| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Kinase Fusion Gene:AKAP13_MUSK |
Kinase Fusion Protein Summary |
 Kinase Fusion gene summary Kinase Fusion gene summary |
| Kinase Fusion partner gene information | Kinase Fusion gene name: AKAP13_MUSK | KinaseFusionDB ID: KFG247 | FusionGDB2.0 ID: KFG247 | Hgene | Tgene | Gene symbol | AKAP13 | MUSK | Gene ID | 11214 | 4593 | |
| Gene name | A-kinase anchoring protein 13 | muscle associated receptor tyrosine kinase | ||||||||||
| Synonyms | AKAP-13|AKAP-Lbc|ARHGEF13|BRX|HA-3|Ht31|LBC|PRKA13|PROTO-LB|PROTO-LBC|c-lbc|p47 | CMS9|FADS|FADS1 | ||||||||||
| Cytomap | 15q25.3 | 9q31.3 | ||||||||||
| Type of gene | protein-coding | protein-coding | ||||||||||
| Description | A-kinase anchor protein 13A kinase (PRKA) anchor protein 13LBC oncogenebreast cancer nuclear receptor-binding auxiliary proteinguanine nucleotide exchange factor Lbchuman thyroid-anchoring protein 31lymphoid blast crisis oncogenenon-oncogenic Rho G | muscle, skeletal receptor tyrosine-protein kinasemuscle, skeletal, receptor tyrosine kinasemuscle-specific kinase receptormuscle-specific tyrosine-protein kinase receptor | ||||||||||
| Modification date | 20240411 | 20240411 | ||||||||||
| UniProtAcc | Q12802 | O15146 | ||||||||||
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000560579, ENST00000361243, ENST00000394510, ENST00000394518, ENST00000560302, | ENST00000189978, ENST00000374448, ENST00000416899, ENST00000374438, ENST00000374439, ENST00000374440, | |||||||||
| Context (manual curation of fusion genes in KinaseFusionDB) | PubMed: AKAP13 [Title/Abstract] AND MUSK [Title/Abstract] AND fusion [Title/Abstract] | |||||||||||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | AKAP13(86228104)-MUSK(113509921), # samples:2 | |||||||||||
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | AKAP13 | GO:0007186 | G protein-coupled receptor signaling pathway | 11546812 |
| Hgene | AKAP13 | GO:0035025 | positive regulation of Rho protein signal transduction | 11546812 |
| Tgene | MUSK | GO:0007528 | neuromuscular junction development | 25537362 |
 Kinase Fusion gene breakpoints across AKAP13 (5'-gene) Kinase Fusion gene breakpoints across AKAP13 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
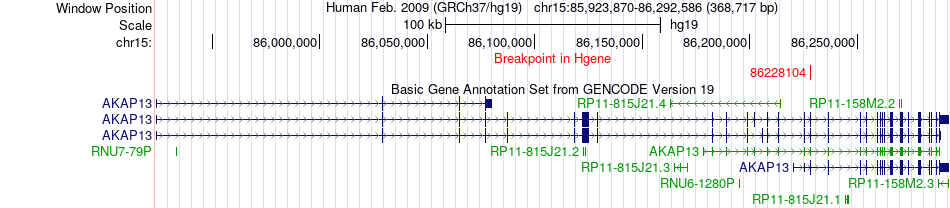 |
 Kinase Fusion gene breakpoints across MUSK (3'-gene) Kinase Fusion gene breakpoints across MUSK (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
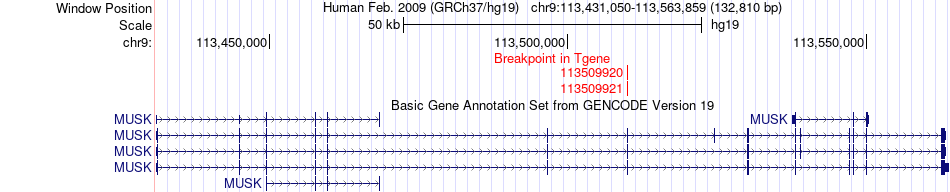 |
Top |
Kinase Fusion Gene Sample Information |
 Kinase Fusion gene information. Kinase Fusion gene information. |
 Kinase Fusion gene information from four resources (ChiTars 5.0, ChimerDB 4.0, COSMIC, and CCLE) Kinase Fusion gene information from four resources (ChiTars 5.0, ChimerDB 4.0, COSMIC, and CCLE)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Sample | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp |
| ChimerDB4 | TCGA-R5-A805-01A | AKAP13 | chr15 | 86228104 | MUSK | chr9 | 113509921 |
| ChimerDB4 | TCGA-R5-A805 | AKAP13 | chr15 | 86228104 | MUSK | chr9 | 113509920 |
Top |
Kinase Fusion ORF Analysis |
 Kinase Fusion information from ORFfinder translation from full-length transcript sequence from KinaseFusionDB. Kinase Fusion information from ORFfinder translation from full-length transcript sequence from KinaseFusionDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp | Seq length (transcript) | Seq length (amino acids) |
| ENST00000394518 | ENST00000189978 | AKAP13 | chr15 | 86228104 | MUSK | chr9 | 113509920 | 7251 | 2381 |
