| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Kinase Fusion Gene:PLK1_PLK1 |
Kinase Fusion Protein Summary |
 Kinase Fusion gene summary Kinase Fusion gene summary |
| Kinase Fusion partner gene information | Kinase Fusion gene name: PLK1_PLK1 | KinaseFusionDB ID: KFG4746 | FusionGDB2.0 ID: KFG4746 | Hgene | Tgene | Gene symbol | PLK1 | PLK1 | Gene ID | 5347 | 5347 | |
| Gene name | polo like kinase 1 | polo like kinase 1 | ||||||||||
| Synonyms | PLK|STPK13 | PLK|STPK13 | ||||||||||
| Cytomap | 16p12.2 | 16p12.2 | ||||||||||
| Type of gene | protein-coding | protein-coding | ||||||||||
| Description | serine/threonine-protein kinase PLK1PLK-1cell cycle regulated protein kinasepolo (Drosophia)-like kinaseserine/threonine-protein kinase 13 | serine/threonine-protein kinase PLK1PLK-1cell cycle regulated protein kinasepolo (Drosophia)-like kinaseserine/threonine-protein kinase 13 | ||||||||||
| Modification date | 20240411 | 20240411 | ||||||||||
| UniProtAcc | P53350 | P53350 | ||||||||||
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000300093, ENST00000564202, | ENST00000564202, ENST00000300093, | |||||||||
| Context (manual curation of fusion genes in KinaseFusionDB) | PubMed: PLK1 [Title/Abstract] AND PLK1 [Title/Abstract] AND fusion [Title/Abstract] | |||||||||||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | PLK1(23695200)-PLK1(23700558), # samples:1 | |||||||||||
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | PLK1 | GO:0000086 | G2/M transition of mitotic cell cycle | 19160488 |
| Hgene | PLK1 | GO:0000132 | establishment of mitotic spindle orientation | 23509069 |
| Hgene | PLK1 | GO:0000278 | mitotic cell cycle | 18615013 |
| Hgene | PLK1 | GO:0000281 | mitotic cytokinesis | 19468302 |
| Hgene | PLK1 | GO:0001578 | microtubule bundle formation | 12939256 |
| Hgene | PLK1 | GO:0006302 | double-strand break repair | 22325354 |
| Hgene | PLK1 | GO:0006468 | protein phosphorylation | 19468300|20679239|22701722|23509069 |
| Hgene | PLK1 | GO:0007095 | mitotic G2 DNA damage checkpoint signaling | 18662541 |
| Hgene | PLK1 | GO:0016567 | protein ubiquitination | 16885022 |
| Hgene | PLK1 | GO:0018105 | peptidyl-serine phosphorylation | 16885022 |
| Hgene | PLK1 | GO:0030071 | regulation of mitotic metaphase/anaphase transition | 23509069 |
| Hgene | PLK1 | GO:0031648 | protein destabilization | 16885022 |
| Hgene | PLK1 | GO:0032465 | regulation of cytokinesis | 17351640 |
| Hgene | PLK1 | GO:0045862 | positive regulation of proteolysis | 16885022 |
| Hgene | PLK1 | GO:0071168 | protein localization to chromatin | 21111234 |
| Hgene | PLK1 | GO:0097681 | double-strand break repair via alternative nonhomologous end joining | 37440612|37674080 |
| Hgene | PLK1 | GO:1901673 | regulation of mitotic spindle assembly | 22621898 |
| Hgene | PLK1 | GO:1904668 | positive regulation of ubiquitin protein ligase activity | 15148369 |
| Hgene | PLK1 | GO:1904776 | regulation of protein localization to cell cortex | 23509069 |
| Hgene | PLK1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination | 28512243 |
| Tgene | PLK1 | GO:0000086 | G2/M transition of mitotic cell cycle | 19160488 |
| Tgene | PLK1 | GO:0000132 | establishment of mitotic spindle orientation | 23509069 |
| Tgene | PLK1 | GO:0000278 | mitotic cell cycle | 18615013 |
| Tgene | PLK1 | GO:0000281 | mitotic cytokinesis | 19468302 |
| Tgene | PLK1 | GO:0001578 | microtubule bundle formation | 12939256 |
| Tgene | PLK1 | GO:0006302 | double-strand break repair | 22325354 |
| Tgene | PLK1 | GO:0006468 | protein phosphorylation | 19468300|20679239|22701722|23509069 |
| Tgene | PLK1 | GO:0007095 | mitotic G2 DNA damage checkpoint signaling | 18662541 |
| Tgene | PLK1 | GO:0016567 | protein ubiquitination | 16885022 |
| Tgene | PLK1 | GO:0018105 | peptidyl-serine phosphorylation | 16885022 |
| Tgene | PLK1 | GO:0030071 | regulation of mitotic metaphase/anaphase transition | 23509069 |
| Tgene | PLK1 | GO:0031648 | protein destabilization | 16885022 |
| Tgene | PLK1 | GO:0032465 | regulation of cytokinesis | 17351640 |
| Tgene | PLK1 | GO:0045862 | positive regulation of proteolysis | 16885022 |
| Tgene | PLK1 | GO:0071168 | protein localization to chromatin | 21111234 |
| Tgene | PLK1 | GO:0097681 | double-strand break repair via alternative nonhomologous end joining | 37440612|37674080 |
| Tgene | PLK1 | GO:1901673 | regulation of mitotic spindle assembly | 22621898 |
| Tgene | PLK1 | GO:1904668 | positive regulation of ubiquitin protein ligase activity | 15148369 |
| Tgene | PLK1 | GO:1904776 | regulation of protein localization to cell cortex | 23509069 |
| Tgene | PLK1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination | 28512243 |
 Kinase Fusion gene breakpoints across PLK1 (5'-gene) Kinase Fusion gene breakpoints across PLK1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
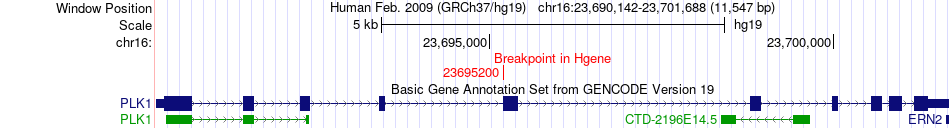 |
 Kinase Fusion gene breakpoints across PLK1 (3'-gene) Kinase Fusion gene breakpoints across PLK1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
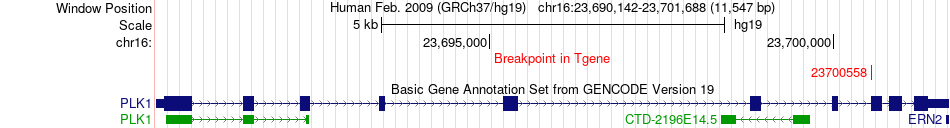 |
Top |
Kinase Fusion Gene Sample Information |
 Kinase Fusion gene information. Kinase Fusion gene information. |
 Kinase Fusion gene information from four resources (ChiTars 5.0, ChimerDB 4.0, COSMIC, and CCLE) Kinase Fusion gene information from four resources (ChiTars 5.0, ChimerDB 4.0, COSMIC, and CCLE)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Sample | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp |
| ChiTaRS5.0 | BI033419 | PLK1 | chr16 | 23695200 | PLK1 | chr16 | 23700558 |
Top |
Kinase Fusion ORF Analysis |
 Kinase Fusion information from ORFfinder translation from full-length transcript sequence from KinaseFusionDB. Kinase Fusion information from ORFfinder translation from full-length transcript sequence from KinaseFusionDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp | Seq length (transcript) | Seq length (amino acids) |
Top |
Kinase Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from KinaseFusionDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from KinaseFusionDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >Henst_Tenst_Hgene_Hchr_Hbp_Tgene_Tchr_Tbp_length(fusion AA)_AAseq |
Multiple Sequence Alignment of All Fusion Protein Isoforms |
Top |
Kinase Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr16:23695200/chr16:23700558) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| PLK1 | PLK1 |
