| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Kinase Fusion Gene:AKAP9_BRAF |
Kinase Fusion Protein Summary |
 Kinase Fusion gene summary Kinase Fusion gene summary |
| Kinase Fusion partner gene information | Kinase Fusion gene name: AKAP9_BRAF | KinaseFusionDB ID: KFG255 | FusionGDB2.0 ID: KFG255 | Hgene | Tgene | Gene symbol | AKAP9 | BRAF | Gene ID | 10142 | 673 | |
| Gene name | A-kinase anchoring protein 9 | B-Raf proto-oncogene, serine/threonine kinase | ||||||||||
| Synonyms | AKAP-9|AKAP350|AKAP450|CG-NAP|HYPERION|LQT11|MU-RMS-40.16A|PPP1R45|PRKA9|YOTIAO | B-RAF1|B-raf|BRAF-1|BRAF1|NS7|RAFB1 | ||||||||||
| Cytomap | 7q21.2 | 7q34 | ||||||||||
| Type of gene | protein-coding | protein-coding | ||||||||||
| Description | A-kinase anchor protein 9A kinase (PRKA) anchor protein (yotiao) 9A kinase (PRKA) anchor protein 9A-kinase anchor protein 350 kDaA-kinase anchor protein 450 kDaAKAP 120-like proteinAKAP9-BRAF fusion proteincentrosome- and Golgi-localized PKN-associ | serine/threonine-protein kinase B-raf94 kDa B-raf proteinB-Raf proto-oncogene serine/threonine-protein kinase (p94)B-Raf serine/threonine-proteinmurine sarcoma viral (v-raf) oncogene homolog B1proto-oncogene B-Rafv-raf murine sarcoma viral oncogene | ||||||||||
| Modification date | 20240407 | 20240416 | ||||||||||
| UniProtAcc | Q99996 | P15056 | ||||||||||
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000356239, ENST00000358100, ENST00000359028, ENST00000394564, ENST00000491695, | ENST00000288602, | |||||||||
| Context (manual curation of fusion genes in KinaseFusionDB) | PubMed: AKAP9 [Title/Abstract] AND BRAF [Title/Abstract] AND fusion [Title/Abstract] | |||||||||||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | AKAP9(91632550)-BRAF(140434354), # samples:1 AKAP9(91632550)-BRAF(140487386), # samples:1 AKAP9(91739875)-BRAF(140513187), # samples:1 | |||||||||||
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | AKAP9 | GO:0031503 | protein-containing complex localization | 19218243 |
| Hgene | AKAP9 | GO:1903358 | regulation of Golgi organization | 27666745 |
| Tgene | BRAF | GO:0000165 | MAPK cascade | 18567582|29433126 |
| Tgene | BRAF | GO:0006468 | protein phosphorylation | 17563371 |
| Tgene | BRAF | GO:0007173 | epidermal growth factor receptor signaling pathway | 18567582 |
| Tgene | BRAF | GO:0010828 | positive regulation of glucose transmembrane transport | 23010278 |
| Tgene | BRAF | GO:0033138 | positive regulation of peptidyl-serine phosphorylation | 19667065 |
| Tgene | BRAF | GO:0043066 | negative regulation of apoptotic process | 19667065 |
| Tgene | BRAF | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 22065586 |
| Tgene | BRAF | GO:0071277 | cellular response to calcium ion | 18567582 |
| Tgene | BRAF | GO:0090150 | establishment of protein localization to membrane | 23010278 |
 Kinase Fusion gene breakpoints across AKAP9 (5'-gene) Kinase Fusion gene breakpoints across AKAP9 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
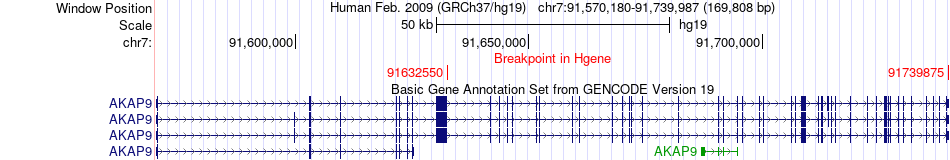 |
 Kinase Fusion gene breakpoints across BRAF (3'-gene) Kinase Fusion gene breakpoints across BRAF (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
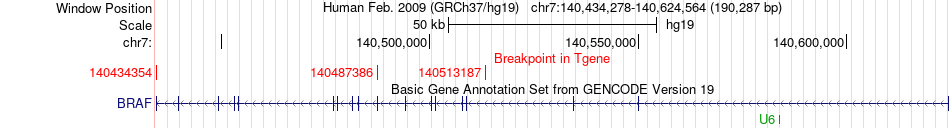 |
Top |
Kinase Fusion Gene Sample Information |
 Kinase Fusion gene information. Kinase Fusion gene information. |
 Kinase Fusion gene information from four resources (ChiTars 5.0, ChimerDB 4.0, COSMIC, and CCLE) Kinase Fusion gene information from four resources (ChiTars 5.0, ChimerDB 4.0, COSMIC, and CCLE)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Sample | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp |
| ChiTaRS5.0 | FN170090 | AKAP9 | chr7 | 91739875 | BRAF | chr7 | 140513187 |