| ENST00000394518 | ENST00000189978 | AKAP13 | chr15 | 86228104 | MUSK | chr9 | 113509921 | 7251 | 2381 |
Top |
Kinase Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from KinaseFusionDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from KinaseFusionDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >Henst_Tenst_Hgene_Hchr_Hbp_Tgene_Tchr_Tbp_length(fusion AA)_AAseq >ENST00000394518_ENST00000189978_AKAP13_chr15_86228104_MUSK_chr9_113509920_length(amino acids)=2381 MKLNPQQAPLYGDCVVTVLLAEEDKAEDDVVFYLVFLGSTLRHCTSTRKVSSDTLETIAPGHDCCETVKVQLCASKEGLPVFVVAEEDFH FVQDEAYDAAQFLATSAGNQQALNFTRFLDQSGPPSGDVNSLDKKLVLAFRHLKLPTEWNVLGTDQSLHDAGPRETLMHFAVRLGLLRLT WFLLQKPGGRGALSIHNQEGATPVSLALERGYHKLHQLLTEENAGEPDSWSSLSYEIPYGDCSVRHHRELDIYTLTSESDSHHEHPFPGD GCTGPIFKLMNIQQQLMKTNLKQMDSLMPLMMTAQDPSSAPETDGQFLPCAPEPTDPQRLSSSEETESTQCCPGSPVAQTESPCDLSSIV EEENTDRSCRKKNKGVERKGEEVEPAPIVDSGTVSDQDSCLQSLPDCGVKGTEGLSSCGNRNEETGTKSSGMPTDQESLSSGDAVLQRDL VMEPGTAQYSSGGELGGISTTNVSTPDTAGEMEHGLMNPDATVWKNVLQGGESTKERFENSNIGTAGASDVHVTSKPVDKISVPNCAPAA SSLDGNKPAESSLAFSNEETSTEKTAETETSRSREESADAPVDQNSVVIPAAAKDKISDGLEPYTLLAAGIGEAMSPSDLALLGLEEDVM PHQNSETNSSHAQSQKGKSSPICSTTGDDKLCADSACQQNTVTSSGDLVAKLCDNIVSESESTTARQPSSQDPPDASHCEDPQAHTVTSD PVRDTQERADFCPFKVVDNKGQRKDVKLDKPLTNMLEVVSHPHPVVPKMEKELVPDQAVISDSTFSLANSPGSESVTKDDALSFVPSQKE KGTATPELHTATDYRDGPDGNSNEPDTRPLEDRAVGLSTSSTAAELQHGMGNTSLTGLGGEHEGPAPPAIPEALNIKGNTDSSLQSVGKA TLALDSVLTEEGKLLVVSESSAAQEQDKDKAVTCSSIKENALSSGTLQEEQRTPPPGQDTQQFHEKSISADCAKDKALQLSNSPGASSAF LKAETEHNKEVAPQVSLLTQGGAAQSLVPPGASLATESRQEALGAEHNSSALLPCLLPDGSDGSDALNCSQPSPLDVGVKNTQSQGKTSA CEVSGDVTVDVTGVNALQGMAEPRRENISHNTQDILIPNVLLSQEKNAVLGLPVALQDKAVTDPQGVGTPEMIPLDWEKGKLEGADHSCT MGDAEEAQIDDEAHPVLLQPVAKELPTDMELSAHDDGAPAGVREVMRAPPSGRERSTPSLPCMVSAQDAPLPKGADLIEEAASRIVDAVI EQVKAAGALLTEGEACHMSLSSPELGPLTKGLESAFTEKVSTFPPGESLPMGSTPEEATGSLAGCFAGREEPEKIILPVQGPEPAAEMPD VKAEDEVDFRASSISEEVAVGSIAATLKMKQGPMTQAINRENWCTIEPCPDAASLLASKQSPECENFLDVGLGRECTSKQGVLKRESGSD SDLFHSPSDDMDSIIFPKPEEEHLACDITGSSSSTDDTASLDRHSSHGSDVSLSQILKPNRSRDRQSLDGFYSHGMGAEGRESESEPADP GDVEEEEMDSITEVPANCSVLRSSMRSLSPFRRHSWGPGKNAASDAEMNHRSSMRVLGDVVRRPPIHRRSFSLEGLTGGAGVGNKPSSSL EVSSANAEELRHPFSGEERVDSLVSLSEEDLESDQREHRMFDQQICHRSKQQGFNYCTSAISSPLTKSISLMTISHPGLDNSRPFHSTFH NTSANLTESITEENYNFLPHSPSKKDSEWKSGTKVSRTFSYIKNKMSSSKKSKVSSGSIQESVKDRVIDSRLQLFITKPGLYTCIATNKH GEKFSTAKAAATISIAAFSKPQKDNKGYCAQYRGEVCNAVLAKDALVFLNTSYADPEEAQELLVHTAWNELKVVSPVCRPAAEALLCNHI FQECSPGVVPTPIPICREYCLAVKELFCAKEWLVMEEKTHRGLYRSEMHLLSVPECSKLPSMHWDPTACARLPHLDYNKENLKTFPPMTS SKPSVDIPNLPSSSSSSFSVSPTYSMTVIISIMSSFAIFVLLTITTLYCCRRRKQWKNKKRESAAVTLTTLPSELLLDRLHPNPMYQRMP LLLNPKLLSLEYPRNNIEYVRDIGEGAFGRVFQARAPGLLPYEPFTMVAVKMLKEEASADMQADFQREAALMAEFDNPNIVKLLGVCAVG KPMCLLFEYMAYGDLNEFLRSMSPHTVCSLSHSDLSMRAQVSSPGPPPLSCAEQLCIARQVAAGMAYLSERKFVHRDLATRNCLVGENMV VKIADFGLSRNIYSADYYKANENDAIPIRWMPPESIFYNRYTTESDVWAYGVVLWEIFSYGLQPYYGMAHEEVIYYVRDGNILSCPENCP -------------------------------------------------------------- >ENST00000394518_ENST00000189978_AKAP13_chr15_86228104_MUSK_chr9_113509921_length(amino acids)=2381 MKLNPQQAPLYGDCVVTVLLAEEDKAEDDVVFYLVFLGSTLRHCTSTRKVSSDTLETIAPGHDCCETVKVQLCASKEGLPVFVVAEEDFH FVQDEAYDAAQFLATSAGNQQALNFTRFLDQSGPPSGDVNSLDKKLVLAFRHLKLPTEWNVLGTDQSLHDAGPRETLMHFAVRLGLLRLT WFLLQKPGGRGALSIHNQEGATPVSLALERGYHKLHQLLTEENAGEPDSWSSLSYEIPYGDCSVRHHRELDIYTLTSESDSHHEHPFPGD GCTGPIFKLMNIQQQLMKTNLKQMDSLMPLMMTAQDPSSAPETDGQFLPCAPEPTDPQRLSSSEETESTQCCPGSPVAQTESPCDLSSIV EEENTDRSCRKKNKGVERKGEEVEPAPIVDSGTVSDQDSCLQSLPDCGVKGTEGLSSCGNRNEETGTKSSGMPTDQESLSSGDAVLQRDL VMEPGTAQYSSGGELGGISTTNVSTPDTAGEMEHGLMNPDATVWKNVLQGGESTKERFENSNIGTAGASDVHVTSKPVDKISVPNCAPAA