| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Kinase Fusion Gene:PDGFRA_USP8 |
Kinase Fusion Protein Summary |
 Kinase Fusion gene summary Kinase Fusion gene summary |
| Kinase Fusion partner gene information | Kinase Fusion gene name: PDGFRA_USP8 | KinaseFusionDB ID: KFG4561 | FusionGDB2.0 ID: KFG4561 | Hgene | Tgene | Gene symbol | PDGFRA | USP8 | Gene ID | 5156 | 9101 | |
| Gene name | platelet derived growth factor receptor alpha | ubiquitin specific peptidase 8 | ||||||||||
| Synonyms | CD140A|PDGFR-2|PDGFR2 | HumORF8|PITA4|SPG59|UBPY | ||||||||||
| Cytomap | 4q12 | 15q21.2 | ||||||||||
| Type of gene | protein-coding | protein-coding | ||||||||||
| Description | platelet-derived growth factor receptor alphaCD140 antigen-like family member ACD140a antigenPDGF-R-alphaalpha-type platelet-derived growth factor receptorplatelet-derived growth factor receptor 2platelet-derived growth factor receptor, alpha polype | ubiquitin carboxyl-terminal hydrolase 8deubiquitinating enzyme 8ubiquitin isopeptidase Yubiquitin thiolesterase 8ubiquitin-specific-processing protease 8 | ||||||||||
| Modification date | 20240416 | 20240407 | ||||||||||
| UniProtAcc | P16234 | P40818 | ||||||||||
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000257290, ENST00000508170, | ENST00000558892, ENST00000307179, ENST00000396444, ENST00000425032, ENST00000433963, | |||||||||
| Context (manual curation of fusion genes in KinaseFusionDB) | PubMed: PDGFRA [Title/Abstract] AND USP8 [Title/Abstract] AND fusion [Title/Abstract] | |||||||||||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | PDGFRA(55156721)-USP8(50751197), # samples:2 | |||||||||||
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | PDGFRA | GO:0008284 | positive regulation of cell population proliferation | 10806482 |
| Hgene | PDGFRA | GO:0010544 | negative regulation of platelet activation | 8188664 |
| Hgene | PDGFRA | GO:0018108 | peptidyl-tyrosine phosphorylation | 2536956|8188664 |
| Hgene | PDGFRA | GO:0030335 | positive regulation of cell migration | 17470632 |
| Hgene | PDGFRA | GO:0034614 | cellular response to reactive oxygen species | 24190966 |
| Hgene | PDGFRA | GO:0038091 | positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway | 17470632 |
| Hgene | PDGFRA | GO:0046777 | protein autophosphorylation | 2536956|8188664 |
| Hgene | PDGFRA | GO:0048008 | platelet-derived growth factor receptor signaling pathway | 2536956|10806482 |
| Hgene | PDGFRA | GO:0048146 | positive regulation of fibroblast proliferation | 10806482 |
| Tgene | USP8 | GO:0070536 | protein K63-linked deubiquitination | 16520378 |
| Tgene | USP8 | GO:0071108 | protein K48-linked deubiquitination | 16520378 |
 Kinase Fusion gene breakpoints across PDGFRA (5'-gene) Kinase Fusion gene breakpoints across PDGFRA (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
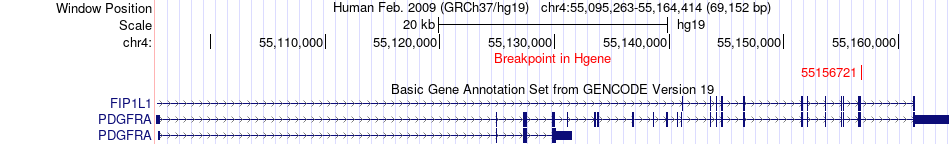 |
 Kinase Fusion gene breakpoints across USP8 (3'-gene) Kinase Fusion gene breakpoints across USP8 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
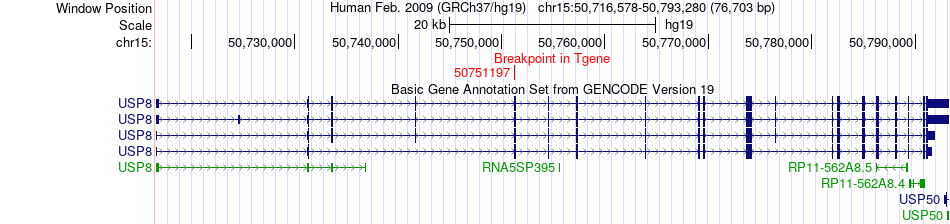 |
Top |
Kinase Fusion Gene Sample Information |
 Kinase Fusion gene information. Kinase Fusion gene information. |
 Kinase Fusion gene information from four resources (ChiTars 5.0, ChimerDB 4.0, COSMIC, and CCLE) Kinase Fusion gene information from four resources (ChiTars 5.0, ChimerDB 4.0, COSMIC, and CCLE)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Sample | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp |
| ChimerDB4 | TCGA-Z4-AAPG-01A | PDGFRA | chr4 | 55156721 | USP8 | chr15 | 50751197 |
Top |
Kinase Fusion ORF Analysis |
 Kinase Fusion information from ORFfinder translation from full-length transcript sequence from KinaseFusionDB. Kinase Fusion information from ORFfinder translation from full-length transcript sequence from KinaseFusionDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp | Seq length (transcript) | Seq length (amino acids) |
| ENST00000257290 | ENST00000307179 | PDGFRA | chr4 | 55156721 | USP8 | chr15 | 50751197 | 7163 | 2103 |
Top |
Kinase Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from KinaseFusionDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from KinaseFusionDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >Henst_Tenst_Hgene_Hchr_Hbp_Tgene_Tchr_Tbp_length(fusion AA)_AAseq >ENST00000257290_ENST00000307179_PDGFRA_chr4_55156721_USP8_chr15_50751197_length(amino acids)=2103 MNQLQNAVFEPITVGATGRETEEETARDHWRPWARSLLHVWDIHCGITSEEKFPRAMGTSHPAFLVLGCLLTGLSLILCQLSLPSILPNE NEKVVQLNSSFSLRCFGESEVSWQYPMSEEESSDVEIRNEENNSGLFVTVLEVSSASAAHTGLYTCYYNHTQTEENELEGRHIYIYVPDP DVAFVPLGMTDYLVIVEDDDSAIIPCRTTDPETPVTLHNSEGVVPASYDSRQGFNGTFTVGPYICEATVKGKKFQTIPFNVYALKATSEL DLEMEALKTVYKSGETIVVTCAVFNNEVVDLQWTYPGEVKGKGITMLEEIKVPSIKLVYTLTVPEATVKDSGDYECAARQATREVKEMKK VTISVHEKGFIEIKPTFSQLEAVNLHEVKHFVVEVRAYPPPRISWLKNNLTLIENLTEITTDVEKIQEIRYRSKLKLIRAKEEDSGHYTI VAQNEDAVKSYTFELLTQVPSSILDLVDDHHGSTGGQTVRCTAEGTPLPDIEWMICKDIKKCNNETSWTILANNVSNIITEIHSRDRSTV EGRVTFAKVEETIAVRCLAKNLLGAENRELKLVAPTLRSELTVAAAVLVLLVIVIISLIVLVVIWKQKPRYEIRWRVIESISPDGHEYIY VDPMQLPYDSRWEFPRDGLVLGRVLGSGAFGKVVEGTAYGLSRSQPVMKVAVKMLKPTARSSEKQALMSELKIMTHLGPHLNIVNLLGAC TKSGPIYIITEYCFYGDLVNYLHKNRDSFLSHHPEKPKKELDIFGLNPADESTRSYVILSFENNGDYMDMKQADTTQYVPMLERKEVSKY SDIQRSLYDRPASYKKKSMLDSEVKNLLSDDNSEGLTLLDLLSFTYQVARGMEFLASKNCVHRDLAARNVLLAQGKIVKICDFGLARDIM HDSNYVSKGSTFLPVKWMAPESIFDNLYTTLSDVWSYGILLWEIFSLGGTPYPGMMVDSTFYNKIKSGYRMAKPDHATSEVYEIMVKCWN SEPEKRPSFYHLSEIVENLLPGQYKKSYEKIHLDFLKSDHPAVARMRVDSDNAYIGVTYKNEEDKLKDWEGGLDEQRLSADSGYIIPLPD IDPVPEEEDLGKRNRHRYEEAEVRKKLEEKDRQEEAQRLQQKRQETGREDGGTLAKGSLENVLDSKDKTQKSNGEKNEKCETKEKGAITA KELYTMMTDKNISLIIMDARRMQDYQDSCILHSLSVPEEAISPGVTASWIEAHLPDDSKDTWKKRGNVEYVVLLDWFSSAKDLQIGTTLR SLKDALFKWESKTVLRNEPLVLEGGYENWLLCYPQYTTNAKVTPPPRRQNEEVSISLDFTYPSLEESIPSKPAAQTPPASIEVDENIELI SGQNERMGPLNISTPVEPVAASKSDVSPIIQPVPSIKNVPQIDRTKKPAVKLPEEHRIKSESTNHEQQSPQSGKVIPDRSTKPVVFSPTL MLTDEEKARIHAETALLMEKNKQEKELRERQQEEQKEKLRKEEQEQKAKKKQEAEENEITEKQQKAKEEMEKKESEQAKKEDKETSAKRG KEITGVKRQSKSEHETSDAKKSVEDRGKRCPTPEIQKKSTGDVPHTSVTGDSGSGKPFKIKGQPESGILRTGTFREDTDDTERNKAQREP LTRARSEEMGRIVPGLPSGWAKFLDPITGTFRYYHSPTNTVHMYPPEMAPSSAPPSTPPTHKAKPQIPAERDREPSKLKRSYSSPDITQA IQEEEKRKPTVTPTVNRENKPTCYPKAEISRLSASQIRNLNPVFGGSGPALTGLRNLGNTCYMNSILQCLCNAPHLADYFNRNCYQDDIN RSNLLGHKGEVAEEFGIIMKALWTGQYRYISPKDFKITIGKINDQFAGYSQQDSQELLLFLMDGLHEDLNKADNRKRYKEENNDHLDDFK AAEHAWQKHKQLNESIIVALFQGQFKSTVQCLTCHKKSRTFEAFMYLSLPLASTSKCTLQDCLRLFSKEEKLTDNNRFYCSHCRARRDSL KKIEIWKLPPVLLVHLKRFSYDGRWKQKLQTSVDFPLENLDLSQYVIGPKNNLKKYNLFSVSNHYGGLDGGHYTAYCKNAARQRWFKFDD -------------------------------------------------------------- |
Multiple Sequence Alignment of All Fusion Protein Isoforms |
Top |
Kinase Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr4:55156721/chr15:50751197) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| PDGFRA | USP8 |
