| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Kinase Fusion Gene:MCM5_LYN |
Kinase Fusion Protein Summary |
 Kinase Fusion gene summary Kinase Fusion gene summary |
| Kinase Fusion partner gene information | Kinase Fusion gene name: MCM5_LYN | KinaseFusionDB ID: KFG3732 | FusionGDB2.0 ID: KFG3732 | Hgene | Tgene | Gene symbol | MCM5 | LYN | Gene ID | 4174 | 4067 | |
| Gene name | minichromosome maintenance complex component 5 | LYN proto-oncogene, Src family tyrosine kinase | ||||||||||
| Synonyms | CDC46|MGORS8|P1-CDC46 | JTK8|SAIDV|p53Lyn|p56Lyn | ||||||||||
| Cytomap | 22q12.3 | 8q12.1 | ||||||||||
| Type of gene | protein-coding | protein-coding | ||||||||||
| Description | DNA replication licensing factor MCM5CDC46 homologMCM5 minichromosome maintenance deficient 5, cell division cycle 46minichromosome maintenance deficient 5 (cell division cycle 46) | tyrosine-protein kinase Lynlck/Yes-related novel protein tyrosine kinasev-yes-1 Yamaguchi sarcoma viral related oncogene homolog | ||||||||||
| Modification date | 20240403 | 20240411 | ||||||||||
| UniProtAcc | P33992 | P07948 | ||||||||||
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000216122, ENST00000382011, ENST00000465557, | ENST00000420292, ENST00000519728, ENST00000520220, | |||||||||
| Context (manual curation of fusion genes in KinaseFusionDB) | PubMed: MCM5 [Title/Abstract] AND LYN [Title/Abstract] AND fusion [Title/Abstract] | |||||||||||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | MCM5(35799535)-LYN(56910905), # samples:3 | |||||||||||
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | MCM5 | GO:0006268 | DNA unwinding involved in DNA replication | 22474384 |
| Tgene | LYN | GO:0000077 | DNA damage checkpoint signaling | 10891478 |
| Tgene | LYN | GO:0006468 | protein phosphorylation | 11517336 |
| Tgene | LYN | GO:0006974 | DNA damage response | 11517336 |
| Tgene | LYN | GO:0018108 | peptidyl-tyrosine phosphorylation | 7682714|11782428 |
| Tgene | LYN | GO:0030222 | eosinophil differentiation | 11823534 |
| Tgene | LYN | GO:0038043 | interleukin-5-mediated signaling pathway | 11823534 |
| Tgene | LYN | GO:0043410 | positive regulation of MAPK cascade | 10891478 |
| Tgene | LYN | GO:0046777 | protein autophosphorylation | 7682714 |
 Kinase Fusion gene breakpoints across MCM5 (5'-gene) Kinase Fusion gene breakpoints across MCM5 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
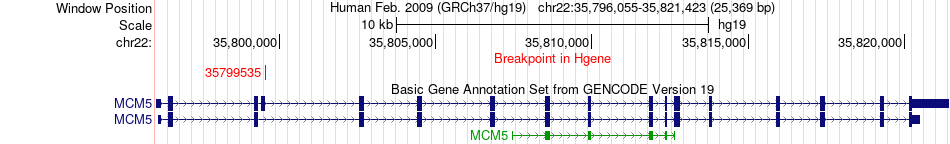 |
 Kinase Fusion gene breakpoints across LYN (3'-gene) Kinase Fusion gene breakpoints across LYN (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
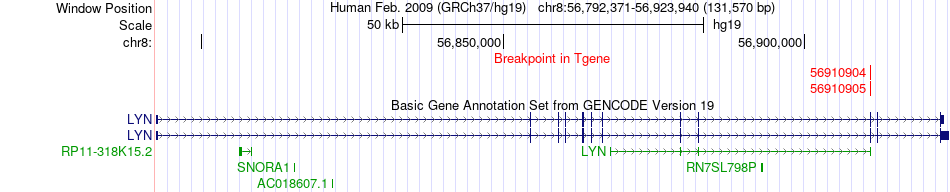 |
Top |
Kinase Fusion Gene Sample Information |
 Kinase Fusion gene information. Kinase Fusion gene information. |
 Kinase Fusion gene information from four resources (ChiTars 5.0, ChimerDB 4.0, COSMIC, and CCLE) Kinase Fusion gene information from four resources (ChiTars 5.0, ChimerDB 4.0, COSMIC, and CCLE)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Sample | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp |
