| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Kinase Fusion Gene:MAPKAPK2_SRGAP2 |
Kinase Fusion Protein Summary |
 Kinase Fusion gene summary Kinase Fusion gene summary |
| Kinase Fusion partner gene information | Kinase Fusion gene name: MAPKAPK2_SRGAP2 | KinaseFusionDB ID: KFG3594 | FusionGDB2.0 ID: KFG3594 | Hgene | Tgene | Gene symbol | MAPKAPK2 | SRGAP2 | Gene ID | 9261 | 23380 | |
| Gene name | MAPK activated protein kinase 2 | SLIT-ROBO Rho GTPase activating protein 2 | ||||||||||
| Synonyms | MAPKAP-K2|MK-2|MK2 | ARHGAP34|FNBP2|SRGAP2A|SRGAP3 | ||||||||||
| Cytomap | 1q32.1 | 1q32.1 | ||||||||||
| Type of gene | protein-coding | protein-coding | ||||||||||
| Description | MAP kinase-activated protein kinase 2MAPKAP kinase 2mitogen-activated protein kinase-activated protein kinase 2 | SLIT-ROBO Rho GTPase-activating protein 2SLIT-ROBO GAP2formin-binding protein 2rho GTPase-activating protein 34 | ||||||||||
| Modification date | 20240305 | 20240403 | ||||||||||
| UniProtAcc | P49137 | O75044 | ||||||||||
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000294981, ENST00000367103, ENST00000479009, | ENST00000419187, ENST00000471256, ENST00000414007, | |||||||||
| Context (manual curation of fusion genes in KinaseFusionDB) | PubMed: MAPKAPK2 [Title/Abstract] AND SRGAP2 [Title/Abstract] AND fusion [Title/Abstract] | |||||||||||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | MAPKAPK2(206858853)-SRGAP2(206589249), # samples:2 SRGAP2(206135293)-MAPKAPK2(206904483), # samples:1 | |||||||||||
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | MAPKAPK2 | GO:0018105 | peptidyl-serine phosphorylation | 15850461 |
| Hgene | MAPKAPK2 | GO:0032680 | regulation of tumor necrosis factor production | 15014438 |
| Hgene | MAPKAPK2 | GO:0034097 | response to cytokine | 8774846 |
| Hgene | MAPKAPK2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization | 14517288|15014438|20932473 |
| Tgene | SRGAP2 | GO:0043547 | positive regulation of GTPase activity | 21148482|27373832 |
| Tgene | SRGAP2 | GO:0046847 | filopodium assembly | 22559944 |
| Tgene | SRGAP2 | GO:0051014 | actin filament severing | 21148482 |
| Tgene | SRGAP2 | GO:0060996 | dendritic spine development | 22559944 |
| Tgene | SRGAP2 | GO:1904861 | excitatory synapse assembly | 27373832 |
| Tgene | SRGAP2 | GO:1904862 | inhibitory synapse assembly | 27373832 |
| Tgene | SRGAP2 | GO:2001223 | negative regulation of neuron migration | 22559944 |
 Kinase Fusion gene breakpoints across MAPKAPK2 (5'-gene) Kinase Fusion gene breakpoints across MAPKAPK2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
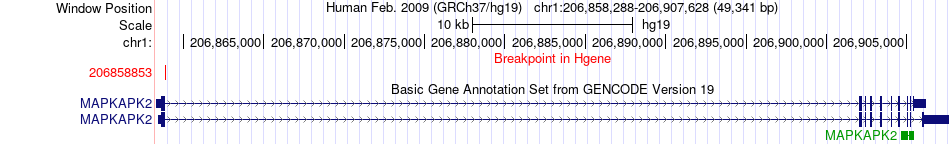 |
 Kinase Fusion gene breakpoints across SRGAP2 (3'-gene) Kinase Fusion gene breakpoints across SRGAP2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
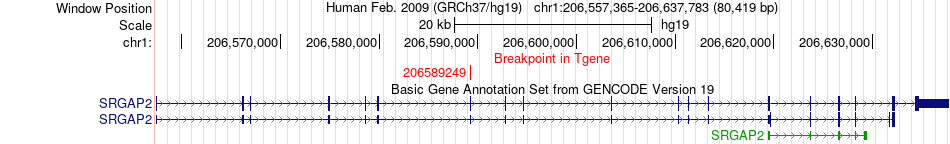 |
Top |
Kinase Fusion Gene Sample Information |
 Kinase Fusion gene information. Kinase Fusion gene information. |