| FUNCTION: Tyrosine-protein kinase that acts as a cell-surface receptor for PDGFA, PDGFB and PDGFC and plays an essential role in the regulation of embryonic development, cell proliferation, survival and chemotaxis. Depending on the context, promotes or inhibits cell proliferation and cell migration. Plays an important role in the differentiation of bone marrow-derived mesenchymal stem cells. Required for normal skeleton development and cephalic closure during embryonic development. Required for normal development of the mucosa lining the gastrointestinal tract, and for recruitment of mesenchymal cells and normal development of intestinal villi. Plays a role in cell migration and chemotaxis in wound healing. Plays a role in platelet activation, secretion of agonists from platelet granules, and in thrombin-induced platelet aggregation. Binding of its cognate ligands - homodimeric PDGFA, homodimeric PDGFB, heterodimers formed by PDGFA and PDGFB or homodimeric PDGFC -leads to the activation of several signaling cascades; the response depends on the nature of the bound ligand and is modulated by the formation of heterodimers between PDGFRA and PDGFRB. Phosphorylates PIK3R1, PLCG1, and PTPN11. Activation of PLCG1 leads to the production of the cellular signaling molecules diacylglycerol and inositol 1,4,5-trisphosphate, mobilization of cytosolic Ca(2+) and the activation of protein kinase C. Phosphorylates PIK3R1, the regulatory subunit of phosphatidylinositol 3-kinase, and thereby mediates activation of the AKT1 signaling pathway. Mediates activation of HRAS and of the MAP kinases MAPK1/ERK2 and/or MAPK3/ERK1. Promotes activation of STAT family members STAT1, STAT3 and STAT5A and/or STAT5B. Receptor signaling is down-regulated by protein phosphatases that dephosphorylate the receptor and its down-stream effectors, and by rapid internalization of the activated receptor. {ECO:0000269|PubMed:10734113, ECO:0000269|PubMed:10947961, ECO:0000269|PubMed:11297552, ECO:0000269|PubMed:12522257, ECO:0000269|PubMed:1646396, ECO:0000269|PubMed:17087943, ECO:0000269|PubMed:1709159, ECO:0000269|PubMed:17141222, ECO:0000269|PubMed:20972453, ECO:0000269|PubMed:21224473, ECO:0000269|PubMed:21596750, ECO:0000269|PubMed:2554309, ECO:0000269|PubMed:8188664, ECO:0000269|PubMed:8760137, ECO:0000269|PubMed:8943348}. | FUNCTION: Hydrolase that can remove conjugated ubiquitin from proteins and therefore plays an important regulatory role at the level of protein turnover by preventing degradation. Converts both 'Lys-48' an 'Lys-63'-linked ubiquitin chains. Catalytic activity is enhanced in the M phase. Involved in cell proliferation. Required to enter into S phase in response to serum stimulation. May regulate T-cell anergy mediated by RNF128 via the formation of a complex containing RNF128 and OTUB1. Probably regulates the stability of STAM2 and RASGRF1. Regulates endosomal ubiquitin dynamics, cargo sorting, membrane traffic at early endosomes, and maintenance of ESCRT-0 stability. The level of protein ubiquitination on endosomes is essential for maintaining the morphology of the organelle. Deubiquitinates EPS15 and controls tyrosine kinase stability. Removes conjugated ubiquitin from EGFR thus regulating EGFR degradation and downstream MAPK signaling. Involved in acrosome biogenesis through interaction with the spermatid ESCRT-0 complex and microtubules. Deubiquitinates BIRC6/bruce and KIF23/MKLP1. Deubiquitinates BACE1 which inhibits BACE1 lysosomal degradation and modulates BACE-mediated APP cleavage and amyloid-beta formation (PubMed:27302062). {ECO:0000269|PubMed:16520378, ECO:0000269|PubMed:17711858, ECO:0000269|PubMed:18329369, ECO:0000269|PubMed:27302062, ECO:0000269|PubMed:9628861}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
