| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Kinase Fusion Gene:MARK2_PLA2G16 |
Kinase Fusion Protein Summary |
 Kinase Fusion gene summary Kinase Fusion gene summary |
| Kinase Fusion partner gene information | Kinase Fusion gene name: MARK2_PLA2G16 | KinaseFusionDB ID: KFG3634 | FusionGDB2.0 ID: KFG3634 | Hgene | Tgene | Gene symbol | MARK2 | PLA2G16 | Gene ID | 2011 | 11145 | |
| Gene name | microtubule affinity regulating kinase 2 | phospholipase A and acyltransferase 3 | ||||||||||
| Synonyms | EMK-1|EMK1|PAR-1|Par-1b|Par1b | AdPLA|FPLD9|H-REV107|H-REV107-1|HRASLS3|HREV107|HREV107-1|HREV107-3|HRSL3|PLA2G16|PLAAT-3 | ||||||||||
| Cytomap | 11q13.1 | 11q12.3-q13.1 | ||||||||||
| Type of gene | protein-coding | protein-coding | ||||||||||
| Description | serine/threonine-protein kinase MARK2ELKL motif kinase 1MAP/microtubule affinity-regulating kinase 2PAR1 homolog bSer/Thr protein kinase PAR-1Bserine/threonine protein kinase EMKtesticular tissue protein Li 117 | phospholipase A and acyltransferase 3Ca-independent phospholipase A1/2H-rev 107 protein homologHRAS-like suppressor 1HRAS-like suppressor 3adipose-specific PLA2adipose-specific phospholipase A2group XVI phospholipase A1/A2group XVI phospholipase A | ||||||||||
| Modification date | 20240407 | 20240305 | ||||||||||
| UniProtAcc | Q7KZI7 | P53816 | ||||||||||
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000315032, ENST00000350490, ENST00000361128, ENST00000377809, ENST00000402010, ENST00000413835, ENST00000502399, ENST00000508192, ENST00000377810, ENST00000408948, ENST00000425897, ENST00000509502, ENST00000513765, | ENST00000394613, ENST00000323646, ENST00000415826, | |||||||||
| Context (manual curation of fusion genes in KinaseFusionDB) | PubMed: MARK2 [Title/Abstract] AND PLA2G16 [Title/Abstract] AND fusion [Title/Abstract] | |||||||||||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | MARK2(63607032)-PLA2G16(63342518), # samples:1 | |||||||||||
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | MARK2 | GO:0006468 | protein phosphorylation | 14976552 |
| Hgene | MARK2 | GO:0010976 | positive regulation of neuron projection development | 12429843 |
| Hgene | MARK2 | GO:0018105 | peptidyl-serine phosphorylation | 10542369|16717194 |
| Hgene | MARK2 | GO:0030010 | establishment of cell polarity | 12429843 |
| Hgene | MARK2 | GO:0035556 | intracellular signal transduction | 14976552 |
| Hgene | MARK2 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity | 15324659 |
| Hgene | MARK2 | GO:0070507 | regulation of microtubule cytoskeleton organization | 10542369 |
| Tgene | PLA2G16 | GO:0006644 | phospholipid metabolic process | 20100577 |
| Tgene | PLA2G16 | GO:0070292 | N-acylphosphatidylethanolamine metabolic process | 22825852 |
 Kinase Fusion gene breakpoints across MARK2 (5'-gene) Kinase Fusion gene breakpoints across MARK2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
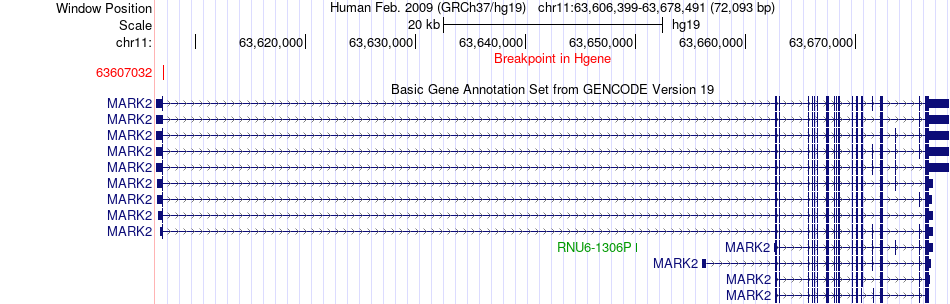 |
 Kinase Fusion gene breakpoints across PLA2G16 (3'-gene) Kinase Fusion gene breakpoints across PLA2G16 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
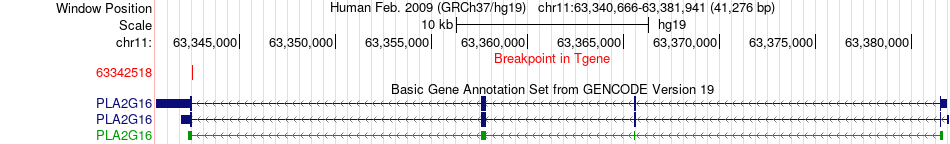 |
Top |
Kinase Fusion Gene Sample Information |
 Kinase Fusion gene information. Kinase Fusion gene information. |
 Kinase Fusion gene information from four resources (ChiTars 5.0, ChimerDB 4.0, COSMIC, and CCLE) Kinase Fusion gene information from four resources (ChiTars 5.0, ChimerDB 4.0, COSMIC, and CCLE)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Sample | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp |
| ChimerDB4 | TCGA-BR-A44T-01A | MARK2 | chr11 | 63607032 | PLA2G16 | chr11 | 63342518 |
Top |
Kinase Fusion ORF Analysis |
 Kinase Fusion information from ORFfinder translation from full-length transcript sequence from KinaseFusionDB. Kinase Fusion information from ORFfinder translation from full-length transcript sequence from KinaseFusionDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp | Seq length (transcript) | Seq length (amino acids) |
Top |
Kinase Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from KinaseFusionDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from KinaseFusionDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >Henst_Tenst_Hgene_Hchr_Hbp_Tgene_Tchr_Tbp_length(fusion AA)_AAseq |
Multiple Sequence Alignment of All Fusion Protein Isoforms |
Top |
Kinase Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr11:63607032/chr11:63342518) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MARK2 | PLA2G16 |
| FUNCTION: Serine/threonine-protein kinase (PubMed:23666762). Involved in cell polarity and microtubule dynamics regulation. Phosphorylates CRTC2/TORC2, DCX, HDAC7, KIF13B, MAP2, MAP4 and RAB11FIP2. Phosphorylates the microtubule-associated protein MAPT/TAU (PubMed:23666762). Plays a key role in cell polarity by phosphorylating the microtubule-associated proteins MAP2, MAP4 and MAPT/TAU at KXGS motifs, causing detachment from microtubules, and their disassembly. Regulates epithelial cell polarity by phosphorylating RAB11FIP2. Involved in the regulation of neuronal migration through its dual activities in regulating cellular polarity and microtubule dynamics, possibly by phosphorylating and regulating DCX. Regulates axogenesis by phosphorylating KIF13B, promoting interaction between KIF13B and 14-3-3 and inhibiting microtubule-dependent accumulation of KIF13B. Also required for neurite outgrowth and establishment of neuronal polarity. Regulates localization and activity of some histone deacetylases by mediating phosphorylation of HDAC7, promoting subsequent interaction between HDAC7 and 14-3-3 and export from the nucleus. Also acts as a positive regulator of the Wnt signaling pathway, probably by mediating phosphorylation of dishevelled proteins (DVL1, DVL2 and/or DVL3). Modulates the developmental decision to build a columnar versus a hepatic epithelial cell apparently by promoting a switch from a direct to a transcytotic mode of apical protein delivery. Essential for the asymmetric development of membrane domains of polarized epithelial cells. {ECO:0000269|PubMed:11433294, ECO:0000269|PubMed:12429843, ECO:0000269|PubMed:14976552, ECO:0000269|PubMed:15158914, ECO:0000269|PubMed:15324659, ECO:0000269|PubMed:15365179, ECO:0000269|PubMed:16775013, ECO:0000269|PubMed:16980613, ECO:0000269|PubMed:18626018, ECO:0000269|PubMed:20194617, ECO:0000269|PubMed:23666762}. | FUNCTION: Exhibits both phospholipase A1/2 and acyltransferase activities (PubMed:19615464, PubMed:19047760, PubMed:22825852, PubMed:22605381, PubMed:26503625). Shows phospholipase A1 (PLA1) and A2 (PLA2) activity, catalyzing the calcium-independent release of fatty acids from the sn-1 or sn-2 position of glycerophospholipids (PubMed:19615464, PubMed:19047760, PubMed:22825852, PubMed:22605381, PubMed:22923616). For most substrates, PLA1 activity is much higher than PLA2 activity (PubMed:19615464). Shows O-acyltransferase activity,catalyzing the transfer of a fatty acyl group from glycerophospholipid to the hydroxyl group of lysophospholipid (PubMed:19615464). Shows N-acyltransferase activity, catalyzing the calcium-independent transfer of a fatty acyl group at the sn-1 position of phosphatidylcholine (PC) and other glycerophospholipids to the primary amine of phosphatidylethanolamine (PE), forming N-acylphosphatidylethanolamine (NAPE), which serves as precursor for N-acylethanolamines (NAEs) (PubMed:19615464, PubMed:19047760, PubMed:22825852, PubMed:22605381). Exhibits high N-acyltransferase activity and low phospholipase A1/2 activity (PubMed:22825852). Required for complete organelle rupture and degradation that occur during eye lens terminal differentiation, when fiber cells that compose the lens degrade all membrane-bound organelles in order to provide lens with transparency to allow the passage of light. Organelle membrane degradation is probably catalyzed by the phospholipase activity (By similarity). {ECO:0000250|UniProtKB:Q8R3U1, ECO:0000269|PubMed:19047760, ECO:0000269|PubMed:19615464, ECO:0000269|PubMed:22605381, ECO:0000269|PubMed:22825852, ECO:0000269|PubMed:22923616, ECO:0000303|PubMed:26503625}.; FUNCTION: (Microbial infection) Acts as a host factor for picornaviruses: required during early infection to promote viral genome release into the cytoplasm (PubMed:28077878). May act as a cellular sensor of membrane damage at sites of virus entry, which relocalizes to sites of membrane rupture upon virus unfection (PubMed:28077878). Facilitates safe passage of the RNA away from LGALS8, enabling viral genome translation by host ribosome (PubMed:28077878). May also be involved in initiating pore formation, increasing pore size or in maintaining pores for genome delivery (PubMed:28077878). The lipid-modifying enzyme activity is required for this process (PubMed:28077878). {ECO:0000269|PubMed:28077878}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
 - Retained domain in the 5'-partner of fusion protein (protein functional feature from UniProt). - Retained domain in the 5'-partner of fusion protein (protein functional feature from UniProt). |
| Partner | Hgeneene | Hbp | Tgeneene | Tbp | ENST | BPexon | TotalExon | Protein feature loci | BPloci | TotalLen | Feature | Note |
 - Retained domain in the 3'-partner of fusion protein (protein functional feature from UniProt). - Retained domain in the 3'-partner of fusion protein (protein functional feature from UniProt). |
| Partner | Hgeneene | Hbp | Tgeneene | Tbp | ENST | BPexon | TotalExon | Protein feature loci | BPloci | TotalLen | Feature | Note |
Top |
Kinase-Substrate Information of MARK2_PLA2G16 |
 Phosphorylation target of the kinase Phosphorylation target of the kinase(phosphosite, 03-17-2024) |
| Kinase | Kinase UniProt Acc | Kinase species | Substrate | Substrate UniProt Acc | Substrate phosphorylated residues | Substrate phosphorylated sites (+/-7AA) | Domain |
| MARK2 | Q7KZI7 | human | KIF13B | Q9NQT8 | S1410 | SNKGRWEsQQDVsQT | |
| MARK2 | Q7KZI7 | human | ARHGEF2 | Q92974 | S186 | NMRNRtLsVEsLIDE | |
| MARK2 | Q7KZI7 | human | RAB11FIP2 | Q7L804 | S227 | QRLsSAHsMsDLSGS | |
| MARK2 | Q7KZI7 | human | MAPT | P10636-8 | S262 | NVKskIGstENLkHQ | Tubulin-binding |
| MARK2 | Q7KZI7 | human | HDAC4 | P56524 | S467 | RPLGRtQsAPLPQNA | |
| MARK2 | Q7KZI7 | human | MAPT | P10636-8 | S324 | kVTskCGsLGNIHHk | Tubulin-binding |