 Kinase Fusion gene information from four resources (ChiTars 5.0, ChimerDB 4.0, COSMIC, and CCLE) Kinase Fusion gene information from four resources (ChiTars 5.0, ChimerDB 4.0, COSMIC, and CCLE)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Sample | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp |
| ChimerDB4 | TCGA-CV-5435 | MAPKAPK2 | chr1 | 206858853 | SRGAP2 | chr1 | 206589249 |
Top |
Kinase Fusion ORF Analysis |
 Kinase Fusion information from ORFfinder translation from full-length transcript sequence from KinaseFusionDB. Kinase Fusion information from ORFfinder translation from full-length transcript sequence from KinaseFusionDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp | Seq length (transcript) | Seq length (amino acids) |
Top |
Kinase Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from KinaseFusionDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from KinaseFusionDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >Henst_Tenst_Hgene_Hchr_Hbp_Tgene_Tchr_Tbp_length(fusion AA)_AAseq |
Multiple Sequence Alignment of All Fusion Protein Isoforms |
Top |
Kinase Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:206858853/chr1:206589249) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MAPKAPK2 | SRGAP2 |
| FUNCTION: Stress-activated serine/threonine-protein kinase involved in cytokine production, endocytosis, reorganization of the cytoskeleton, cell migration, cell cycle control, chromatin remodeling, DNA damage response and transcriptional regulation. Following stress, it is phosphorylated and activated by MAP kinase p38-alpha/MAPK14, leading to phosphorylation of substrates. Phosphorylates serine in the peptide sequence, Hyd-X-R-X(2)-S, where Hyd is a large hydrophobic residue. Phosphorylates ALOX5, CDC25B, CDC25C, CEP131, ELAVL1, HNRNPA0, HSP27/HSPB1, KRT18, KRT20, LIMK1, LSP1, PABPC1, PARN, PDE4A, RCSD1, RPS6KA3, TAB3 and TTP/ZFP36. Phosphorylates HSF1; leading to the interaction with HSP90 proteins and inhibiting HSF1 homotrimerization, DNA-binding and transactivation activities (PubMed:16278218). Mediates phosphorylation of HSP27/HSPB1 in response to stress, leading to the dissociation of HSP27/HSPB1 from large small heat-shock protein (sHsps) oligomers and impairment of their chaperone activities and ability to protect against oxidative stress effectively. Involved in inflammatory response by regulating tumor necrosis factor (TNF) and IL6 production post-transcriptionally: acts by phosphorylating AU-rich elements (AREs)-binding proteins ELAVL1, HNRNPA0, PABPC1 and TTP/ZFP36, leading to the regulation of the stability and translation of TNF and IL6 mRNAs. Phosphorylation of TTP/ZFP36, a major post-transcriptional regulator of TNF, promotes its binding to 14-3-3 proteins and reduces its ARE mRNA affinity, leading to inhibition of dependent degradation of ARE-containing transcripts. Phosphorylates CEP131 in response to cellular stress induced by ultraviolet irradiation