|
Home |
Download |
Statistics |
Examples |
Help |
Contact |
Terms of Use |
Navigation
1. Overview of KinaseFusionDB.2. Understanding of KinaseFusionDB's Annotation Category.
- Kinase Fusion Gene Search Result Page.
- Kinase Fusion Gene Annotation Result Page.
-- 1) Kinase Fusion Gene Summary.
-- 2) Kinase Fusion Gene Sample Information.
-- 3) Kinase Fusion ORF Analysis.
-- 4) Kinase Fusion Amino Acid Sequences.
-- 5) Multiple Sequence Alignment of All Fusion Protein Isoforms.
-- 6) Kinase Fusion Protein Functional Features.
-- 7) Kinase Fusion Protein Structures.
-- 8) Comparison of Fusion Protein Isoforms.
-- 9) Comparison of Fusion Protein Sequences/Structures with Known Sequences/Structures from PDB.
-- 10) pLDDT Scores and Difference Analysis of pLDDT Scores Between the Active Sites (Best) and Non-Active Sites.
-- 11) Ramachandran Plot of Kinase Fusion Protein Structures.
-- 12) Potential Active Site Information.
-- 13) Virtual Screening Results.
-- 14) Kinase-Substrate Information.
-- 15) Related Drugs with This Kinase Fusion Protein.
-- 16) Related Disease with This Kinase Fusion Protein.
3. Download Data and Contact Us.
1. Overview of KinaseFusionDB.
Kinase fusion genes were the most targeted fusion gene group. Kinase inhibitors block the uncontrolled signal transduction in diverse mechanisms relating to the amplified kinase function context in cancer (i.e., amplified copy number variation of kinase gene, loss of kinase protein degradation, fusion between strong promoter gene and kinase gene). To develop effective personalized therapeutics and next-generation kinase inhibitors, we must determine the detailed mechanisms of action and contexts of individual kinase inhibitors in kinase fusion oncoproteins. Currently, we have ~7000 kinase fusion genes from FusionGDB2 (1), CCLE (2), and COSMIC (3). ORF annotation of the breakpoints and gene isoforms found ~1400 kinase fusion proteins. Kinase domain retention search, fusion protein 3D structure prediction, virtual screening, and PubMed search for all reported drugs of all human kinase fusion oncoproteins will provide novel knowledge for prioritizing the kinase inhibitors by using the whole 3D structures of kinase fusion proteins, not relying on the partial structure-based drug screening approach (KinaseFusionDB).2. Understanding KinaseFusionDB's Annotation Categories
Search page, example: ABL1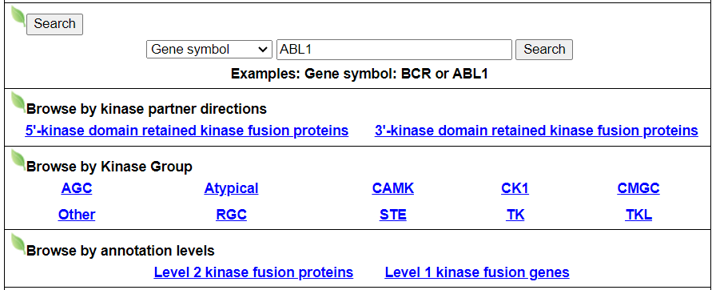
Input query
- Official HUGO gene symbol.
FusionGene Search Result Page
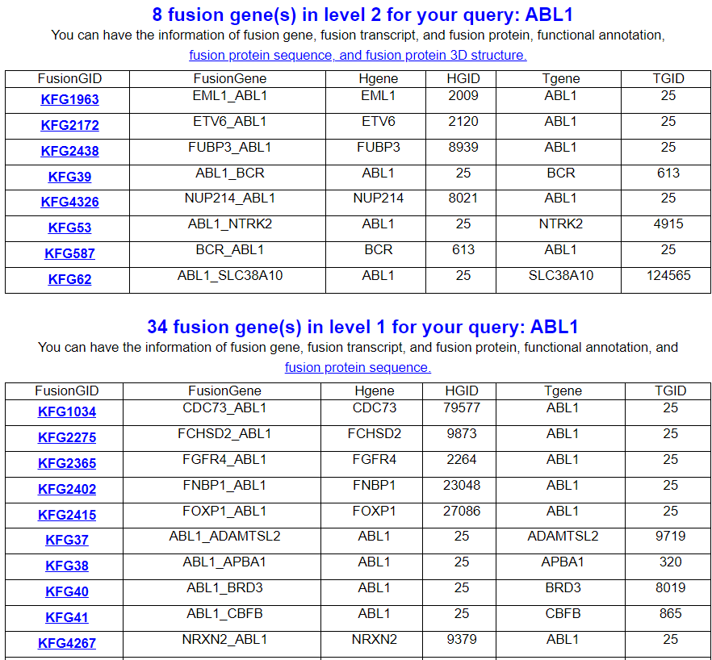
Select your kinase fusion gene from the fusion gene list.
We provide a classification in the first query search results into two Levels.
- Level 2 provides fusion protein sequences with multiple sequence alignment, fusion protein functional feature retention search, fusion protein 3D structures, comparison of fusion protein isoforms, comparison of fusion protein sequences/structures with known sequences/structures from PDB, active sites, confident structure selection through pLDDT distributions between active vs non-active sites, Ramachandran plots, virtual screening against all FDA-approved kinase inhibitors, top 20 bindings, interacting kinase inhibitor information, and PubMed search of drugs/diseases.
- Level 1 provides the fusion gene summary, functional annotation, ORF annotation, full-length fusion transcript sequences, and kinase-substrate information of ~ 7000 kinase fusion genes.
Kinase Fusion Gene Annotation Result Page

These are KinaseFusionDB's annotation categories for your query with links to their corresponding annotation parts.
1) Kinase Fusion Protein Summary.