SSLDGNKPAESSLAFSNEETSTEKTAETETSRSREESADAPVDQNSVVIPAAAKDKISDGLEPYTLLAAGIGEAMSPSDLALLGLEEDVM PHQNSETNSSHAQSQKGKSSPICSTTGDDKLCADSACQQNTVTSSGDLVAKLCDNIVSESESTTARQPSSQDPPDASHCEDPQAHTVTSD PVRDTQERADFCPFKVVDNKGQRKDVKLDKPLTNMLEVVSHPHPVVPKMEKELVPDQAVISDSTFSLANSPGSESVTKDDALSFVPSQKE KGTATPELHTATDYRDGPDGNSNEPDTRPLEDRAVGLSTSSTAAELQHGMGNTSLTGLGGEHEGPAPPAIPEALNIKGNTDSSLQSVGKA TLALDSVLTEEGKLLVVSESSAAQEQDKDKAVTCSSIKENALSSGTLQEEQRTPPPGQDTQQFHEKSISADCAKDKALQLSNSPGASSAF LKAETEHNKEVAPQVSLLTQGGAAQSLVPPGASLATESRQEALGAEHNSSALLPCLLPDGSDGSDALNCSQPSPLDVGVKNTQSQGKTSA CEVSGDVTVDVTGVNALQGMAEPRRENISHNTQDILIPNVLLSQEKNAVLGLPVALQDKAVTDPQGVGTPEMIPLDWEKGKLEGADHSCT MGDAEEAQIDDEAHPVLLQPVAKELPTDMELSAHDDGAPAGVREVMRAPPSGRERSTPSLPCMVSAQDAPLPKGADLIEEAASRIVDAVI EQVKAAGALLTEGEACHMSLSSPELGPLTKGLESAFTEKVSTFPPGESLPMGSTPEEATGSLAGCFAGREEPEKIILPVQGPEPAAEMPD VKAEDEVDFRASSISEEVAVGSIAATLKMKQGPMTQAINRENWCTIEPCPDAASLLASKQSPECENFLDVGLGRECTSKQGVLKRESGSD SDLFHSPSDDMDSIIFPKPEEEHLACDITGSSSSTDDTASLDRHSSHGSDVSLSQILKPNRSRDRQSLDGFYSHGMGAEGRESESEPADP GDVEEEEMDSITEVPANCSVLRSSMRSLSPFRRHSWGPGKNAASDAEMNHRSSMRVLGDVVRRPPIHRRSFSLEGLTGGAGVGNKPSSSL EVSSANAEELRHPFSGEERVDSLVSLSEEDLESDQREHRMFDQQICHRSKQQGFNYCTSAISSPLTKSISLMTISHPGLDNSRPFHSTFH NTSANLTESITEENYNFLPHSPSKKDSEWKSGTKVSRTFSYIKNKMSSSKKSKVSSGSIQESVKDRVIDSRLQLFITKPGLYTCIATNKH GEKFSTAKAAATISIAAFSKPQKDNKGYCAQYRGEVCNAVLAKDALVFLNTSYADPEEAQELLVHTAWNELKVVSPVCRPAAEALLCNHI FQECSPGVVPTPIPICREYCLAVKELFCAKEWLVMEEKTHRGLYRSEMHLLSVPECSKLPSMHWDPTACARLPHLDYNKENLKTFPPMTS SKPSVDIPNLPSSSSSSFSVSPTYSMTVIISIMSSFAIFVLLTITTLYCCRRRKQWKNKKRESAAVTLTTLPSELLLDRLHPNPMYQRMP LLLNPKLLSLEYPRNNIEYVRDIGEGAFGRVFQARAPGLLPYEPFTMVAVKMLKEEASADMQADFQREAALMAEFDNPNIVKLLGVCAVG KPMCLLFEYMAYGDLNEFLRSMSPHTVCSLSHSDLSMRAQVSSPGPPPLSCAEQLCIARQVAAGMAYLSERKFVHRDLATRNCLVGENMV VKIADFGLSRNIYSADYYKANENDAIPIRWMPPESIFYNRYTTESDVWAYGVVLWEIFSYGLQPYYGMAHEEVIYYVRDGNILSCPENCP -------------------------------------------------------------- |
Multiple Sequence Alignment of All Fusion Protein Isoforms |
Top |
Kinase Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr15:86228104/chr9:113509921) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| AKAP13 | MUSK |
| FUNCTION: Scaffold protein that plays an important role in assembling signaling complexes downstream of several types of G protein-coupled receptors. Activates RHOA in response to signaling via G protein-coupled receptors via its function as Rho guanine nucleotide exchange factor (PubMed:11546812, PubMed:15229649, PubMed:23090968, PubMed:25186459, PubMed:24993829). May also activate other Rho family members (PubMed:11546812). Part of a kinase signaling complex that links ADRA1A and ADRA1B adrenergic receptor signaling to the activation of downstream p38 MAP kinases, such as MAPK11 and MAPK14 (PubMed:17537920, PubMed:23716597, PubMed:21224381). Part of a signaling complex that links ADRA1B signaling to the activation of RHOA and IKBKB/IKKB, leading to increased NF-kappa-B transcriptional activity (PubMed:23090968). Part of a RHOA-dependent signaling cascade that mediates responses to lysophosphatidic acid (LPA), a signaling molecule that activates G-protein coupled receptors and potentiates transcriptional activation of the glucocorticoid receptor NR3C1 (PubMed:16469733). Part of a signaling cascade that stimulates MEF2C-dependent gene expression in response to lysophosphatidic acid (LPA) (By similarity). Part of a signaling pathway that activates MAPK11 and/or MAPK14 and leads to increased transcription activation of the estrogen receptors ESR1 and ESR2 (PubMed:9627117, PubMed:11579095). Part