 - Retained domain in the 5'-partner of fusion protein (protein functional feature from UniProt). - Retained domain in the 5'-partner of fusion protein (protein functional feature from UniProt). |
| Partner | Hgeneene | Hbp | Tgeneene | Tbp | ENST | BPexon | TotalExon | Protein feature loci | BPloci | TotalLen | Feature | Note |
| Hgene | PDGFRA | 55156721 | USP8 | 50751197 | ENST00000257290 | 22 | 23 | 24_113 | 10401 | 1090 | Domain | Note=Ig-like C2-type 1 |
| Hgene | PDGFRA | 55156721 | USP8 | 50751197 | ENST00000257290 | 22 | 23 | 117_201 | 10401 | 1090 | Domain | Note=Ig-like C2-type 2 |
| Hgene | PDGFRA | 55156721 | USP8 | 50751197 | ENST00000257290 | 22 | 23 | 202_306 | 10401 | 1090 | Domain | Note=Ig-like C2-type 3 |
| Hgene | PDGFRA | 55156721 | USP8 | 50751197 | ENST00000257290 | 22 | 23 | 319_410 | 10401 | 1090 | Domain | Note=Ig-like C2-type 4 |
| Hgene | PDGFRA | 55156721 | USP8 | 50751197 | ENST00000257290 | 22 | 23 | 414_517 | 10401 | 1090 | Domain | Note=Ig-like C2-type 5 |
| Hgene | PDGFRA | 55156721 | USP8 | 50751197 | ENST00000257290 | 22 | 23 | 593_954 | 10401 | 1090 | Domain | Note=Protein kinase;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00159 |
 - Retained domain in the 3'-partner of fusion protein (protein functional feature from UniProt). - Retained domain in the 3'-partner of fusion protein (protein functional feature from UniProt). |
| Partner | Hgeneene | Hbp | Tgeneene | Tbp | ENST | BPexon | TotalExon | Protein feature loci | BPloci | TotalLen | Feature | Note |
Top |
Kinase Fusion Protein Structures |
 CIF files of the predicted kinase fusion proteins CIF files of the predicted kinase fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Kinase Fusion protein CIF link (fusion AA seq ID in KinaseFusionDB) | Henst | Tenst | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp | AA seq | Len(AA seq) |
| PDB file >>>193_PDGFRA_USP8 | ENST00000257290 | ENST00000307179 | PDGFRA | chr4 | 55156721 | USP8 | chr15 | 50751197 | MNQLQNAVFEPITVGATGRETEEETARDHWRPWARSLLHVWDIHCGITSEEKFPRAMGTSHPAFLVLGCLLTGLSLILCQLSLPSILPNE NEKVVQLNSSFSLRCFGESEVSWQYPMSEEESSDVEIRNEENNSGLFVTVLEVSSASAAHTGLYTCYYNHTQTEENELEGRHIYIYVPDP DVAFVPLGMTDYLVIVEDDDSAIIPCRTTDPETPVTLHNSEGVVPASYDSRQGFNGTFTVGPYICEATVKGKKFQTIPFNVYALKATSEL DLEMEALKTVYKSGETIVVTCAVFNNEVVDLQWTYPGEVKGKGITMLEEIKVPSIKLVYTLTVPEATVKDSGDYECAARQATREVKEMKK VTISVHEKGFIEIKPTFSQLEAVNLHEVKHFVVEVRAYPPPRISWLKNNLTLIENLTEITTDVEKIQEIRYRSKLKLIRAKEEDSGHYTI VAQNEDAVKSYTFELLTQVPSSILDLVDDHHGSTGGQTVRCTAEGTPLPDIEWMICKDIKKCNNETSWTILANNVSNIITEIHSRDRSTV EGRVTFAKVEETIAVRCLAKNLLGAENRELKLVAPTLRSELTVAAAVLVLLVIVIISLIVLVVIWKQKPRYEIRWRVIESISPDGHEYIY VDPMQLPYDSRWEFPRDGLVLGRVLGSGAFGKVVEGTAYGLSRSQPVMKVAVKMLKPTARSSEKQALMSELKIMTHLGPHLNIVNLLGAC TKSGPIYIITEYCFYGDLVNYLHKNRDSFLSHHPEKPKKELDIFGLNPADESTRSYVILSFENNGDYMDMKQADTTQYVPMLERKEVSKY SDIQRSLYDRPASYKKKSMLDSEVKNLLSDDNSEGLTLLDLLSFTYQVARGMEFLASKNCVHRDLAARNVLLAQGKIVKICDFGLARDIM HDSNYVSKGSTFLPVKWMAPESIFDNLYTTLSDVWSYGILLWEIFSLGGTPYPGMMVDSTFYNKIKSGYRMAKPDHATSEVYEIMVKCWN