acid starvation | 1.01e-04 |
| MTOR | GO:0036294 | cellular response to decreased oxygen levels | 1.05e-04 |
| MTOR | GO:0071375 | cellular response to peptide hormone stimulus | 1.05e-04 |
| MTOR | GO:0097191 | extrinsic apoptotic signaling pathway | 1.05e-04 |
| MTOR | GO:1901655 | cellular response to ketone | 1.18e-04 |
| MTOR | GO:0006383 | transcription by RNA polymerase III | 1.61e-04 |
| MTOR | GO:0071453 | cellular response to oxygen levels | 1.79e-04 |
| MTOR | GO:0062012 | regulation of small molecule metabolic process | 1.86e-04 |
| MTOR | GO:0019216 | regulation of lipid metabolic process | 2.04e-04 |
| MTOR | GO:0008361 | regulation of cell size | 2.62e-04 |
| MTOR | GO:0046626 | regulation of insulin receptor signaling pathway | 2.62e-04 |
| MTOR | GO:0032868 | response to insulin | 2.81e-04 |
| MTOR | GO:1900076 | regulation of cellular response to insulin stimulus | 2.87e-04 |
| MTOR | GO:0008286 | insulin receptor signaling pathway | 2.90e-04 |
| MTOR | GO:0043200 | response to amino acid | 2.97e-04 |
| MTOR | GO:0046777 | protein autophosphorylation | 3.48e-04 |
| MTOR | GO:1901653 | cellular response to peptide | 3.49e-04 |
| MTOR | GO:0010827 | regulation of glucose transmembrane transport | 4.09e-04 |
| MTOR | GO:0098781 | ncRNA transcription | 4.23e-04 |
Top |
Related Drugs to CHD1_MTOR |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
 Distribution of the number of studies mentioning CHD1-MTOR and kinase inhibitors the PubMed Abstract (04-01-2024) Distribution of the number of studies mentioning CHD1-MTOR and kinase inhibitors the PubMed Abstract (04-01-2024) |
| Fusion gene - drug pair 1 | Fusion gene - drug pair 2 | PMID | Publication date | DOI | Study title |
Top |
Related Diseases to CHD1_MTOR |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Related diseases from the literature mentioned this fusion gene and drug. Related diseases from the literature mentioned this fusion gene and drug. (PubMed, 04-01-2024) |
| MeSH ID | MeSH term |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
Top |
Clinical Trials of the Found Drugs/Small Molecules |
 Statistics of the Clinical Trials of the Found Kinase Inibitors from clinicaltrials.gov (06-17-2024) Statistics of the Clinical Trials of the Found Kinase Inibitors from clinicaltrials.gov (06-17-2024) |
 Clinical Trials from clinicaltrials.gov (06-17-2024) Clinical Trials from clinicaltrials.gov (06-17-2024) |
| Fusion Gene | Kinase Inhibitor | NCT ID | Study Status | Phases | Disease | # Enrolment | Date |