of a signaling cascade that links cAMP and EGFR signaling to BRAF signaling and to PKA-mediated phosphorylation of KSR1, leading to the activation of downstream MAP kinases, such as MAPK1 or MAPK3 (PubMed:21102438). Functions as a scaffold protein that anchors cAMP-dependent protein kinase (PKA) and PRKD1. This promotes activation of PRKD1, leading to increased phosphorylation of HDAC5 and ultimately cardiomyocyte hypertrophy (By similarity). Has no guanine nucleotide exchange activity on CDC42, Ras or Rac (PubMed:11546812). Required for normal embryonic heart development, and in particular for normal sarcomere formation in the developing cardiomyocytes (By similarity). Plays a role in cardiomyocyte growth and cardiac hypertrophy in response to activation of the beta-adrenergic receptor by phenylephrine or isoproterenol (PubMed:17537920, PubMed:23090968). Required for normal adaptive cardiac hypertrophy in response to pressure overload (PubMed:23716597). Plays a role in osteogenesis (By similarity). {ECO:0000250|UniProtKB:E9Q394, ECO:0000269|PubMed:11546812, ECO:0000269|PubMed:11579095, ECO:0000269|PubMed:17537920, ECO:0000269|PubMed:21224381, ECO:0000269|PubMed:23716597, ECO:0000269|PubMed:24993829, ECO:0000269|PubMed:25186459, ECO:0000269|PubMed:9627117, ECO:0000269|PubMed:9891067}. | FUNCTION: Receptor tyrosine kinase which plays a central role in the formation and the maintenance of the neuromuscular junction (NMJ), the synapse between the motor neuron and the skeletal muscle (PubMed:25537362). Recruitment of AGRIN by LRP4 to the MUSK signaling complex induces phosphorylation and activation of MUSK, the kinase of the complex. The activation of MUSK in myotubes regulates the formation of NMJs through the regulation of different processes including the specific expression of genes in subsynaptic nuclei, the reorganization of the actin cytoskeleton and the clustering of the acetylcholine receptors (AChR) in the postsynaptic membrane. May regulate AChR phosphorylation and clustering through activation of ABL1 and Src family kinases which in turn regulate MUSK. DVL1 and PAK1 that form a ternary complex with MUSK are also important for MUSK-dependent regulation of AChR clustering. May positively regulate Rho family GTPases through FNTA. Mediates the phosphorylation of FNTA which promotes prenylation, recruitment to membranes and activation of RAC1 a regulator of the actin cytoskeleton and of gene expression. Other effectors of the MUSK signaling include DNAJA3 which functions downstream of MUSK. May also play a role within the central nervous system by mediating cholinergic responses, synaptic plasticity and memory formation (By similarity). {ECO:0000250, ECO:0000269|PubMed:25537362}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
 - Retained domain in the 5'-partner of fusion protein (protein functional feature from UniProt). - Retained domain in the 5'-partner of fusion protein (protein functional feature from UniProt). |
| Partner | Hgeneene | Hbp | Tgeneene | Tbp | ENST | BPexon | TotalExon | Protein feature loci | BPloci | TotalLen | Feature | Note |
 - Retained domain in the 3'-partner of fusion protein (protein functional feature from UniProt). - Retained domain in the 3'-partner of fusion protein (protein functional feature from UniProt). |
| Partner | Hgeneene | Hbp | Tgeneene | Tbp | ENST | BPexon | TotalExon | Protein feature loci | BPloci | TotalLen | Feature | Note |
| Tgene | AKAP13 | 86228104 | MUSK | 113509920 | ENST00000394518 | 5 | 15 | 312_450 | 251 | 870 | Domain | Note=FZ;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00090 |