| FUNCTION: Serine/threonine-protein kinase that performs several important functions throughout M phase of the cell cycle, including the regulation of centrosome maturation and spindle assembly, the removal of cohesins from chromosome arms, the inactivation of anaphase-promoting complex/cyclosome (APC/C) inhibitors, and the regulation of mitotic exit and cytokinesis (PubMed:8991084, PubMed:11202906, PubMed:12207013, PubMed:12447691, PubMed:12524548, PubMed:12738781, PubMed:12852856, PubMed:12939256, PubMed:14532005, PubMed:14734534, PubMed:15070733, PubMed:15148369, PubMed:15469984, PubMed:16198290, PubMed:16247472, PubMed:16980960, PubMed:17081991, PubMed:17351640, PubMed:17376779, PubMed:17617734, PubMed:18174154, PubMed:18331714, PubMed:18418051, PubMed:18477460, PubMed:18521620, PubMed:18615013, PubMed:19160488, PubMed:19351716, PubMed:19468300, PubMed:19468302, PubMed:19473992, PubMed:19509060, PubMed:19597481, PubMed:23455478, PubMed:23509069). Polo-like kinase proteins act by binding and phosphorylating proteins that are already phosphorylated on a specific motif recognized by the POLO box domains (PubMed:8991084, PubMed:11202906, PubMed:12207013, PubMed:12447691, PubMed:12524548, PubMed:12738781, PubMed:12852856, PubMed:12939256, PubMed:14532005, PubMed:14734534, PubMed:15070733, PubMed:15148369, PubMed:15469984, PubMed:16198290, PubMed:16247472, PubMed:16980960, PubMed:17081991, PubMed:17351640, PubMed:17376779, PubMed:17617734, PubMed:18174154, PubMed:18331714, PubMed:18418051, PubMed:18477460, PubMed:18521620, PubMed:18615013, PubMed:19160488, PubMed:19351716, PubMed:19468300, PubMed:19468302, PubMed:19473992, PubMed:19509060, PubMed:19597481, PubMed:23455478, PubMed:23509069). Phosphorylates BORA, BUB1B/BUBR1, CCNB1, CDC25C, CEP55, ECT2, ERCC6L, FBXO5/EMI1, FOXM1, KIF20A/MKLP2, CENPU, NEDD1, NINL, NPM1, NUDC, PKMYT1/MYT1, KIZ, PPP1R12A/MYPT1, POLQ, PRC1, RACGAP1/CYK4, RAD51, RHNO1, SGO1, STAG2/SA2, TEX14, TOPORS, p73/TP73, TPT1, WEE1 and HNRNPU (PubMed:8991084, PubMed:11202906, PubMed:12207013, PubMed:12447691, PubMed:12524548, PubMed:12738781, PubMed:12852856, PubMed:12939256, PubMed:14532005, PubMed:14734534, PubMed:15070733, PubMed:15148369, PubMed:15469984, PubMed:16198290, PubMed:16247472, PubMed:16980960, PubMed:17081991, PubMed:17218258, PubMed:17351640, PubMed:17376779, PubMed:17617734, PubMed:18174154, PubMed:18331714, PubMed:18418051, PubMed:18477460, PubMed:18521620, PubMed:18615013, PubMed:19160488, PubMed:19351716, PubMed:19468300, PubMed:19468302, PubMed:19473992, PubMed:19509060, PubMed:22325354, PubMed:19597481, PubMed:23455478, PubMed:23509069, PubMed:25986610, PubMed:26811421, PubMed:37674080, PubMed:37440612). Plays a key role in centrosome functions and the assembly of bipolar spindles by phosphorylating KIZ, NEDD1 and NINL (PubMed:16980960, PubMed:19509060). NEDD1 phosphorylation promotes subsequent targeting of the gamma-tubulin ring complex (gTuRC) to the centrosome, an important step for spindle formation (PubMed:19509060). Phosphorylation of NINL component of the centrosome leads to NINL dissociation from other centrosomal proteins (PubMed:12852856). Involved in mitosis exit and cytokinesis by phosphorylating CEP55, ECT2, KIF20A/MKLP2, CENPU, PRC1 and RACGAP1 (PubMed:12939256, PubMed:16247472, PubMed:17351640, PubMed:19468300, PubMed:19468302). Recruited at the central spindle by phosphorylating and docking PRC1 and KIF20A/MKLP2; creates its own docking sites on PRC1 and KIF20A/MKLP2 by mediating phosphorylation of sites subsequently recognized by the POLO box domains (PubMed:12939256, PubMed:17351640). Phosphorylates RACGAP1, thereby creating a docking site for the Rho GTP exchange factor ECT2 that is essential for the cleavage furrow formation (PubMed:19468300, PubMed:19468302). Promotes the central spindle recruitment of ECT2 (PubMed:16247472). Plays a central role in G2/M transition of mitotic cell cycle by phosphorylating CCNB1, CDC25C, FOXM1, CENPU, PKMYT1/MYT1, PPP1R12A/MYPT1 and WEE1 (PubMed:11202906, PubMed:12447691, PubMed:12524548, PubMed:19160488). Part of a regulatory circuit that promotes the activation of CDK1 by phosphorylating the positive regulator CDC25C and inhibiting the negative regulators WEE1 and PKMYT1/MYT1 (PubMed:11202906). Also acts by mediating phosphorylation of cyclin-B1 (CCNB1) on centrosomes in prophase (PubMed:12447691, PubMed:12524548). Phosphorylates FOXM1, a key mitotic transcription regulator, leading to enhance FOXM1 transcriptional activity (PubMed:19160488). Involved in kinetochore functions and sister chromatid cohesion by phosphorylating BUB1B/BUBR1, FBXO5/EMI1 and STAG2/SA2 (PubMed:15469984, PubMed:15148369, PubMed:17376779, PubMed:18331714). PLK1 is high on non-attached kinetochores suggesting a role of PLK1 in kinetochore attachment or in spindle assembly checkpoint (SAC) regulation (PubMed:17617734). Required for kinetochore localization of BUB1B (PubMed:17376779). Regulates the dissociation of cohesin from chromosomes by phosphorylating cohesin subunits such as STAG2/SA2 (By similarity). Phosphorylates SGO1: required for spindle pole localization of isoform 3 of SGO1 and plays a role in regulating its centriole cohesion function (PubMed:18331714). Mediates phosphorylation of FBXO5/EMI1, a negative regulator of the APC/C complex during prophase, leading to FBXO5/EMI1 ubiquitination and degradation by the proteasome (PubMed:15469984, PubMed:15148369). Acts as a negative regulator of p53 family members: phosphorylates TOPORS, leading to inhibit the sumoylation of p53/TP53 and simultaneously enhance the ubiquitination and subsequent degradation of p53/TP53 (PubMed:19473992). Phosphorylates the transactivation domain of the transcription factor p73/TP73, leading to inhibit p73/TP73-mediated transcriptional activation and pro-apoptotic functions. Phosphorylates BORA, and thereby promotes the degradation of BORA (PubMed:18521620). Contributes to the regulation of AURKA function (PubMed:18662541, PubMed:18615013). Also required for recovery after DNA damage checkpoint and entry into mitosis (PubMed:18662541, PubMed:18615013). Phosphorylates MISP, leading to stabilization of cortical and astral microtubule attachments required for proper spindle positioning (PubMed:23509069). Together with MEIKIN, acts as a regulator of kinetochore function during meiosis I: required both for mono-orientation of kinetochores on sister chromosomes and protection of centromeric cohesin from separase-mediated cleavage (By similarity). Phosphorylates CEP68 and is required for its degradation (PubMed:25503564). Regulates nuclear envelope breakdown during prophase by phosphorylating DCTN1 resulting in its