| ChimerDB4 | AY803272 | AKAP9 | chr7 | 91632550 | BRAF | chr7 | 140434354 |
| ChimerKB3 | . | AKAP9 | chr7 | 91632549 | BRAF | chr7 | 140487384 |
| ChiTaRS5.0 | AY803272 | AKAP9 | chr7 | 91632550 | BRAF | chr7 | 140487386 |
| ChimerKB3 | . | AKAP9 | chr7 | 91470486 | BRAF | chr7 | 140080822 |
| COSMIC | 900219 | AKAP9 | chr7 | 91632913 | BRAF | chr7 | 140489178 |
Top |
Kinase Fusion ORF Analysis |
 Kinase Fusion information from ORFfinder translation from full-length transcript sequence from KinaseFusionDB. Kinase Fusion information from ORFfinder translation from full-length transcript sequence from KinaseFusionDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp | Seq length (transcript) | Seq length (amino acids) |
| ENST00000356239 | ENST00000288602 | AKAP9 | chr7 | 91632550 | BRAF | chr7 | 140487386 | 4830 | 1502 |
| ENST00000356239 | ENST00000288602 | AKAP9 | chr7 | 91632549 | BRAF | chr7 | 140487384 | 4830 | 1502 |
Top |
Kinase Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from KinaseFusionDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from KinaseFusionDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >Henst_Tenst_Hgene_Hchr_Hbp_Tgene_Tchr_Tbp_length(fusion AA)_AAseq >ENST00000356239_ENST00000288602_AKAP9_chr7_91632550_BRAF_chr7_140487386_length(amino acids)=1502 MFSPAFLAEAMEDEERQKKLEAGKAKLAQFRQRKAQSDGQSPSKKQKKKRKTSSSKHDVSAHHDLNIDQSQCNEMYINSSQRVESTVIPE STIMRTLHSGEITSHEQGFSVELESEISTTADDCSSEVNGCSFVMRTGKPTNLLREEEFGVDDSYSEQGAQDSPTHLEMMESELAGKQHE IEELNRELEEMRVTYGTEGLQQLQEFEAAIKQRDGIITQLTANLQQARREKDETMREFLELTEQSQKLQIQFQQLQASETLRNSTHSSTA ADLLQAKQQILTHQQQLEEQDHLLEDYQKKKEDFTMQISFLQEKIKVYEMEQDKKVENSNKEEIQEKETIIEELNTKIIEEEKKTLELKD KLTTADKLLGELQEQIVQKNQEIKNMKLELTNSKQKERQSSEEIKQLMGTVEELQKRNHKDSQFETDIVQRMEQETQRKLEQLRAELDEM YGQQIVQMKQELIRQHMAQMEEMKTRHKGEMENALRSYSNITVNEDQIKLMNVAINELNIKLQDTNSQKEKLKEELGLILEEKCALQRQL EDLVEELSFSREQIQRARQTIAEQESKLNEAHKSLSTVEDLKAEIVSASESRKELELKHEAEVTNYKIKLEMLEKEKNAVLDRMAESQEA ELERLRTQLLFSHEEELSKLKEDLEIEHRINIEKLKDNLGIHYKQQIDGLQNEMSQKIETMQFEKDNLITKQNQLILEISKLKDLQQSLV NSKSEEMTLQINELQKEIEILRQEEKEKGTLEQEVQELQLKTELLEKQMKEKENDLQEKFAQLEAENSILKDEKKTLEDMLKIHTPVSQE ERLIFLDSIKSKSKDSVWEKEIEILIEENEDLKQQCIQLNEEIEKQRNTFSFAEKNFEVNYQELQEEYACLLKVKDDLEDSKNKQELEYK SKLKALNEELHLQRINPTTVKMKSSVFDEDKTFVAETLEMGEVVEKDTTELMEKLEVTKREKLELSQRLSDLSEQLKQKHGEISFLNEEV KSLKQEKEQVSLRCRELEIIINHNRAENVQSCDTQVSSLLDGVVTMTSRGAEGSVSKVNKSFGEESKIMVEDKVSFENMTVGEESKQEQL ILDHLPSVTKESSLRATQPSENDKLQKELNVLKSEQDLIRDQGFRGDGGSTTGLSATPPASLPGSLTNVKALQKSPGPQRERKSSSSSED RNRMKTLGRRDSSDDWEIPDGQITVGQRIGSGSFGTVYKGKWHGDVAVKMLNVTAPTPQQLQAFKNEVGVLRKTRHVNILLFMGYSTKPQ LAIVTQWCEGSSLYHHLHIIETKFEMIKLIDIARQTAQGMDYLHAKSIIHRDLKSNNIFLHEDLTVKIGDFGLATVKSRWSGSHQFEQLS GSILWMAPEVIRMQDKNPYSFQSDVYAFGIVLYELMTGQLPYSNINNRDQIIFMVGRGYLSPDLSKVRSNCPKAMKRLMAECLKKKRDER -------------------------------------------------------------- >ENST00000356239_ENST00000288602_AKAP9_chr7_91632549_BRAF_chr7_140487384_length(amino acids)=1502 MFSPAFLAEAMEDEERQKKLEAGKAKLAQFRQRKAQSDGQSPSKKQKKKRKTSSSKHDVSAHHDLNIDQSQCNEMYINSSQRVESTVIPE STIMRTLHSGEITSHEQGFSVELESEISTTADDCSSEVNGCSFVMRTGKPTNLLREEEFGVDDSYSEQGAQDSPTHLEMMESELAGKQHE IEELNRELEEMRVTYGTEGLQQLQEFEAAIKQRDGIITQLTANLQQARREKDETMREFLELTEQSQKLQIQFQQLQASETLRNSTHSSTA ADLLQAKQQILTHQQQLEEQDHLLEDYQKKKEDFTMQISFLQEKIKVYEMEQDKKVENSNKEEIQEKETIIEELNTKIIEEEKKTLELKD KLTTADKLLGELQEQIVQKNQEIKNMKLELTNSKQKERQSSEEIKQLMGTVEELQKRNHKDSQFETDIVQRMEQETQRKLEQLRAELDEM YGQQIVQMKQELIRQHMAQMEEMKTRHKGEMENALRSYSNITVNEDQIKLMNVAINELNIKLQDTNSQKEKLKEELGLILEEKCALQRQL EDLVEELSFSREQIQRARQTIAEQESKLNEAHKSLSTVEDLKAEIVSASESRKELELKHEAEVTNYKIKLEMLEKEKNAVLDRMAESQEA ELERLRTQLLFSHEEELSKLKEDLEIEHRINIEKLKDNLGIHYKQQIDGLQNEMSQKIETMQFEKDNLITKQNQLILEISKLKDLQQSLV NSKSEEMTLQINELQKEIEILRQEEKEKGTLEQEVQELQLKTELLEKQMKEKENDLQEKFAQLEAENSILKDEKKTLEDMLKIHTPVSQE