which promotes binding of CEP131 to 14-3-3 proteins and inhibits formation of novel centriolar satellites (PubMed:26616734). Also involved in late G2/M checkpoint following DNA damage through a process of post-transcriptional mRNA stabilization: following DNA damage, relocalizes from nucleus to cytoplasm and phosphorylates HNRNPA0 and PARN, leading to stabilization of GADD45A mRNA. Involved in toll-like receptor signaling pathway (TLR) in dendritic cells: required for acute TLR-induced macropinocytosis by phosphorylating and activating RPS6KA3. {ECO:0000269|PubMed:10383393, ECO:0000269|PubMed:11844797, ECO:0000269|PubMed:12456657, ECO:0000269|PubMed:12565831, ECO:0000269|PubMed:14499342, ECO:0000269|PubMed:14517288, ECO:0000269|PubMed:15014438, ECO:0000269|PubMed:15629715, ECO:0000269|PubMed:16278218, ECO:0000269|PubMed:16456544, ECO:0000269|PubMed:17481585, ECO:0000269|PubMed:18021073, ECO:0000269|PubMed:20932473, ECO:0000269|PubMed:26616734, ECO:0000269|PubMed:8093612, ECO:0000269|PubMed:8280084, ECO:0000269|PubMed:8774846}. | FUNCTION: Postsynaptic RAC1 GTPase activating protein (GAP) that plays a key role in neuronal morphogenesis and migration mainly during development of the cerebral cortex (PubMed:20810653, PubMed:27373832, PubMed:28333212). Regulates excitatory and inhibitory synapse maturation and density in cortical pyramidal neurons (PubMed:22559944, PubMed:27373832). SRGAP2/SRGAP2A limits excitatory and inhibitory synapse density through its RAC1-specific GTPase activating activity, while it promotes maturation of both excitatory and inhibitory synapses through its ability to bind to the postsynaptic scaffolding protein HOMER1 at excitatory synapses, and the postsynaptic protein GPHN at inhibitory synapses (By similarity). Mechanistically, acts by binding and deforming membranes, thereby regulating actin dynamics to regulate cell migration and differentiation (PubMed:27373832). Promotes cell repulsion and contact inhibition of locomotion: localizes to protrusions with curved edges and controls the duration of RAC1 activity in contact protrusions (By similarity). In non-neuronal cells, may also play a role in cell migration by regulating the formation of lamellipodia and filopodia (PubMed:20810653, PubMed:21148482). {ECO:0000250|UniProtKB:Q91Z67, ECO:0000269|PubMed:20810653, ECO:0000269|PubMed:21148482, ECO:0000269|PubMed:22559944, ECO:0000269|PubMed:27373832, ECO:0000269|PubMed:28333212}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
 - Retained domain in the 5'-partner of fusion protein (protein functional feature from UniProt). - Retained domain in the 5'-partner of fusion protein (protein functional feature from UniProt). |
| Partner | Hgeneene | Hbp | Tgeneene | Tbp | ENST | BPexon | TotalExon | Protein feature loci | BPloci | TotalLen | Feature | Note |
 - Retained domain in the 3'-partner of fusion protein (protein functional feature from UniProt). - Retained domain in the 3'-partner of fusion protein (protein functional feature from UniProt). |