1) The Kinase Fusion Gene Summary category shows a basic fusion gene summary following multiple annotations for each fusion partner gene. It also provides the two gene assessment scores to provide the impact of each gene across pan-cancer fusion genes. It provides the functional gene groups to assign potential loss-of-functional effects with ORF annotation and the breakpoints loci on the gene structures of individual partners using the UCSC genome browser.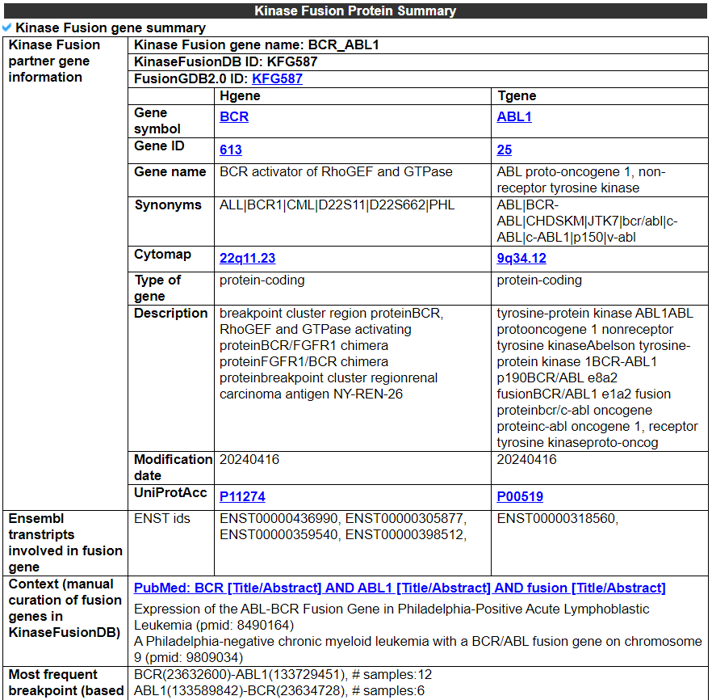
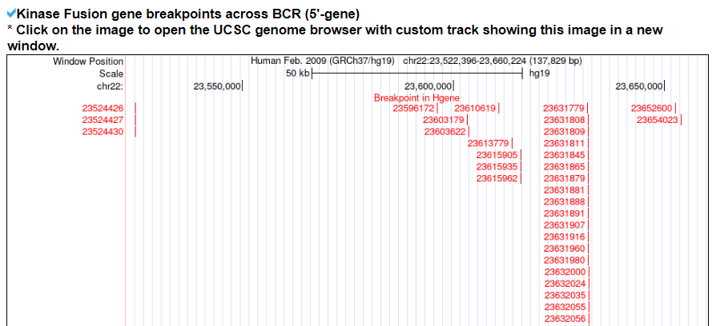
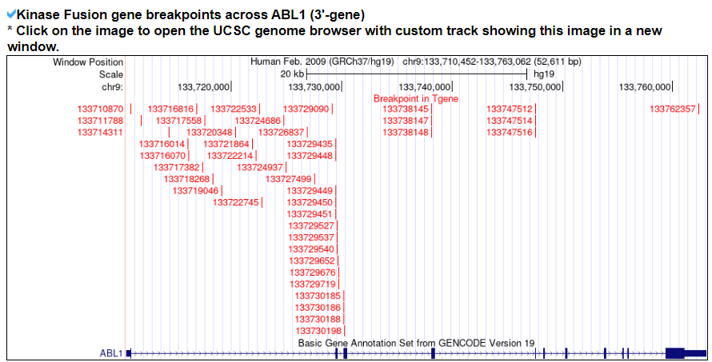
2) Kinase Fusion Gene Sample Information.
This Kinase Fusion Gene Sample Information category shows the cell-line or patient sample information, which expressed individual kinase fusion genes.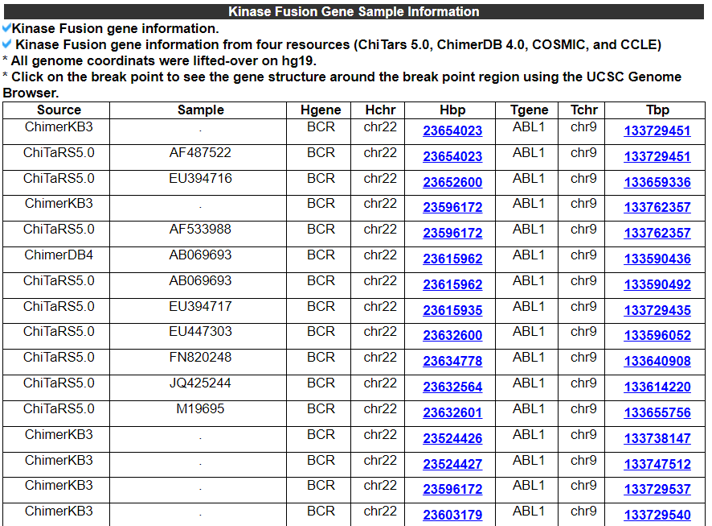
3) Kinase Fusion Gene ORF analysis.
The Kinase Fusion ORF Analysis category provides the in-frame fusion gene information and the length of full-length fusion mRNA and fusion amino acid sequences.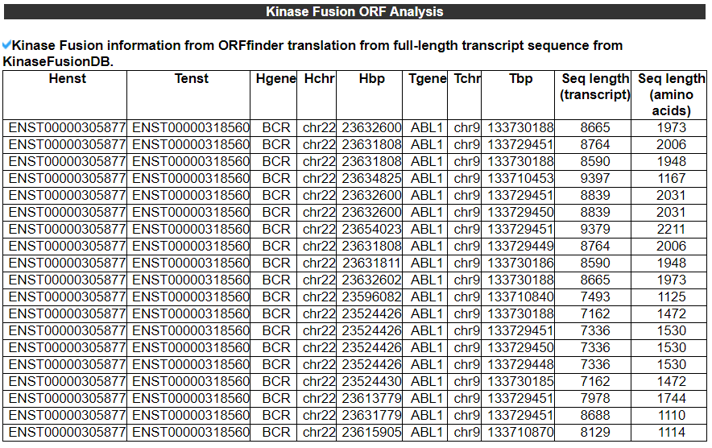
4) Kinase Fusion Amino Acid Sequences.
The Kinase Fusion Amino Acid Sequences category provides amino acid sequences of all fusion protein isoforms.
5) Multiple Sequence Alignment.