ERLIFLDSIKSKSKDSVWEKEIEILIEENEDLKQQCIQLNEEIEKQRNTFSFAEKNFEVNYQELQEEYACLLKVKDDLEDSKNKQELEYK SKLKALNEELHLQRINPTTVKMKSSVFDEDKTFVAETLEMGEVVEKDTTELMEKLEVTKREKLELSQRLSDLSEQLKQKHGEISFLNEEV KSLKQEKEQVSLRCRELEIIINHNRAENVQSCDTQVSSLLDGVVTMTSRGAEGSVSKVNKSFGEESKIMVEDKVSFENMTVGEESKQEQL ILDHLPSVTKESSLRATQPSENDKLQKELNVLKSEQDLIRDQGFRGDGGSTTGLSATPPASLPGSLTNVKALQKSPGPQRERKSSSSSED RNRMKTLGRRDSSDDWEIPDGQITVGQRIGSGSFGTVYKGKWHGDVAVKMLNVTAPTPQQLQAFKNEVGVLRKTRHVNILLFMGYSTKPQ LAIVTQWCEGSSLYHHLHIIETKFEMIKLIDIARQTAQGMDYLHAKSIIHRDLKSNNIFLHEDLTVKIGDFGLATVKSRWSGSHQFEQLS GSILWMAPEVIRMQDKNPYSFQSDVYAFGIVLYELMTGQLPYSNINNRDQIIFMVGRGYLSPDLSKVRSNCPKAMKRLMAECLKKKRDER -------------------------------------------------------------- |
Multiple Sequence Alignment of All Fusion Protein Isoforms |
Top |
Kinase Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr7:91632550/chr7:140434354) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| AKAP9 | BRAF |
| FUNCTION: Scaffolding protein that assembles several protein kinases and phosphatases on the centrosome and Golgi apparatus. Required to maintain the integrity of the Golgi apparatus (PubMed:10202149, PubMed:15047863). Required for microtubule nucleation at the cis-side of the Golgi apparatus (PubMed:15047863, PubMed:19242490). Required for association of the centrosomes with the poles of the bipolar mitotic spindle during metaphase (PubMed:25657325). In complex with PDE4DIP isoform 13/MMG8/SMYLE, recruits CAMSAP2 to the Golgi apparatus and tethers non-centrosomal minus-end microtubules to the Golgi, an important step for polarized cell movement (PubMed:27666745, PubMed:28814570). In complex with PDE4DIP isoform 13/MMG8/SMYLE, EB1/MAPRE1 and CDK5RAP2, contributes to microtubules nucleation and extension also from the centrosome to the cell periphery (PubMed:29162697). {ECO:0000269|PubMed:10202149, ECO:0000269|PubMed:15047863, ECO:0000269|PubMed:19242490, ECO:0000269|PubMed:25657325, ECO:0000269|PubMed:27666745, ECO:0000269|PubMed:28814570, ECO:0000269|PubMed:29162697}.; FUNCTION: [Isoform 4]: Associated with the N-methyl-D-aspartate receptor and is specifically found in the neuromuscular junction (NMJ) as well as in neuronal synapses, suggesting a role in the organization of postsynaptic specializations. {ECO:0000269|PubMed:9482789}. | FUNCTION: Protein kinase involved in the transduction of mitogenic signals from the cell membrane to the nucleus (Probable). Phosphorylates MAP2K1, and thereby activates the MAP kinase signal transduction pathway (PubMed:21441910, PubMed:29433126). Phosphorylates PFKFB2 (PubMed:36402789). May play a role in the postsynaptic responses of hippocampal neurons (PubMed:1508179). {ECO:0000269|PubMed:1508179, ECO:0000269|PubMed:21441910, ECO:0000269|PubMed:29433126, ECO:0000269|PubMed:36402789, ECO:0000305}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
 - Retained domain in the 5'-partner of fusion protein (protein functional feature from UniProt). - Retained domain in the 5'-partner of fusion protein (protein functional feature from UniProt). |
| Partner | Hgeneene | Hbp | Tgeneene | Tbp | ENST | BPexon | TotalExon | Protein feature loci | BPloci | TotalLen | Feature | Note |
 - Retained domain in the 3'-partner of fusion protein (protein functional feature from UniProt). - Retained domain in the 3'-partner of fusion protein (protein functional feature from UniProt). |
| Partner | Hgeneene | Hbp | Tgeneene | Tbp | ENST | BPexon | TotalExon | Protein feature loci | BPloci | TotalLen | Feature | Note |
| Tgene | AKAP9 | 91632549 | BRAF | 140487384 | ENST00000356239 | 7 | 18 | 457_717 | 380 | 767 | Domain | Note=Protein kinase;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00159 |