| Partner | Hgeneene | Hbp | Tgeneene | Tbp | ENST | BPexon | TotalExon | Protein feature loci | BPloci | TotalLen | Feature | Note |
Top |
Kinase-Substrate Information of MAPKAPK2_SRGAP2 |
 Phosphorylation target of the kinase Phosphorylation target of the kinase(phosphosite, 03-17-2024) |
| Kinase | Kinase UniProt Acc | Kinase species | Substrate | Substrate UniProt Acc | Substrate phosphorylated residues | Substrate phosphorylated sites (+/-7AA) | Domain |
| MAPKAPK2 | P49137 | human | TAB3 | Q8N5C8 | S506 | HKYQRsSsSGSDDyA | |
| MAPKAPK2 | P49137 | human | CDC25B | P30305 | S169 | VLRNItNsQAPDGRR | M-inducer_phosp |
| MAPKAPK2 | P49137 | human | POU5F1 | Q01860 | S93 | PQGGLEtsQPEGEAG | |
| MAPKAPK2 | P49137 | human | HNRNPA0 | Q13151 | S84 | VELkRAVsREDsARP | |
| MAPKAPK2 | P49137 | human | GJA1 | P17302 | S255 | HAtsGALsPAkDCGs | |
| MAPKAPK2 | P49137 | human | HSPB1 | P04792 | S78 | PAysRALsRQLssGV | |
| MAPKAPK2 | P49137 | human | LIMK1 | P53667 | S323 | KDLGrSEsLRVVCRP | |
| MAPKAPK2 | P49137 | human | WNK4 | Q96J92 | S575 | PFLFRHAsYSSTTSD | |
| MAPKAPK2 | P49137 | human | CREB1 | P16220 | S119 | EILsRRPsyRkILND | pKID |
| MAPKAPK2 | P49137 | human | RCSD1 | Q6JBY9 | S179 | RRFRRsQsDCGELGD | CAP-ZIP_m |
| MAPKAPK2 | P49137 | human | MRTFA | Q969V6 | S333 | CGLARQNstsLtGKP | |
| MAPKAPK2 | P49137 | human | YWHAZ | P63104 | S58 | VVGARRssWRVVssI | 14-3-3 |
| MAPKAPK2 | P49137 | human | AGO2 | Q9UKV8 | S387 | SkLMRsAsFNtDPyV | ArgoL2 |
| MAPKAPK2 | P49137 | human | BAG2 | O95816 | S20 | GRFCRSSsMADRSSR | |
| MAPKAPK2 | P49137 | human | POU5F1 | Q01860 | S111 | EsNsDGAsPEPCtVt | |
| MAPKAPK2 | P49137 | human | MDM2 | Q00987 | S157 | SHLVSRPstSsRRRA | |
| MAPKAPK2 | P49137 | human | RBM7 | Q9Y580 | S204 | QrkVrMNsyPYLADr | |
| MAPKAPK2 | P49137 | human | MDM2 | Q00987 | S166 | SsRRRAIsEtEENsD | |
| MAPKAPK2 | P49137 | human | EEF2K | O00418 | S377 | PPLLRPLsENsGDEN | |
| MAPKAPK2 | P49137 | human | TH | P07101-4 | S44 | RFIGRRQsLIEDARK | TOH_N |
| MAPKAPK2 | P49137 | human | KRT18 | P05783 | S53 | IsVsrstsFrGGMGs | |
| MAPKAPK2 | P49137 | human | NELFE | P18615 | S51 | GGVkRsLsEQPVMDt | |
| MAPKAPK2 | P49137 | human | RBM7 | Q9Y580 | S136 | QIIQRsFssPENFQr | |
| MAPKAPK2 | P49137 | human | MAPKAPK2 | P49137 | T338 | VPQtPLHtsRVLKED | |
| MAPKAPK2 | P49137 | human | ZFP36L1 | Q07352 | S92 | RFRDRsFsEGGERLL | Tis11B_N |
| MAPKAPK2 | P49137 | human | RTN4 | Q9NQC3-2 | S107 | VAPERQPsWDPSPVS | |
| MAPKAPK2 | P49137 | human | PLK1 | P53350 | S326 | LTIPPRFsIAPssLD | |
| MAPKAPK2 | P49137 | human | CEP131 | Q9UPN4 | S47 | kPIVRsVsVVTGsEQ | |
| MAPKAPK2 | P49137 | human | RCSD1 | Q6JBY9 | S244 | PPLRRsPsRTEKQEE | RCSD |
| MAPKAPK2 | P49137 | human | AKT1 | P31749 | S473 | RPHFPQFsysAsGtA | Pkinase_C |
| MAPKAPK2 | P49137 | human | ARPC5 | O15511 | S77 | AVKDrAGsIVLKVLI | P16-Arc |
| MAPKAPK2 | P49137 | human | TP53 | P04637 | S20 | PLsQEtFsDLWkLLP | P53_TAD |