The Multiple Sequence Alignment of All Fusion Protein Isoforms category provides the multiple sequence alignment of all fusion protein isoforms.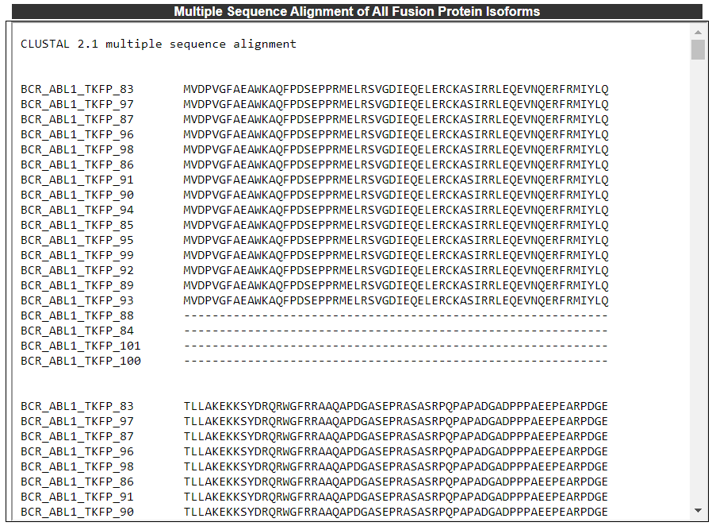
6) Kinase Fusion Protein Functional Features.
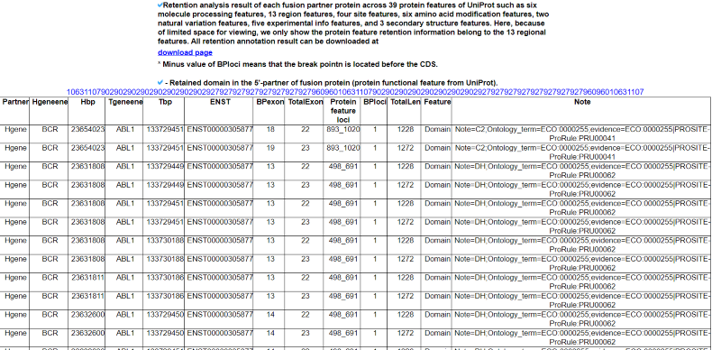
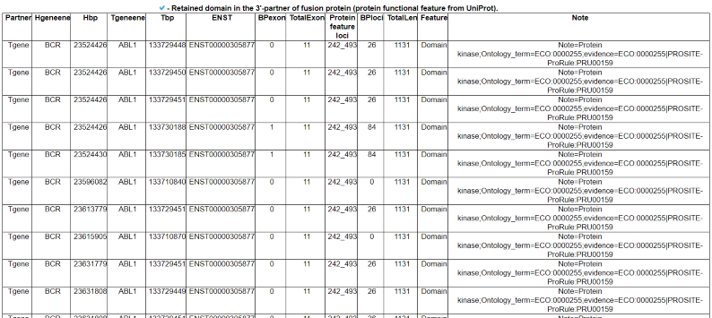
The Kinase Fusion Protein Functional Features category provides the retention information of 39 protein features due to the formation of fusion proteins with a link to an in-house visualization web tool for functional features of fusion genes, FGviewer. When we investigated the major domain features, the 3’-partner’s fusion protein part retained more protein functional domains than the ones of the 5’-partners.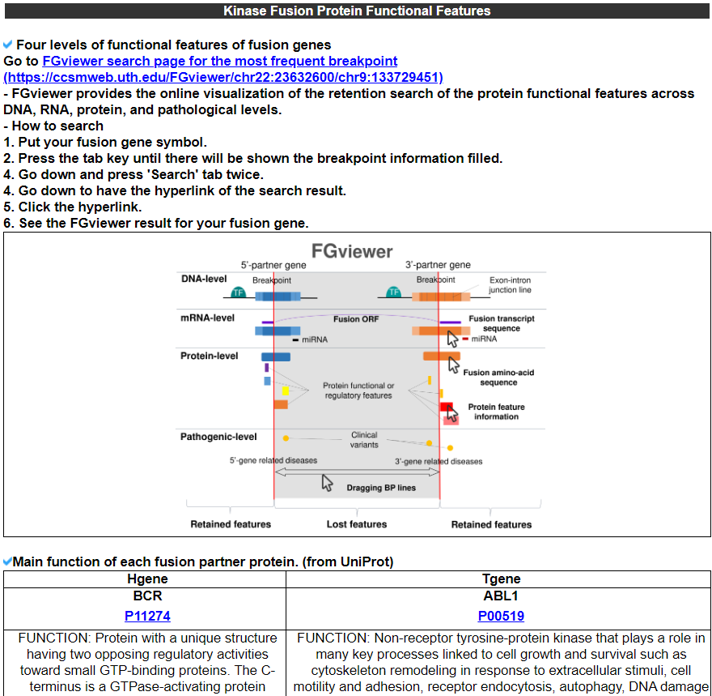
7) Kinase Fusion Protein Structures.
The Kinase Fusion Protein Structure category provides the 3D structures of all fusion protein isoforms predicted by AlphaFold2. The 3D views of fusion protein structures were implemented with Molstar with per-residue confidence scores (pLDDT) colored.Kinase Fusion Protein Structure category provides the 3D structures of all fusion protein isoforms predicted by AlphaFold2. The 3D views of fusion protein structures were implemented with Molstar with per-residue confidence scores (pLDDT) colored.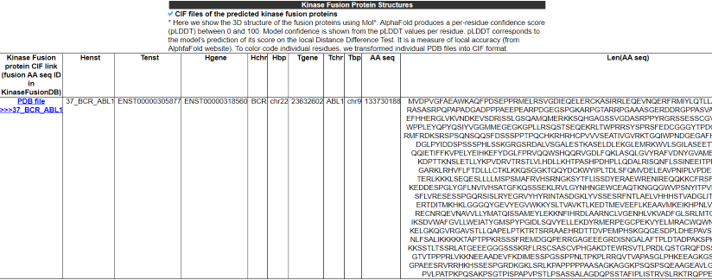

8) Comparison of the Fusion Protein Isoforms.