| Tgene | AKAP9 | 91632550 | BRAF | 140487386 | ENST00000356239 | 7 | 18 | 457_717 | 380 | 767 | Domain | Note=Protein kinase;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00159 |
Top |
Kinase Fusion Protein Structures |
 CIF files of the predicted kinase fusion proteins CIF files of the predicted kinase fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Kinase Fusion protein CIF link (fusion AA seq ID in KinaseFusionDB) | Henst | Tenst | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp | AA seq | Len(AA seq) |
| PDB file >>>19_AKAP9_BRAF | ENST00000356239 | ENST00000288602 | AKAP9 | chr7 | 91632549 | BRAF | chr7 | 140487384 | MFSPAFLAEAMEDEERQKKLEAGKAKLAQFRQRKAQSDGQSPSKKQKKKRKTSSSKHDVSAHHDLNIDQSQCNEMYINSSQRVESTVIPE STIMRTLHSGEITSHEQGFSVELESEISTTADDCSSEVNGCSFVMRTGKPTNLLREEEFGVDDSYSEQGAQDSPTHLEMMESELAGKQHE IEELNRELEEMRVTYGTEGLQQLQEFEAAIKQRDGIITQLTANLQQARREKDETMREFLELTEQSQKLQIQFQQLQASETLRNSTHSSTA ADLLQAKQQILTHQQQLEEQDHLLEDYQKKKEDFTMQISFLQEKIKVYEMEQDKKVENSNKEEIQEKETIIEELNTKIIEEEKKTLELKD KLTTADKLLGELQEQIVQKNQEIKNMKLELTNSKQKERQSSEEIKQLMGTVEELQKRNHKDSQFETDIVQRMEQETQRKLEQLRAELDEM YGQQIVQMKQELIRQHMAQMEEMKTRHKGEMENALRSYSNITVNEDQIKLMNVAINELNIKLQDTNSQKEKLKEELGLILEEKCALQRQL EDLVEELSFSREQIQRARQTIAEQESKLNEAHKSLSTVEDLKAEIVSASESRKELELKHEAEVTNYKIKLEMLEKEKNAVLDRMAESQEA ELERLRTQLLFSHEEELSKLKEDLEIEHRINIEKLKDNLGIHYKQQIDGLQNEMSQKIETMQFEKDNLITKQNQLILEISKLKDLQQSLV NSKSEEMTLQINELQKEIEILRQEEKEKGTLEQEVQELQLKTELLEKQMKEKENDLQEKFAQLEAENSILKDEKKTLEDMLKIHTPVSQE ERLIFLDSIKSKSKDSVWEKEIEILIEENEDLKQQCIQLNEEIEKQRNTFSFAEKNFEVNYQELQEEYACLLKVKDDLEDSKNKQELEYK SKLKALNEELHLQRINPTTVKMKSSVFDEDKTFVAETLEMGEVVEKDTTELMEKLEVTKREKLELSQRLSDLSEQLKQKHGEISFLNEEV KSLKQEKEQVSLRCRELEIIINHNRAENVQSCDTQVSSLLDGVVTMTSRGAEGSVSKVNKSFGEESKIMVEDKVSFENMTVGEESKQEQL ILDHLPSVTKESSLRATQPSENDKLQKELNVLKSEQDLIRDQGFRGDGGSTTGLSATPPASLPGSLTNVKALQKSPGPQRERKSSSSSED RNRMKTLGRRDSSDDWEIPDGQITVGQRIGSGSFGTVYKGKWHGDVAVKMLNVTAPTPQQLQAFKNEVGVLRKTRHVNILLFMGYSTKPQ LAIVTQWCEGSSLYHHLHIIETKFEMIKLIDIARQTAQGMDYLHAKSIIHRDLKSNNIFLHEDLTVKIGDFGLATVKSRWSGSHQFEQLS GSILWMAPEVIRMQDKNPYSFQSDVYAFGIVLYELMTGQLPYSNINNRDQIIFMVGRGYLSPDLSKVRSNCPKAMKRLMAECLKKKRDER | 1502 |
| 3D view using mol* of 19_AKAP9_BRAF | ||||||||||
| PDB file >>>TKFP_32_AKAP9_BRAF | ENST00000356239 | ENST00000288602 | AKAP9 | chr7 | 91632550 | BRAF | chr7 | 140487386 | MFSPAFLAEAMEDEERQKKLEAGKAKLAQFRQRKAQSDGQSPSKKQKKKRKTSSSKHDVSAHHDLNIDQSQCNEMYINSSQRVESTVIPE STIMRTLHSGEITSHEQGFSVELESEISTTADDCSSEVNGCSFVMRTGKPTNLLREEEFGVDDSYSEQGAQDSPTHLEMMESELAGKQHE IEELNRELEEMRVTYGTEGLQQLQEFEAAIKQRDGIITQLTANLQQARREKDETMREFLELTEQSQKLQIQFQQLQASETLRNSTHSSTA ADLLQAKQQILTHQQQLEEQDHLLEDYQKKKEDFTMQISFLQEKIKVYEMEQDKKVENSNKEEIQEKETIIEELNTKIIEEEKKTLELKD KLTTADKLLGELQEQIVQKNQEIKNMKLELTNSKQKERQSSEEIKQLMGTVEELQKRNHKDSQFETDIVQRMEQETQRKLEQLRAELDEM YGQQIVQMKQELIRQHMAQMEEMKTRHKGEMENALRSYSNITVNEDQIKLMNVAINELNIKLQDTNSQKEKLKEELGLILEEKCALQRQL EDLVEELSFSREQIQRARQTIAEQESKLNEAHKSLSTVEDLKAEIVSASESRKELELKHEAEVTNYKIKLEMLEKEKNAVLDRMAESQEA ELERLRTQLLFSHEEELSKLKEDLEIEHRINIEKLKDNLGIHYKQQIDGLQNEMSQKIETMQFEKDNLITKQNQLILEISKLKDLQQSLV NSKSEEMTLQINELQKEIEILRQEEKEKGTLEQEVQELQLKTELLEKQMKEKENDLQEKFAQLEAENSILKDEKKTLEDMLKIHTPVSQE ERLIFLDSIKSKSKDSVWEKEIEILIEENEDLKQQCIQLNEEIEKQRNTFSFAEKNFEVNYQELQEEYACLLKVKDDLEDSKNKQELEYK SKLKALNEELHLQRINPTTVKMKSSVFDEDKTFVAETLEMGEVVEKDTTELMEKLEVTKREKLELSQRLSDLSEQLKQKHGEISFLNEEV KSLKQEKEQVSLRCRELEIIINHNRAENVQSCDTQVSSLLDGVVTMTSRGAEGSVSKVNKSFGEESKIMVEDKVSFENMTVGEESKQEQL ILDHLPSVTKESSLRATQPSENDKLQKELNVLKSEQDLIRDQGFRGDGGSTTGLSATPPASLPGSLTNVKALQKSPGPQRERKSSSSSED RNRMKTLGRRDSSDDWEIPDGQITVGQRIGSGSFGTVYKGKWHGDVAVKMLNVTAPTPQQLQAFKNEVGVLRKTRHVNILLFMGYSTKPQ LAIVTQWCEGSSLYHHLHIIETKFEMIKLIDIARQTAQGMDYLHAKSIIHRDLKSNNIFLHEDLTVKIGDFGLATVKSRWSGSHQFEQLS GSILWMAPEVIRMQDKNPYSFQSDVYAFGIVLYELMTGQLPYSNINNRDQIIFMVGRGYLSPDLSKVRSNCPKAMKRLMAECLKKKRDER | 1502_AKAP9_BRAF |