| ChimerDB4 | TCGA-25-1329-01A | MCM5 | chr22 | 35799535 | LYN | chr8 | 56910905 |
| ChimerDB4 | TCGA-25-1329 | MCM5 | chr22 | 35799535 | LYN | chr8 | 56910904 |
Top |
Kinase Fusion ORF Analysis |
 Kinase Fusion information from ORFfinder translation from full-length transcript sequence from KinaseFusionDB. Kinase Fusion information from ORFfinder translation from full-length transcript sequence from KinaseFusionDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp | Seq length (transcript) | Seq length (amino acids) |
Top |
Kinase Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from KinaseFusionDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from KinaseFusionDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >Henst_Tenst_Hgene_Hchr_Hbp_Tgene_Tchr_Tbp_length(fusion AA)_AAseq |
Multiple Sequence Alignment of All Fusion Protein Isoforms |
Top |
Kinase Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr22:35799535/chr8:56910905) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MCM5 | LYN |
| FUNCTION: Acts as a component of the MCM2-7 complex (MCM complex) which is the replicative helicase essential for 'once per cell cycle' DNA replication initiation and elongation in eukaryotic cells. Core component of CDC45-MCM-GINS (CMG) helicase, the molecular machine that unwinds template DNA during replication, and around which the replisome is built (PubMed:32453425, PubMed:34694004, PubMed:34700328, PubMed:35585232, PubMed:16899510). The active ATPase sites in the MCM2-7 ring are formed through the interaction surfaces of two neighboring subunits such that a critical structure of a conserved arginine finger motif is provided in trans relative to the ATP-binding site of the Walker A box of the adjacent subunit. The six ATPase active sites, however, are likely to contribute differentially to the complex helicase activity (PubMed:32453425). {ECO:0000269|PubMed:16899510, ECO:0000269|PubMed:32453425, ECO:0000269|PubMed:34694004, ECO:0000269|PubMed:34700328, ECO:0000269|PubMed:35585232}. | FUNCTION: Non-receptor tyrosine-protein kinase that transmits signals from cell surface receptors and plays an important role in the regulation of innate and adaptive immune responses, hematopoiesis, responses to growth factors and cytokines, integrin signaling, but also responses to DNA damage and genotoxic agents. Functions primarily as negative regulator, but can also function as activator, depending on the context. Required for the initiation of the B-cell response, but also for its down-regulation and termination. Plays an important role in the regulation of B-cell differentiation, proliferation, survival and apoptosis, and is important for immune self-tolerance. Acts downstream of several immune receptors, including the B-cell receptor, CD79A, CD79B, CD5, CD19, CD22, FCER1, FCGR2, FCGR1A, TLR2 and TLR4. Plays a role in the inflammatory response to bacterial lipopolysaccharide. Mediates the responses to cytokines and growth factors in hematopoietic progenitors, platelets, erythrocytes, and in mature myeloid cells, such as dendritic cells, neutrophils and eosinophils. Acts downstream of EPOR, KIT, MPL, the