| MAPKAPK2 | P49137 | human | TRIM29 | Q14134 | S550 | GyPsLMRsQsPkAQP | |
| MAPKAPK2 | P49137 | human | HSPA1L | P34931 | S241 | DFDNRLVsHFVEEFK | HSP70 |
| MAPKAPK2 | P49137 | human | RPS6KA3 | P51812 | S386 | HQLFRGFsFVAItsD | Pkinase_C |
| MAPKAPK2 | P49137 | human | CDC25B | P30305 | S323 | QRLFRsPsMPCsVIR | M-inducer_phosp |
| MAPKAPK2 | P49137 | human | ZFP36L1 | Q07352 | S203 | PRLQHsFsFAGFPSA | |
| MAPKAPK2 | P49137 | human | TH | P07101-3 | S19 | KGFRRAVsELDAKQA | TOH_N |
| MAPKAPK2 | P49137 | human | PLK1 | P53350 | S383 | DMLQQLHsVNAskPS | |
| MAPKAPK2 | P49137 | human | PARN | O95453 | S557 | NHyYRNNsFTAPsTV | |
| MAPKAPK2 | P49137 | human | DDX5 | P17844 | S198 | CRACRLkstCIyGGA | DEAD |
| MAPKAPK2 | P49137 | human | TH | P07101-2 | S19 | KGFRRAVsELDAKQA | TOH_N |
| MAPKAPK2 | P49137 | human | TH | P07101-2 | S67 | RFIGRRQsLIEDARK | TOH_N |
| MAPKAPK2 | P49137 | human | RPS6KA1 | Q15418 | S380 | HQLFRGFsFVAtGLM | Pkinase_C |
| MAPKAPK2 | P49137 | human | GJA1 | P17302 | S279 | sSPtAPLsPMsPPGy | |
| MAPKAPK2 | P49137 | human | CEP131 | Q9UPN4 | S78 | NNLRRsNsttQVSQP | |
| MAPKAPK2 | P49137 | human | DAZL | Q92904 | S65 | SFFARYGsVkEVKII | RRM_1 |
| MAPKAPK2 | P49137 | human | RIPK1 | Q13546 | S320 | AVVkRMQsLQLdCVA | |
| MAPKAPK2 | P49137 | human | ZFP36L1 | Q07352 | S54 | GGFPRRHsVtLPSSk | Tis11B_N |
| MAPKAPK2 | P49137 | human | NELFE | P18615 | S251 | GPFRRsDsFPERRAP | |
| MAPKAPK2 | P49137 | human | COPS5 | Q92905 | S177 | IDPTRTIsAGkVNLG | |
| MAPKAPK2 | P49137 | human | TH | P07101-4 | S19 | KGFRRAVsELDAKQA | TOH_N |
| MAPKAPK2 | P49137 | human | SRF | P11831 | S103 | RGLKRsLsEMEIGMV | |
| MAPKAPK2 | P49137 | human | TRIM28 | Q13263 | S473 | sGVkRsRsGEGEVsG | |
| MAPKAPK2 | P49137 | human | GJA1 | P17302 | S282 | tAPLsPMsPPGykLV | |
| MAPKAPK2 | P49137 | human | TH | P07101 | S71 | RFIGRRQsLIEDARK | TOH_N |
| MAPKAPK2 | P49137 | human | NELFE | P18615 | S115 | QPFQRsIsADDDLQE | |
| MAPKAPK2 | P49137 | human | LSP1 | P33241 | S252 | PKLARQAsIELPsMA | Caldesmon |
| MAPKAPK2 | P49137 | human | PDE4D | Q08499-6 | S133 | sFLYRSDsDYDLSPk | |
| MAPKAPK2 | P49137 | human | PIAS1 | O75925 | S522 | DTSYINTsLIQDYRH | |
| MAPKAPK2 | P49137 | human | TH | P07101-3 | S404 | GELLHCLsEEPEIRA | Biopterin_H |
| MAPKAPK2 | P49137 | human | ALOX5 | P09917 | S272 | CsLERQLsLEQEVQQ | Lipoxygenase |
| MAPKAPK2 | P49137 | human | MRTFA | Q969V6 | S312 | tPPVRsLstTNSsSS | |
| MAPKAPK2 | P49137 | human | AATF | Q9NY61 | T366 | FTVYRNRtLQkWHDK | AATF-Che1 |
| MAPKAPK2 | P49137 | human | HSPB1 | P04792 | S15 | FsLLrGPsWDPFrDW | |
| MAPKAPK2 | P49137 | human | CDC25B | P30305 | S375 | ARVLRsksLCHDEIE | M-inducer_phosp |
| MAPKAPK2 | P49137 | human | TRIM29 | Q14134 | S552 | PsLMRsQsPkAQPQT | |
| MAPKAPK2 | P49137 | human | HSF1 | Q00613 | S121 | NIkRkVtsVSTLksE | |
| MAPKAPK2 | P49137 | human | BCL2L1 | Q07817 | S62 | PSWHLADsPAVNGAT | |
| MAPKAPK2 | P49137 | human | PFKFB3 | Q16875 | S461 | NPLMRRNsVtPLAsP | |