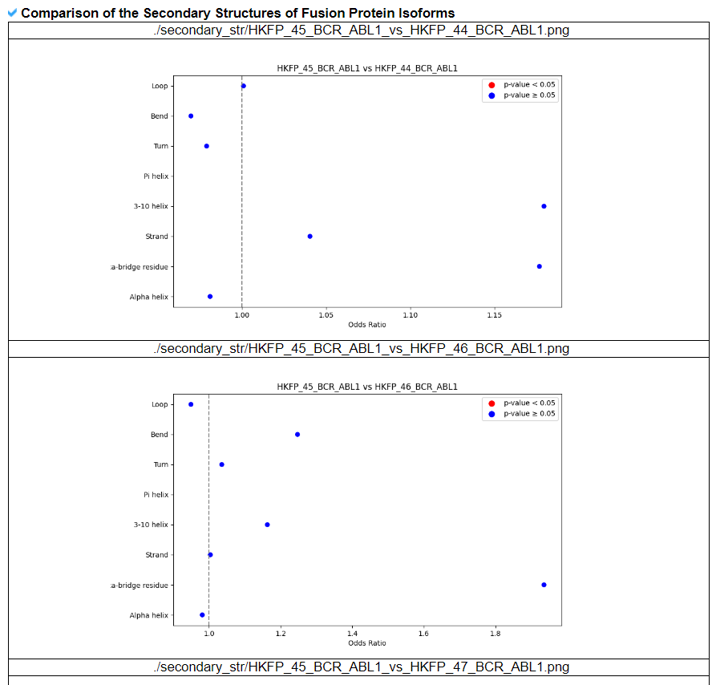
The Comparison of the Fusion Protein Isoforms category provides the superimpose among different kinase fusion protein isoforms.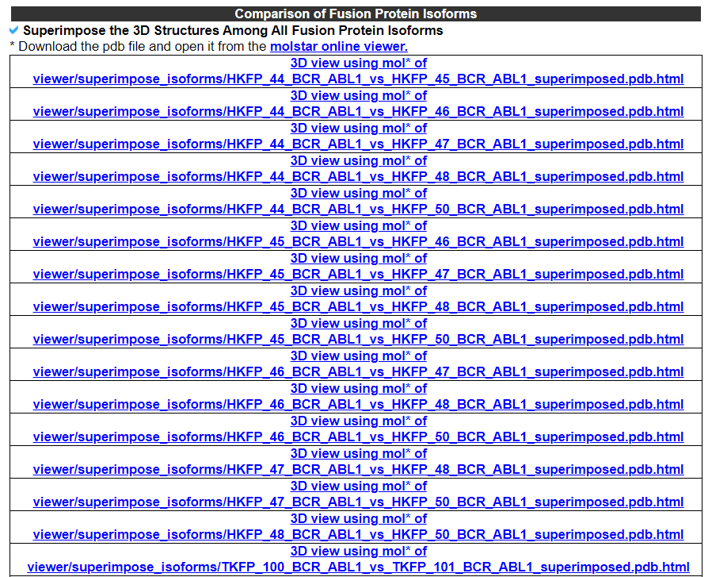
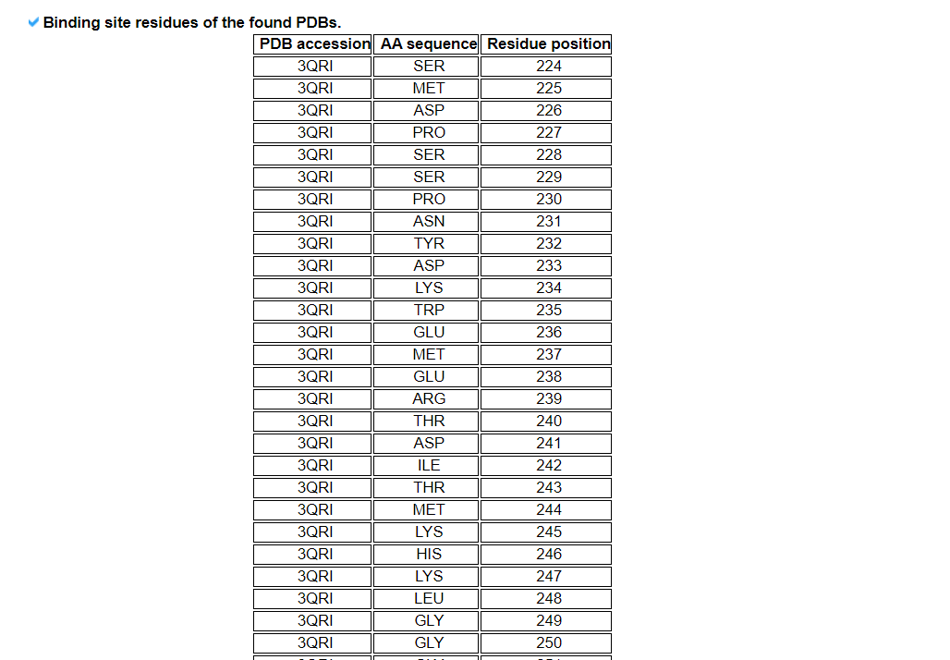
9) Comparison of Fusion Protein Sequences/Structures with Known Sequences/Structures from the PDB.
The Comparison of Fusion Protein Sequences/Structures with Known Sequences/Structures from the PDB category provides the structural comparison between our fusion proteins and known ones from the PDB. We provide multiple sequence alignments and superimpose among these.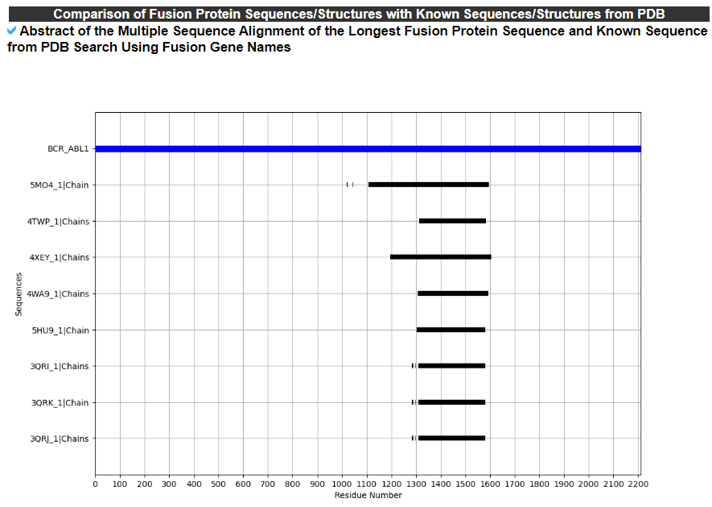


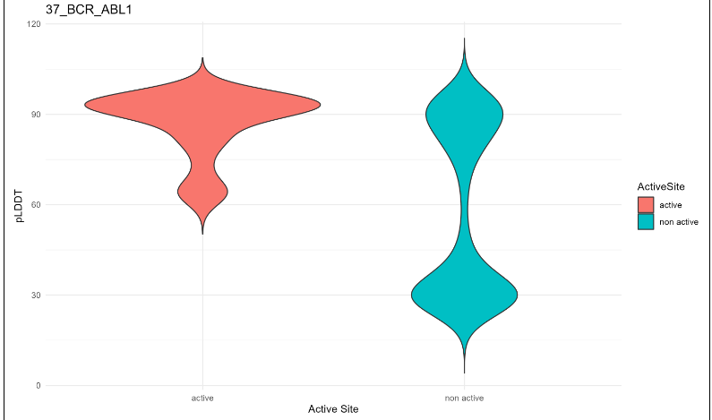
10) pLDDT Scores and Difference Analysis of pLDDT Scores Between the Active Sites (Best) and Non-Active Sites.