| PDB file >>>TKFP_33_AKAP9_BRAF | ENST00000356239 | ENST00000288602 | AKAP9 | chr7 | 91632549 | BRAF | chr7 | 140487384 | MFSPAFLAEAMEDEERQKKLEAGKAKLAQFRQRKAQSDGQSPSKKQKKKRKTSSSKHDVSAHHDLNIDQSQCNEMYINSSQRVESTVIPE STIMRTLHSGEITSHEQGFSVELESEISTTADDCSSEVNGCSFVMRTGKPTNLLREEEFGVDDSYSEQGAQDSPTHLEMMESELAGKQHE IEELNRELEEMRVTYGTEGLQQLQEFEAAIKQRDGIITQLTANLQQARREKDETMREFLELTEQSQKLQIQFQQLQASETLRNSTHSSTA ADLLQAKQQILTHQQQLEEQDHLLEDYQKKKEDFTMQISFLQEKIKVYEMEQDKKVENSNKEEIQEKETIIEELNTKIIEEEKKTLELKD KLTTADKLLGELQEQIVQKNQEIKNMKLELTNSKQKERQSSEEIKQLMGTVEELQKRNHKDSQFETDIVQRMEQETQRKLEQLRAELDEM YGQQIVQMKQELIRQHMAQMEEMKTRHKGEMENALRSYSNITVNEDQIKLMNVAINELNIKLQDTNSQKEKLKEELGLILEEKCALQRQL EDLVEELSFSREQIQRARQTIAEQESKLNEAHKSLSTVEDLKAEIVSASESRKELELKHEAEVTNYKIKLEMLEKEKNAVLDRMAESQEA ELERLRTQLLFSHEEELSKLKEDLEIEHRINIEKLKDNLGIHYKQQIDGLQNEMSQKIETMQFEKDNLITKQNQLILEISKLKDLQQSLV NSKSEEMTLQINELQKEIEILRQEEKEKGTLEQEVQELQLKTELLEKQMKEKENDLQEKFAQLEAENSILKDEKKTLEDMLKIHTPVSQE ERLIFLDSIKSKSKDSVWEKEIEILIEENEDLKQQCIQLNEEIEKQRNTFSFAEKNFEVNYQELQEEYACLLKVKDDLEDSKNKQELEYK SKLKALNEELHLQRINPTTVKMKSSVFDEDKTFVAETLEMGEVVEKDTTELMEKLEVTKREKLELSQRLSDLSEQLKQKHGEISFLNEEV KSLKQEKEQVSLRCRELEIIINHNRAENVQSCDTQVSSLLDGVVTMTSRGAEGSVSKVNKSFGEESKIMVEDKVSFENMTVGEESKQEQL ILDHLPSVTKESSLRATQPSENDKLQKELNVLKSEQDLIRDQGFRGDGGSTTGLSATPPASLPGSLTNVKALQKSPGPQRERKSSSSSED RNRMKTLGRRDSSDDWEIPDGQITVGQRIGSGSFGTVYKGKWHGDVAVKMLNVTAPTPQQLQAFKNEVGVLRKTRHVNILLFMGYSTKPQ LAIVTQWCEGSSLYHHLHIIETKFEMIKLIDIARQTAQGMDYLHAKSIIHRDLKSNNIFLHEDLTVKIGDFGLATVKSRWSGSHQFEQLS GSILWMAPEVIRMQDKNPYSFQSDVYAFGIVLYELMTGQLPYSNINNRDQIIFMVGRGYLSPDLSKVRSNCPKAMKRLMAECLKKKRDER | 1502_AKAP9_BRAF |
Top |
Comparison of Fusion Protein Isoforms |
 Superimpose the 3D Structures Among All Fusion Protein Isoforms Superimpose the 3D Structures Among All Fusion Protein Isoforms * Download the pdb file and open it from the molstar online viewer. |
 Comparison of the Secondary Structures of Fusion Protein Isoforms Comparison of the Secondary Structures of Fusion Protein Isoforms |
Top |
Comparison of Fusion Protein Sequences/Structures with Known Sequences/Structures from PDB |
Top |
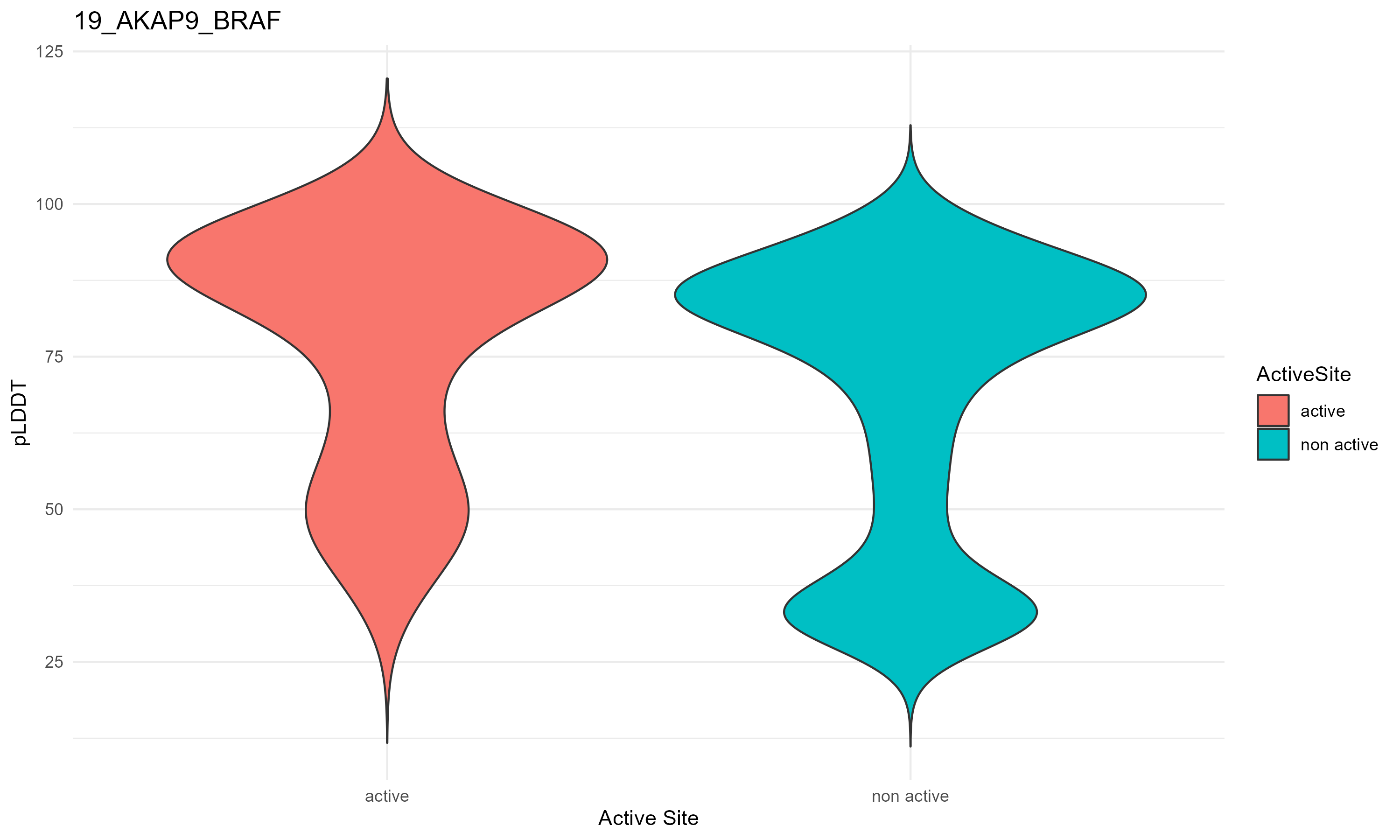
pLDDT score distribution |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. * The blue color at the bottom marks the best active site residues. |
| 19_AKAP9_BRAF.png |
 |
| 19_AKAP9_BRAF.png |
 |
Top |
Potential Active Site Information |
 The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. |
| Kinase Fusion AA seq ID in KinaseFusionDB | Site score | Size | Dscore | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
Top |
Ramachandran Plot of Kinase Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| 19_AKAP9_BRAF_ramachandran.png |
 |
Top |
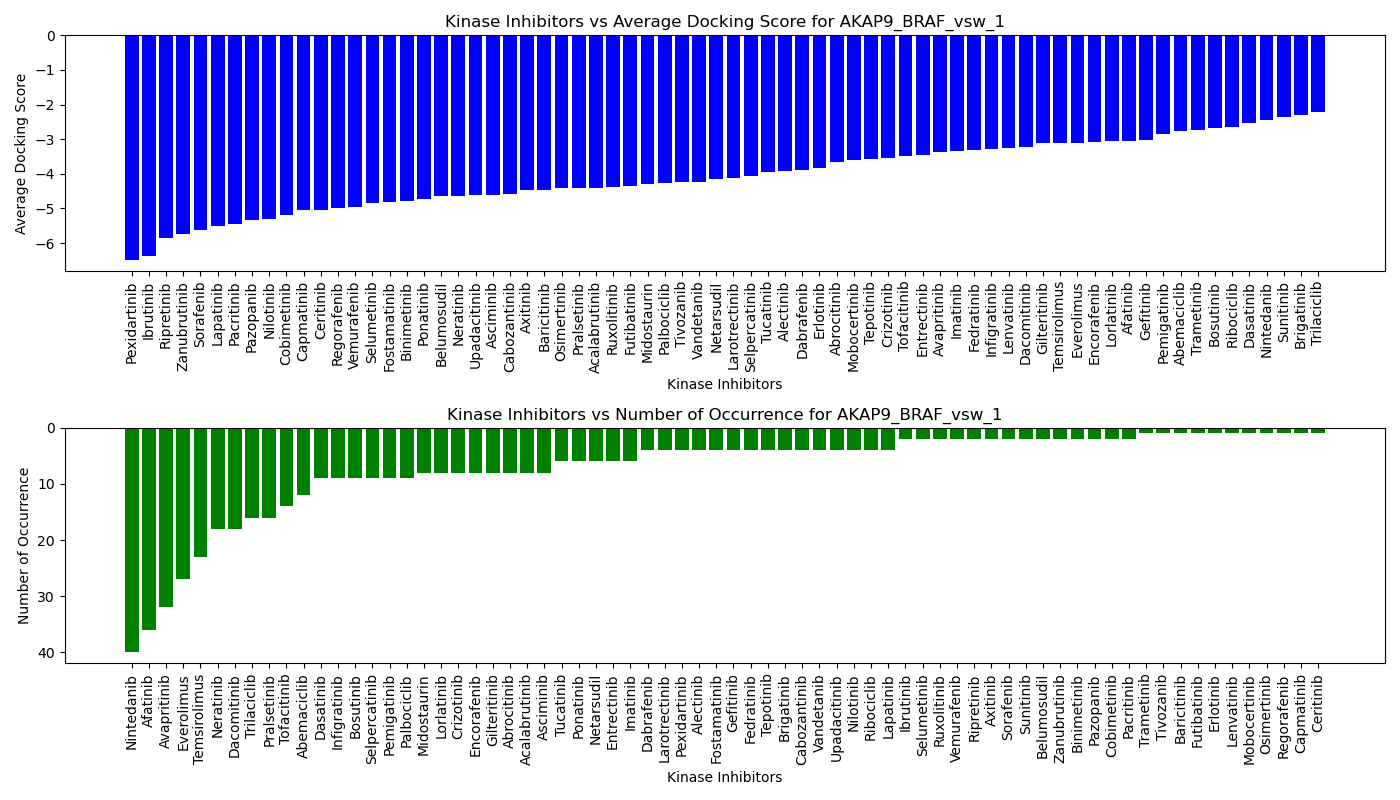
Virtual Screening Results |
 Distribution of the average docking score across all approved kinase inhibitors. Distribution of the average docking score across all approved kinase inhibitors.Distribution of the number of occurrence across all approved kinase inhibitors. |
| 5'-kinase fusion protein case |
| 3'-kinase fusion protein case |
 |
Top |
 Drug information from DrugBank of the top 20 interacting small molecules. Drug information from DrugBank of the top 20 interacting small molecules.* The detailed information of individual kinase inhibitors are available in the download page. |
| Fusion gene name info | Drug | Docking score | Glide g score | Glide energy |
| 19_AKAP9_BRAF_vsw_1-DOCK_HTVS_1-001 | Neratinib | -8.1484 | -8.3307 | -47.0437 |
| 19_AKAP9_BRAF_vsw_1-DOCK_HTVS_1-001 | Neratinib | -8.1484 | -8.3307 | -47.0437 |
| 19_AKAP9_BRAF_vsw_1-DOCK_HTVS_1-001 | Acalabrutinib | -7.941789999999999 | -7.955889999999999 | -43.7382 |
