| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Kinase Fusion Gene:IPO7_WEE1 |
Kinase Fusion Protein Summary |
 Kinase Fusion gene summary Kinase Fusion gene summary |
| Kinase Fusion partner gene information | Kinase Fusion gene name: IPO7_WEE1 | KinaseFusionDB ID: KFG2892 | FusionGDB2.0 ID: KFG2892 | Hgene | Tgene | Gene symbol | IPO7 | WEE1 | Gene ID | 10527 | 7465 | |
| Gene name | importin 7 | WEE1 G2 checkpoint kinase | ||||||||||
| Synonyms | Imp7|RANBP7 | WEE1A|WEE1hu | ||||||||||
| Cytomap | 11p15.4 | 11p15.4 | ||||||||||
| Type of gene | protein-coding | protein-coding | ||||||||||
| Description | importin-7RAN-binding protein 7 | wee1-like protein kinaseWEE1 homologWEE1+ homologprotein kinasewee1A kinase | ||||||||||
| Modification date | 20240407 | 20240407 | ||||||||||
| UniProtAcc | O95373 | P30291 | ||||||||||
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000379719, ENST00000533680, | ENST00000299613, ENST00000450114, | |||||||||
| Context (manual curation of fusion genes in KinaseFusionDB) | PubMed: IPO7 [Title/Abstract] AND WEE1 [Title/Abstract] AND fusion [Title/Abstract] | |||||||||||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | IPO7(9406394)-WEE1(9597435), # samples:1 IPO7(9406394)-WEE1(9597434), # samples:1 | |||||||||||
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | WEE1 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle | 7743995|8348613|8428596 |
 Kinase Fusion gene breakpoints across IPO7 (5'-gene) Kinase Fusion gene breakpoints across IPO7 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
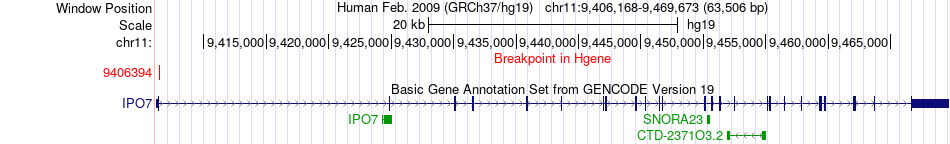 |
 Kinase Fusion gene breakpoints across WEE1 (3'-gene) Kinase Fusion gene breakpoints across WEE1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
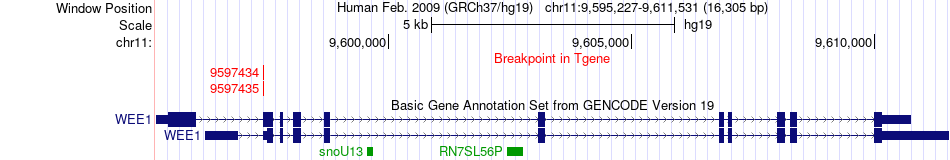 |
Top |
Kinase Fusion Gene Sample Information |
 Kinase Fusion gene information. Kinase Fusion gene information. |
 Kinase Fusion gene information from four resources (ChiTars 5.0, ChimerDB 4.0, COSMIC, and CCLE) Kinase Fusion gene information from four resources (ChiTars 5.0, ChimerDB 4.0, COSMIC, and CCLE)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Sample | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp |
| ChimerDB4 | TCGA-22-4593-01A | IPO7 | chr11 | 9406394 | WEE1 | chr11 | 9597435 |
| ChimerDB4 | TCGA-22-4593 | IPO7 | chr11 | 9406394 | WEE1 | chr11 | 9597434 |
Top |
Kinase Fusion ORF Analysis |
 Kinase Fusion information from ORFfinder translation from full-length transcript sequence from KinaseFusionDB. Kinase Fusion information from ORFfinder translation from full-length transcript sequence from KinaseFusionDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp | Seq length (transcript) | Seq length (amino acids) |
| ENST00000379719 | ENST00000450114 | IPO7 | chr11 | 9406394 | WEE1 | chr11 | 9597435 | 2196 | 482 |
| ENST00000379719 | ENST00000450114 | IPO7 | chr11 | 9406394 | WEE1 | chr11 | 9597434 | 2196 | 482 |
Top |