chemokine receptor CXCR4, as well as the receptors for IL3, IL5 and CSF2. Plays an important role in integrin signaling. Regulates cell proliferation, survival, differentiation, migration, adhesion, degranulation, and cytokine release. Down-regulates signaling pathways by phosphorylation of immunoreceptor tyrosine-based inhibitory motifs (ITIM), that then serve as binding sites for phosphatases, such as PTPN6/SHP-1, PTPN11/SHP-2 and INPP5D/SHIP-1, that modulate signaling by dephosphorylation of kinases and their substrates. Phosphorylates LIME1 in response to CD22 activation. Phosphorylates BTK, CBL, CD5, CD19, CD72, CD79A, CD79B, CSF2RB, DOK1, HCLS1, LILRB3/PIR-B, MS4A2/FCER1B, SYK and TEC. Promotes phosphorylation of SIRPA, PTPN6/SHP-1, PTPN11/SHP-2 and INPP5D/SHIP-1. Mediates phosphorylation of the BCR-ABL fusion protein. Required for rapid phosphorylation of FER in response to FCER1 activation. Mediates KIT phosphorylation. Acts as an effector of EPOR (erythropoietin receptor) in controlling KIT expression and may play a role in erythroid differentiation during the switch between proliferation and maturation. Depending on the context, activates or inhibits several signaling cascades. Regulates phosphatidylinositol 3-kinase activity and AKT1 activation. Regulates activation of the MAP kinase signaling cascade, including activation of MAP2K1/MEK1, MAPK1/ERK2, MAPK3/ERK1, MAPK8/JNK1 and MAPK9/JNK2. Mediates activation of STAT5A and/or STAT5B. Phosphorylates LPXN on 'Tyr-72'. Kinase activity facilitates TLR4-TLR6 heterodimerization and signal initiation. Phosphorylates SCIMP on 'Tyr-107'; this enhances binding of SCIMP to TLR4, promoting the phosphorylation of TLR4, and a selective cytokine response to lipopolysaccharide in macrophages (By similarity). Phosphorylates CLNK (By similarity). Phosphorylates BCAR1/CAS and NEDD9/HEF1 (PubMed:9020138). {ECO:0000250|UniProtKB:P25911, ECO:0000269|PubMed:10574931, ECO:0000269|PubMed:10748115, ECO:0000269|PubMed:10891478, ECO:0000269|PubMed:11435302, ECO:0000269|PubMed:11517336, ECO:0000269|PubMed:11825908, ECO:0000269|PubMed:14726379, ECO:0000269|PubMed:15795233, ECO:0000269|PubMed:16467205, ECO:0000269|PubMed:17640867, ECO:0000269|PubMed:17977829, ECO:0000269|PubMed:18056483, ECO:0000269|PubMed:18070987, ECO:0000269|PubMed:18235045, ECO:0000269|PubMed:18577747, ECO:0000269|PubMed:18802065, ECO:0000269|PubMed:19290919, ECO:0000269|PubMed:20037584, ECO:0000269|PubMed:7687428, ECO:0000269|PubMed:9020138}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
 - Retained domain in the 5'-partner of fusion protein (protein functional feature from UniProt). - Retained domain in the 5'-partner of fusion protein (protein functional feature from UniProt). |
| Partner | Hgeneene | Hbp | Tgeneene | Tbp | ENST | BPexon | TotalExon | Protein feature loci | BPloci | TotalLen | Feature | Note |
 - Retained domain in the 3'-partner of fusion protein (protein functional feature from UniProt). - Retained domain in the 3'-partner of fusion protein (protein functional feature from UniProt). |