| MAPKAPK2 | P49137 | human | TSC2 | P49815 | S1254 | TALyksLsVPAASTA | |
| MAPKAPK2 | P49137 | human | TH | P07101 | S19 | KGFRRAVsELDAKQA | TOH_N |
| MAPKAPK2 | P49137 | human | CDC25C | P30307 | S216 | sGLyRsPsMPENLNR | M-inducer_phosp |
| MAPKAPK2 | P49137 | human | TH | P07101-3 | S40 | RFIGRRQsLIEDARK | TOH_N |
| MAPKAPK2 | P49137 | human | BECN1 | Q14457 | S90 | IPPARMMstEsANsF | |
| MAPKAPK2 | P49137 | human | KRT20 | P35900 | S13 | RSFHRsLsSSLQAPV | |
| MAPKAPK2 | P49137 | human | HSPB1 | P04792 | S82 | RALsRQLssGVsEIr | |
| MAPKAPK2 | P49137 | human | CDC25B | P30305 | S353 | VQNkRRRsVtPPEEQ | M-inducer_phosp |
 Biological Network Integration of This Kinase and Substrates Biological Network Integration of This Kinase and Substrates (GeneMANIA website) |
 Enriched GO biological processes of the phosphorylation target genes of the kinase Enriched GO biological processes of the phosphorylation target genes of the kinase |
| Kinase | GOID | GO term | P.adjust |
| MAPKAPK2 | ID | Description | 0.00e+00 |
| MAPKAPK2 | GO:0031330 | negative regulation of cellular catabolic process | 9.29e-06 |
| MAPKAPK2 | GO:0010586 | miRNA metabolic process | 1.44e-05 |
| MAPKAPK2 | GO:0031667 | response to nutrient levels | 3.85e-05 |
| MAPKAPK2 | GO:0034655 | nucleobase-containing compound catabolic process | 3.85e-05 |
| MAPKAPK2 | GO:0036293 | response to decreased oxygen levels | 5.86e-05 |
| MAPKAPK2 | GO:0062197 | cellular response to chemical stress | 5.86e-05 |
| MAPKAPK2 | GO:0070482 | response to oxygen levels | 9.77e-05 |
| MAPKAPK2 | GO:0009895 | negative regulation of catabolic process | 9.89e-05 |
| MAPKAPK2 | GO:1903311 | regulation of mRNA metabolic process | 1.19e-04 |
| MAPKAPK2 | GO:0006401 | RNA catabolic process | 1.21e-04 |
| MAPKAPK2 | GO:0071480 | cellular response to gamma radiation | 1.63e-04 |
| MAPKAPK2 | GO:0061614 | miRNA transcription | 2.51e-04 |
| MAPKAPK2 | GO:0016242 | negative regulation of macroautophagy | 2.51e-04 |
| MAPKAPK2 | GO:0006402 | mRNA catabolic process | 2.51e-04 |
| MAPKAPK2 | GO:0034504 | protein localization to nucleus | 2.51e-04 |
| MAPKAPK2 | GO:0051348 | negative regulation of transferase activity | 3.26e-04 |
| MAPKAPK2 | GO:0043488 | regulation of mRNA stability | 4.10e-04 |
| MAPKAPK2 | GO:0070918 | regulatory ncRNA processing | 4.37e-04 |
| MAPKAPK2 | GO:0043487 | regulation of RNA stability | 4.80e-04 |
| MAPKAPK2 | GO:0010507 | negative regulation of autophagy | 4.80e-04 |
| MAPKAPK2 | GO:0031669 | cellular response to nutrient levels | 4.80e-04 |
| MAPKAPK2 | GO:0043086 | negative regulation of catalytic activity | 4.80e-04 |
| MAPKAPK2 | GO:0061013 | regulation of mRNA catabolic process | 4.80e-04 |
| MAPKAPK2 | GO:0034599 | cellular response to oxidative stress | 4.97e-04 |
| MAPKAPK2 | GO:1901653 | cellular response to peptide | 6.03e-04 |
| MAPKAPK2 | GO:0035196 | miRNA processing | 6.72e-04 |
| MAPKAPK2 | GO:0001701 | in utero embryonic development | 7.84e-04 |