localization in the nuclear envelope (PubMed:20679239). Phosphorylates the heat shock transcription factor HSF1, promoting HSF1 nuclear translocation upon heat shock (PubMed:15661742). Phosphorylates HSF1 also in the early mitotic period; this phosphorylation regulates HSF1 localization to the spindle pole, the recruitment of the SCF(BTRC) ubiquitin ligase complex induicing HSF1 degradation, and hence mitotic progression (PubMed:18794143). Regulates mitotic progression by phosphorylating RIOK2 (PubMed:21880710). Through the phosphorylation of DZIP1 regulates the localization during mitosis of the BBSome, a ciliary protein complex involved in cilium biogenesis (PubMed:27979967). Regulates DNA repair during mitosis by mediating phosphorylation of POLQ and RHNO1, thereby promoting POLQ recruitment to DNA damage sites (PubMed:37674080, PubMed:37440612). {ECO:0000250|UniProtKB:P70032, ECO:0000250|UniProtKB:Q5F2C3, ECO:0000269|PubMed:11202906, ECO:0000269|PubMed:12207013, ECO:0000269|PubMed:12447691, ECO:0000269|PubMed:12524548, ECO:0000269|PubMed:12738781, ECO:0000269|PubMed:12852856, ECO:0000269|PubMed:12939256, ECO:0000269|PubMed:14532005, ECO:0000269|PubMed:14734534, ECO:0000269|PubMed:15070733, ECO:0000269|PubMed:15148369, ECO:0000269|PubMed:15469984, ECO:0000269|PubMed:15661742, ECO:0000269|PubMed:16198290, ECO:0000269|PubMed:16247472, ECO:0000269|PubMed:16980960, ECO:0000269|PubMed:17081991, ECO:0000269|PubMed:17218258, ECO:0000269|PubMed:17351640, ECO:0000269|PubMed:17376779, ECO:0000269|PubMed:17617734, ECO:0000269|PubMed:18174154, ECO:0000269|PubMed:18331714, ECO:0000269|PubMed:18418051, ECO:0000269|PubMed:18477460, ECO:0000269|PubMed:18521620, ECO:0000269|PubMed:18615013, ECO:0000269|PubMed:18662541, ECO:0000269|PubMed:18794143, ECO:0000269|PubMed:19160488, ECO:0000269|PubMed:19351716, ECO:0000269|PubMed:19468300, ECO:0000269|PubMed:19468302, ECO:0000269|PubMed:19473992, ECO:0000269|PubMed:19509060, ECO:0000269|PubMed:19597481, ECO:0000269|PubMed:20679239, ECO:0000269|PubMed:21880710, ECO:0000269|PubMed:22325354, ECO:0000269|PubMed:23455478, ECO:0000269|PubMed:23509069, ECO:0000269|PubMed:25503564, ECO:0000269|PubMed:25986610, ECO:0000269|PubMed:26811421, ECO:0000269|PubMed:27979967, ECO:0000269|PubMed:37440612, ECO:0000269|PubMed:37674080, ECO:0000269|PubMed:8991084}. | FUNCTION: Serine/threonine-protein kinase that performs several important functions throughout M phase of the cell cycle, including the regulation of centrosome maturation and spindle assembly, the removal of cohesins from chromosome arms, the inactivation of anaphase-promoting complex/cyclosome (APC/C) inhibitors, and the regulation of mitotic exit and cytokinesis (PubMed:8991084, PubMed:11202906, PubMed:12207013, PubMed:12447691, PubMed:12524548, PubMed:12738781, PubMed:12852856, PubMed:12939256, PubMed:14532005, PubMed:14734534, PubMed:15070733, PubMed:15148369, PubMed:15469984, PubMed:16198290, PubMed:16247472, PubMed:16980960, PubMed:17081991, PubMed:17351640, PubMed:17376779, PubMed:17617734, PubMed:18174154, PubMed:18331714, PubMed:18418051, PubMed:18477460, PubMed:18521620, PubMed:18615013, PubMed:19160488, PubMed:19351716, PubMed:19468300, PubMed:19468302, PubMed:19473992, PubMed:19509060, PubMed:19597481, PubMed:23455478, PubMed:23509069). Polo-like kinase proteins act by binding and phosphorylating proteins that are already phosphorylated on a specific motif recognized by the POLO box domains (PubMed:8991084, PubMed:11202906, PubMed:12207013, PubMed:12447691, PubMed:12524548, PubMed:12738781, PubMed:12852856, PubMed:12939256, PubMed:14532005, PubMed:14734534, PubMed:15070733, PubMed:15148369, PubMed:15469984, PubMed:16198290, PubMed:16247472, PubMed:16980960, PubMed:17081991, PubMed:17351640, PubMed:17376779, PubMed:17617734, PubMed:18174154, PubMed:18331714, PubMed:18418051, PubMed:18477460, PubMed:18521620, PubMed:18615013, PubMed:19160488, PubMed:19351716, PubMed:19468300, PubMed:19468302, PubMed:19473992, PubMed:19509060, PubMed:19597481, PubMed:23455478, PubMed:23509069). Phosphorylates BORA, BUB1B/BUBR1, CCNB1, CDC25C, CEP55, ECT2, ERCC6L, FBXO5/EMI1, FOXM1, KIF20A/MKLP2, CENPU, NEDD1, NINL, NPM1, NUDC, PKMYT1/MYT1, KIZ, PPP1R12A/MYPT1, POLQ, PRC1, RACGAP1/CYK4, RAD51, RHNO1, SGO1, STAG2/SA2, TEX14, TOPORS, p73/TP73, TPT1, WEE1 and HNRNPU (PubMed:8991084, PubMed:11202906, PubMed:12207013, PubMed:12447691, PubMed:12524548, PubMed:12738781, PubMed:12852856, PubMed:12939256, PubMed:14532005, PubMed:14734534, PubMed:15070733, PubMed:15148369, PubMed:15469984, PubMed:16198290, PubMed:16247472, PubMed:16980960, PubMed:17081991, PubMed:17218258, PubMed:17351640, PubMed:17376779, PubMed:17617734, PubMed:18174154, PubMed:18331714, PubMed:18418051, PubMed:18477460, PubMed:18521620, PubMed:18615013, PubMed:19160488, PubMed:19351716, PubMed:19468300, PubMed:19468302, PubMed:19473992, PubMed:19509060, PubMed:22325354, PubMed:19597481, PubMed:23455478, PubMed:23509069, PubMed:25986610, PubMed:26811421, PubMed:37674080, PubMed:37440612). Plays a key role in centrosome functions and the assembly of bipolar spindles by phosphorylating KIZ, NEDD1 and NINL (PubMed:16980960, PubMed:19509060). NEDD1 phosphorylation promotes subsequent targeting of the gamma-tubulin ring complex (gTuRC) to the centrosome, an important step for spindle formation (PubMed:19509060). Phosphorylation of NINL component of the centrosome leads to NINL dissociation from other centrosomal proteins (PubMed:12852856). Involved in mitosis exit and cytokinesis by phosphorylating CEP55, ECT2, KIF20A/MKLP2, CENPU, PRC1 and RACGAP1 (PubMed:12939256, PubMed:16247472, PubMed:17351640, PubMed:19468300, PubMed:19468302). Recruited at the central spindle by phosphorylating and docking PRC1 and KIF20A/MKLP2; creates its own docking sites on PRC1 and KIF20A/MKLP2 by mediating phosphorylation of sites subsequently recognized by the POLO box domains (PubMed:12939256, PubMed:17351640). Phosphorylates RACGAP1, thereby creating a docking site for the Rho GTP exchange factor ECT2 that is essential for the cleavage furrow formation (PubMed:19468300, PubMed:19468302). Promotes the central spindle recruitment of ECT2 (PubMed:16247472). Plays a central role in G2/M transition of mitotic cell cycle by phosphorylating CCNB1, CDC25C, FOXM1, CENPU, PKMYT1/MYT1, PPP1R12A/MYPT1 and WEE1 (PubMed:11202906, PubMed:12447691, PubMed:12524548, PubMed:19160488). Part of a regulatory circuit that promotes the activation of CDK1 by phosphorylating the positive regulator CDC25C and inhibiting the negative regulators WEE1 and PKMYT1/MYT1 (PubMed:11202906). Also