| Partner | Hgeneene | Hbp | Tgeneene | Tbp | ENST | BPexon | TotalExon | Protein feature loci | BPloci | TotalLen | Feature | Note |
Top |
Kinase-Substrate Information of MCM5_LYN |
 Phosphorylation target of the kinase Phosphorylation target of the kinase(phosphosite, 03-17-2024) |
| Kinase | Kinase UniProt Acc | Kinase species | Substrate | Substrate UniProt Acc | Substrate phosphorylated residues | Substrate phosphorylated sites (+/-7AA) | Domain |
| LYN | P07948 | human | NMT1 | P30419 | Y180 | YTLLNENyVEDDDNM | NMT |
| LYN | P07948 | human | ABL1 | P00519 | Y393 | RLMtGDtytAHAGAk | PK_Tyr_Ser-Thr |
| LYN | P07948 | human | YY1 | P25490 | Y8 | MASGDTLyIATDGSE | |
| LYN | P07948 | human | AKAP8 | O43823 | Y53 | TTGATySyGPASWEA | |
| LYN | P07948 | human | GCSAM | Q8N6F7 | Y106 | sGNsAEEyyENVPCk | HGAL |
| LYN | P07948 | human | ACLY | P53396 | Y682 | sRtTDGVyEGVAIGG | Ligase_CoA |
| LYN | P07948 | human | RGS16 | O15492 | Y177 | RFLkSPAyRDLAAQA | RGS |
| LYN | P07948 | human | SLC4A1 | P02730 | Y21 | ENLEQEEyEDPDIPE | |
| LYN | P07948 | human | SLC4A1 | P02730 | Y8 | MEELQDDyEDMMEEN | |
| LYN | P07948 | human | PTPN6 | P29350 | Y564 | sKHkEDVyENLHTkN | |
| LYN | P07948 | human | FCGR2A | P12318 | Y304 | tDDDKNIyLTLPPND | |
| LYN | P07948 | human | ACLY | P53396 | Y131 | YATREGDyVLFHHEG | |
| LYN | P07948 | human | CSF2RB | P32927 | Y882 | KALKQQDyLSLPPWE | |
| LYN | P07948 | human | RMDN3 | Q96TC7 | Y176 | DAESEGGytTANAEs | |
| LYN | P07948 | human | CSF2RB | P32927 | Y822 | VLQQVGDyCFLPGLG | |
| LYN | P07948 | human | TRIM28 | Q13263 | Y517 | PGsTtEDyNLIVIER | |
| LYN | P07948 | human | CASP8 | Q14790 | Y380 | tDsEEQPyLEMDLSs | Peptidase_C14 |
| LYN | P07948 | human | SYK | P43405 | Y323 | sTVsFNPyEPELAPW | |
| LYN | P07948 | human | CSF2RB | P32927 | Y466 | ALRFCGIyGyRLRRk | |
| LYN | P07948 | human | CASP8 | Q14790 | Y448 | TILTEVNyEVSNKDD | Peptidase_C14 |
| LYN | P07948 | human | CD19 | P15391 | Y500 | TSLGsQsyEDMRGIL | |
| LYN | P07948 | human | IRF5 | Q13568 | Y335 | QLQGQDLyAIRLCQC | IRF-3 |
| LYN | P07948 | human | LY96 | Q9Y6Y9 | Y131 | IKFSKGKyKCVVEAI | E1_DerP2_DerF2 |
| LYN | P07948 | human | YY1 | P25490 | Y383 | IHtGDrPyVCPFDGC | zf-C2H2 |
| LYN | P07948 | human | SLC4A1 | P02730 | Y359 | AKPDssFyKGLDLNG | |
| LYN | P07948 | human | ABL1 | P00519 | Y128 | SLEKHSWyHGPVSRN | SH2 |
| LYN | P07948 | human | AKAP8 | O43823 | Y154 | RPsySyDyEFDLGSD | |
| LYN | P07948 | human | CDKN1B | P46527 | Y88 | kGsLPEFyyRPPRPP | |
| LYN | P07948 | human | FCGR2B | P31994 | Y292 | GAENTITysLLMHPD | |
| LYN | P07948 | human | TRIM28 | Q13263 | Y449 | PMEVQEGyGFGsGDD | |
| LYN | P07948 | human | AKAP8 | O43823 | Y539 | IVkMLEkyLKGEDPF | AKAP95 |
| LYN | P07948 | human | PTEN | P60484 | Y240 | RREDKFMyFEFPQPL | PTEN_C2 |
| LYN | P07948 | human | LAT2 | Q9GZY6 | Y95 | EDPASSRyQNFSkGs | LAT2 |
| LYN | P07948 | human | CBL | P22681 | Y371 | TQEQyELyCEMGSTF | |
| LYN | P07948 | human | ABL1 | P00519 | Y226 | KRNKPtVyGVsPNyD | |
| LYN | P07948 | human | LY96 | Q9Y6Y9 | Y22 | FTEAQKQyWVCNSSD | |
| LYN | P07948 | human | ABL1 | P00519 | Y115 | QGWVPsNyItPVNSL | |
| LYN | P07948 | human | GCSAM | Q8N6F7 | Y128 | LGGTEtEySLLHMPS | HGAL |