| Tgene | AKAP13 | 86228104 | MUSK | 113509921 | ENST00000394518 | 5 | 15 | 312_450 | 251 | 870 | Domain | Note=FZ;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00090 |
| Tgene | AKAP13 | 86228104 | MUSK | 113509920 | ENST00000394518 | 5 | 15 | 575_856 | 251 | 870 | Domain | Note=Protein kinase;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00159 |
| Tgene | AKAP13 | 86228104 | MUSK | 113509921 | ENST00000394518 | 5 | 15 | 575_856 | 251 | 870 | Domain | Note=Protein kinase;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00159 |
Top |
Kinase Fusion Protein Structures |
 CIF files of the predicted kinase fusion proteins CIF files of the predicted kinase fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Kinase Fusion protein CIF link (fusion AA seq ID in KinaseFusionDB) | Henst | Tenst | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp | AA seq | Len(AA seq) |
| PDB file >>>15_AKAP13_MUSK | ENST00000394518 | ENST00000189978 | AKAP13 | chr15 | 86228104 | MUSK | chr9 | 113509921 | MKLNPQQAPLYGDCVVTVLLAEEDKAEDDVVFYLVFLGSTLRHCTSTRKVSSDTLETIAPGHDCCETVKVQLCASKEGLPVFVVAEEDFH FVQDEAYDAAQFLATSAGNQQALNFTRFLDQSGPPSGDVNSLDKKLVLAFRHLKLPTEWNVLGTDQSLHDAGPRETLMHFAVRLGLLRLT WFLLQKPGGRGALSIHNQEGATPVSLALERGYHKLHQLLTEENAGEPDSWSSLSYEIPYGDCSVRHHRELDIYTLTSESDSHHEHPFPGD GCTGPIFKLMNIQQQLMKTNLKQMDSLMPLMMTAQDPSSAPETDGQFLPCAPEPTDPQRLSSSEETESTQCCPGSPVAQTESPCDLSSIV EEENTDRSCRKKNKGVERKGEEVEPAPIVDSGTVSDQDSCLQSLPDCGVKGTEGLSSCGNRNEETGTKSSGMPTDQESLSSGDAVLQRDL VMEPGTAQYSSGGELGGISTTNVSTPDTAGEMEHGLMNPDATVWKNVLQGGESTKERFENSNIGTAGASDVHVTSKPVDKISVPNCAPAA SSLDGNKPAESSLAFSNEETSTEKTAETETSRSREESADAPVDQNSVVIPAAAKDKISDGLEPYTLLAAGIGEAMSPSDLALLGLEEDVM PHQNSETNSSHAQSQKGKSSPICSTTGDDKLCADSACQQNTVTSSGDLVAKLCDNIVSESESTTARQPSSQDPPDASHCEDPQAHTVTSD PVRDTQERADFCPFKVVDNKGQRKDVKLDKPLTNMLEVVSHPHPVVPKMEKELVPDQAVISDSTFSLANSPGSESVTKDDALSFVPSQKE KGTATPELHTATDYRDGPDGNSNEPDTRPLEDRAVGLSTSSTAAELQHGMGNTSLTGLGGEHEGPAPPAIPEALNIKGNTDSSLQSVGKA TLALDSVLTEEGKLLVVSESSAAQEQDKDKAVTCSSIKENALSSGTLQEEQRTPPPGQDTQQFHEKSISADCAKDKALQLSNSPGASSAF LKAETEHNKEVAPQVSLLTQGGAAQSLVPPGASLATESRQEALGAEHNSSALLPCLLPDGSDGSDALNCSQPSPLDVGVKNTQSQGKTSA CEVSGDVTVDVTGVNALQGMAEPRRENISHNTQDILIPNVLLSQEKNAVLGLPVALQDKAVTDPQGVGTPEMIPLDWEKGKLEGADHSCT MGDAEEAQIDDEAHPVLLQPVAKELPTDMELSAHDDGAPAGVREVMRAPPSGRERSTPSLPCMVSAQDAPLPKGADLIEEAASRIVDAVI EQVKAAGALLTEGEACHMSLSSPELGPLTKGLESAFTEKVSTFPPGESLPMGSTPEEATGSLAGCFAGREEPEKIILPVQGPEPAAEMPD VKAEDEVDFRASSISEEVAVGSIAATLKMKQGPMTQAINRENWCTIEPCPDAASLLASKQSPECENFLDVGLGRECTSKQGVLKRESGSD SDLFHSPSDDMDSIIFPKPEEEHLACDITGSSSSTDDTASLDRHSSHGSDVSLSQILKPNRSRDRQSLDGFYSHGMGAEGRESESEPADP GDVEEEEMDSITEVPANCSVLRSSMRSLSPFRRHSWGPGKNAASDAEMNHRSSMRVLGDVVRRPPIHRRSFSLEGLTGGAGVGNKPSSSL EVSSANAEELRHPFSGEERVDSLVSLSEEDLESDQREHRMFDQQICHRSKQQGFNYCTSAISSPLTKSISLMTISHPGLDNSRPFHSTFH NTSANLTESITEENYNFLPHSPSKKDSEWKSGTKVSRTFSYIKNKMSSSKKSKVSSGSIQESVKDRVIDSRLQLFITKPGLYTCIATNKH GEKFSTAKAAATISIAAFSKPQKDNKGYCAQYRGEVCNAVLAKDALVFLNTSYADPEEAQELLVHTAWNELKVVSPVCRPAAEALLCNHI FQECSPGVVPTPIPICREYCLAVKELFCAKEWLVMEEKTHRGLYRSEMHLLSVPECSKLPSMHWDPTACARLPHLDYNKENLKTFPPMTS SKPSVDIPNLPSSSSSSFSVSPTYSMTVIISIMSSFAIFVLLTITTLYCCRRRKQWKNKKRESAAVTLTTLPSELLLDRLHPNPMYQRMP LLLNPKLLSLEYPRNNIEYVRDIGEGAFGRVFQARAPGLLPYEPFTMVAVKMLKEEASADMQADFQREAALMAEFDNPNIVKLLGVCAVG KPMCLLFEYMAYGDLNEFLRSMSPHTVCSLSHSDLSMRAQVSSPGPPPLSCAEQLCIARQVAAGMAYLSERKFVHRDLATRNCLVGENMV VKIADFGLSRNIYSADYYKANENDAIPIRWMPPESIFYNRYTTESDVWAYGVVLWEIFSYGLQPYYGMAHEEVIYYVRDGNILSCPENCP | 2381 |
| 3D view using mol* of 15_AKAP13_MUSK | ||||||||||