acts by mediating phosphorylation of cyclin-B1 (CCNB1) on centrosomes in prophase (PubMed:12447691, PubMed:12524548). Phosphorylates FOXM1, a key mitotic transcription regulator, leading to enhance FOXM1 transcriptional activity (PubMed:19160488). Involved in kinetochore functions and sister chromatid cohesion by phosphorylating BUB1B/BUBR1, FBXO5/EMI1 and STAG2/SA2 (PubMed:15469984, PubMed:15148369, PubMed:17376779, PubMed:18331714). PLK1 is high on non-attached kinetochores suggesting a role of PLK1 in kinetochore attachment or in spindle assembly checkpoint (SAC) regulation (PubMed:17617734). Required for kinetochore localization of BUB1B (PubMed:17376779). Regulates the dissociation of cohesin from chromosomes by phosphorylating cohesin subunits such as STAG2/SA2 (By similarity). Phosphorylates SGO1: required for spindle pole localization of isoform 3 of SGO1 and plays a role in regulating its centriole cohesion function (PubMed:18331714). Mediates phosphorylation of FBXO5/EMI1, a negative regulator of the APC/C complex during prophase, leading to FBXO5/EMI1 ubiquitination and degradation by the proteasome (PubMed:15469984, PubMed:15148369). Acts as a negative regulator of p53 family members: phosphorylates TOPORS, leading to inhibit the sumoylation of p53/TP53 and simultaneously enhance the ubiquitination and subsequent degradation of p53/TP53 (PubMed:19473992). Phosphorylates the transactivation domain of the transcription factor p73/TP73, leading to inhibit p73/TP73-mediated transcriptional activation and pro-apoptotic functions. Phosphorylates BORA, and thereby promotes the degradation of BORA (PubMed:18521620). Contributes to the regulation of AURKA function (PubMed:18662541, PubMed:18615013). Also required for recovery after DNA damage checkpoint and entry into mitosis (PubMed:18662541, PubMed:18615013). Phosphorylates MISP, leading to stabilization of cortical and astral microtubule attachments required for proper spindle positioning (PubMed:23509069). Together with MEIKIN, acts as a regulator of kinetochore function during meiosis I: required both for mono-orientation of kinetochores on sister chromosomes and protection of centromeric cohesin from separase-mediated cleavage (By similarity). Phosphorylates CEP68 and is required for its degradation (PubMed:25503564). Regulates nuclear envelope breakdown during prophase by phosphorylating DCTN1 resulting in its localization in the nuclear envelope (PubMed:20679239). Phosphorylates the heat shock transcription factor HSF1, promoting HSF1 nuclear translocation upon heat shock (PubMed:15661742). Phosphorylates HSF1 also in the early mitotic period; this phosphorylation regulates HSF1 localization to the spindle pole, the recruitment of the SCF(BTRC) ubiquitin ligase complex induicing HSF1 degradation, and hence mitotic progression (PubMed:18794143). Regulates mitotic progression by phosphorylating RIOK2 (PubMed:21880710). Through the phosphorylation of DZIP1 regulates the localization during mitosis of the BBSome, a ciliary protein complex involved in cilium biogenesis (PubMed:27979967). Regulates DNA repair during mitosis by mediating phosphorylation of POLQ and RHNO1, thereby promoting POLQ recruitment to DNA damage sites (PubMed:37674080, PubMed:37440612). {ECO:0000250|UniProtKB:P70032, ECO:0000250|UniProtKB:Q5F2C3, ECO:0000269|PubMed:11202906, ECO:0000269|PubMed:12207013, ECO:0000269|PubMed:12447691, ECO:0000269|PubMed:12524548, ECO:0000269|PubMed:12738781, ECO:0000269|PubMed:12852856, ECO:0000269|PubMed:12939256, ECO:0000269|PubMed:14532005, ECO:0000269|PubMed:14734534, ECO:0000269|PubMed:15070733, ECO:0000269|PubMed:15148369, ECO:0000269|PubMed:15469984, ECO:0000269|PubMed:15661742, ECO:0000269|PubMed:16198290, ECO:0000269|PubMed:16247472, ECO:0000269|PubMed:16980960, ECO:0000269|PubMed:17081991, ECO:0000269|PubMed:17218258, ECO:0000269|PubMed:17351640, ECO:0000269|PubMed:17376779, ECO:0000269|PubMed:17617734, ECO:0000269|PubMed:18174154, ECO:0000269|PubMed:18331714, ECO:0000269|PubMed:18418051, ECO:0000269|PubMed:18477460, ECO:0000269|PubMed:18521620, ECO:0000269|PubMed:18615013, ECO:0000269|PubMed:18662541, ECO:0000269|PubMed:18794143, ECO:0000269|PubMed:19160488, ECO:0000269|PubMed:19351716, ECO:0000269|PubMed:19468300, ECO:0000269|PubMed:19468302, ECO:0000269|PubMed:19473992, ECO:0000269|PubMed:19509060, ECO:0000269|PubMed:19597481, ECO:0000269|PubMed:20679239, ECO:0000269|PubMed:21880710, ECO:0000269|PubMed:22325354, ECO:0000269|PubMed:23455478, ECO:0000269|PubMed:23509069, ECO:0000269|PubMed:25503564, ECO:0000269|PubMed:25986610, ECO:0000269|PubMed:26811421, ECO:0000269|PubMed:27979967, ECO:0000269|PubMed:37440612, ECO:0000269|PubMed:37674080, ECO:0000269|PubMed:8991084}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
 - Retained domain in the 5'-partner of fusion protein (protein functional feature from UniProt). - Retained domain in the 5'-partner of fusion protein (protein functional feature from UniProt). |
| Partner | Hgeneene | Hbp | Tgeneene | Tbp | ENST | BPexon | TotalExon | Protein feature loci | BPloci | TotalLen | Feature | Note |
 - Retained domain in the 3'-partner of fusion protein (protein functional feature from UniProt). - Retained domain in the 3'-partner of fusion protein (protein functional feature from UniProt). |
| Partner | Hgeneene | Hbp | Tgeneene | Tbp | ENST | BPexon | TotalExon | Protein feature loci | BPloci | TotalLen | Feature | Note |
Top |
Kinase-Substrate Information of PLK1_PLK1 |
 Phosphorylation target of the kinase Phosphorylation target of the kinase(phosphosite, 03-17-2024) |
| Kinase | Kinase UniProt Acc | Kinase species | Substrate | Substrate UniProt Acc | Substrate phosphorylated residues | Substrate phosphorylated sites (+/-7AA) | Domain |
| PLK1 | P53350 | human | DCTN1 | Q14203 | S179 | sASAGELsSSEPSTP | |
| PLK1 | P53350 | human | CCNT1 | O60563 | S564 | KTYSLSSsFSSSSsT | |
| PLK1 | P53350 | human | NINL | Q9Y2I6 | S88 | RPSDEDssSLESAAS | |
| PLK1 | P53350 | human | PTEN | P60484 | S380 | EPDHyRYsDttDsDP | |
| PLK1 | P53350 | human | PINX1 | Q96BK5 | S226 | ATGKDVEsyLQPkAK | |
| PLK1 | P53350 | human | CLASP2 | O75122 | S1092 | MKLtQEEsFSVWDEH | |
| PLK1 | P53350 | human | ANAPC1 | Q9H1A4 | T520 | GLPAPsLtMsNtMPR | |
| PLK1 | P53350 | human | MAP9 | Q49MG5 | S289 | SDENKENsFsADHVT | |
| PLK1 | P53350 | human | PCNT | O95613 | T1221 | ALPELDRtLSECAEM | |
| PLK1 | P53350 | human | CDC25C | P30307 | S75 | PkRCLDLsNLSSGEI | |
| PLK1 | P53350 | human | PRC1 | O43663 | T616 | TSGILNstNIQs___ | |
| PLK1 | P53350 | human | KIF2C | Q99661 | S715 | MQLEEQAsRQIsSKK | |