| LYN | P07948 | human | SYK | P43405 | Y348 | LPMDtEVyEsPyADP | |
| LYN | P07948 | human | GCSAM | Q8N6F7 | Y107 | GNsAEEyyENVPCkA | HGAL |
| LYN | P07948 | human | IKBKB | O14920 | Y188 | sFVGTLQyLAPELLE | Pkinase |
| LYN | P07948 | human | ACLY | P53396 | Y252 | EAyPEEAyIADLDAk | Citrate_bind |
| LYN | P07948 | human | ABL1 | P00519 | Y215 | GLITTLHyPAPKRNK | |
| LYN | P07948 | human | ABL1 | P00519 | Y70 | PNLFVALyDFVAsGD | SH3_1 |
| LYN | P07948 | human | PEAK1 | Q9H792 | Y635 | IVINPNAyDNLAIyK | |
| LYN | P07948 | human | CDK4 | P11802 | Y17 | AEIGVGAyGTVYkAR | Pkinase |
| LYN | P07948 | human | AKAP8 | O43823 | Y146 | CLPEHNPyRPsySyD | |
| LYN | P07948 | human | AKAP8 | O43823 | Y170 | NGsFGGQySECRDPA | |
| LYN | P07948 | human | SYK | P43405 | Y526 | LRADENyykAQtHGK | PK_Tyr_Ser-Thr |
| LYN | P07948 | human | SOCS1 | O15524 | Y80 | LLDACGFyWGPLSVH | SH2 |
| LYN | P07948 | human | LYN | P07948 | Y508 | ytAtEGQyQQQP___ | |
| LYN | P07948 | human | PTGS2 | P35354 | Y120 | PPTYNADyGYKSWEA | |
| LYN | P07948 | human | GLO1 | Q04760 | Y136 | GIAVPDVysACkRFE | Glyoxalase |
| LYN | P07948 | human | LAT2 | Q9GZY6 | Y119 | DPIAMEyyNWGRFSk | LAT2 |
| LYN | P07948 | human | SLC4A1 | P02730 | Y904 | EEEGRDEyDEVAMPV | |
| LYN | P07948 | human | FCGR2C | P31995 | Y310 | tDDDKNIyLTLPPND | |
| LYN | P07948 | human | BCR | P11274 | Y177 | ADAEKPFyVNVEFHH | |
| LYN | P07948 | human | ACLY | P53396 | Y659 | yRPGsVAyVSRsGGM | |
| LYN | P07948 | human | BTK | Q06187 | Y223 | LKKVVALyDyMPMNA | SH3_1 |
| LYN | P07948 | human | AKAP8 | O43823 | Y51 | SVTTGATySyGPASW | |
| LYN | P07948 | human | CD19 | P15391 | Y531 | HEEDADsyENMDNPD | |
| LYN | P07948 | human | ABL1 | P00519 | Y139 | VSRNAAEyLLsSGIN | SH2 |
| LYN | P07948 | human | LAT2 | Q9GZY6 | Y110 | RHGsEEAyIDPIAME | LAT2 |
| LYN | P07948 | human | IRF5 | Q13568 | Y313 | PSDkQRFyTNQLLDV | IRF-3 |
| LYN | P07948 | human | LYN | P07948 | Y397 | RVIEDNEytAREGAk | PK_Tyr_Ser-Thr |
| LYN | P07948 | human | AKAP8 | O43823 | Y152 | PyRPsySyDyEFDLG | |
| LYN | P07948 | human | AKAP8 | O43823 | Y150 | HNPyRPsySyDyEFD | |
| LYN | P07948 | human | PTPN6 | P29350 | Y536 | QkGQEsEyGNItyPP | |
| LYN | P07948 | human | SLAMF1 | Q13291 | Y327 | ETNSITVyASVTLPE | |
| LYN | P07948 | human | GCSAM | Q8N6F7 | Y86 | tysEELCytLINHRV | HGAL |
| LYN | P07948 | human | RGS16 | O15492 | Y168 | TLMEKDSyPRFLkSP | RGS |
| LYN | P07948 | human | ABL1 | P00519 | Y172 | LRyEGRVyHyRINTA | SH2 |
| LYN | P07948 | human | AKAP8 | O43823 | Y80 | PAMHMASyGPEPCTD | |
| LYN | P07948 | human | LAT2 | Q9GZY6 | Y136 | EDDDANsyENVLICK | LAT2 |
| LYN | P07948 | human | LAT2 | Q9GZY6 | Y118 | IDPIAMEyyNWGRFS | LAT2 |
| LYN | P07948 | human | SYK | P43405 | Y352 | tEVyEsPyADPEEIR | |
| LYN | P07948 | human | ACLY | P53396 | Y384 | VRrGGPNyQEGLrVM | Citrate_bind |
| LYN | P07948 | human | ACLY | P53396 | Y227 | KVDATADyICkVkWG | |
| LYN | P07948 | human | HCLS1 | P14317 | Y222 | MEAPttAykkTTPIE | |
| LYN | P07948 | human | BTK | Q06187 | Y551 | RYVLDDEytsSVGSk | PK_Tyr_Ser-Thr |
| LYN | P07948 | human | ABL1 | P00519 | Y185 | TAsDGKLyVsSESRF | SH2 |