The pLDDT Scores and Difference Analysis of pLDDT scores Between the Active Sites and Non-Active Sites category provides confident kinase fusion protein 3D structures.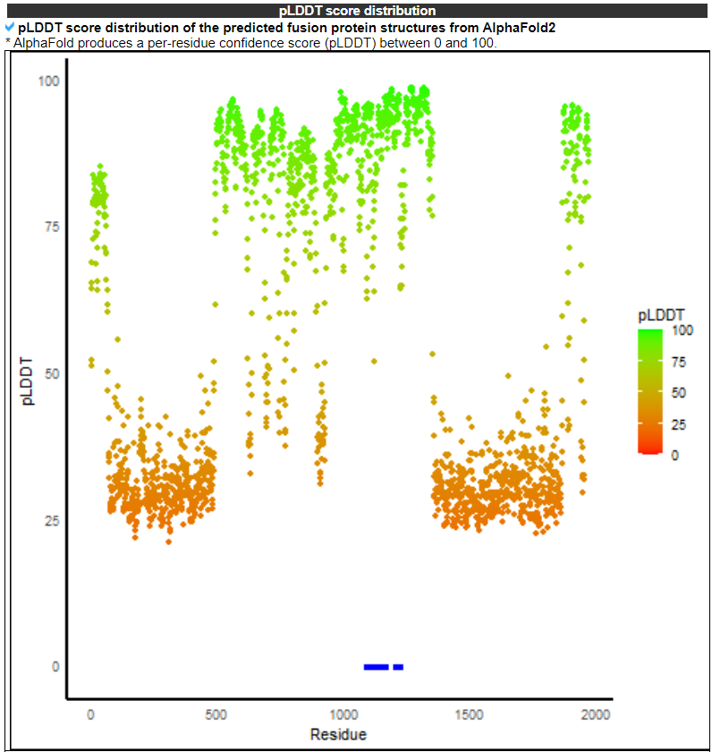
11) Ramachandra plot of Kinase Fusion Protein Structures.
The Ramachandran Plot of the Kinase Fusion Protein Structure category provides the Ramachandran plots of all kinase fusion protein isoforms.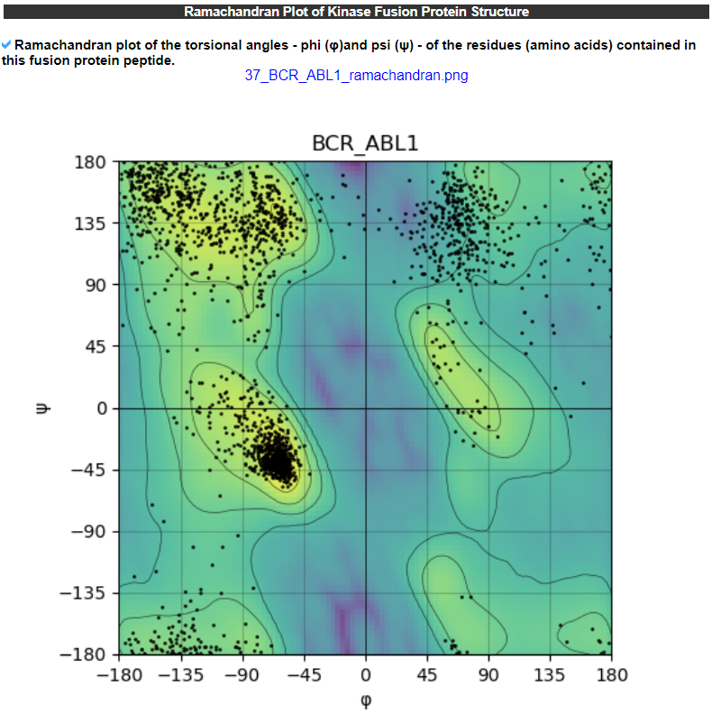
12) Potential Active Site Information.
The Potential Active Site Information category provides the best active site information predicted from the whole 3D structures of the fusion proteins using SiteMap.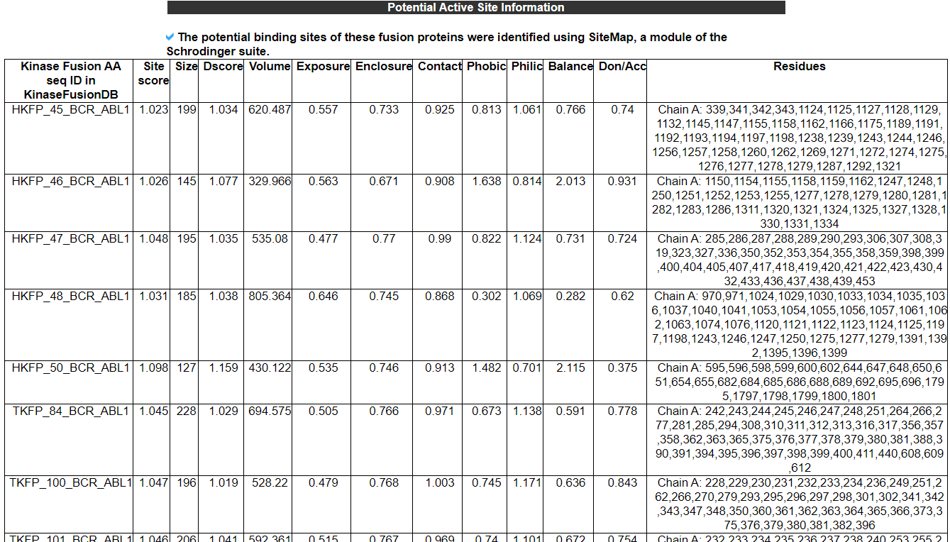
13) Virtual Screening Results.