| PDB file >>>TKFP_25_AKAP13_MUSK | ENST00000394518 | ENST00000189978 | AKAP13 | chr15 | 86228104 | MUSK | chr9 | 113509920 | MKLNPQQAPLYGDCVVTVLLAEEDKAEDDVVFYLVFLGSTLRHCTSTRKVSSDTLETIAPGHDCCETVKVQLCASKEGLPVFVVAEEDFH FVQDEAYDAAQFLATSAGNQQALNFTRFLDQSGPPSGDVNSLDKKLVLAFRHLKLPTEWNVLGTDQSLHDAGPRETLMHFAVRLGLLRLT WFLLQKPGGRGALSIHNQEGATPVSLALERGYHKLHQLLTEENAGEPDSWSSLSYEIPYGDCSVRHHRELDIYTLTSESDSHHEHPFPGD GCTGPIFKLMNIQQQLMKTNLKQMDSLMPLMMTAQDPSSAPETDGQFLPCAPEPTDPQRLSSSEETESTQCCPGSPVAQTESPCDLSSIV EEENTDRSCRKKNKGVERKGEEVEPAPIVDSGTVSDQDSCLQSLPDCGVKGTEGLSSCGNRNEETGTKSSGMPTDQESLSSGDAVLQRDL VMEPGTAQYSSGGELGGISTTNVSTPDTAGEMEHGLMNPDATVWKNVLQGGESTKERFENSNIGTAGASDVHVTSKPVDKISVPNCAPAA SSLDGNKPAESSLAFSNEETSTEKTAETETSRSREESADAPVDQNSVVIPAAAKDKISDGLEPYTLLAAGIGEAMSPSDLALLGLEEDVM PHQNSETNSSHAQSQKGKSSPICSTTGDDKLCADSACQQNTVTSSGDLVAKLCDNIVSESESTTARQPSSQDPPDASHCEDPQAHTVTSD PVRDTQERADFCPFKVVDNKGQRKDVKLDKPLTNMLEVVSHPHPVVPKMEKELVPDQAVISDSTFSLANSPGSESVTKDDALSFVPSQKE KGTATPELHTATDYRDGPDGNSNEPDTRPLEDRAVGLSTSSTAAELQHGMGNTSLTGLGGEHEGPAPPAIPEALNIKGNTDSSLQSVGKA TLALDSVLTEEGKLLVVSESSAAQEQDKDKAVTCSSIKENALSSGTLQEEQRTPPPGQDTQQFHEKSISADCAKDKALQLSNSPGASSAF LKAETEHNKEVAPQVSLLTQGGAAQSLVPPGASLATESRQEALGAEHNSSALLPCLLPDGSDGSDALNCSQPSPLDVGVKNTQSQGKTSA CEVSGDVTVDVTGVNALQGMAEPRRENISHNTQDILIPNVLLSQEKNAVLGLPVALQDKAVTDPQGVGTPEMIPLDWEKGKLEGADHSCT MGDAEEAQIDDEAHPVLLQPVAKELPTDMELSAHDDGAPAGVREVMRAPPSGRERSTPSLPCMVSAQDAPLPKGADLIEEAASRIVDAVI EQVKAAGALLTEGEACHMSLSSPELGPLTKGLESAFTEKVSTFPPGESLPMGSTPEEATGSLAGCFAGREEPEKIILPVQGPEPAAEMPD VKAEDEVDFRASSISEEVAVGSIAATLKMKQGPMTQAINRENWCTIEPCPDAASLLASKQSPECENFLDVGLGRECTSKQGVLKRESGSD SDLFHSPSDDMDSIIFPKPEEEHLACDITGSSSSTDDTASLDRHSSHGSDVSLSQILKPNRSRDRQSLDGFYSHGMGAEGRESESEPADP GDVEEEEMDSITEVPANCSVLRSSMRSLSPFRRHSWGPGKNAASDAEMNHRSSMRVLGDVVRRPPIHRRSFSLEGLTGGAGVGNKPSSSL EVSSANAEELRHPFSGEERVDSLVSLSEEDLESDQREHRMFDQQICHRSKQQGFNYCTSAISSPLTKSISLMTISHPGLDNSRPFHSTFH NTSANLTESITEENYNFLPHSPSKKDSEWKSGTKVSRTFSYIKNKMSSSKKSKVSSGSIQESVKDRVIDSRLQLFITKPGLYTCIATNKH GEKFSTAKAAATISIAAFSKPQKDNKGYCAQYRGEVCNAVLAKDALVFLNTSYADPEEAQELLVHTAWNELKVVSPVCRPAAEALLCNHI FQECSPGVVPTPIPICREYCLAVKELFCAKEWLVMEEKTHRGLYRSEMHLLSVPECSKLPSMHWDPTACARLPHLDYNKENLKTFPPMTS SKPSVDIPNLPSSSSSSFSVSPTYSMTVIISIMSSFAIFVLLTITTLYCCRRRKQWKNKKRESAAVTLTTLPSELLLDRLHPNPMYQRMP LLLNPKLLSLEYPRNNIEYVRDIGEGAFGRVFQARAPGLLPYEPFTMVAVKMLKEEASADMQADFQREAALMAEFDNPNIVKLLGVCAVG KPMCLLFEYMAYGDLNEFLRSMSPHTVCSLSHSDLSMRAQVSSPGPPPLSCAEQLCIARQVAAGMAYLSERKFVHRDLATRNCLVGENMV VKIADFGLSRNIYSADYYKANENDAIPIRWMPPESIFYNRYTTESDVWAYGVVLWEIFSYGLQPYYGMAHEEVIYYVRDGNILSCPENCP | 2381_AKAP13_MUSK |
| PDB file >>>TKFP_26_AKAP13_MUSK | ENST00000394518 | ENST00000189978 | AKAP13 | chr15 | 86228104 | MUSK | chr9 | 113509921 | MKLNPQQAPLYGDCVVTVLLAEEDKAEDDVVFYLVFLGSTLRHCTSTRKVSSDTLETIAPGHDCCETVKVQLCASKEGLPVFVVAEEDFH FVQDEAYDAAQFLATSAGNQQALNFTRFLDQSGPPSGDVNSLDKKLVLAFRHLKLPTEWNVLGTDQSLHDAGPRETLMHFAVRLGLLRLT WFLLQKPGGRGALSIHNQEGATPVSLALERGYHKLHQLLTEENAGEPDSWSSLSYEIPYGDCSVRHHRELDIYTLTSESDSHHEHPFPGD GCTGPIFKLMNIQQQLMKTNLKQMDSLMPLMMTAQDPSSAPETDGQFLPCAPEPTDPQRLSSSEETESTQCCPGSPVAQTESPCDLSSIV EEENTDRSCRKKNKGVERKGEEVEPAPIVDSGTVSDQDSCLQSLPDCGVKGTEGLSSCGNRNEETGTKSSGMPTDQESLSSGDAVLQRDL VMEPGTAQYSSGGELGGISTTNVSTPDTAGEMEHGLMNPDATVWKNVLQGGESTKERFENSNIGTAGASDVHVTSKPVDKISVPNCAPAA SSLDGNKPAESSLAFSNEETSTEKTAETETSRSREESADAPVDQNSVVIPAAAKDKISDGLEPYTLLAAGIGEAMSPSDLALLGLEEDVM PHQNSETNSSHAQSQKGKSSPICSTTGDDKLCADSACQQNTVTSSGDLVAKLCDNIVSESESTTARQPSSQDPPDASHCEDPQAHTVTSD PVRDTQERADFCPFKVVDNKGQRKDVKLDKPLTNMLEVVSHPHPVVPKMEKELVPDQAVISDSTFSLANSPGSESVTKDDALSFVPSQKE KGTATPELHTATDYRDGPDGNSNEPDTRPLEDRAVGLSTSSTAAELQHGMGNTSLTGLGGEHEGPAPPAIPEALNIKGNTDSSLQSVGKA