Kinase Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from KinaseFusionDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from KinaseFusionDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >Henst_Tenst_Hgene_Hchr_Hbp_Tgene_Tchr_Tbp_length(fusion AA)_AAseq >ENST00000379719_ENST00000450114_IPO7_chr11_9406394_WEE1_chr11_9597435_length(amino acids)=482 MDPNTIIEALRGTMDPALREAAERQLNESLLSKARGIDSSSVKLRGSSLFMDTEKSGKREFDVRQTPQVNINPFTPDSLLLHSSGQCRRR KRTYWNDSCGEDMEASDYELEDETRPAKRITITESNMKSRYTTEFHELEKIGSGEFGSVFKCVKRLDGCIYAIKRSKKPLAGSVDEQNAL REVYAHAVLGQHSHVVRYFSAWAEDDHMLIQNEYCNGGSLADAISENYRIMSYFKEAELKDLLLQVGRGLRYIHSMSLVHMDIKPSNIFI SRTSIPNAASEEGDEDDWASNKVMFKIGDLGHVTRISSPQVEEGDSRFLANEVLQENYTHLPKADIFALALTVVCAAGAEPLPRNGDQWH EIRQGRLPRIPQVLSQEFTELLKVMIHPDPERRPSAMALVKHSVLLSASRKSAEQLRIELNAEKFKNSLLQKELKKAQMAKAAAEERALF -------------------------------------------------------------- >ENST00000379719_ENST00000450114_IPO7_chr11_9406394_WEE1_chr11_9597434_length(amino acids)=482 MDPNTIIEALRGTMDPALREAAERQLNESLLSKARGIDSSSVKLRGSSLFMDTEKSGKREFDVRQTPQVNINPFTPDSLLLHSSGQCRRR KRTYWNDSCGEDMEASDYELEDETRPAKRITITESNMKSRYTTEFHELEKIGSGEFGSVFKCVKRLDGCIYAIKRSKKPLAGSVDEQNAL REVYAHAVLGQHSHVVRYFSAWAEDDHMLIQNEYCNGGSLADAISENYRIMSYFKEAELKDLLLQVGRGLRYIHSMSLVHMDIKPSNIFI SRTSIPNAASEEGDEDDWASNKVMFKIGDLGHVTRISSPQVEEGDSRFLANEVLQENYTHLPKADIFALALTVVCAAGAEPLPRNGDQWH EIRQGRLPRIPQVLSQEFTELLKVMIHPDPERRPSAMALVKHSVLLSASRKSAEQLRIELNAEKFKNSLLQKELKKAQMAKAAAEERALF -------------------------------------------------------------- |
Multiple Sequence Alignment of All Fusion Protein Isoforms |
Top |
Kinase Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr11:9406394/chr11:9597435) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| IPO7 | WEE1 |
| FUNCTION: Functions in nuclear protein import, either by acting as autonomous nuclear transport receptor or as an adapter-like protein in association with the importin-beta subunit KPNB1. Acting autonomously, is thought to serve itself as receptor for nuclear localization signals (NLS) and to promote translocation of import substrates through the nuclear pore complex (NPC) by an energy requiring, Ran-dependent mechanism. At the nucleoplasmic side of the NPC, Ran binds to importin, the importin/substrate complex dissociates and importin is re-exported from the nucleus to the cytoplasm where GTP hydrolysis releases Ran. The directionality of nuclear import is thought to be conferred by an asymmetric distribution of the GTP- and GDP-bound forms of Ran between the cytoplasm and nucleus. Mediates autonomously the nuclear import of ribosomal proteins RPL23A, RPS7 and RPL5 (PubMed:11682607). In association with KPNB1 mediates the nuclear import of H1 histone and the Ran-binding site of IPO7 is not required but synergizes with that of KPNB1 in importin/substrate complex dissociation. Promotes odontoblast differentiation via promoting nuclear translocation of DLX3, KLF4, SMAD2, thereby facilitating the transcription of target genes that play a role in odontoblast differentiation (By similarity). Facilitates BMP4-induced translocation of SMAD1 to the nucleus and recruitment to the MSX1 gene promoter, thereby promotes the expression of the odontogenic regulator