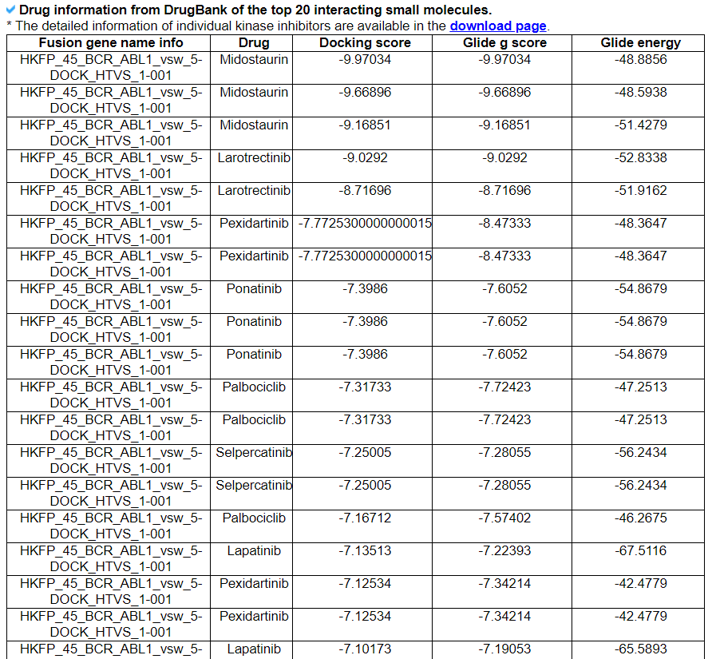
The Virtual Screening Results category provides the list of virtual screening results. The screening table will consist of the top 20 docking results and the GLIDE score for each fusion protein target.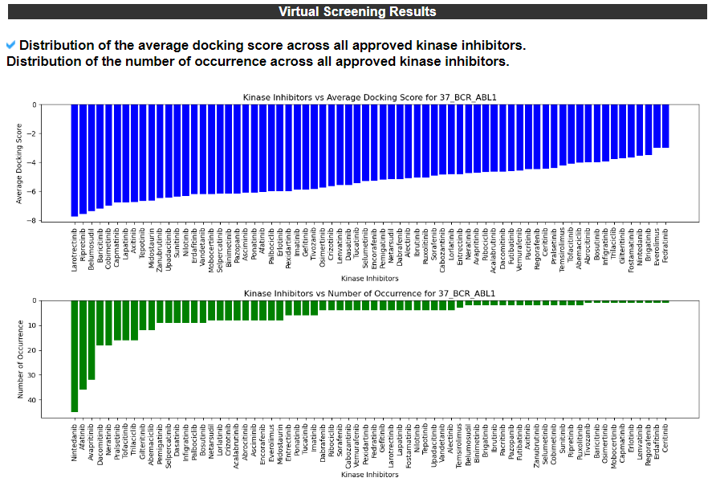
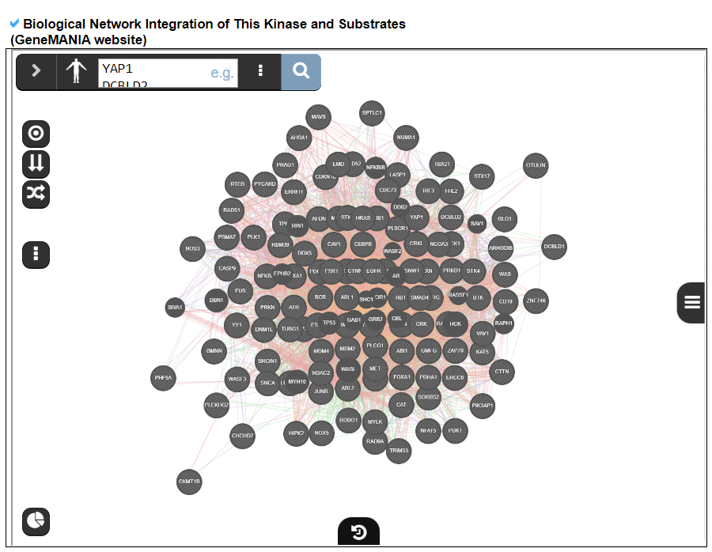
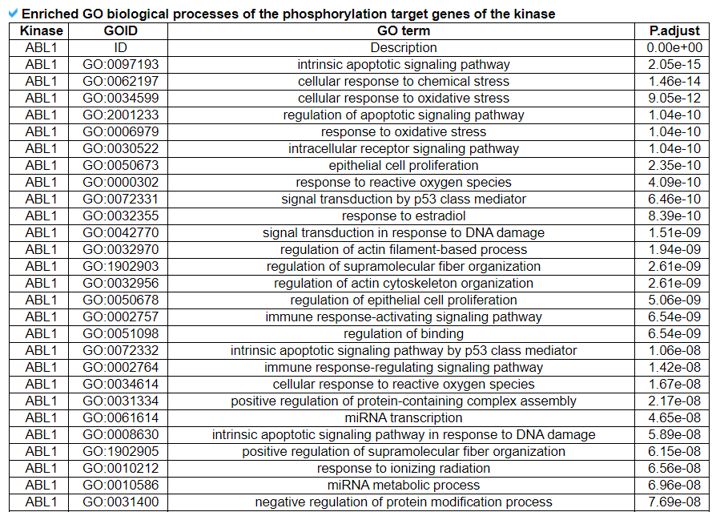
14) Kinase-Substrate Information.
The Kinase-Substrate Information category provides the phosphorylation target gene information and their enriched biological processes.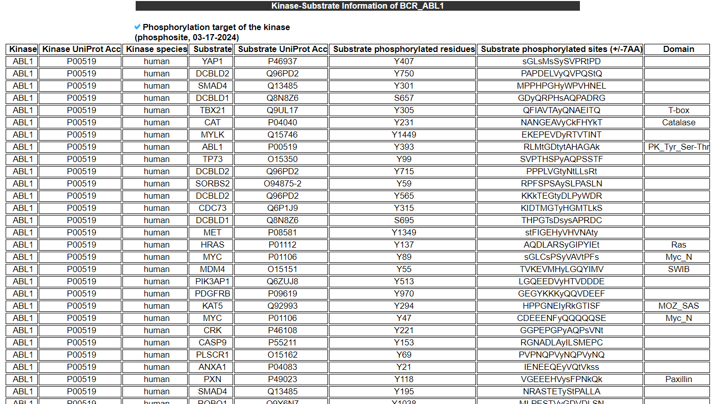
15) Related Drugs.