| PLK1 | P53350 | human | KMT2E | Q8IZD2 | S861 | sPLLLNDsCSLPDLT | |
| PLK1 | P53350 | human | MAD1L1 | Q9Y6D9 | T680 | SKMQLLEtEFSHTVG | MAD |
| PLK1 | P53350 | human | MDC1 | Q14676 | S20 | EEEEtEQssESLRCN | |
| PLK1 | P53350 | human | VCP | P55072 | T76 | CIVLsDDtCSDEkIR | CDC48_N |
| PLK1 | P53350 | human | CDC27 | P30260 | T209 | LtEtPQDtIELNRLN | |
| PLK1 | P53350 | human | BORA | Q6PGQ7 | S497 | SSNIQMDsGYNtQNC | |
| PLK1 | P53350 | human | TNKS | O95271 | T982 | SLIsPAStPSCLsAA | |
| PLK1 | P53350 | human | KNL1 | Q8NG31-2 | T875 | DKNDMDItKSYTIEI | MELT |
| PLK1 | P53350 | human | TTK | P33981 | T46 | NKISADttDNsGTVN | |
| PLK1 | P53350 | human | CENPQ | Q7L2Z9 | T123 | RLLQQCEtLKVPPKk | CENP-Q |
| PLK1 | P53350 | human | PCNT | O95613 | T1209 | TAPALEEtWSDVALP | |
| PLK1 | P53350 | human | STK38 | Q15208 | T407 | EIKSIDDtSNFDEFP | Pkinase_C |
| PLK1 | P53350 | human | PPP6R2 | O75170 | T10 | WKFDLNTtSHVDkLL | |
| PLK1 | P53350 | human | STK38 | Q15208 | T7 | _MAMtGstPCssMsN | |
| PLK1 | P53350 | human | CDC20 | Q12834 | S170 | KtCRYIPsLPDrILD | |
| PLK1 | P53350 | human | TOP2A | P11388 | T1343 | FsDFDEKtDDEDFVP | |
| PLK1 | P53350 | human | TTK | P33981 | T33 | kFkNEDLtDELsLNK | |
| PLK1 | P53350 | human | NEDD1 | Q8NHV4 | S637 | HSLLERysVNEGLVA | |
| PLK1 | P53350 | human | TOP2A | P11388 | S1525 | PIKyLEEsDEDDLF_ | |
| PLK1 | P53350 | human | CDC16 | Q13042 | S112 | EkYLkDEsGFkDPSS | |
| PLK1 | P53350 | human | KIF2B | Q8N4N8 | T125 | MIPQKNQtAsGDsLD | |
| PLK1 | P53350 | human | HSPA1B | P0DMV8 | S633 | GPkGGsGsGPtIEEV | |
| PLK1 | P53350 | human | DVL2 | O14641 | T206 | MTSELEStSLGDsDE | Dishevelled |
| PLK1 | P53350 | human | CLASP2 | O75122 | S1053 | DQFPDDLsLDHsDLV | |
| PLK1 | P53350 | human | PTPN1 | P18031 | S393 | sPAKGEPsLPEKDED | |
| PLK1 | P53350 | human | MDM2 | Q00987 | S260 | sLDsEDYsLsEEGQE | |
| PLK1 | P53350 | human | HNRNPU | Q00839 | S59 | AMEPGNGsLDLGGDs | |
| PLK1 | P53350 | human | TOPORS | Q9NS56 | S718 | kDRDGYEsSYRRRTL | |
| PLK1 | P53350 | human | CENPQ | Q7L2Z9 | S138 | MEDLtNVssLLNMER | CENP-Q |
| PLK1 | P53350 | human | CTNNB1 | P35222 | S311 | LAYGNQEsKLIILAS | |
| PLK1 | P53350 | human | TTK | P33981 | T594 | RLYDYEItDQYIYMV | Pkinase |
| PLK1 | P53350 | human | KLF4 | O43474 | S234 | GkFVLkAsLsAPGSE | |
| PLK1 | P53350 | human | CENPQ | Q7L2Z9 | T256 | sQMKsMstFIEEAYK | CENP-Q |
| PLK1 | P53350 | human | TTK | P33981 | S321 | SkPsGNDsCELRNLk | |
| PLK1 | P53350 | human | BRCA1 | P38398 | S1164 | GEIKEDTsFAENDIk | |
| PLK1 | P53350 | human | ANAPC1 | Q9H1A4 | S377 | NIssHNQsPkRHSIS | |
| PLK1 | P53350 | human | STK3 | Q13188 | T384 | GtMKRNAtsPQVQRP | |
| PLK1 | P53350 | human | LRRK1 | Q38SD2 | S1817 | GDSIADVsIMYSEEL | |
| PLK1 | P53350 | human | TTK | P33981 | S108 | DkYGQNEsFARIQVR | |
| PLK1 | P53350 | human | CENPQ | Q7L2Z9 | S253 | HNssQMKsMstFIEE | CENP-Q |
| PLK1 | P53350 | human | ANAPC7 | Q9UJX3 | S23 | HsNVRLLssLLLtMs | |
| PLK1 | P53350 | human | TTK | P33981 | T458 | CktPsSNtLDDyMsC | |
| PLK1 | P53350 | human | CEP68 | Q76N32 | S332 | ADPVLQDsGVDLDSF | |
| PLK1 | P53350 | human | PPP1R7 | Q15435 | S44 | sGIVADLsEQsLkDG | |
| PLK1 | P53350 | human | TTK | P33981 | T12 | DLsGRELtIDsIMNk | |
| PLK1 | P53350 | human | NEDD1 | Q8NHV4 | S426 | VNKGsDEsIGkGDGF | |
| PLK1 | P53350 | human | TTK | P33981 | S333 | NLkSVQNsHFkEPLV | |
| PLK1 | P53350 | human | MAD1L1 | Q9Y6D9 | S22 | RsLNNFIsQRVEGGs | |
| PLK1 | P53350 | human | PPP1R7 | Q15435 | S27 | RRVEsEEsGDEEGKk | |
| PLK1 | P53350 | human | CASP8 | Q14790 | S305 | IYQLMDHsNMDCFIC | Peptidase_C14 |
| PLK1 | P53350 | human | USP16 | Q9Y5T5 | S386 | HESFLDLsLPVLDDQ | UCH |
| PLK1 | P53350 | human | NCAPH2 | Q6IBW4 | S288 | EsRsPQQsAALPRRY | |
| PLK1 | P53350 | human | PPP1R7 | Q15435 | S24 | EVDRRVEsEEsGDEE | |
| PLK1 | P53350 | human | FOXM1 | Q08050 | S739 | SKILLDIsFPGLDED | |
| PLK1 | P53350 | human | MAP2K1 | Q02750 | S222 | LIDsMANsFVGtRSY | Pkinase |
| PLK1 | P53350 | human | PCNT | O95613 | S1235 | MSSVAEIsSHMREsF | |
| PLK1 | P53350 | human | PRC1 | O43663 | S615 | ATSGILNstNIQs__ | |
| PLK1 | P53350 | human | CDC6 | Q99741 | T37 | SDAkLEPtNVQTVtC | |
| PLK1 | P53350 | human | CENPQ | Q7L2Z9 | S249 | LDILHNssQMKsMst | CENP-Q |
| PLK1 | P53350 | human | VIM | P08670 | T336 | EVdALkGtNEsLERQ | Filament |
| PLK1 | P53350 | human | CCNB1 | P14635 | S126 | PILVDTAsPsPMETs | |
| PLK1 | P53350 | human | RACGAP1 | Q9H0H5 | S164 | sFDKtDEsLDWDssL | |
| PLK1 | P53350 | human | FOXM1 | Q08050-2 | S724 | SKILLDIsFPGLDED | |
| PLK1 | P53350 | human | NCAPD3 | P42695 | S1419 | AIstPEKsISDVtFG | |
| PLK1 | P53350 | human | TTK | P33981 | S291 | CDVktDDsVVPCFMK | |
| PLK1 | P53350 | human | PAX3/FOXO1 | AAC50053 | S503 | PRTSSNAsTISGRLs | |
| PLK1 | P53350 | human | AXIN2 | Q9Y2T1 | S311 | SDALTDDsMSMTDSS | |
| PLK1 | P53350 | human | RAD21 | O60216 | S175 | REIMREGsAFEDDDM | |
| PLK1 | P53350 | human | MDC1 | Q14676 | T4 | ____MEDtQAIDWDV | |
| PLK1 | P53350 | human | TTK | P33981 | T371 | LLAkLEEtkEyQEPE | |
| PLK1 | P53350 | human | BRCA2 | P51587 | S205 | LATPPtLsstVLIVR | |
| PLK1 | P53350 | human | BRF1 | Q92994 | S450 | GDGELDLsGIDDLEI | BRF1 |
| PLK1 | P53350 | human | TNKS | O95271 | T930 | LAHGADPtMkNQEGQ | Ank_5 |
| PLK1 | P53350 | human | YY1 | P25490 | T39 | PVETIETtVVGEEEE | |
| PLK1 | P53350 | human | WDCP | Q9H6R7 | S686 | RsDVFRDsFsHsPGA | |
| PLK1 | P53350 | human | CDC25B | P30305 | S465 | PLERDAEsFLLksPI | Rhodanese |
| PLK1 | P53350 | human | NUAK1 | O60285 | S480 | QREsGYYsSPERSES | |
| PLK1 | P53350 | human | TP53BP1 | Q12888 | S1618 | LtkAADIsLDNLVEG | |
| PLK1 | P53350 | human | VIM | P08670 | S83 | GVRLLQDsVdFsLAd | Filament_head |
| PLK1 | P53350 | human | CDC23 | Q9UJX2 | T562 | QLRNQGEtPttEVPA | |
| PLK1 | P53350 | human | SUZ12 | Q15022 | S539 | RPKRTkAsMsEFLEs | |
| PLK1 | P53350 | human | MAP2K1 | Q02750 | S218 | VsGQLIDsMANsFVG | Pkinase |
| PLK1 | P53350 | human | BRCA2 | P51587 | S193 | AEVDPDMsWSSSLAT | |
| PLK1 | P53350 | human | STK3 | Q13188 | S15 | KSKLKkLsEDsLTkQ | |
| PLK1 | P53350 | human | CENPU | Q71F23 | S77 | TFDPPLHstAIyADE | |
| PLK1 | P53350 | human | CDC27 | P30260 | T430 | INDsLEItkLDssII | |
| PLK1 | P53350 | human | CHEK2 | O96017 | T205 | VFVFFDLtVDDQsVY | |
| PLK1 | P53350 | human | KIF2C | Q99661 | S632 | EELSsQMssFNEAMT | |
| PLK1 | P53350 | human | WEE1 | P30291 | S53 | GHstGEDsAFQEPDS | |
| PLK1 | P53350 | human | VIM | P08670 | S459 | GQVINEtsQHHDDLE | |
| PLK1 | P53350 | human | KDM1A | O60341 | S126 | EYREMDEsLANLsED | |
| PLK1 | P53350 | human | CDC23 | Q9UJX2 | T565 | NQGEtPttEVPAPFF | |