MSX1 in dental mesenchymal cells (By similarity). Also promotes odontoblast differentiation by facilitating the nuclear translocation of HDAC6 and subsequent repression of RUNX2 expression (By similarity). Inhibits osteoblast differentiation by inhibiting nuclear translocation of RUNX2 and therefore inhibition of RUNX2 target gene transcription (By similarity). In vitro, mediates nuclear import of H2A, H2B, H3 and H4 histones. {ECO:0000250|UniProtKB:Q9EPL8, ECO:0000269|PubMed:10228156, ECO:0000269|PubMed:11682607, ECO:0000269|PubMed:9687515}.; FUNCTION: (Microbial infection) Mediates the nuclear import of HIV-1 reverse transcription complex (RTC) integrase. Binds and mediates the nuclear import of HIV-1 Rev. {ECO:0000269|PubMed:12853482, ECO:0000269|PubMed:16704975}. | FUNCTION: Acts as a negative regulator of entry into mitosis (G2 to M transition) by protecting the nucleus from cytoplasmically activated cyclin B1-complexed CDK1 before the onset of mitosis by mediating phosphorylation of CDK1 on 'Tyr-15'. Specifically phosphorylates and inactivates cyclin B1-complexed CDK1 reaching a maximum during G2 phase and a minimum as cells enter M phase. Phosphorylation of cyclin B1-CDK1 occurs exclusively on 'Tyr-15' and phosphorylation of monomeric CDK1 does not occur. Its activity increases during S and G2 phases and decreases at M phase when it is hyperphosphorylated. A correlated decrease in protein level occurs at M/G1 phase, probably due to its degradation. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
 - Retained domain in the 5'-partner of fusion protein (protein functional feature from UniProt). - Retained domain in the 5'-partner of fusion protein (protein functional feature from UniProt). |
| Partner | Hgeneene | Hbp | Tgeneene | Tbp | ENST | BPexon | TotalExon | Protein feature loci | BPloci | TotalLen | Feature | Note |
 - Retained domain in the 3'-partner of fusion protein (protein functional feature from UniProt). - Retained domain in the 3'-partner of fusion protein (protein functional feature from UniProt). |
| Partner | Hgeneene | Hbp | Tgeneene | Tbp | ENST | BPexon | TotalExon | Protein feature loci | BPloci | TotalLen | Feature | Note |
| Tgene | IPO7 | 9406394 | WEE1 | 9597434 | ENST00000379719 | 0 | 11 | 299_569 | 0 | 433 | Domain | Note=Protein kinase;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00159 |
| Tgene | IPO7 | 9406394 | WEE1 | 9597434 | ENST00000379719 | 0 | 11 | 299_569 | 192 | 647 | Domain | Note=Protein kinase;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00159 |
| Tgene | IPO7 | 9406394 | WEE1 | 9597435 | ENST00000379719 | 0 | 11 | 299_569 | 0 | 433 | Domain | Note=Protein kinase;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00159 |
| Tgene | IPO7 | 9406394 | WEE1 | 9597435 | ENST00000379719 | 0 | 11 | 299_569 | 192 | 647 | Domain | Note=Protein kinase;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00159 |
Top |
Kinase Fusion Protein Structures |