SEPEKRPSFYHLSEIVENLLPGQYKKSYEKIHLDFLKSDHPAVARMRVDSDNAYIGVTYKNEEDKLKDWEGGLDEQRLSADSGYIIPLPD IDPVPEEEDLGKRNRHRYEEAEVRKKLEEKDRQEEAQRLQQKRQETGREDGGTLAKGSLENVLDSKDKTQKSNGEKNEKCETKEKGAITA KELYTMMTDKNISLIIMDARRMQDYQDSCILHSLSVPEEAISPGVTASWIEAHLPDDSKDTWKKRGNVEYVVLLDWFSSAKDLQIGTTLR SLKDALFKWESKTVLRNEPLVLEGGYENWLLCYPQYTTNAKVTPPPRRQNEEVSISLDFTYPSLEESIPSKPAAQTPPASIEVDENIELI SGQNERMGPLNISTPVEPVAASKSDVSPIIQPVPSIKNVPQIDRTKKPAVKLPEEHRIKSESTNHEQQSPQSGKVIPDRSTKPVVFSPTL MLTDEEKARIHAETALLMEKNKQEKELRERQQEEQKEKLRKEEQEQKAKKKQEAEENEITEKQQKAKEEMEKKESEQAKKEDKETSAKRG KEITGVKRQSKSEHETSDAKKSVEDRGKRCPTPEIQKKSTGDVPHTSVTGDSGSGKPFKIKGQPESGILRTGTFREDTDDTERNKAQREP LTRARSEEMGRIVPGLPSGWAKFLDPITGTFRYYHSPTNTVHMYPPEMAPSSAPPSTPPTHKAKPQIPAERDREPSKLKRSYSSPDITQA IQEEEKRKPTVTPTVNRENKPTCYPKAEISRLSASQIRNLNPVFGGSGPALTGLRNLGNTCYMNSILQCLCNAPHLADYFNRNCYQDDIN RSNLLGHKGEVAEEFGIIMKALWTGQYRYISPKDFKITIGKINDQFAGYSQQDSQELLLFLMDGLHEDLNKADNRKRYKEENNDHLDDFK AAEHAWQKHKQLNESIIVALFQGQFKSTVQCLTCHKKSRTFEAFMYLSLPLASTSKCTLQDCLRLFSKEEKLTDNNRFYCSHCRARRDSL KKIEIWKLPPVLLVHLKRFSYDGRWKQKLQTSVDFPLENLDLSQYVIGPKNNLKKYNLFSVSNHYGGLDGGHYTAYCKNAARQRWFKFDD | 2103 |
| 3D view using mol* of 193_PDGFRA_USP8 | ||||||||||
| PDB file >>>HKFP_280_PDGFRA_USP8 | ENST00000257290 | ENST00000307179 | PDGFRA | chr4 | 55156721 | USP8 | chr15 | 50751197 | MNQLQNAVFEPITVGATGRETEEETARDHWRPWARSLLHVWDIHCGITSEEKFPRAMGTSHPAFLVLGCLLTGLSLILCQLSLPSILPNE NEKVVQLNSSFSLRCFGESEVSWQYPMSEEESSDVEIRNEENNSGLFVTVLEVSSASAAHTGLYTCYYNHTQTEENELEGRHIYIYVPDP DVAFVPLGMTDYLVIVEDDDSAIIPCRTTDPETPVTLHNSEGVVPASYDSRQGFNGTFTVGPYICEATVKGKKFQTIPFNVYALKATSEL DLEMEALKTVYKSGETIVVTCAVFNNEVVDLQWTYPGEVKGKGITMLEEIKVPSIKLVYTLTVPEATVKDSGDYECAARQATREVKEMKK VTISVHEKGFIEIKPTFSQLEAVNLHEVKHFVVEVRAYPPPRISWLKNNLTLIENLTEITTDVEKIQEIRYRSKLKLIRAKEEDSGHYTI VAQNEDAVKSYTFELLTQVPSSILDLVDDHHGSTGGQTVRCTAEGTPLPDIEWMICKDIKKCNNETSWTILANNVSNIITEIHSRDRSTV EGRVTFAKVEETIAVRCLAKNLLGAENRELKLVAPTLRSELTVAAAVLVLLVIVIISLIVLVVIWKQKPRYEIRWRVIESISPDGHEYIY VDPMQLPYDSRWEFPRDGLVLGRVLGSGAFGKVVEGTAYGLSRSQPVMKVAVKMLKPTARSSEKQALMSELKIMTHLGPHLNIVNLLGAC TKSGPIYIITEYCFYGDLVNYLHKNRDSFLSHHPEKPKKELDIFGLNPADESTRSYVILSFENNGDYMDMKQADTTQYVPMLERKEVSKY SDIQRSLYDRPASYKKKSMLDSEVKNLLSDDNSEGLTLLDLLSFTYQVARGMEFLASKNCVHRDLAARNVLLAQGKIVKICDFGLARDIM HDSNYVSKGSTFLPVKWMAPESIFDNLYTTLSDVWSYGILLWEIFSLGGTPYPGMMVDSTFYNKIKSGYRMAKPDHATSEVYEIMVKCWN SEPEKRPSFYHLSEIVENLLPGQYKKSYEKIHLDFLKSDHPAVARMRVDSDNAYIGVTYKNEEDKLKDWEGGLDEQRLSADSGYIIPLPD IDPVPEEEDLGKRNRHRYEEAEVRKKLEEKDRQEEAQRLQQKRQETGREDGGTLAKGSLENVLDSKDKTQKSNGEKNEKCETKEKGAITA KELYTMMTDKNISLIIMDARRMQDYQDSCILHSLSVPEEAISPGVTASWIEAHLPDDSKDTWKKRGNVEYVVLLDWFSSAKDLQIGTTLR SLKDALFKWESKTVLRNEPLVLEGGYENWLLCYPQYTTNAKVTPPPRRQNEEVSISLDFTYPSLEESIPSKPAAQTPPASIEVDENIELI SGQNERMGPLNISTPVEPVAASKSDVSPIIQPVPSIKNVPQIDRTKKPAVKLPEEHRIKSESTNHEQQSPQSGKVIPDRSTKPVVFSPTL MLTDEEKARIHAETALLMEKNKQEKELRERQQEEQKEKLRKEEQEQKAKKKQEAEENEITEKQQKAKEEMEKKESEQAKKEDKETSAKRG KEITGVKRQSKSEHETSDAKKSVEDRGKRCPTPEIQKKSTGDVPHTSVTGDSGSGKPFKIKGQPESGILRTGTFREDTDDTERNKAQREP LTRARSEEMGRIVPGLPSGWAKFLDPITGTFRYYHSPTNTVHMYPPEMAPSSAPPSTPPTHKAKPQIPAERDREPSKLKRSYSSPDITQA IQEEEKRKPTVTPTVNRENKPTCYPKAEISRLSASQIRNLNPVFGGSGPALTGLRNLGNTCYMNSILQCLCNAPHLADYFNRNCYQDDIN RSNLLGHKGEVAEEFGIIMKALWTGQYRYISPKDFKITIGKINDQFAGYSQQDSQELLLFLMDGLHEDLNKADNRKRYKEENNDHLDDFK AAEHAWQKHKQLNESIIVALFQGQFKSTVQCLTCHKKSRTFEAFMYLSLPLASTSKCTLQDCLRLFSKEEKLTDNNRFYCSHCRARRDSL KKIEIWKLPPVLLVHLKRFSYDGRWKQKLQTSVDFPLENLDLSQYVIGPKNNLKKYNLFSVSNHYGGLDGGHYTAYCKNAARQRWFKFDD | 2103_PDGFRA_USP8 |