The Related Drugs category provides the manually curated results of used drugs targeting curated kinase fusion genes from PubMed literature and MyCancerGenome. Using the systematic PubMed abstract search of all combinations of kinase fusion gene names and 72 kinase inhibitors, we found 2911 unique PMIDs mentioning 90 kinase fusion genes and 60 kinase inhibitors in the abstracts.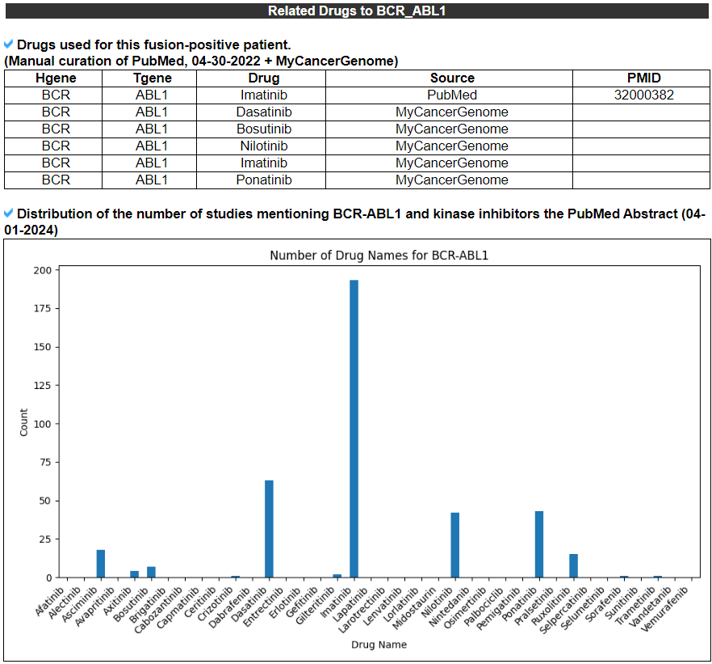
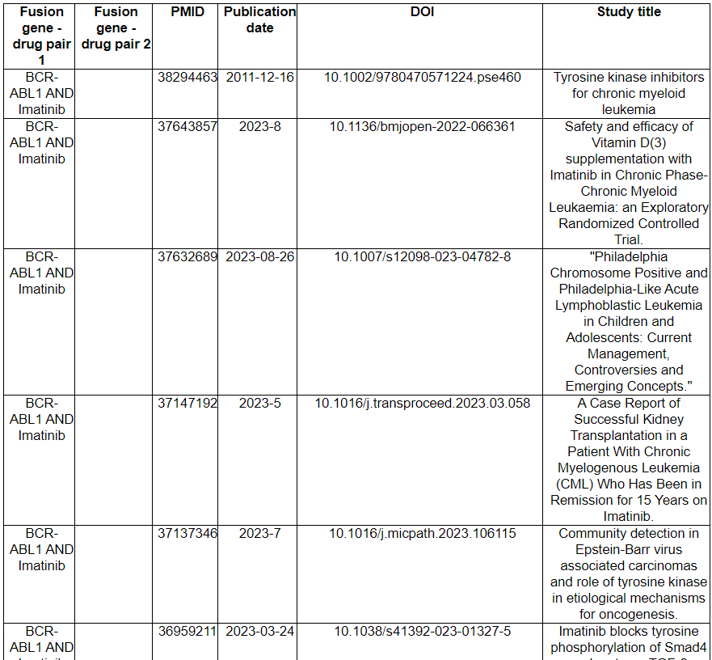
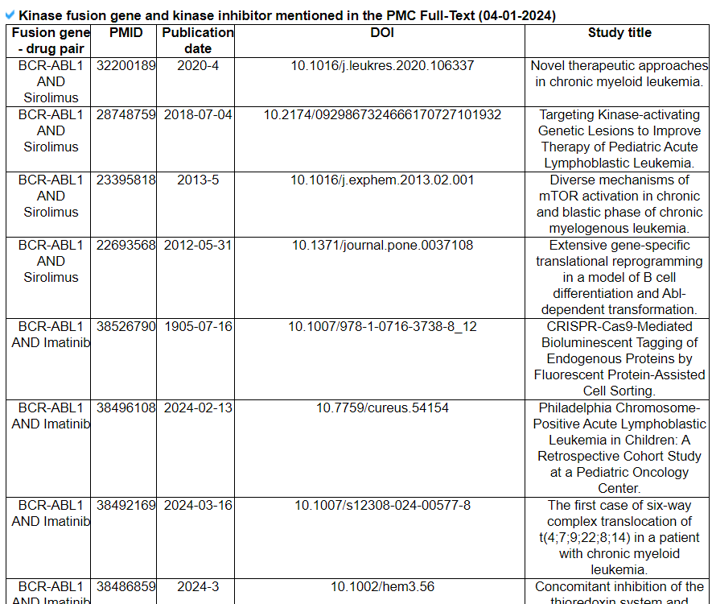
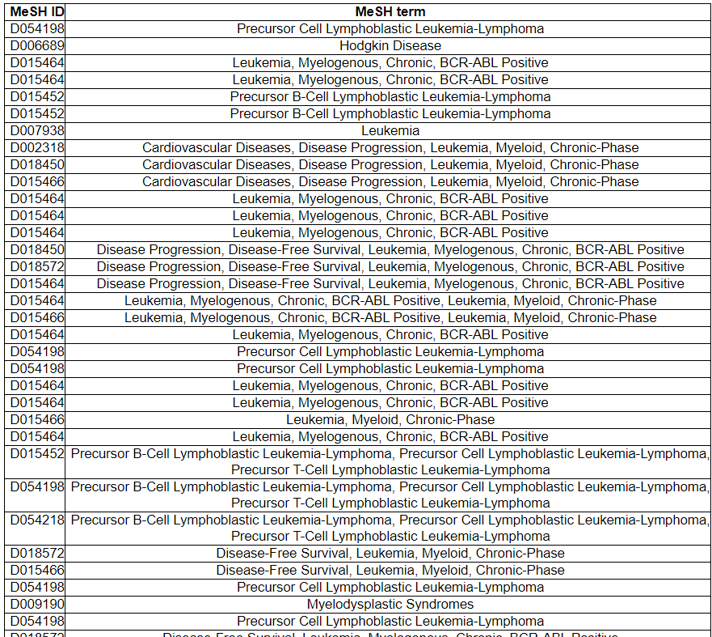
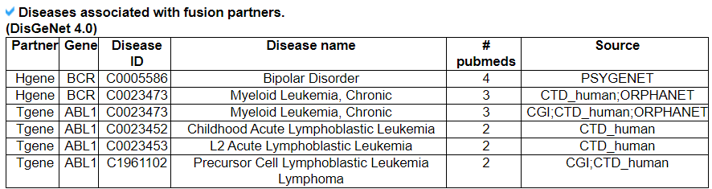
16) Related Diseases
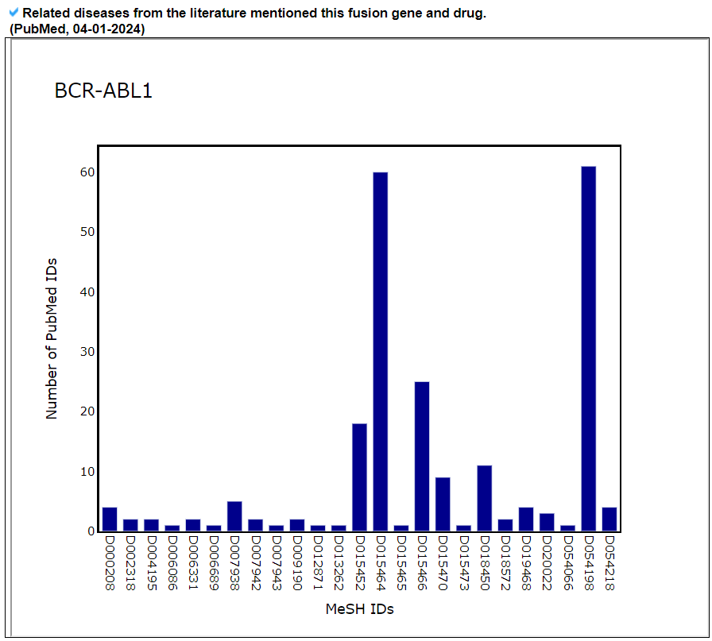