 CIF files of the predicted kinase fusion proteins CIF files of the predicted kinase fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Kinase Fusion protein CIF link (fusion AA seq ID in KinaseFusionDB) | Henst | Tenst | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp | AA seq | Len(AA seq) |
| PDB file >>>TKFP_433_IPO7_WEE1 | ENST00000379719 | ENST00000450114 | IPO7 | chr11 | 9406394 | WEE1 | chr11 | 9597435 | MDPNTIIEALRGTMDPALREAAERQLNESLLSKARGIDSSSVKLRGSSLFMDTEKSGKREFDVRQTPQVNINPFTPDSLLLHSSGQCRRR KRTYWNDSCGEDMEASDYELEDETRPAKRITITESNMKSRYTTEFHELEKIGSGEFGSVFKCVKRLDGCIYAIKRSKKPLAGSVDEQNAL REVYAHAVLGQHSHVVRYFSAWAEDDHMLIQNEYCNGGSLADAISENYRIMSYFKEAELKDLLLQVGRGLRYIHSMSLVHMDIKPSNIFI SRTSIPNAASEEGDEDDWASNKVMFKIGDLGHVTRISSPQVEEGDSRFLANEVLQENYTHLPKADIFALALTVVCAAGAEPLPRNGDQWH EIRQGRLPRIPQVLSQEFTELLKVMIHPDPERRPSAMALVKHSVLLSASRKSAEQLRIELNAEKFKNSLLQKELKKAQMAKAAAEERALF | 482_IPO7_WEE1 |
| 3D view using mol* of TKFP_433_IPO7_WEE1 | ||||||||||
| PDB file >>>TKFP_434_IPO7_WEE1 | ENST00000379719 | ENST00000450114 | IPO7 | chr11 | 9406394 | WEE1 | chr11 | 9597434 | MDPNTIIEALRGTMDPALREAAERQLNESLLSKARGIDSSSVKLRGSSLFMDTEKSGKREFDVRQTPQVNINPFTPDSLLLHSSGQCRRR KRTYWNDSCGEDMEASDYELEDETRPAKRITITESNMKSRYTTEFHELEKIGSGEFGSVFKCVKRLDGCIYAIKRSKKPLAGSVDEQNAL REVYAHAVLGQHSHVVRYFSAWAEDDHMLIQNEYCNGGSLADAISENYRIMSYFKEAELKDLLLQVGRGLRYIHSMSLVHMDIKPSNIFI SRTSIPNAASEEGDEDDWASNKVMFKIGDLGHVTRISSPQVEEGDSRFLANEVLQENYTHLPKADIFALALTVVCAAGAEPLPRNGDQWH EIRQGRLPRIPQVLSQEFTELLKVMIHPDPERRPSAMALVKHSVLLSASRKSAEQLRIELNAEKFKNSLLQKELKKAQMAKAAAEERALF | 482_IPO7_WEE1 |
Top |
Comparison of Fusion Protein Isoforms |
 Superimpose the 3D Structures Among All Fusion Protein Isoforms Superimpose the 3D Structures Among All Fusion Protein Isoforms * Download the pdb file and open it from the molstar online viewer. |
 Comparison of the Secondary Structures of Fusion Protein Isoforms Comparison of the Secondary Structures of Fusion Protein Isoforms |
Top |
Comparison of Fusion Protein Sequences/Structures with Known Sequences/Structures from PDB |
Top |
pLDDT score distribution |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. * The blue color at the bottom marks the best active site residues. |
| TKFP_433_IPO7_WEE1.png |
 |
Top |
Potential Active Site Information |
 The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. |
| Kinase Fusion AA seq ID in KinaseFusionDB | Site score | Size | Dscore | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
| TKFP_433_IPO7_WEE1 | 1.062 | 134 | 1.094 | 329.966 | 0.485 | 0.755 | 0.987 | 0.831 | 0.918 | 0.905 | 0.755 | Chain A: 141,142,143,144,145,146,147,149,162,164,1 82,186,196,212,213,214,215,218,219,222,262,264,266 ,267,269,298,299 |
Top |
Ramachandran Plot of Kinase Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| TKFP_433_IPO7_WEE1_ramachandran.png |
 |
Top |
Virtual Screening Results |
 Distribution of the average docking score across all approved kinase inhibitors. Distribution of the average docking score across all approved kinase inhibitors.Distribution of the number of occurrence across all approved kinase inhibitors. |
| 5'-kinase fusion protein case |
| 3'-kinase fusion protein case |
Top |