| 19_AKAP9_BRAF_vsw_1-DOCK_HTVS_1-001 | Acalabrutinib | -7.941789999999999 | -7.955889999999999 | -43.7382 |
| 19_AKAP9_BRAF_vsw_1-DOCK_HTVS_1-001 | Pexidartinib | -7.915210000000001 | -8.616010000000001 | -41.0086 |
| 19_AKAP9_BRAF_vsw_1-DOCK_HTVS_1-001 | Pexidartinib | -7.915210000000001 | -8.616010000000001 | -41.0086 |
| 19_AKAP9_BRAF_vsw_1-DOCK_HTVS_1-001 | Midostaurin | -7.58125 | -7.58125 | -49.4291 |
| 19_AKAP9_BRAF_vsw_1-DOCK_HTVS_1-001 | Neratinib | -7.467860000000001 | -7.653760000000001 | -44.7525 |
| 19_AKAP9_BRAF_vsw_1-DOCK_HTVS_1-001 | Zanubrutinib | -7.46563 | -7.46563 | -41.9919 |
| 19_AKAP9_BRAF_vsw_1-DOCK_HTVS_1-001 | Ibrutinib | -7.266660000000001 | -7.266660000000001 | -48.1168 |
| 19_AKAP9_BRAF_vsw_1-DOCK_HTVS_1-001 | Acalabrutinib | -6.72611 | -6.74021 | -46.118 |
| 19_AKAP9_BRAF_vsw_1-DOCK_HTVS_1-001 | Acalabrutinib | -6.72611 | -6.74021 | -46.118 |
| 19_AKAP9_BRAF_vsw_1-DOCK_HTVS_1-001 | Midostaurin | -6.6803 | -6.6803 | -48.7289 |
| 19_AKAP9_BRAF_vsw_1-DOCK_HTVS_1-001 | Midostaurin | -6.2609900000000005 | -6.2609900000000005 | -50.1829 |
| 19_AKAP9_BRAF_vsw_1-DOCK_HTVS_1-001 | Netarsudil | -6.12105 | -6.13215 | -47.983999999999995 |
| 19_AKAP9_BRAF_vsw_1-DOCK_HTVS_1-001 | Netarsudil | -6.12105 | -6.13215 | -47.983999999999995 |
| 19_AKAP9_BRAF_vsw_1-DOCK_HTVS_1-001 | Axitinib | -6.0109699999999995 | -6.01417 | -41.2245 |
| 19_AKAP9_BRAF_vsw_1-DOCK_HTVS_1-001 | Lapatinib | -5.9949699999999995 | -6.0837699999999995 | -52.9724 |
| 19_AKAP9_BRAF_vsw_1-DOCK_HTVS_1-001 | Lapatinib | -5.94945 | -6.03825 | -52.824 |
| 19_AKAP9_BRAF_vsw_1-DOCK_HTVS_1-001 | Pralsetinib | -5.8501199999999995 | -5.9416199999999995 | -46.9968 |
Top |
Kinase-Substrate Information of AKAP9_BRAF |
 Phosphorylation target of the kinase Phosphorylation target of the kinase(phosphosite, 03-17-2024) |
| Kinase | Kinase UniProt Acc | Kinase species | Substrate | Substrate UniProt Acc | Substrate phosphorylated residues | Substrate phosphorylated sites (+/-7AA) | Domain |
| BRAF | P15056 | human | SLC9A1 | P19634 | T653 | LRsYNRHtLVADPyE | NEXCaM_BD |
| BRAF | P15056 | human | EEF1A1 | P68104 | T88 | EtSkyyVtIIDAPGH | GTP_EFTU |
| BRAF | P15056 | human | BRAF | P15056 | S465 | tVGQRIGsGsFGtVY | PK_Tyr_Ser-Thr |
| BRAF | P15056 | human | MAP2K1 | Q02750 | S222 | LIDsMANsFVGtRSY | Pkinase |
| BRAF | P15056 | human | EEF1A2 | Q05639 | S21 | GHVDSGkstttGHLI | GTP_EFTU |
| BRAF | P15056 | human | MAP2K1 | Q02750 | S218 | VsGQLIDsMANsFVG | Pkinase |
| BRAF | P15056 | human | TTK | P33981 | S281 | VPVNLLNsPDCDVkt | |
| BRAF | P15056 | human | BAD | Q92934 | S99 | PFrGrsRsAPPNLWA | Bcl-2_BAD |
| BRAF | P15056 | human | MAP2K2 | P36507 | S222 | VsGQLIDsMANsFVG | Pkinase |
| BRAF | P15056 | human | BAD | Q92934 | S75 | EIRsRHssyPAGtED | Bcl-2_BAD |
| BRAF | P15056 | human | NANOG | Q9H9S0 | S68 | LIQDsPDsStsPKGK | |
| BRAF | P15056 | human | EEF1A1 | P68104 | S21 | GHVDSGkstttGHLI | GTP_EFTU |
| BRAF | P15056 | human | BRAF | P15056 | S614 | SHQFEQLsGSILWMA | PK_Tyr_Ser-Thr |
| BRAF | P15056 | human | BAD | Q92934 | S118 | GRELRRMsDEFVDsF | Bcl-2_BAD |
| BRAF | P15056 | human | BRAF | P15056 | S467 | GQRIGsGsFGtVYkG | PK_Tyr_Ser-Thr |
| BRAF | P15056 | human | BRAF | P15056 | T373 | APNVHINtIEPVNID | |
| BRAF | P15056 | human | MAP2K2 | P36507 | S226 | LIDsMANsFVGtRSY | Pkinase |
| BRAF | P15056 | human | TTK | P33981 | S436 | VFSVskQsPPIsTSk | |
| BRAF | P15056 | human | BAD | Q92934 | S134 | KGLPRPKsAGtAtQM | Bcl-2_BAD |
 Biological Network Integration of This Kinase and Substrates Biological Network Integration of This Kinase and Substrates (GeneMANIA website) |
 Enriched GO biological processes of the phosphorylation target genes of the kinase Enriched GO biological processes of the phosphorylation target genes of the kinase |