| LYN | P07948 | human | GCSAM | Q8N6F7 | Y80 | QDNVDQtysEELCyt | HGAL |
| LYN | P07948 | human | SYK | P43405 | Y525 | ALRADENyykAQtHG | PK_Tyr_Ser-Thr |
| LYN | P07948 | human | IKBKB | O14920 | Y199 | ELLEQQKyTVTVDYW | Pkinase |
| LYN | P07948 | human | CSF2RB | P32927 | Y468 | RFCGIyGyRLRRkWE | |
| LYN | P07948 | human | LASP1 | Q14847 | Y171 | IPtsAPVyQQPQQQP | |
| LYN | P07948 | human | YY1 | P25490 | Y254 | sPPDySEyMTGkKLP | |
| LYN | P07948 | human | WAS | P42768 | Y291 | AEtsKLIyDFIEDQG | PBD |
| LYN | P07948 | human | SLCO1B1 | Q9Y6L6 | Y645 | VLyIILIyAMKKKYQ | TM |
| LYN | P07948 | human | MCM7 | P33993 | Y600 | WASkDAtyTSARtLL | MCM_lid |
| LYN | P07948 | human | DAPP1 | Q9UN19 | Y139 | kVEEPsIyEsVRVHT | |
| LYN | P07948 | human | TRIM28 | Q13263 | Y458 | FGsGDDPyssAEPHV | |
| LYN | P07948 | human | AKAP8 | O43823 | Y311 | KRkQFQLyEEPDTkL | |
| LYN | P07948 | human | AKAP8 | O43823 | Y436 | TVEFLQEyIVNRNKK | AKAP95 |
| LYN | P07948 | human | PPP1R8 | Q12972 | Y335 | NEPKKKKyAKEAWPG | |
| LYN | P07948 | human | IKBKG | Q9Y6K9 | Y374 | PLPPAPAyLssPLAL |
 Biological Network Integration of This Kinase and Substrates Biological Network Integration of This Kinase and Substrates (GeneMANIA website) |
 Enriched GO biological processes of the phosphorylation target genes of the kinase Enriched GO biological processes of the phosphorylation target genes of the kinase |
| Kinase | GOID | GO term | P.adjust |
| LYN | ID | Description | 0.00e+00 |
| LYN | GO:0050853 | B cell receptor signaling pathway | 1.46e-10 |
| LYN | GO:0050851 | antigen receptor-mediated signaling pathway | 5.71e-10 |
| LYN | GO:0002429 | immune response-activating cell surface receptor signaling pathway | 1.89e-09 |
| LYN | GO:0002768 | immune response-regulating cell surface receptor signaling pathway | 3.80e-09 |
| LYN | GO:0002237 | response to molecule of bacterial origin | 6.44e-09 |
| LYN | GO:0002366 | leukocyte activation involved in immune response | 1.06e-08 |
| LYN | GO:0002275 | myeloid cell activation involved in immune response | 1.06e-08 |
| LYN | GO:0002263 | cell activation involved in immune response | 1.06e-08 |
| LYN | GO:0002443 | leukocyte mediated immunity | 5.29e-08 |
| LYN | GO:0002757 | immune response-activating signaling pathway | 5.65e-08 |
| LYN | GO:0042113 | B cell activation | 6.82e-08 |
| LYN | GO:0043299 | leukocyte degranulation | 7.74e-08 |
| LYN | GO:0002764 | immune response-regulating signaling pathway | 8.18e-08 |
| LYN | GO:0002274 | myeloid leukocyte activation | 2.95e-07 |
| LYN | GO:0002279 | mast cell activation involved in immune response | 2.95e-07 |
| LYN | GO:0038093 | Fc receptor signaling pathway | 2.95e-07 |
| LYN | GO:0032496 | response to lipopolysaccharide | 3.87e-07 |
| LYN | GO:0002444 | myeloid leukocyte mediated immunity | 4.51e-07 |
| LYN | GO:0071216 | cellular response to biotic stimulus | 5.48e-07 |
| LYN | GO:0050854 | regulation of antigen receptor-mediated signaling pathway | 6.50e-07 |
| LYN | GO:0045576 | mast cell activation | 8.13e-07 |
| LYN | GO:1902105 | regulation of leukocyte differentiation | 2.49e-06 |