| PLK1 | P53350 | human | TTK | P33981 | T676 | NQMQPDttsVVkDSQ | Pkinase |
| PLK1 | P53350 | human | RIOK2 | Q9BVS4 | S548 | KSSLEAAsFWGE___ | |
| PLK1 | P53350 | human | RAPGEF2 | Q17RH5 | S794 | RECKNFNsMFAIISG | RasGEF |
| PLK1 | P53350 | human | TP73 | O15350 | T27 | SSLEPDStyFDLPQS | |
| PLK1 | P53350 | human | TSC1 | Q92574 | S579 | CQTSLETsIFTPsPC | Hamartin |
| PLK1 | P53350 | human | RUVBL1 | Q9Y265 | T239 | kEIIQDVtLHDLDVA | TIP49 |
| PLK1 | P53350 | human | CEP55 | Q53EZ4 | S436 | PtAALNEsLVECPkC | |
| PLK1 | P53350 | human | CDC25B | P30305 | S375 | ARVLRsksLCHDEIE | M-inducer_phosp |
| PLK1 | P53350 | human | STK38 | Q15208 | T183 | TLLMKKDtLTEEETQ | Pkinase |
| PLK1 | P53350 | human | RACGAP1 | Q9H0H5 | T260 | QPWNsDstLNSRQLE | |
| PLK1 | P53350 | human | BCL2L1 | Q07817 | S62 | PSWHLADsPAVNGAT | |
| PLK1 | P53350 | human | TERF1 | P54274 | S435 | KKLKLIssDsED___ | |
| PLK1 | P53350 | human | ERCC6L | Q2NKX8 | T1063 | VKQFDAstPkNDIsP | |
| PLK1 | P53350 | human | NUDC | Q9Y266 | S326 | QHPEMDFskAkFN__ | |
| PLK1 | P53350 | human | VIM | P08670 | T327 | RRQVQsLtCEVdALk | Filament |
| PLK1 | P53350 | human | SNCB | Q16143 | S118 | LMEPEGEsYEDPPQE | Synuclein |
| PLK1 | P53350 | human | TNKS | O95271 | T839 | DTQGRNStPLHLAAG | Ank_4 |
| PLK1 | P53350 | human | KIZ | Q2M2Z5 | T379 | WSTSSDLtISISEDD | |
| PLK1 | P53350 | human | BRCA2 | P51587 | S206 | ATPPtLsstVLIVRN | |
| PLK1 | P53350 | human | RAPGEF2 | Q17RH5 | S1164 | DERRQRHsVSIVETN | |
| PLK1 | P53350 | human | RBBP8 | Q99708 | T693 | ETVDMDCtLVSETVL | |
| PLK1 | P53350 | human | PINX1 | Q96BK5 | S110 | SDKKEKKsFSLEEKs | |
| PLK1 | P53350 | human | SUZ12 | Q15022 | S546 | sMsEFLEsEDGEVEQ | VEFS-Box |
| PLK1 | P53350 | human | MAD1L1 | Q9Y6D9 | S29 | sQRVEGGsGLDISTS | |
| PLK1 | P53350 | human | HSF1 | Q00613 | S419 | SALLDLFsPSVTVPD | Vert_HS_TF |
| PLK1 | P53350 | human | MAP2K2 | P36507 | S222 | VsGQLIDsMANsFVG | Pkinase |
| PLK1 | P53350 | human | NINL | Q9Y2I6 | S87 | VRPSDEDssSLESAA | |
| PLK1 | P53350 | human | CDC25B | P30305 | S397 | RELIGDYskAFLLQt | |
| PLK1 | P53350 | human | RAPGEF2 | Q17RH5 | S921 | KKWRSLGsLSQGSTN | |
| PLK1 | P53350 | human | PPP1R7 | Q15435 | T277 | LENNNKLtMLDIASN | LRR_9 |
| PLK1 | P53350 | human | CLASP2 | O75122 | S1027 | NPyNysDsIsPFNKs | |
| PLK1 | P53350 | human | CLIP1 | P30622 | S195 | LtKtAsEsIsNLsEA | |
| PLK1 | P53350 | human | CDC27 | P30260 | S435 | EItkLDssIIsEGkI | |
| PLK1 | P53350 | human | YAP1 | P46937 | T77 | NAVMNPktANVPQtV | |
| PLK1 | P53350 | human | TEX14 | Q8IWB6 | S437 | QKAATVKsDIYSFSM | PK_Tyr_Ser-Thr |
| PLK1 | P53350 | human | CCNB1 | P14635 | S147 | EDLCQAFsDVILAVN | |
| PLK1 | P53350 | human | SNCA | P37840 | S129 | NEAyEMPsEEGyQDy | Synuclein |
| PLK1 | P53350 | human | RACGAP1 | Q9H0H5 | S157 | GsILsDIsFDKtDEs | |
| PLK1 | P53350 | human | CENPQ | Q7L2Z9 | S139 | EDLtNVssLLNMERA | CENP-Q |
| PLK1 | P53350 | human | FIRRM | Q9NSG2 | S744 | ETKNKVVsFLEkTGF | |
| PLK1 | P53350 | human | CDC25B | P30305 | S353 | VQNkRRRsVtPPEEQ | M-inducer_phosp |
| PLK1 | P53350 | human | NOTCH1 | P46531 | S1791 | REPLGEDsVGLkPLK | |
| PLK1 | P53350 | human | CDC25B | P30305 | T127 | DPHMAEQtFEQAIQA | M-inducer_phosp |
| PLK1 | P53350 | human | TTK | P33981 | T210 | LSASTVLtAQEsFsG | |
| PLK1 | P53350 | human | TTK | P33981 | S214 | TVLtAQEsFsGsLGH | |
| PLK1 | P53350 | human | CDC25B | P30305 | T265 | LTPAEGDtEEDDGFV | M-inducer_phosp |
| PLK1 | P53350 | human | KIF2C | Q99661 | S633 | ELSsQMssFNEAMTQ | |
| PLK1 | P53350 | human | IRS1 | P35568 | S24 | GYLRKPKsMHKRFFV | PH |
| PLK1 | P53350 | human | CENPQ | Q7L2Z9 | S255 | ssQMKsMstFIEEAY | CENP-Q |
| PLK1 | P53350 | human | PCNT | O95613 | S1241 | IsSHMREsFLMsPES | |
| PLK1 | P53350 | human | TTK | P33981 | S362 | TLKNktEssLLAkLE | |
| PLK1 | P53350 | human | RB1 | P06400 | S758 | SIIVFYNsVFMQRLK | |
| PLK1 | P53350 | human | RBBP8 | Q99708 | S723 | EERKMNDsLEDMFDR | |
| PLK1 | P53350 | human | SUZ12 | Q15022 | S541 | KRTkAsMsEFLEsED | |
| PLK1 | P53350 | human | RIOK2 | Q9BVS4 | S335 | TKEGsEFsFsDGEVA | |
| PLK1 | P53350 | human | MRE11 | P49959 | S688 | sKGVDFEssEDDDDD | |
| PLK1 | P53350 | human | KIF2A | O00139 | T554 | ANRVKELtVDPTAAG | |
| PLK1 | P53350 | human | OPTN | Q96CV9 | S177 | ssGssEDsFVEIRMA | |
| PLK1 | P53350 | human | ANAPC1 | Q9H1A4 | S373 | VQRFNIssHNQsPkR | |
| PLK1 | P53350 | human | CLSPN | Q9HAW4 | S30 | EADSPSDsGQGsYET | |
| PLK1 | P53350 | human | TTK | P33981 | S345 | PLVSDEKsSELIItD | |
| PLK1 | P53350 | human | PIN1 | Q13526 | S65 | sHLLVkHsQSRRPss | Rotamase |
| PLK1 | P53350 | human | ESPL1 | Q14674 | S1399 | KVNFsDDsDLEDPVS | |
| PLK1 | P53350 | human | FBXO5 | Q9UKT4 | S149 | yEDsGYSsFSLQSGL | |
| PLK1 | P53350 | human | ANAPC7 | Q9UJX3 | S17 | MAAAGLHsNVRLLss | |
| PLK1 | P53350 | human | RAP1GAP | P47736 | S525 | AGQKtPDsGHVSQEP | |
| PLK1 | P53350 | human | ANAPC1 | Q9H1A4 | S547 | PLsKLLGsLDEVVLL | |
| PLK1 | P53350 | human | ANAPC1 | Q9H1A4 | S688 | FDFEGsLsPVIAPkk | Apc1_MidN |
| PLK1 | P53350 | human | CIP2A | Q8TCG1 | S904 | NPETVNLsI______ | |
| PLK1 | P53350 | human | RAPGEF2 | Q17RH5 | T632 | AIREFAVtATPDQYS | RA |
| PLK1 | P53350 | human | TNKS | O95271 | S978 | VVSASLIsPAStPSC | |
| PLK1 | P53350 | human | BIRC5 | O15392 | S20 | FLkDHRIstFkNWPF | BIR |
| PLK1 | P53350 | human | IKBKB | O14920 | S740 | sFtALDWsWLQTEEE | IKKbetaNEMObind |
| PLK1 | P53350 | human | GTSE1 | Q9NYZ3 | S454 | RsIRRRDsCLNsKTK | |
| PLK1 | P53350 | human | ZMYM2 | Q9UBW7 | S303 | QkQPGVDsLsPVAsL | |
| PLK1 | P53350 | human | CDC27 | P30260 | T205 | PEtVLtEtPQDtIEL | |
| PLK1 | P53350 | human | PTPN1 | P18031 | S286 | KFIMGDSsVQDQWKE | |
| PLK1 | P53350 | human | USP16 | Q9Y5T5 | S330 | ILKAFGNsTEkLDEE | UCH |
| PLK1 | P53350 | human | RAPGEF2 | Q17RH5 | S1010 | KPVKSETsPVAPRAG | |
| PLK1 | P53350 | human | PLEKHG6 | Q3KR16 | T574 | HLVVTEDtDEDAPLV | |
| PLK1 | P53350 | human | CENPQ | Q7L2Z9 | S248 | DLDILHNssQMKsMs | CENP-Q |
| PLK1 | P53350 | human | TRIOBP | Q9H2D6-5 | T457 | QAEEREHtLRRCQQE | |
| PLK1 | P53350 | human | WDCP | Q9H6R7 | S695 | sHsPGAVssLKVFTG | |
| PLK1 | P53350 | human | KIF2B | Q8N4N8 | S204 | HLDssKIsVLEPPQE | |
| PLK1 | P53350 | human | PRKN | O60260 | S378 | AyHEGECsAVFEASG | |
| PLK1 | P53350 | human | RBBP8 | Q99708 | T731 | LEDMFDRtTHEEYES | |
| PLK1 | P53350 | human | CTNNB1 | P35222 | S718 | QDDPsyRsFHsGGyG | |
| PLK1 | P53350 | human | SUN1 | O94901-9 | S138 | RPPVLDEsWIREQTT | MRP |
| PLK1 | P53350 | human | NOTCH1 | P46531 | S2439 | SFLSGEPsQADVQPL | |
| PLK1 | P53350 | human | RIOK2 | Q9BVS4 | S380 | PEQIkEdsLsEEsAD | |