TLALDSVLTEEGKLLVVSESSAAQEQDKDKAVTCSSIKENALSSGTLQEEQRTPPPGQDTQQFHEKSISADCAKDKALQLSNSPGASSAF LKAETEHNKEVAPQVSLLTQGGAAQSLVPPGASLATESRQEALGAEHNSSALLPCLLPDGSDGSDALNCSQPSPLDVGVKNTQSQGKTSA CEVSGDVTVDVTGVNALQGMAEPRRENISHNTQDILIPNVLLSQEKNAVLGLPVALQDKAVTDPQGVGTPEMIPLDWEKGKLEGADHSCT MGDAEEAQIDDEAHPVLLQPVAKELPTDMELSAHDDGAPAGVREVMRAPPSGRERSTPSLPCMVSAQDAPLPKGADLIEEAASRIVDAVI EQVKAAGALLTEGEACHMSLSSPELGPLTKGLESAFTEKVSTFPPGESLPMGSTPEEATGSLAGCFAGREEPEKIILPVQGPEPAAEMPD VKAEDEVDFRASSISEEVAVGSIAATLKMKQGPMTQAINRENWCTIEPCPDAASLLASKQSPECENFLDVGLGRECTSKQGVLKRESGSD SDLFHSPSDDMDSIIFPKPEEEHLACDITGSSSSTDDTASLDRHSSHGSDVSLSQILKPNRSRDRQSLDGFYSHGMGAEGRESESEPADP GDVEEEEMDSITEVPANCSVLRSSMRSLSPFRRHSWGPGKNAASDAEMNHRSSMRVLGDVVRRPPIHRRSFSLEGLTGGAGVGNKPSSSL EVSSANAEELRHPFSGEERVDSLVSLSEEDLESDQREHRMFDQQICHRSKQQGFNYCTSAISSPLTKSISLMTISHPGLDNSRPFHSTFH NTSANLTESITEENYNFLPHSPSKKDSEWKSGTKVSRTFSYIKNKMSSSKKSKVSSGSIQESVKDRVIDSRLQLFITKPGLYTCIATNKH GEKFSTAKAAATISIAAFSKPQKDNKGYCAQYRGEVCNAVLAKDALVFLNTSYADPEEAQELLVHTAWNELKVVSPVCRPAAEALLCNHI FQECSPGVVPTPIPICREYCLAVKELFCAKEWLVMEEKTHRGLYRSEMHLLSVPECSKLPSMHWDPTACARLPHLDYNKENLKTFPPMTS SKPSVDIPNLPSSSSSSFSVSPTYSMTVIISIMSSFAIFVLLTITTLYCCRRRKQWKNKKRESAAVTLTTLPSELLLDRLHPNPMYQRMP LLLNPKLLSLEYPRNNIEYVRDIGEGAFGRVFQARAPGLLPYEPFTMVAVKMLKEEASADMQADFQREAALMAEFDNPNIVKLLGVCAVG KPMCLLFEYMAYGDLNEFLRSMSPHTVCSLSHSDLSMRAQVSSPGPPPLSCAEQLCIARQVAAGMAYLSERKFVHRDLATRNCLVGENMV VKIADFGLSRNIYSADYYKANENDAIPIRWMPPESIFYNRYTTESDVWAYGVVLWEIFSYGLQPYYGMAHEEVIYYVRDGNILSCPENCP | 2381_AKAP13_MUSK |
Top |
Comparison of Fusion Protein Isoforms |
 Superimpose the 3D Structures Among All Fusion Protein Isoforms Superimpose the 3D Structures Among All Fusion Protein Isoforms * Download the pdb file and open it from the molstar online viewer. |
 Comparison of the Secondary Structures of Fusion Protein Isoforms Comparison of the Secondary Structures of Fusion Protein Isoforms |
Top |
Comparison of Fusion Protein Sequences/Structures with Known Sequences/Structures from PDB |
Top |
pLDDT score distribution |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. * The blue color at the bottom marks the best active site residues. |
| 15_AKAP13_MUSK.png |
 |
| 15_AKAP13_MUSK.png |
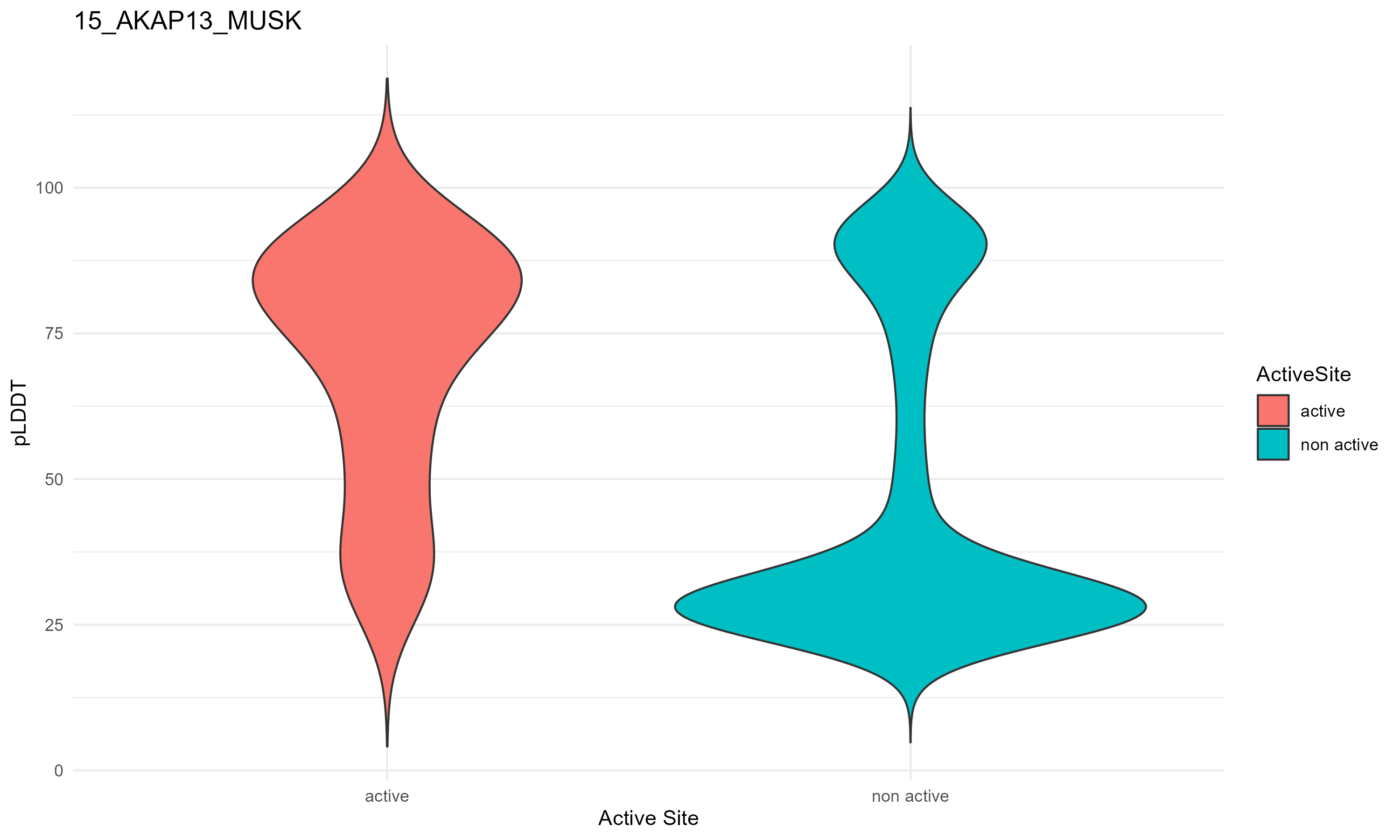 |
Top |
Potential Active Site Information |
 The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. |
| Kinase Fusion AA seq ID in KinaseFusionDB | Site score | Size | Dscore | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
Top |
Ramachandran Plot of Kinase Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| 15_AKAP13_MUSK_ramachandran.png |
 |
Top |
Virtual Screening Results |
 Distribution of the average docking score across all approved kinase inhibitors. Distribution of the average docking score across all approved kinase inhibitors.Distribution of the number of occurrence across all approved kinase inhibitors. |
| 5'-kinase fusion protein case |
| 3'-kinase fusion protein case |
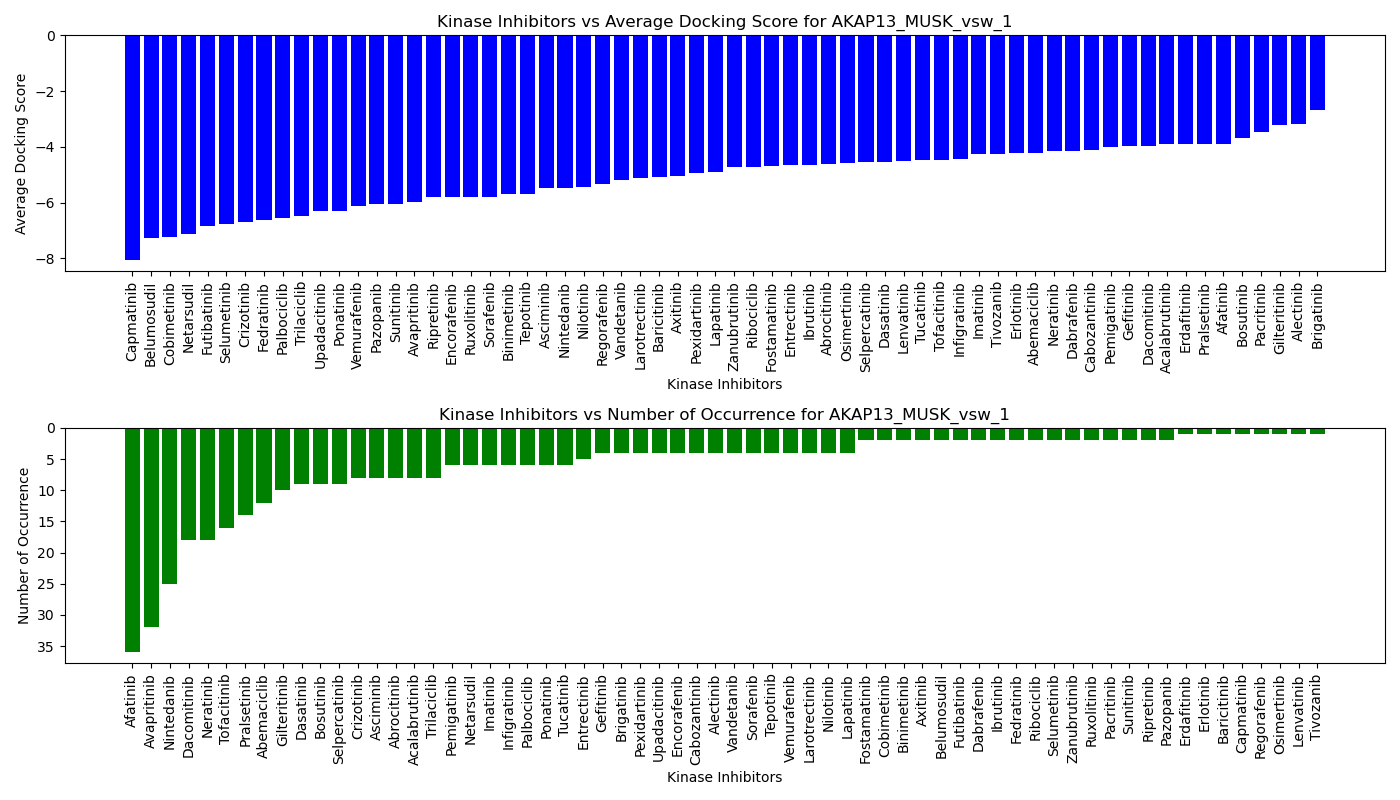 |
Top |
 Drug information from DrugBank of the top 20 interacting small molecules. Drug information from DrugBank of the top 20 interacting small molecules.* The detailed information of individual kinase inhibitors are available in the download page. |
| Fusion gene name info | Drug | Docking score | Glide g score | Glide energy |
| 15_AKAP13_MUSK_vsw_1-DOCK_HTVS_1-001 | Netarsudil | -8.73225 | -8.74335 | -54.9457 |
| 15_AKAP13_MUSK_vsw_1-DOCK_HTVS_1-001 | Netarsudil | -8.73225 | -8.74335 | -54.9457 |
| 15_AKAP13_MUSK_vsw_1-DOCK_HTVS_1-001 | Capmatinib | -8.04547 | -8.05107 | -46.534 |