| Kinase | GOID | GO term | P.adjust |
| BRAF | ID | Description | 0.00e+00 |
| BRAF | GO:0048679 | regulation of axon regeneration | 5.62e-05 |
| BRAF | GO:0030878 | thyroid gland development | 5.62e-05 |
| BRAF | GO:0035270 | endocrine system development | 5.62e-05 |
| BRAF | GO:0070570 | regulation of neuron projection regeneration | 5.62e-05 |
| BRAF | GO:0048538 | thymus development | 1.42e-04 |
| BRAF | GO:0031103 | axon regeneration | 1.42e-04 |
| BRAF | GO:0060324 | face development | 1.42e-04 |
| BRAF | GO:0031102 | neuron projection regeneration | 1.74e-04 |
| BRAF | GO:0050772 | positive regulation of axonogenesis | 3.79e-04 |
| BRAF | GO:0048678 | response to axon injury | 4.26e-04 |
| BRAF | GO:0060502 | epithelial cell proliferation involved in lung morphogenesis | 5.02e-04 |
| BRAF | GO:0060439 | trachea morphogenesis | 5.09e-04 |
| BRAF | GO:0048534 | hematopoietic or lymphoid organ development | 5.09e-04 |
| BRAF | GO:0033674 | positive regulation of kinase activity | 5.09e-04 |
| BRAF | GO:0048308 | organelle inheritance | 7.57e-04 |
| BRAF | GO:0048313 | Golgi inheritance | 7.57e-04 |
| BRAF | GO:0061684 | chaperone-mediated autophagy | 7.57e-04 |
| BRAF | GO:1903358 | regulation of Golgi organization | 8.17e-04 |
| BRAF | GO:0051347 | positive regulation of transferase activity | 8.44e-04 |
| BRAF | GO:0060438 | trachea development | 9.59e-04 |
| BRAF | GO:2000641 | regulation of early endosome to late endosome transport | 9.59e-04 |
| BRAF | GO:0010720 | positive regulation of cell development | 9.59e-04 |
| BRAF | GO:0050770 | regulation of axonogenesis | 1.21e-03 |
| BRAF | GO:1903034 | regulation of response to wounding | 1.55e-03 |
| BRAF | GO:0044342 | type B pancreatic cell proliferation | 1.85e-03 |
| BRAF | GO:0031099 | regeneration | 1.92e-03 |
| BRAF | GO:0002520 | immune system development | 2.09e-03 |
| BRAF | GO:1903649 | regulation of cytoplasmic transport | 2.16e-03 |
| BRAF | GO:0043550 | regulation of lipid kinase activity | 3.11e-03 |
| BRAF | GO:0050769 | positive regulation of neurogenesis | 3.35e-03 |
| BRAF | GO:0045022 | early endosome to late endosome transport | 3.60e-03 |
| BRAF | GO:0060428 | lung epithelium development | 3.60e-03 |
| BRAF | GO:0035987 | endodermal cell differentiation | 4.20e-03 |
| BRAF | GO:0098927 | vesicle-mediated transport between endosomal compartments | 4.20e-03 |
| BRAF | GO:0048009 | insulin-like growth factor receptor signaling pathway | 4.60e-03 |
| BRAF | GO:0051962 | positive regulation of nervous system development | 4.69e-03 |
| BRAF | GO:0060425 | lung morphogenesis | 4.69e-03 |
| BRAF | GO:0018105 | peptidyl-serine phosphorylation | 4.74e-03 |
| BRAF | GO:0001706 | endoderm formation | 5.11e-03 |
| BRAF | GO:0018209 | peptidyl-serine modification | 5.11e-03 |
| BRAF | GO:0046902 | regulation of mitochondrial membrane permeability | 5.41e-03 |
| BRAF | GO:0070371 | ERK1 and ERK2 cascade | 6.22e-03 |
| BRAF | GO:0048645 | animal organ formation | 6.22e-03 |
| BRAF | GO:0035019 | somatic stem cell population maintenance | 6.45e-03 |
| BRAF | GO:0006414 | translational elongation | 6.46e-03 |
| BRAF | GO:0071260 | cellular response to mechanical stimulus | 6.46e-03 |
| BRAF | GO:0010506 | regulation of autophagy | 6.46e-03 |
| BRAF | GO:0031346 | positive regulation of cell projection organization | 6.46e-03 |
| BRAF | GO:0090559 | regulation of membrane permeability | 6.84e-03 |
Top |
Related Drugs to AKAP9_BRAF |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
 Distribution of the number of studies mentioning AKAP9-BRAF and kinase inhibitors the PubMed Abstract (04-01-2024) Distribution of the number of studies mentioning AKAP9-BRAF and kinase inhibitors the PubMed Abstract (04-01-2024) |