 Drug information from DrugBank of the top 20 interacting small molecules. Drug information from DrugBank of the top 20 interacting small molecules.* The detailed information of individual kinase inhibitors are available in the download page. |
| Fusion gene name info | Drug | Docking score | Glide g score | Glide energy |
Top |
Kinase-Substrate Information of IPO7_WEE1 |
 Phosphorylation target of the kinase Phosphorylation target of the kinase(phosphosite, 03-17-2024) |
| Kinase | Kinase UniProt Acc | Kinase species | Substrate | Substrate UniProt Acc | Substrate phosphorylated residues | Substrate phosphorylated sites (+/-7AA) | Domain |
| WEE1 | P30291 | human | HSP90AA1 | P07900 | Y38 | SLIINtFySNkEIFL | |
| WEE1 | P30291 | human | CDK2 | P24941 | Y15 | EkIGEGtyGVVykAR | Pkinase |
| WEE1 | P30291 | human | CDK1 | P06493 | Y15 | EkIGEGtyGVVykGR | Pkinase |
| WEE1 | P30291 | human | ERG | P11308 | Y183 | FQRLtPSyNADILLS | SAM_PNT |
 Biological Network Integration of This Kinase and Substrates Biological Network Integration of This Kinase and Substrates (GeneMANIA website) |
 Enriched GO biological processes of the phosphorylation target genes of the kinase Enriched GO biological processes of the phosphorylation target genes of the kinase |
| Kinase | GOID | GO term | P.adjust |
| WEE1 | ID | Description | 0.00e+00 |
| WEE1 | GO:0018105 | peptidyl-serine phosphorylation | 1.79e-03 |
| WEE1 | GO:0051054 | positive regulation of DNA metabolic process | 1.79e-03 |
| WEE1 | GO:0018209 | peptidyl-serine modification | 1.79e-03 |
| WEE1 | GO:0045740 | positive regulation of DNA replication | 1.79e-03 |
| WEE1 | GO:0042307 | positive regulation of protein import into nucleus | 1.79e-03 |
| WEE1 | GO:0042306 | regulation of protein import into nucleus | 3.09e-03 |
| WEE1 | GO:0046824 | positive regulation of nucleocytoplasmic transport | 3.09e-03 |
| WEE1 | GO:0044773 | mitotic DNA damage checkpoint signaling | 4.75e-03 |
| WEE1 | GO:0044774 | mitotic DNA integrity checkpoint signaling | 4.75e-03 |
| WEE1 | GO:1900182 | positive regulation of protein localization to nucleus | 4.88e-03 |
| WEE1 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle | 5.16e-03 |
| WEE1 | GO:0046822 | regulation of nucleocytoplasmic transport | 5.16e-03 |
| WEE1 | GO:2001252 | positive regulation of chromosome organization | 5.16e-03 |
| WEE1 | GO:1902749 | regulation of cell cycle G2/M phase transition | 5.43e-03 |
| WEE1 | GO:0006275 | regulation of DNA replication | 5.50e-03 |
| WEE1 | GO:0000077 | DNA damage checkpoint signaling | 5.50e-03 |
| WEE1 | GO:0031570 | DNA integrity checkpoint signaling | 5.50e-03 |
| WEE1 | GO:0007098 | centrosome cycle | 5.50e-03 |
| WEE1 | GO:0000086 | G2/M transition of mitotic cell cycle | 5.50e-03 |
| WEE1 | GO:0007093 | mitotic cell cycle checkpoint signaling | 5.50e-03 |
| WEE1 | GO:1900180 | regulation of protein localization to nucleus | 5.50e-03 |
| WEE1 | GO:0031023 | microtubule organizing center organization | 5.85e-03 |
| WEE1 | GO:0044839 | cell cycle G2/M phase transition | 5.85e-03 |
| WEE1 | GO:0090316 | positive regulation of intracellular protein transport | 5.85e-03 |