Top |
Comparison of Fusion Protein Isoforms |
 Superimpose the 3D Structures Among All Fusion Protein Isoforms Superimpose the 3D Structures Among All Fusion Protein Isoforms * Download the pdb file and open it from the molstar online viewer. |
 Comparison of the Secondary Structures of Fusion Protein Isoforms Comparison of the Secondary Structures of Fusion Protein Isoforms |
Top |
Comparison of Fusion Protein Sequences/Structures with Known Sequences/Structures from PDB |
Top |
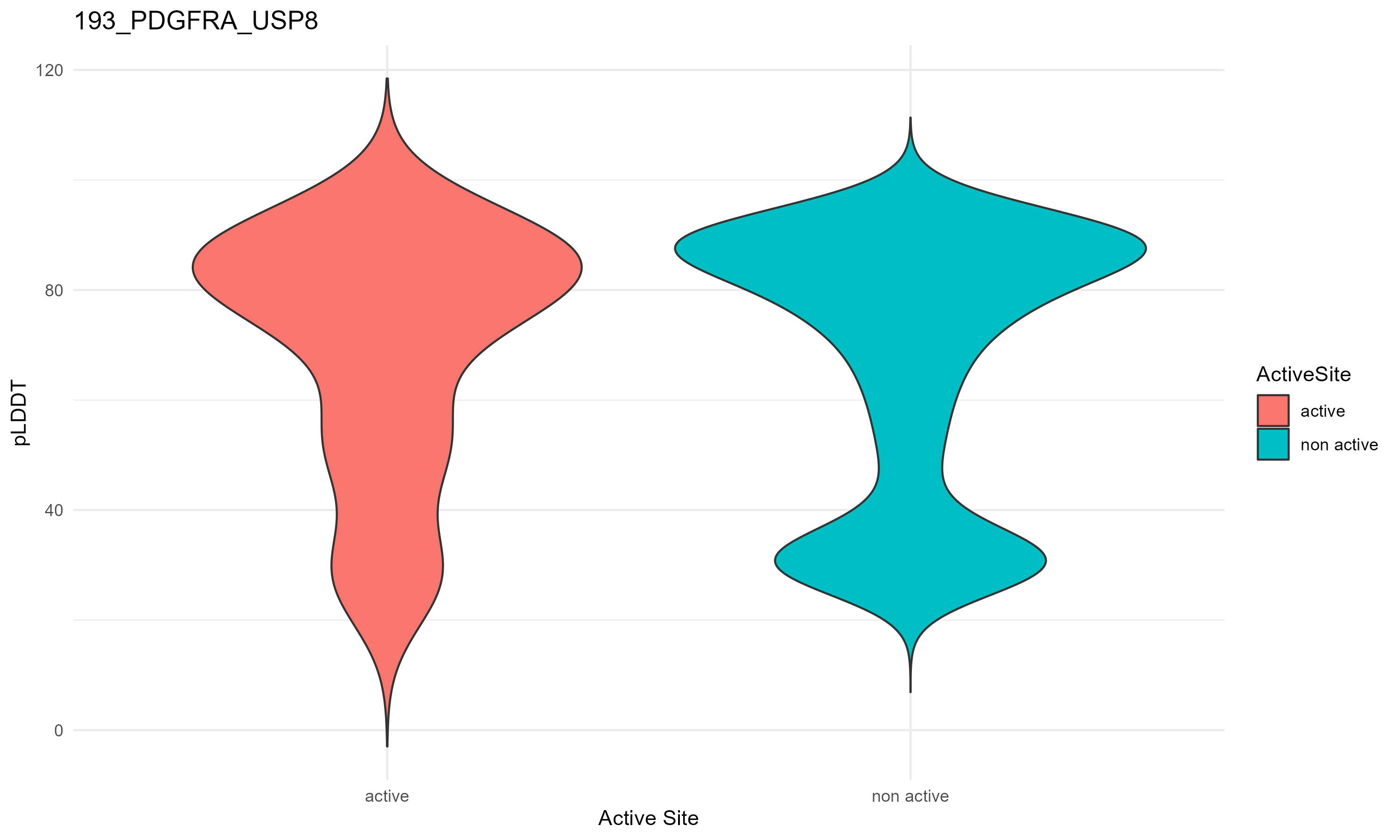
pLDDT score distribution |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. * The blue color at the bottom marks the best active site residues. |
| 193_PDGFRA_USP8.png |
 |
| 193_PDGFRA_USP8.png |
 |
Top |
Potential Active Site Information |
 The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. |
| Kinase Fusion AA seq ID in KinaseFusionDB | Site score | Size | Dscore | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
Top |
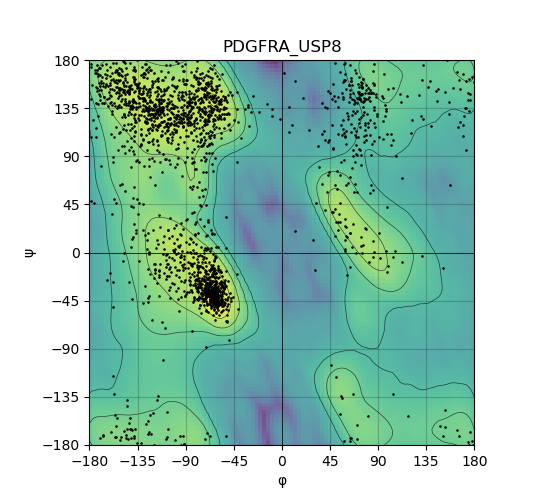
Ramachandran Plot of Kinase Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| 193_PDGFRA_USP8_ramachandran.png |
 |
Top |
Virtual Screening Results |
 Distribution of the average docking score across all approved kinase inhibitors. Distribution of the average docking score across all approved kinase inhibitors.Distribution of the number of occurrence across all approved kinase inhibitors. |
| 5'-kinase fusion protein case |
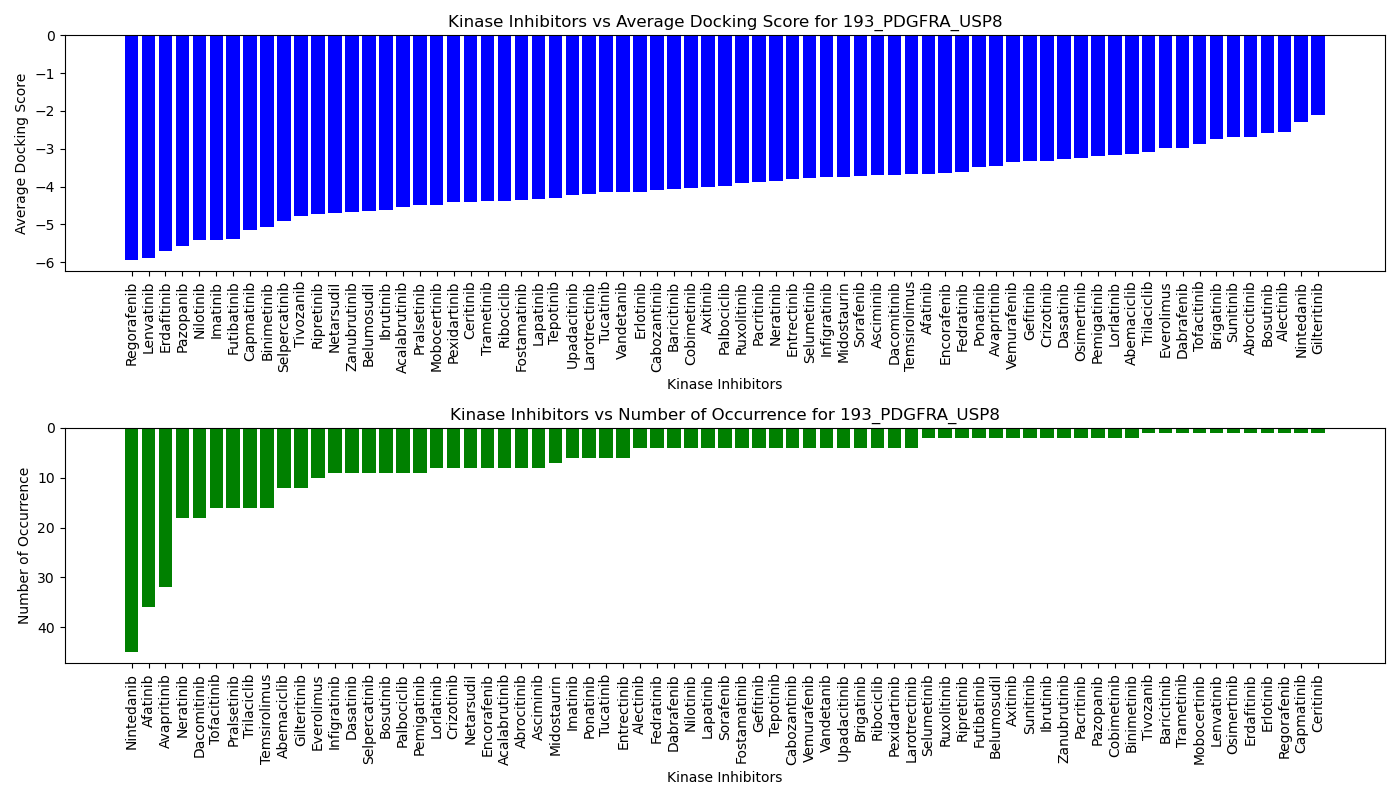 |
| 3'-kinase fusion protein case |
Top |
 Drug information from DrugBank of the top 20 interacting small molecules. Drug information from DrugBank of the top 20 interacting small molecules.* The detailed information of individual kinase inhibitors are available in the download page. |
| Fusion gene name info | Drug | Docking score | Glide g score | Glide energy |
| 193_PDGFRA_USP8-DOCK_HTVS_1-001 | Pralsetinib | -7.321039999999999 | -7.412539999999999 | -54.7896 |
| 193_PDGFRA_USP8-DOCK_HTVS_1-001 | Pralsetinib | -6.92358 | -7.01508 | -56.156000000000006 |
| 193_PDGFRA_USP8-DOCK_HTVS_1-001 | Pralsetinib | -6.73038 | -6.82188 | -55.8496 |
| 193_PDGFRA_USP8-DOCK_HTVS_1-001 | Netarsudil | -6.679710000000001 | -6.690810000000001 | -52.9302 |
| 193_PDGFRA_USP8-DOCK_HTVS_1-001 | Netarsudil | -6.679710000000001 | -6.690810000000001 | -52.9302 |
| 193_PDGFRA_USP8-DOCK_HTVS_1-001 | Nilotinib | -6.3756699999999995 | -6.51527 | -56.8545 |
| 193_PDGFRA_USP8-DOCK_HTVS_1-001 | Nilotinib | -6.3756699999999995 | -6.51527 | -56.8545 |
| 193_PDGFRA_USP8-DOCK_HTVS_1-001 | Selpercatinib | -6.30575 | -6.33625 | -49.3693 |
| 193_PDGFRA_USP8-DOCK_HTVS_1-001 | Imatinib | -6.14816 | -6.35476 | -56.5003 |
| 193_PDGFRA_USP8-DOCK_HTVS_1-001 | Imatinib | -6.14816 | -6.35476 | -56.5003 |
| 193_PDGFRA_USP8-DOCK_HTVS_1-001 | Imatinib | -6.14816 | -6.35476 | -56.5003 |
| 193_PDGFRA_USP8-DOCK_HTVS_1-001 | Futibatinib | -6.14741 | -6.14741 | -44.6274 |
| 193_PDGFRA_USP8-DOCK_HTVS_1-001 | Selpercatinib | -6.04637 | -6.0768699999999995 | -51.5937 |
| 193_PDGFRA_USP8-DOCK_HTVS_1-001 | Selpercatinib | -6.04637 | -6.0768699999999995 | -51.5937 |
| 193_PDGFRA_USP8-DOCK_HTVS_1-001 | Afatinib | -6.01775 | -6.20005 | -55.9055 |
| 193_PDGFRA_USP8-DOCK_HTVS_1-001 | Afatinib | -6.01775 | -6.20005 | -55.9055 |
| 193_PDGFRA_USP8-DOCK_HTVS_1-001 | Afatinib | -6.01635 | -6.20005 | -55.9055 |
| 193_PDGFRA_USP8-DOCK_HTVS_1-001 | Tepotinib | -5.98554 | -5.98664 | -49.9192 |