| MARK2 | Q7KZI7 | human | DLG4 | P78352 | S561 | EFPDKFGsCVPHTTR | Guanylate_kin |
| MARK2 | Q7KZI7 | human | STK3 | Q13188 | S15 | KSKLKkLsEDsLTkQ | |
| MARK2 | Q7KZI7 | human | ARHGEF2 | Q92974-2 | S885 | PVDPRRRsLPAGDAL | |
| MARK2 | Q7KZI7 | human | PINK1 | Q9BXM7 | T313 | EGLGHGRtLFLVMkN | Pkinase |
| MARK2 | Q7KZI7 | human | UTRN | P46939 | S1258 | TLEERMKsTEVLPEk | |
| MARK2 | Q7KZI7 | human | ARHGEF2 | Q92974 | S143 | GsRRGRssLsLAksV | |
| MARK2 | Q7KZI7 | human | RAB11FIP1 | Q6WKZ4-3 | S234 | QKTPLSQsMSVLPTS | |
| MARK2 | Q7KZI7 | human | HDAC7 | Q8WUI4 | S155 | FPLRKtVsEPNLkLR | |
| MARK2 | Q7KZI7 | human | HDAC4 | P56524 | S246 | FPLRkTAsEPNLKLR | |
| MARK2 | Q7KZI7 | human | PARD3 | Q8TEW0 | S873 | VDDQKAGsPsRDVGP | |
| MARK2 | Q7KZI7 | human | MARK2 | Q7KZI7 | S400 | HkVQRsVsANPKQRR | |
| MARK2 | Q7KZI7 | human | PTK2 | Q05397 | Y397 | sVsEtDDyAEIIDEE | |
| MARK2 | Q7KZI7 | human | ARHGEF2 | Q92974-2 | S959 | SRLSPPHsPRDFTRM | |
| MARK2 | Q7KZI7 | human | MAPT | P10636-8 | S356 | rVQskIGsLDNItHV | Tubulin-binding |
| MARK2 | Q7KZI7 | human | FEZ1 | Q99689 | S58 | SEIISFKsMEDLVNE | FEZ |
| MARK2 | Q7KZI7 | human | RNF41 | Q9H4P4 | S254 | IENAHERsWPQGLAT | USP8_interact |
| MARK2 | Q7KZI7 | human | MYL9 | P24844 | S20 | KRPQRAtsNVFAMFD | |
| MARK2 | Q7KZI7 | human | BAIAP2 | Q9UQB8 | S366 | ktLPRsssMAAGLER | |
| MARK2 | Q7KZI7 | human | ARHGEF2 | Q92974 | S172 | LGLRRILsQstDsLN | |
| MARK2 | Q7KZI7 | human | HDAC5 | Q9UQL6 | S259 | FPLRkTAsEPNLKVR | |
| MARK2 | Q7KZI7 | human | PARD3 | Q8TEW0 | S144 | PLHVRRssDPALIGL | |
| MARK2 | Q7KZI7 | human | EIF2S1 | P05198 | S52 | MILLsELsRRRIRsI | S1 |
| MARK2 | Q7KZI7 | human | KIF13B | Q9NQT8 | S1381 | KLSRRsIssPNVNRL |
 Biological Network Integration of This Kinase and Substrates Biological Network Integration of This Kinase and Substrates (GeneMANIA website) |
 Enriched GO biological processes of the phosphorylation target genes of the kinase Enriched GO biological processes of the phosphorylation target genes of the kinase |
| Kinase | GOID | GO term | P.adjust |
| MARK2 | ID | Description | 0.00e+00 |
| MARK2 | GO:0007163 | establishment or maintenance of cell polarity | 9.52e-05 |
| MARK2 | GO:0007409 | axonogenesis | 1.17e-04 |
| MARK2 | GO:0030010 | establishment of cell polarity | 1.64e-04 |
| MARK2 | GO:0016242 | negative regulation of macroautophagy | 1.20e-03 |
| MARK2 | GO:0043393 | regulation of protein binding | 2.17e-03 |
| MARK2 | GO:0051646 | mitochondrion localization | 2.17e-03 |
| MARK2 | GO:0010823 | negative regulation of mitochondrion organization | 2.17e-03 |
| MARK2 | GO:0033674 | positive regulation of kinase activity | 2.17e-03 |
| MARK2 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity | 2.17e-03 |
| MARK2 | GO:0031346 | positive regulation of cell projection organization | 2.17e-03 |
| MARK2 | GO:0035088 | establishment or maintenance of apical/basal cell polarity | 2.17e-03 |
| MARK2 | GO:0061245 | establishment or maintenance of bipolar cell polarity | 2.17e-03 |
| MARK2 | GO:0006476 | protein deacetylation | 2.17e-03 |
| MARK2 | GO:0010639 | negative regulation of organelle organization | 2.17e-03 |
| MARK2 | GO:0035601 | protein deacylation | 3.02e-03 |
| MARK2 | GO:0098732 | macromolecule deacylation | 3.23e-03 |
| MARK2 | GO:0051347 | positive regulation of transferase activity | 3.35e-03 |
| MARK2 | GO:0035418 | protein localization to synapse | 3.97e-03 |
| MARK2 | GO:1902902 | negative regulation of autophagosome assembly | 4.22e-03 |