| PLK1 | P53350 | human | NPM1 | P06748 | S4 | ____MEDsMDMDMsP | |
| PLK1 | P53350 | human | MDC1 | Q14676 | T847 | PERQTDVtGEEELTK | |
| PLK1 | P53350 | human | AURKB | Q96GD4 | T236 | RRktMCGtLDyLPPE | Pkinase |
| PLK1 | P53350 | human | STK3 | Q13188 | T336 | TSVEsVGtMRATSTM | |
| PLK1 | P53350 | human | FADD | Q13158 | S194 | QNRsGAMsPMsWNsD | |
| PLK1 | P53350 | human | CDC25B | P30305 | S513 | RAVNDYPsLYYPEMY | Rhodanese |
| PLK1 | P53350 | human | USP16 | Q9Y5T5 | S486 | SEYEAEMsLQGEVNI | UCH |
| PLK1 | P53350 | human | KIF2B | Q8N4N8 | S201 | YRRHLDssKIsVLEP | |
| PLK1 | P53350 | human | BRF1 | Q92994 | T270 | RLTEFEDtPTSQLTI | TFIIB |
| PLK1 | P53350 | human | RACGAP1 | Q9H0H5 | S149 | RLstIDEsGsILsDI | |
| PLK1 | P53350 | human | CDC25C | P30307 | S198 | sDELMEFsLKDQEAK | M-inducer_phosp |
| PLK1 | P53350 | human | GORASP1 | Q9BQQ3 | S189 | AAWGGEGsLGCGIGY | GRASP55_65 |
| PLK1 | P53350 | human | BRCA2 | P51587 | T207 | TPPtLsstVLIVRNE | |
| PLK1 | P53350 | human | PLK1 | P53350 | T210 | YDGERKktLCGtPNy | Pkinase |
| PLK1 | P53350 | human | ESPL1 | Q14674 | T1363 | AGPHVPFtVFEEVCP | |
| PLK1 | P53350 | human | ATXN10 | Q9UBB4 | T82 | ASsLQLItECFRCLR | |
| PLK1 | P53350 | human | RACGAP1 | Q9H0H5 | S170 | EsLDWDssLVkTFKL | |
| PLK1 | P53350 | human | CDC25B | P30305 | S291 | AVPPGMEsLISAPLV | M-inducer_phosp |
| PLK1 | P53350 | human | KAT7 | O95251 | S57 | sQssQDssPVRNLQs | |
| PLK1 | P53350 | human | GTSE1 | Q9NYZ3 | S242 | LLLPRAAsVRGRsIP | |
| PLK1 | P53350 | human | REST | Q13127 | S1030 | MsEGsDDsGLHGARP | |
| PLK1 | P53350 | human | CLIP1 | P30622 | S1364 | DDLNNYDsDDQEKQS | |
| PLK1 | P53350 | human | MTHFR | P42898 | T549 | NVKGENItNAPELQP | |
| PLK1 | P53350 | human | STIL | Q15468 | S1108 | DRSTVGLsLIsPNNM | |
| PLK1 | P53350 | human | ANAPC4 | Q9UJX5 | S779 | kEEVLsEsEAENQQA | |
| PLK1 | P53350 | human | TSC1 | Q92574 | S468 | GFLGDLAsEEDsIEK | Hamartin |
| PLK1 | P53350 | human | BUB1B | O60566 | T1008 | LNANDEAtVSVLGEL | |
| PLK1 | P53350 | human | ANAPC1 | Q9H1A4 | T291 | kFsEQGGtPQNVATS | |
| PLK1 | P53350 | human | CCNB1 | P14635 | S133 | sPsPMETsGCAPAEE | |
| PLK1 | P53350 | human | TNKS | O95271 | T1128 | VEEEMQStIREHRDG | PARP |
| PLK1 | P53350 | human | CENPU | Q71F23 | T78 | FDPPLHstAIyADEE | |
| PLK1 | P53350 | human | PINX1 | Q96BK5 | S117 | sFSLEEKsKISKNRV | |
| PLK1 | P53350 | human | NINL | Q9Y2I6 | T161 | SDEEAEStKEAQNEL | |
| PLK1 | P53350 | human | NEDD1 | Q8NHV4 | T382 | PRsINTDtLSkEtDs | |
| PLK1 | P53350 | human | STK3 | Q13188 | S18 | LKkLsEDsLTkQPEE | |
| PLK1 | P53350 | human | NUDC | Q9Y266 | S274 | kkINPENskLsDLDs | |
| PLK1 | P53350 | human | IKBKB | O14920 | S750 | QTEEEEHsCLEQAS_ | |
| PLK1 | P53350 | human | ANAPC7 | Q9UJX3 | S30 | ssLLLtMsNNNPELF | |
| PLK1 | P53350 | human | CEP192 | Q8TEP8 | T44 | GLPVAVStLARDRSS | |
| PLK1 | P53350 | human | PKMYT1 | Q99640 | T495 | LLSLFEDtLDPT___ | |
| PLK1 | P53350 | human | FOXM1 | Q08050 | S730 | VLDTMNDsLSKILLD | |
| PLK1 | P53350 | human | PINX1 | Q96BK5 | T141 | DLSSRSKtDLDCIFG | |
| PLK1 | P53350 | human | WDR62 | O43379 | S897 | KPESLENsILDSLEP | |
| PLK1 | P53350 | human | TTK | P33981 | T564 | LEEADNQtLDSYRNE | Pkinase |
| PLK1 | P53350 | human | TOP2A | P11388 | S1337 | LDsDEDFsDFDEKtD | |
| PLK1 | P53350 | human | CDC25B | P30305 | T404 | skAFLLQtVDGkHQD | |
| PLK1 | P53350 | human | ZMYM2 | Q9UBW7 | S309 | DsLsPVAsLPkQIFQ | |
| PLK1 | P53350 | human | CTNNB1 | P35222 | S60 | EEEDVDTsQVLyEWE | |
| PLK1 | P53350 | human | MYC | P01106 | S77 | LLPtPPLsPsRRsGL | Myc_N |
| PLK1 | P53350 | human | ANAPC1 | Q9H1A4 | S355 | AALsrAHsPALGVHs | |
| PLK1 | P53350 | human | TTK | P33981 | S37 | EDLtDELsLNKISAD | |
| PLK1 | P53350 | human | PARP10 | Q53GL7 | T601 | EVRELLAtLEGLDLD | |
| PLK1 | P53350 | human | PRKDC | P78527 | S3205 | tPLPEDNsMNVDQDG | FAT |
| PLK1 | P53350 | human | PPP1R7 | Q15435 | S47 | VADLsEQsLkDGEEr | |
| PLK1 | P53350 | human | HSPA1B | P0DMV8 | S631 | AQGPkGGsGsGPtIE | |
| PLK1 | P53350 | human | CHEK2 | O96017 | T68 | SsLEtVstQELYsIP | |
| PLK1 | P53350 | human | FBXO5 | Q9UKT4 | S145 | TSRLyEDsGYSsFSL | |
| PLK1 | P53350 | human | KIF2B | Q8N4N8 | S200 | EYRRHLDssKIsVLE | |
| PLK1 | P53350 | human | CDC27 | P30260 | S434 | LEItkLDssIIsEGk | |
| PLK1 | P53350 | human | CHEK2 | O96017 | S164 | IAYIEDHsGNGTFVN | FHA |
| PLK1 | P53350 | human | CENPQ | Q7L2Z9 | T135 | PKkMEDLtNVssLLN | CENP-Q |
| PLK1 | P53350 | human | TTK | P33981 | S363 | LKNktEssLLAkLEE | |
| PLK1 | P53350 | human | SVIL | O95425 | S238 | sFSGRDssFtEVPRs | |
| PLK1 | P53350 | human | CDC25B | P30305 | S50 | PVRAAASsPVttLtQ | |
| PLK1 | P53350 | human | CCNB1 | P14635 | S128 | LVDTAsPsPMETsGC | |
| PLK1 | P53350 | human | BRCA2 | P51587 | T203 | SSLATPPtLsstVLI | |
| PLK1 | P53350 | human | BUB1B | O60566 | T792 | PRNSAELtVIKVsSQ | |
| PLK1 | P53350 | human | CDCA5 | Q96FF9 | T151 | RsysRLEtLGsASts | Sororin |
| PLK1 | P53350 | human | PPP6R2 | O75170 | S289 | GtEGLVDsFsQGLER | SAPS |
| PLK1 | P53350 | human | PRKAA2 | P54646 | T172 | sDGEFLRtsCGsPNy | Pkinase |
| PLK1 | P53350 | human | PKMYT1 | Q99640 | S426 | CSLLLDSsLSSNWDD | |
| PLK1 | P53350 | human | VIM | P08670 | S339 | ALkGtNEsLERQMRE | Filament |
| PLK1 | P53350 | human | ZMYM2 | Q9UBW7 | S305 | QPGVDsLsPVAsLPk | |
| PLK1 | P53350 | human | RAD21 | O60216 | S454 | EPsRLQEsVMEAsRT | |
| PLK1 | P53350 | human | RACGAP1 | Q9H0H5 | S214 | AVDQGNEsIVAKTTV | |
| PLK1 | P53350 | human | ATXN10 | Q9UBB4 | S77 | QVENLASsLQLItEC | |
| PLK1 | P53350 | human | CDC25B | P30305 | T167 | sPVLRNItNsQAPDG | M-inducer_phosp |
| PLK1 | P53350 | human | RAN | P62826 | S135 | DRkVKAksIVFHRkk | Ras |
| PLK1 | P53350 | human | SUGT1 | Q9Y2Z0 | S331 | VkRAMNksFMEsGGt | SGS |
| PLK1 | P53350 | human | KIF2C | Q99661 | S621 | ALIPGNLsKEEEELS | |
| PLK1 | P53350 | human | CDCA5 | Q96FF9 | T115 | VPAtPtStPVPNPEA | Sororin |
| PLK1 | P53350 | human | BUB1B | O60566 | T680 | IEDsREAtHSsGFSG | |
| PLK1 | P53350 | human | HSF1 | Q00613 | S216 | IPLMLNDsGsAHSMP | |
| PLK1 | P53350 | human | MAP2K2 | P36507 | S226 | LIDsMANsFVGtRSY | Pkinase |
| PLK1 | P53350 | human | PTEN | P60484 | T383 | HyRYsDttDsDPENE | |
| PLK1 | P53350 | human | STK3 | Q13188 | S385 | tMKRNAtsPQVQRPs | |
| PLK1 | P53350 | human | SRI | P30626 | T155 | YstNGKItFDDYIAC | |
| PLK1 | P53350 | human | NUAK1 | O60285 | S476 | KKTQQREsGYYsSPE | |
| PLK1 | P53350 | human | STIL | Q15468 | S1116 | LIsPNNMsFAtkKYM | |
| PLK1 | P53350 | human | CLASP2 | O75122 | S1034 | sIsPFNKsALKEAMF | |
| PLK1 | P53350 | human | CDC27 | P30260 | T446 | EGkIStItPQIQAFN | |