| MAPKAPK2 | GO:0031668 | cellular response to extracellular stimulus | 7.84e-04 |
| MAPKAPK2 | GO:2001233 | regulation of apoptotic signaling pathway | 7.84e-04 |
| MAPKAPK2 | GO:1901873 | regulation of post-translational protein modification | 7.84e-04 |
| MAPKAPK2 | GO:0010332 | response to gamma radiation | 7.84e-04 |
| MAPKAPK2 | GO:0010823 | negative regulation of mitochondrion organization | 8.14e-04 |
| MAPKAPK2 | GO:2001242 | regulation of intrinsic apoptotic signaling pathway | 8.14e-04 |
| MAPKAPK2 | GO:0018105 | peptidyl-serine phosphorylation | 9.06e-04 |
| MAPKAPK2 | GO:0001666 | response to hypoxia | 9.24e-04 |
| MAPKAPK2 | GO:0001558 | regulation of cell growth | 9.24e-04 |
| MAPKAPK2 | GO:1901800 | positive regulation of proteasomal protein catabolic process | 9.46e-04 |
| MAPKAPK2 | GO:0071248 | cellular response to metal ion | 9.46e-04 |
| MAPKAPK2 | GO:0035821 | modulation of process of another organism | 1.01e-03 |
| MAPKAPK2 | GO:0018209 | peptidyl-serine modification | 1.01e-03 |
| MAPKAPK2 | GO:0043434 | response to peptide hormone | 1.01e-03 |
| MAPKAPK2 | GO:0002763 | positive regulation of myeloid leukocyte differentiation | 1.11e-03 |
| MAPKAPK2 | GO:0097193 | intrinsic apoptotic signaling pathway | 1.19e-03 |
| MAPKAPK2 | GO:0045931 | positive regulation of mitotic cell cycle | 1.25e-03 |
| MAPKAPK2 | GO:0070301 | cellular response to hydrogen peroxide | 1.56e-03 |
| MAPKAPK2 | GO:0098781 | ncRNA transcription | 1.56e-03 |
| MAPKAPK2 | GO:1904375 | regulation of protein localization to cell periphery | 1.56e-03 |
| MAPKAPK2 | GO:0071241 | cellular response to inorganic substance | 1.56e-03 |
| MAPKAPK2 | GO:0044772 | mitotic cell cycle phase transition | 1.56e-03 |
Top |
Related Drugs to MAPKAPK2_SRGAP2 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
 Distribution of the number of studies mentioning MAPKAPK2-SRGAP2 and kinase inhibitors the PubMed Abstract (04-01-2024) Distribution of the number of studies mentioning MAPKAPK2-SRGAP2 and kinase inhibitors the PubMed Abstract (04-01-2024) |
| Fusion gene - drug pair 1 | Fusion gene - drug pair 2 | PMID | Publication date | DOI | Study title |
Top |
Related Diseases to MAPKAPK2_SRGAP2 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Related diseases from the literature mentioned this fusion gene and drug. Related diseases from the literature mentioned this fusion gene and drug. (PubMed, 04-01-2024) |
| MeSH ID | MeSH term |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
Top |
Clinical Trials of the Found Drugs/Small Molecules |
 Statistics of the Clinical Trials of the Found Kinase Inibitors from clinicaltrials.gov (06-17-2024) Statistics of the Clinical Trials of the Found Kinase Inibitors from clinicaltrials.gov (06-17-2024) |
 Clinical Trials from clinicaltrials.gov (06-17-2024) Clinical Trials from clinicaltrials.gov (06-17-2024) |
| Fusion Gene | Kinase Inhibitor | NCT ID | Study Status | Phases | Disease | # Enrolment | Date |