| MARK2 | GO:0010975 | regulation of neuron projection development | 4.36e-03 |
| MARK2 | GO:0051091 | positive regulation of DNA-binding transcription factor activity | 4.48e-03 |
| MARK2 | GO:0010507 | negative regulation of autophagy | 4.48e-03 |
| MARK2 | GO:0032273 | positive regulation of protein polymerization | 4.48e-03 |
| MARK2 | GO:0022604 | regulation of cell morphogenesis | 4.48e-03 |
| MARK2 | GO:0051896 | regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction | 4.48e-03 |
| MARK2 | GO:0010832 | negative regulation of myotube differentiation | 4.48e-03 |
| MARK2 | GO:1901524 | regulation of mitophagy | 4.48e-03 |
| MARK2 | GO:0061912 | selective autophagy | 4.48e-03 |
| MARK2 | GO:0032886 | regulation of microtubule-based process | 4.48e-03 |
| MARK2 | GO:0000422 | autophagy of mitochondrion | 4.48e-03 |
| MARK2 | GO:0061726 | mitochondrion disassembly | 4.48e-03 |
| MARK2 | GO:0051098 | regulation of binding | 4.48e-03 |
| MARK2 | GO:0019896 | axonal transport of mitochondrion | 4.76e-03 |
| MARK2 | GO:0008356 | asymmetric cell division | 5.16e-03 |
| MARK2 | GO:1902414 | protein localization to cell junction | 5.28e-03 |
| MARK2 | GO:0045860 | positive regulation of protein kinase activity | 5.28e-03 |
| MARK2 | GO:0043491 | phosphatidylinositol 3-kinase/protein kinase B signal transduction | 5.72e-03 |
| MARK2 | GO:0031644 | regulation of nervous system process | 6.59e-03 |
| MARK2 | GO:0062197 | cellular response to chemical stress | 7.62e-03 |
| MARK2 | GO:0034643 | establishment of mitochondrion localizatio | 3.42e-04 |
| MARK2 | GO:0016236 | macroautophagy | 8.34e-03 |
| MARK2 | GO:0008360 | regulation of cell shape | 9.38e-03 |
| MARK2 | GO:0009895 | negative regulation of catabolic process | 9.46e-03 |
| MARK2 | GO:0051654 | establishment of mitochondrion localization | 9.46e-03 |
| MARK2 | GO:0090140 | regulation of mitochondrial fission | 9.46e-03 |
| MARK2 | GO:1903008 | organelle disassembly | 9.46e-03 |
| MARK2 | GO:0010506 | regulation of autophagy | 9.46e-03 |
| MARK2 | GO:2000377 | regulation of reactive oxygen species metabolic process | 9.46e-03 |
| MARK2 | GO:0034063 | stress granule assembly | 9.46e-03 |
Top |
Related Drugs to MARK2_PLA2G16 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
 Distribution of the number of studies mentioning MARK2-PLA2G16 and kinase inhibitors the PubMed Abstract (04-01-2024) Distribution of the number of studies mentioning MARK2-PLA2G16 and kinase inhibitors the PubMed Abstract (04-01-2024) |
| Fusion gene - drug pair 1 | Fusion gene - drug pair 2 | PMID | Publication date | DOI | Study title |
Top |
Related Diseases to MARK2_PLA2G16 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Related diseases from the literature mentioned this fusion gene and drug. Related diseases from the literature mentioned this fusion gene and drug. (PubMed, 04-01-2024) |
| MeSH ID | MeSH term |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
Top |
Clinical Trials of the Found Drugs/Small Molecules |
 Statistics of the Clinical Trials of the Found Kinase Inibitors from clinicaltrials.gov (06-17-2024) Statistics of the Clinical Trials of the Found Kinase Inibitors from clinicaltrials.gov (06-17-2024) |
 Clinical Trials from clinicaltrials.gov (06-17-2024) Clinical Trials from clinicaltrials.gov (06-17-2024) |
| Fusion Gene | Kinase Inhibitor | NCT ID | Study Status | Phases | Disease | # Enrolment | Date |