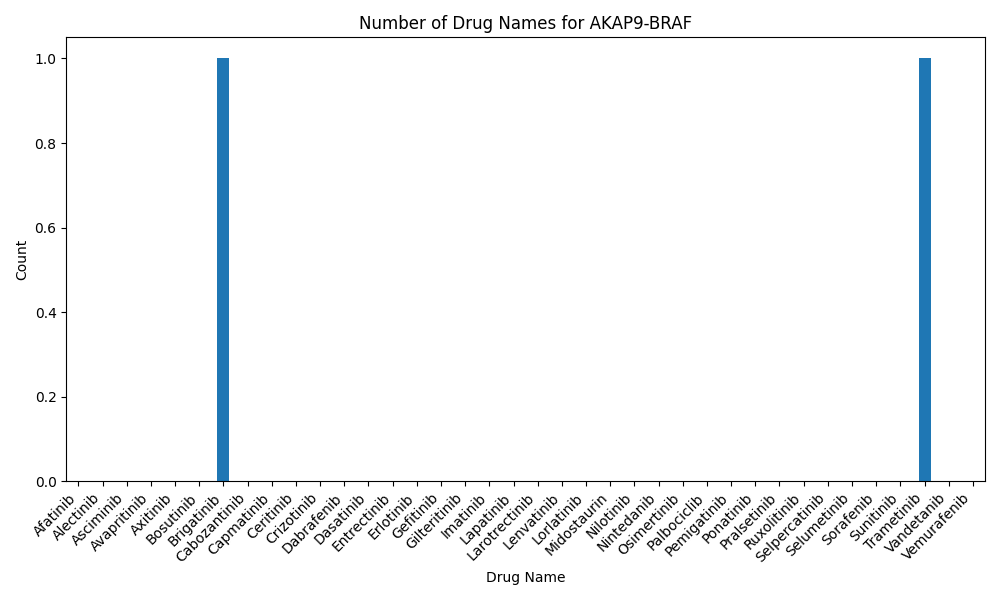 |
| Fusion gene - drug pair 1 | Fusion gene - drug pair 2 | PMID | Publication date | DOI | Study title |
| AKAP9-BRAF AND Trametinib | 30792255 | 2019-03-01 | 10.1542/peds.2018-2469 | Giant Congenital Melanocytic Nevus Treated With Trametinib. | |
| AKAP9-BRAF AND Brigatinib | HIP1-ALK AND Brigatinib | 34687488 | 2021-11 | 10.1002/gcc.23005 | Clinicopathological features and resistance mechanisms in HIP1-ALK-rearranged lung cancer: A multicenter study. |
Top |
Related Diseases to AKAP9_BRAF |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
| AKAP9 | BRAF | Thyroid Gland Papillary Carcinoma | MyCancerGenome | |
| AKAP9 | BRAF | Low-Grade Glioma | MyCancerGenome | |
| AKAP9 | BRAF | Nos | MyCancerGenome | |
| AKAP9 | BRAF | Pancreatic Adenocarcinoma | MyCancerGenome | |
| AKAP9 | BRAF | Pilocytic Astrocytoma | MyCancerGenome | |
| AKAP9 | BRAF | Astrocytoma | MyCancerGenome |
 Related diseases from the literature mentioned this fusion gene and drug. Related diseases from the literature mentioned this fusion gene and drug. (PubMed, 04-01-2024) |
| MeSH ID | MeSH term |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | AKAP9 | C2678483 | Long Qt Syndrome 11 | 4 | CLINGEN;CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Tgene | BRAF | C0025202 | melanoma | 24 | CGI;CTD_human;UNIPROT |
| Tgene | BRAF | C1275081 | Cardio-facio-cutaneous syndrome | 14 | CLINGEN;CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Tgene | BRAF | C0009402 | Colorectal Carcinoma | 8 | CTD_human;UNIPROT |
| Tgene | BRAF | C0028326 | Noonan Syndrome | 8 | CLINGEN;CTD_human;GENOMICS_ENGLAND;ORPHANET |
| Tgene | BRAF | C0238463 | Papillary thyroid carcinoma | 8 | CTD_human;ORPHANET |
| Tgene | BRAF | C0040136 | Thyroid Neoplasm | 6 | CGI;CTD_human |
| Tgene | BRAF | C0151468 | Thyroid Gland Follicular Adenoma | 6 | CTD_human |
| Tgene | BRAF | C0175704 | LEOPARD Syndrome | 6 | CLINGEN;GENOMICS_ENGLAND |
| Tgene | BRAF | C0549473 | Thyroid carcinoma | 6 | CGI;CTD_human |
| Tgene | BRAF | C3150970 | NOONAN SYNDROME 7 | 5 | CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Tgene | BRAF | C0009404 | Colorectal Neoplasms | 4 | CTD_human |
| Tgene | BRAF | C3150971 | LEOPARD SYNDROME 3 | 4 | CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Tgene | BRAF | C1519086 | Pilomyxoid astrocytoma | 3 | ORPHANET |
| Tgene | BRAF | C0004565 | Melanoma, B16 | 2 | CTD_human |
| Tgene | BRAF | C0009075 | Melanoma, Cloudman S91 | 2 | CTD_human |
| Tgene | BRAF | C0018598 | Melanoma, Harding-Passey | 2 | CTD_human |
| Tgene | BRAF | C0023443 | Hairy Cell Leukemia | 2 | CGI;ORPHANET |
| Tgene | BRAF | C0025205 | Melanoma, Experimental | 2 | CTD_human |
| Tgene | BRAF | C0033578 | Prostatic Neoplasms | 2 | CTD_human |
| Tgene | BRAF | C0152013 | Adenocarcinoma of lung (disorder) | 2 | CGI;CTD_human |
| Tgene | BRAF | C0376358 | Malignant neoplasm of prostate | 2 | CTD_human |
| Tgene | BRAF | C0587248 | Costello syndrome (disorder) | 2 | CLINGEN;CTD_human |
| Tgene | BRAF | C3501843 | Nonmedullary Thyroid Carcinoma | 2 | CTD_human |
| Tgene | BRAF | C3501844 | Familial Nonmedullary Thyroid Cancer | 2 | CTD_human |
Top |
Clinical Trials of the Found Drugs/Small Molecules |
 Statistics of the Clinical Trials of the Found Kinase Inibitors from clinicaltrials.gov (06-17-2024) Statistics of the Clinical Trials of the Found Kinase Inibitors from clinicaltrials.gov (06-17-2024) |
 Clinical Trials from clinicaltrials.gov (06-17-2024) Clinical Trials from clinicaltrials.gov (06-17-2024) |
| Fusion Gene | Kinase Inhibitor | NCT ID | Study Status | Phases | Disease | # Enrolment | Date |