| WEE1 | GO:0006606 | protein import into nucleus | 5.85e-03 |
| WEE1 | GO:0051170 | import into nucleus | 5.98e-03 |
| WEE1 | GO:0042770 | signal transduction in response to DNA damage | 7.43e-03 |
| WEE1 | GO:0000075 | cell cycle checkpoint signaling | 7.43e-03 |
| WEE1 | GO:1901991 | negative regulation of mitotic cell cycle phase transition | 7.47e-03 |
| WEE1 | GO:0032388 | positive regulation of intracellular transport | 7.59e-03 |
| WEE1 | GO:0033157 | regulation of intracellular protein transport | 9.28e-03 |
| WEE1 | GO:0033044 | regulation of chromosome organization | 1.05e-02 |
| WEE1 | GO:0045930 | negative regulation of mitotic cell cycle | 1.05e-02 |
| WEE1 | GO:0000082 | G1/S transition of mitotic cell cycle | 1.05e-02 |
| WEE1 | GO:1901988 | negative regulation of cell cycle phase transition | 1.18e-02 |
| WEE1 | GO:0006260 | DNA replication | 1.18e-02 |
| WEE1 | GO:0044843 | cell cycle G1/S phase transition | 1.18e-02 |
| WEE1 | GO:0051222 | positive regulation of protein transport | 1.38e-02 |
| WEE1 | GO:0034504 | protein localization to nucleus | 1.38e-02 |
| WEE1 | GO:0010948 | negative regulation of cell cycle process | 1.42e-02 |
| WEE1 | GO:1904951 | positive regulation of establishment of protein localization | 1.42e-02 |
| WEE1 | GO:0006913 | nucleocytoplasmic transport | 1.42e-02 |
| WEE1 | GO:0051169 | nuclear transport | 1.42e-02 |
| WEE1 | GO:0032386 | regulation of intracellular transport | 1.44e-02 |
| WEE1 | GO:1901990 | regulation of mitotic cell cycle phase transition | 1.56e-02 |
| WEE1 | GO:0055015 | ventricular cardiac muscle cell development | 1.57e-02 |
| WEE1 | GO:0042176 | regulation of protein catabolic process | 1.57e-02 |
| WEE1 | GO:0045793 | positive regulation of cell size | 1.64e-02 |
| WEE1 | GO:0045786 | negative regulation of cell cycle | 1.64e-02 |
Top |
Related Drugs to IPO7_WEE1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
 Distribution of the number of studies mentioning IPO7-WEE1 and kinase inhibitors the PubMed Abstract (04-01-2024) Distribution of the number of studies mentioning IPO7-WEE1 and kinase inhibitors the PubMed Abstract (04-01-2024) |
| Fusion gene - drug pair 1 | Fusion gene - drug pair 2 | PMID | Publication date | DOI | Study title |
Top |
Related Diseases to IPO7_WEE1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Related diseases from the literature mentioned this fusion gene and drug. Related diseases from the literature mentioned this fusion gene and drug. (PubMed, 04-01-2024) |
| MeSH ID | MeSH term |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
Top |
Clinical Trials of the Found Drugs/Small Molecules |
 Statistics of the Clinical Trials of the Found Kinase Inibitors from clinicaltrials.gov (06-17-2024) Statistics of the Clinical Trials of the Found Kinase Inibitors from clinicaltrials.gov (06-17-2024) |
 Clinical Trials from clinicaltrials.gov (06-17-2024) Clinical Trials from clinicaltrials.gov (06-17-2024) |
| Fusion Gene | Kinase Inhibitor | NCT ID | Study Status | Phases | Disease | # Enrolment | Date |