| 193_PDGFRA_USP8-DOCK_HTVS_1-001 | Selpercatinib | -5.97808 | -6.00858 | -44.0647 |
| 193_PDGFRA_USP8-DOCK_HTVS_1-001 | Regorafenib | -5.93821 | -5.93821 | -47.8743 |
Top |
Kinase-Substrate Information of PDGFRA_USP8 |
 Phosphorylation target of the kinase Phosphorylation target of the kinase(phosphosite, 03-17-2024) |
| Kinase | Kinase UniProt Acc | Kinase species | Substrate | Substrate UniProt Acc | Substrate phosphorylated residues | Substrate phosphorylated sites (+/-7AA) | Domain |
| PDGFRA | P16234 | human | PDGFRA | P16234 | Y1018 | RLSADsGyIIPLPDI | |
| PDGFRA | P16234 | human | PDGFRA | P16234 | Y572 | IsPDGHEyIyVDPMQ | |
| PDGFRA | P16234 | human | PDGFRA | P16234 | Y988 | RVDSDNAyIGVtyKN | |
| PDGFRA | P16234 | human | CCDC88C | Q9P219 | Y2025 | PQTVWyEyGCV____ | |
| PDGFRA | P16234 | human | SRC | P12931 | Y419 | RLIEDNEytARQGAk | PK_Tyr_Ser-Thr |
| PDGFRA | P16234 | human | PDGFRA | P16234 | Y574 | PDGHEyIyVDPMQLP |
 Biological Network Integration of This Kinase and Substrates Biological Network Integration of This Kinase and Substrates (GeneMANIA website) |
 Enriched GO biological processes of the phosphorylation target genes of the kinase Enriched GO biological processes of the phosphorylation target genes of the kinase |
| Kinase | GOID | GO term | P.adjust |
| PDGFRA | ID | Description | 0.00e+00 |
| PDGFRA | GO:0031648 | protein destabilization | 4.83e-03 |
| PDGFRA | GO:0022602 | ovulation cycle process | 4.83e-03 |
| PDGFRA | GO:0048008 | platelet-derived growth factor receptor signaling pathway | 4.83e-03 |
| PDGFRA | GO:0042698 | ovulation cycle | 5.44e-03 |
| PDGFRA | GO:0008585 | female gonad development | 7.79e-03 |
| PDGFRA | GO:0046545 | development of primary female sexual characteristics | 7.79e-03 |
| PDGFRA | GO:0046660 | female sex differentiation | 7.79e-03 |
| PDGFRA | GO:0042476 | odontogenesis | 7.79e-03 |
| PDGFRA | GO:0030168 | platelet activation | 7.79e-03 |
| PDGFRA | GO:0048565 | digestive tract development | 7.79e-03 |
| PDGFRA | GO:0055123 | digestive system development | 8.41e-03 |
| PDGFRA | GO:0034614 | cellular response to reactive oxygen species | 8.57e-03 |
| PDGFRA | GO:0051897 | positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction | 9.76e-03 |
| PDGFRA | GO:0046777 | protein autophosphorylation | 1.07e-02 |
| PDGFRA | GO:0000302 | response to reactive oxygen species | 1.07e-02 |
| PDGFRA | GO:0031032 | actomyosin structure organization | 1.07e-02 |
| PDGFRA | GO:0002064 | epithelial cell development | 1.07e-02 |
| PDGFRA | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 1.07e-02 |
| PDGFRA | GO:0007596 | blood coagulation | 1.07e-02 |
| PDGFRA | GO:0050817 | coagulation | 1.07e-02 |
| PDGFRA | GO:0007599 | hemostasis | 1.07e-02 |
| PDGFRA | GO:0008406 | gonad development | 1.07e-02 |
| PDGFRA | GO:0045137 | development of primary sexual characteristics | 1.07e-02 |
| PDGFRA | GO:0051896 | regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction | 1.11e-02 |
| PDGFRA | GO:0034599 | cellular response to oxidative stress | 1.13e-02 |
| PDGFRA | GO:0060828 | regulation of canonical Wnt signaling pathway | 1.14e-02 |
| PDGFRA | GO:0018108 | peptidyl-tyrosine phosphorylation | 1.20e-02 |
| PDGFRA | GO:0018212 | peptidyl-tyrosine modification | 1.20e-02 |
| PDGFRA | GO:0007548 | sex differentiation | 1.21e-02 |