| LYN | GO:0030183 | B cell differentiation | 3.50e-06 |
| LYN | GO:0070661 | leukocyte proliferation | 4.79e-06 |
| LYN | GO:0002703 | regulation of leukocyte mediated immunity | 4.97e-06 |
| LYN | GO:1903131 | mononuclear cell differentiation | 5.42e-06 |
| LYN | GO:0045936 | negative regulation of phosphate metabolic process | 7.73e-06 |
| LYN | GO:0010563 | negative regulation of phosphorus metabolic process | 7.73e-06 |
| LYN | GO:0043303 | mast cell degranulation | 8.10e-06 |
| LYN | GO:0018108 | peptidyl-tyrosine phosphorylation | 9.04e-06 |
| LYN | GO:0002697 | regulation of immune effector process | 9.04e-06 |
| LYN | GO:0018212 | peptidyl-tyrosine modification | 9.04e-06 |
| LYN | GO:0002448 | mast cell mediated immunity | 9.38e-06 |
| LYN | GO:0050855 | regulation of B cell receptor signaling pathway | 1.01e-05 |
| LYN | GO:0002886 | regulation of myeloid leukocyte mediated immunity | 1.05e-05 |
| LYN | GO:0038095 | Fc-epsilon receptor signaling pathway | 1.36e-05 |
| LYN | GO:1903706 | regulation of hemopoiesis | 1.37e-05 |
| LYN | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis | 1.48e-05 |
| LYN | GO:0038096 | Fc-gamma receptor signaling pathway involved in phagocytosis | 1.48e-05 |
| LYN | GO:0046651 | lymphocyte proliferation | 1.54e-05 |
| LYN | GO:0030098 | lymphocyte differentiation | 1.57e-05 |
| LYN | GO:0032943 | mononuclear cell proliferation | 1.74e-05 |
| LYN | GO:0031400 | negative regulation of protein modification process | 1.75e-05 |
| LYN | GO:0050864 | regulation of B cell activation | 1.75e-05 |
| LYN | GO:0045055 | regulated exocytosis | 2.63e-05 |
| LYN | GO:0050730 | regulation of peptidyl-tyrosine phosphorylation | 2.89e-05 |
| LYN | GO:0002431 | Fc receptor mediated stimulatory signaling pathway | 2.92e-05 |
| LYN | GO:0038094 | Fc-gamma receptor signaling pathway | 3.22e-05 |
| LYN | GO:0006909 | phagocytosis | 3.22e-05 |
Top |
Related Drugs to MCM5_LYN |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
 Distribution of the number of studies mentioning MCM5-LYN and kinase inhibitors the PubMed Abstract (04-01-2024) Distribution of the number of studies mentioning MCM5-LYN and kinase inhibitors the PubMed Abstract (04-01-2024) |
| Fusion gene - drug pair 1 | Fusion gene - drug pair 2 | PMID | Publication date | DOI | Study title |
Top |
Related Diseases to MCM5_LYN |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Related diseases from the literature mentioned this fusion gene and drug. Related diseases from the literature mentioned this fusion gene and drug. (PubMed, 04-01-2024) |
| MeSH ID | MeSH term |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
Top |
Clinical Trials of the Found Drugs/Small Molecules |
 Statistics of the Clinical Trials of the Found Kinase Inibitors from clinicaltrials.gov (06-17-2024) Statistics of the Clinical Trials of the Found Kinase Inibitors from clinicaltrials.gov (06-17-2024) |
 Clinical Trials from clinicaltrials.gov (06-17-2024) Clinical Trials from clinicaltrials.gov (06-17-2024) |
| Fusion Gene | Kinase Inhibitor | NCT ID | Study Status | Phases | Disease | # Enrolment | Date |