The Related Diseases category provides the manually curated results of used drugs targeting curated kinase fusion genes from PubMed literature and MyCancerGenome. From the 2911 abstracts found in the Related Drugs category, we found 71 MeSH IDs across 71 diseases.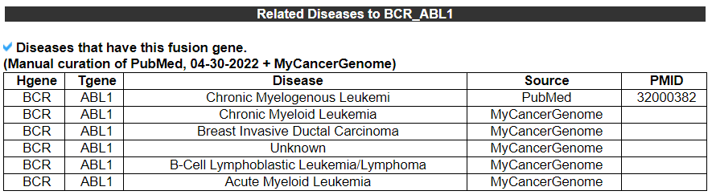
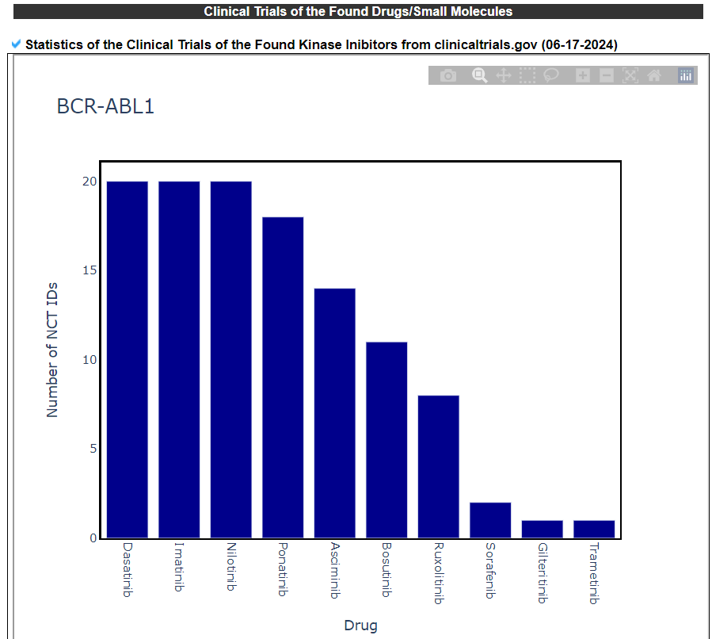
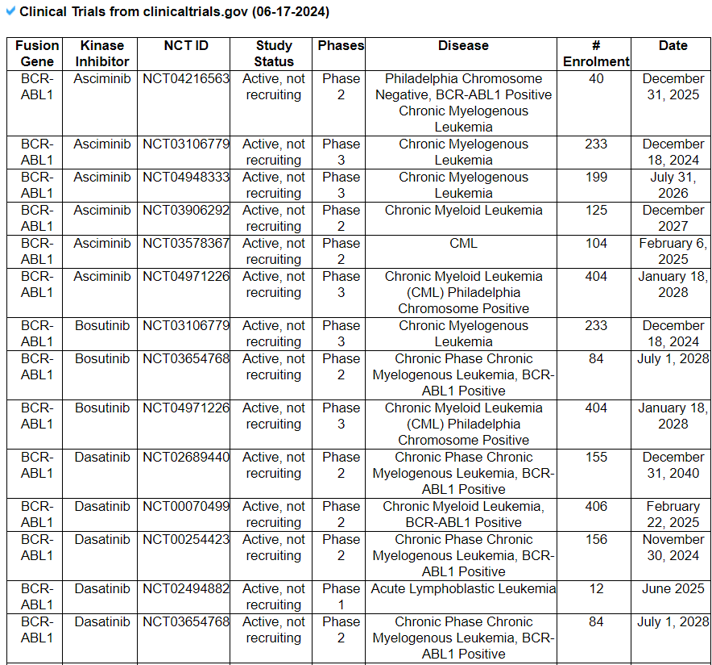
17) Clinical Trials of the Found Drugs/Small Moleculess
The Clinical Trails of the Found Drugs/Small Molecules category provides the previous or current clinical trial records of fusion gene-positive patients and our found drugs from the PubMed search