| PDGFRA | GO:0043491 | phosphatidylinositol 3-kinase/protein kinase B signal transduction | 1.21e-02 |
| PDGFRA | GO:0048511 | rhythmic process | 1.21e-02 |
| PDGFRA | GO:0048608 | reproductive structure development | 1.21e-02 |
| PDGFRA | GO:0061458 | reproductive system development | 1.21e-02 |
| PDGFRA | GO:0070372 | regulation of ERK1 and ERK2 cascade | 1.21e-02 |
| PDGFRA | GO:0060070 | canonical Wnt signaling pathway | 1.21e-02 |
| PDGFRA | GO:0062197 | cellular response to chemical stress | 1.21e-02 |
| PDGFRA | GO:0031647 | regulation of protein stability | 1.24e-02 |
| PDGFRA | GO:0070371 | ERK1 and ERK2 cascade | 1.24e-02 |
| PDGFRA | GO:0033674 | positive regulation of kinase activity | 1.24e-02 |
| PDGFRA | GO:0030111 | regulation of Wnt signaling pathway | 1.24e-02 |
| PDGFRA | GO:0050878 | regulation of body fluid levels | 1.40e-02 |
| PDGFRA | GO:0006979 | response to oxidative stress | 1.61e-02 |
| PDGFRA | GO:0140694 | non-membrane-bounded organelle assembly | 1.61e-02 |
| PDGFRA | GO:0051347 | positive regulation of transferase activity | 1.61e-02 |
| PDGFRA | GO:0042060 | wound healing | 1.61e-02 |
| PDGFRA | GO:0035332 | positive regulation of hippo signaling | 1.61e-02 |
| PDGFRA | GO:0036093 | germ cell proliferation | 1.61e-02 |
| PDGFRA | GO:0071803 | positive regulation of podosome assembly | 1.61e-02 |
| PDGFRA | GO:0048732 | gland development | 1.61e-02 |
Top |
Related Drugs to PDGFRA_USP8 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
 Distribution of the number of studies mentioning PDGFRA-USP8 and kinase inhibitors the PubMed Abstract (04-01-2024) Distribution of the number of studies mentioning PDGFRA-USP8 and kinase inhibitors the PubMed Abstract (04-01-2024) |
| Fusion gene - drug pair 1 | Fusion gene - drug pair 2 | PMID | Publication date | DOI | Study title |
Top |
Related Diseases to PDGFRA_USP8 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Related diseases from the literature mentioned this fusion gene and drug. Related diseases from the literature mentioned this fusion gene and drug. (PubMed, 04-01-2024) |
| MeSH ID | MeSH term |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | PDGFRA | C0238198 | Gastrointestinal Stromal Tumors | 10 | CGI;CLINGEN;CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Hgene | PDGFRA | C3179349 | Gastrointestinal Stromal Sarcoma | 9 | CLINGEN;CTD_human;ORPHANET |
| Hgene | PDGFRA | C0346421 | Chronic eosinophilic leukemia | 4 | ORPHANET |
| Hgene | PDGFRA | C0206141 | Idiopathic Hypereosinophilic Syndrome | 3 | CTD_human;GENOMICS_ENGLAND |
| Hgene | PDGFRA | C0006413 | Burkitt Lymphoma | 2 | ORPHANET |
| Hgene | PDGFRA | C0206142 | Eosinophilic leukemia | 2 | CTD_human |
| Hgene | PDGFRA | C0206143 | Loeffler's Endocarditis | 2 | CTD_human |
| Hgene | PDGFRA | C1292769 | Precursor B-cell lymphoblastic leukemia | 2 | ORPHANET |
| Hgene | PDGFRA | C1540912 | Hypereosinophilic syndrome | 2 | CGI;CTD_human |
Top |
Clinical Trials of the Found Drugs/Small Molecules |
 Statistics of the Clinical Trials of the Found Kinase Inibitors from clinicaltrials.gov (06-17-2024) Statistics of the Clinical Trials of the Found Kinase Inibitors from clinicaltrials.gov (06-17-2024) |
 Clinical Trials from clinicaltrials.gov (06-17-2024) Clinical Trials from clinicaltrials.gov (06-17-2024) |
| Fusion Gene | Kinase Inhibitor | NCT ID | Study Status | Phases | Disease | # Enrolment | Date |