| 15_AKAP13_MUSK_vsw_1-DOCK_HTVS_1-001 | Futibatinib | -7.882339999999999 | -7.882339999999999 | -38.6326 |
| 15_AKAP13_MUSK_vsw_1-DOCK_HTVS_1-001 | Netarsudil | -7.774769999999999 | -7.78587 | -48.2866 |
| 15_AKAP13_MUSK_vsw_1-DOCK_HTVS_1-001 | Netarsudil | -7.774769999999999 | -7.78587 | -48.2866 |
| 15_AKAP13_MUSK_vsw_1-DOCK_HTVS_1-001 | Lapatinib | -7.65195 | -7.74075 | -42.5148 |
| 15_AKAP13_MUSK_vsw_1-DOCK_HTVS_1-001 | Cobimetinib | -7.548369999999999 | -7.55117 | -49.8308 |
| 15_AKAP13_MUSK_vsw_1-DOCK_HTVS_1-001 | Selpercatinib | -7.37475 | -7.40525 | -53.3699 |
| 15_AKAP13_MUSK_vsw_1-DOCK_HTVS_1-001 | Belumosudil | -7.31888 | -7.32658 | -45.6389 |
| 15_AKAP13_MUSK_vsw_1-DOCK_HTVS_1-001 | Palbociclib | -7.23032 | -7.69982 | -48.2918 |
| 15_AKAP13_MUSK_vsw_1-DOCK_HTVS_1-001 | Palbociclib | -7.23032 | -7.69982 | -48.2918 |
| 15_AKAP13_MUSK_vsw_1-DOCK_HTVS_1-001 | Palbociclib | -7.23032 | -7.69982 | -48.2918 |
| 15_AKAP13_MUSK_vsw_1-DOCK_HTVS_1-001 | Ruxolitinib | -7.22553 | -7.22553 | -31.2441 |
| 15_AKAP13_MUSK_vsw_1-DOCK_HTVS_1-001 | Belumosudil | -7.182460000000001 | -7.1901600000000006 | -44.9358 |
| 15_AKAP13_MUSK_vsw_1-DOCK_HTVS_1-001 | Avapritinib | -7.163989999999999 | -7.9614899999999995 | -53.864 |
| 15_AKAP13_MUSK_vsw_1-DOCK_HTVS_1-001 | Avapritinib | -7.163989999999999 | -7.9614899999999995 | -53.864 |
| 15_AKAP13_MUSK_vsw_1-DOCK_HTVS_1-001 | Avapritinib | -7.163989999999999 | -7.9614899999999995 | -53.864 |
| 15_AKAP13_MUSK_vsw_1-DOCK_HTVS_1-001 | Avapritinib | -7.163989999999999 | -7.9614899999999995 | -53.864 |
| 15_AKAP13_MUSK_vsw_1-DOCK_HTVS_1-001 | Tepotinib | -7.12648 | -7.12758 | -45.8429 |
Top |
Kinase-Substrate Information of AKAP13_MUSK |
 Phosphorylation target of the kinase Phosphorylation target of the kinase(phosphosite, 03-17-2024) |
| Kinase | Kinase UniProt Acc | Kinase species | Substrate | Substrate UniProt Acc | Substrate phosphorylated residues | Substrate phosphorylated sites (+/-7AA) | Domain |
 Biological Network Integration of This Kinase and Substrates Biological Network Integration of This Kinase and Substrates (GeneMANIA website) |
 Enriched GO biological processes of the phosphorylation target genes of the kinase Enriched GO biological processes of the phosphorylation target genes of the kinase |
| Kinase | GOID | GO term | P.adjust |
Top |
Related Drugs to AKAP13_MUSK |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
 Distribution of the number of studies mentioning AKAP13-MUSK and kinase inhibitors the PubMed Abstract (04-01-2024) Distribution of the number of studies mentioning AKAP13-MUSK and kinase inhibitors the PubMed Abstract (04-01-2024) |
| Fusion gene - drug pair 1 | Fusion gene - drug pair 2 | PMID | Publication date | DOI | Study title |
Top |
Related Diseases to AKAP13_MUSK |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Related diseases from the literature mentioned this fusion gene and drug. Related diseases from the literature mentioned this fusion gene and drug. (PubMed, 04-01-2024) |
| MeSH ID | MeSH term |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
Top |
Clinical Trials of the Found Drugs/Small Molecules |
 Statistics of the Clinical Trials of the Found Kinase Inibitors from clinicaltrials.gov (06-17-2024) Statistics of the Clinical Trials of the Found Kinase Inibitors from clinicaltrials.gov (06-17-2024) |
 Clinical Trials from clinicaltrials.gov (06-17-2024) Clinical Trials from clinicaltrials.gov (06-17-2024) |
| Fusion Gene | Kinase Inhibitor | NCT ID | Study Status | Phases | Disease | # Enrolment | Date |