| PLK1 | P53350 | human | MRE11 | P49959 | S649 | EVIEVDEsDVEEDIF | |
| PLK1 | P53350 | human | CLIP1 | P30622 | S312 | AsLKRsPsAssLsSM | |
| PLK1 | P53350 | human | STK3 | Q13188 | S316 | LEEEEENsDEDELDs | |
| PLK1 | P53350 | human | BUB1B | O60566 | S676 | LsPIIEDsREAtHSs | |
| PLK1 | P53350 | human | ANAPC1 | Q9H1A4 | T701 | kkARPsEtGsDDDWE | Apc1_MidN |
| PLK1 | P53350 | human | ANAPC1 | Q9H1A4 | T530 | NtMPRPstPLDGVst | |
| PLK1 | P53350 | human | PTEN | P60484 | T382 | DHyRYsDttDsDPEN | |
| PLK1 | P53350 | human | BORA | Q6PGQ7 | T501 | QMDsGYNtQNCGSNI | |
| PLK1 | P53350 | human | CDC25B | P30305 | T58 | PVttLtQtMHDLAGL | |
| PLK1 | P53350 | human | TPT1 | P13693 | S46 | tEGNIDDsLIGGNAs | TCTP |
| PLK1 | P53350 | human | CDC25B | P30305 | S209 | PWkPTHPsSTHALAE | M-inducer_phosp |
| PLK1 | P53350 | human | FIRRM | Q9NSG2 | S43 | SQARGLSsQNLEIQT | |
| PLK1 | P53350 | human | NEDD1 | Q8NHV4 | S397 | GkNQDFssFDDtGKs | |
| PLK1 | P53350 | human | SMAD4 | Q13485 | T197 | ASTETyStPALLAPS | |
| PLK1 | P53350 | human | RAD51 | Q06609 | S14 | LEANADtsVEEESFG | |
| PLK1 | P53350 | human | MAD2L1BP | Q15013 | S102 | kHFyRkPsPQAEEML | p31comet |
| PLK1 | P53350 | human | CHEK2 | O96017 | S210 | DLtVDDQsVYPKALR | |
| PLK1 | P53350 | human | RSF1 | Q96T23 | S1359 | ENVGKVGsPLDysLV | |
| PLK1 | P53350 | human | CDC27 | P30260 | S426 | TQPNINDsLEItkLD | |
| PLK1 | P53350 | human | NEK9 | Q8TD19 | T210 | SEYsMAEtLVGtPYY | Pkinase |
| PLK1 | P53350 | human | IKBKB | O14920 | S733 | TVREQDQsFtALDWs | IKKbetaNEMObind |
| PLK1 | P53350 | human | NINL | Q9Y2I6 | S686 | LEELHEKsQEVIWGL | |
| PLK1 | P53350 | human | ORC2 | Q13416 | S188 | SEYSASNsEDDEGVA | |
| PLK1 | P53350 | human | VIM | P08670 | S56 | srsLyAssPGGVyAt | Filament_head |
| PLK1 | P53350 | human | BIRC5 | O15392 | T21 | LkDHRIstFkNWPFL | BIR |
| PLK1 | P53350 | human | PINX1 | Q96BK5 | T317 | EDATLEEtLVKKKKK |
 Biological Network Integration of This Kinase and Substrates Biological Network Integration of This Kinase and Substrates (GeneMANIA website) |
 Enriched GO biological processes of the phosphorylation target genes of the kinase Enriched GO biological processes of the phosphorylation target genes of the kinase |
| Kinase | GOID | GO term | P.adjust |
| PLK1 | ID | Description | 0.00e+00 |
| PLK1 | GO:0007059 | chromosome segregation | 4.81e-45 |
| PLK1 | GO:0098813 | nuclear chromosome segregation | 1.58e-44 |
| PLK1 | GO:0000819 | sister chromatid segregation | 1.09e-41 |
| PLK1 | GO:0000280 | nuclear division | 4.20e-39 |
| PLK1 | GO:0048285 | organelle fission | 1.83e-38 |
| PLK1 | GO:0140014 | mitotic nuclear division | 7.18e-37 |
| PLK1 | GO:0000070 | mitotic sister chromatid segregation | 7.74e-36 |
| PLK1 | GO:1901987 | regulation of cell cycle phase transition | 1.10e-34 |
| PLK1 | GO:0033044 | regulation of chromosome organization | 2.54e-34 |
| PLK1 | GO:0044772 | mitotic cell cycle phase transition | 3.33e-34 |
| PLK1 | GO:1901990 | regulation of mitotic cell cycle phase transition | 2.78e-31 |
| PLK1 | GO:0051983 | regulation of chromosome segregation | 3.49e-31 |
| PLK1 | GO:0010948 | negative regulation of cell cycle process | 2.52e-30 |
| PLK1 | GO:0045786 | negative regulation of cell cycle | 7.35e-28 |
| PLK1 | GO:0033045 | regulation of sister chromatid segregation | 2.04e-27 |
| PLK1 | GO:0007051 | spindle organization | 3.61e-27 |
| PLK1 | GO:1901988 | negative regulation of cell cycle phase transition | 3.82e-27 |
| PLK1 | GO:0007091 | metaphase/anaphase transition of mitotic cell cycle | 6.24e-27 |
| PLK1 | GO:0044784 | metaphase/anaphase transition of cell cycle | 1.32e-26 |
| PLK1 | GO:0007052 | mitotic spindle organization | 5.15e-25 |
| PLK1 | GO:1901991 | negative regulation of mitotic cell cycle phase transition | 8.98e-25 |
| PLK1 | GO:0045930 | negative regulation of mitotic cell cycle | 1.17e-24 |
| PLK1 | GO:1905818 | regulation of chromosome separation | 2.67e-24 |
| PLK1 | GO:0051306 | mitotic sister chromatid separation | 3.15e-24 |
| PLK1 | GO:0030071 | regulation of mitotic metaphase/anaphase transition | 4.07e-24 |
| PLK1 | GO:1902850 | microtubule cytoskeleton organization involved in mitosis | 4.11e-24 |
| PLK1 | GO:1902099 | regulation of metaphase/anaphase transition of cell cycle | 8.05e-24 |
| PLK1 | GO:0051304 | chromosome separation | 1.68e-23 |
| PLK1 | GO:0007088 | regulation of mitotic nuclear division | 4.60e-23 |
| PLK1 | GO:0010965 | regulation of mitotic sister chromatid separation | 6.04e-23 |
| PLK1 | GO:0000075 | cell cycle checkpoint signaling | 5.76e-21 |
| PLK1 | GO:0051783 | regulation of nuclear division | 5.83e-21 |
| PLK1 | GO:2001251 | negative regulation of chromosome organization | 1.59e-20 |
| PLK1 | GO:0090068 | positive regulation of cell cycle process | 2.97e-20 |
| PLK1 | GO:0010639 | negative regulation of organelle organization | 4.18e-20 |
| PLK1 | GO:0045787 | positive regulation of cell cycle | 6.16e-20 |
| PLK1 | GO:0007093 | mitotic cell cycle checkpoint signaling | 7.73e-20 |
| PLK1 | GO:0051321 | meiotic cell cycle | 2.76e-19 |
| PLK1 | GO:0051303 | establishment of chromosome localization | 7.43e-18 |
| PLK1 | GO:0050000 | chromosome localization | 2.79e-17 |
| PLK1 | GO:0033047 | regulation of mitotic sister chromatid segregation | 1.25e-16 |
| PLK1 | GO:0045931 | positive regulation of mitotic cell cycle | 1.66e-16 |
| PLK1 | GO:0051656 | establishment of organelle localization | 4.48e-16 |
| PLK1 | GO:0051310 | metaphase chromosome alignment | 7.97e-16 |
| PLK1 | GO:0033046 | negative regulation of sister chromatid segregation | 9.84e-16 |
| PLK1 | GO:0033048 | negative regulation of mitotic sister chromatid segregation | 9.84e-16 |
| PLK1 | GO:0045841 | negative regulation of mitotic metaphase/anaphase transition | 9.84e-16 |
| PLK1 | GO:2000816 | negative regulation of mitotic sister chromatid separation | 9.84e-16 |
| PLK1 | GO:1901989 | positive regulation of cell cycle phase transition | 9.92e-16 |
Top |
Related Drugs to PLK1_PLK1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
 Distribution of the number of studies mentioning PLK1-PLK1 and kinase inhibitors the PubMed Abstract (04-01-2024) Distribution of the number of studies mentioning PLK1-PLK1 and kinase inhibitors the PubMed Abstract (04-01-2024) |
| Fusion gene - drug pair 1 | Fusion gene - drug pair 2 | PMID | Publication date | DOI | Study title |
Top |
Related Diseases to PLK1_PLK1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Related diseases from the literature mentioned this fusion gene and drug. Related diseases from the literature mentioned this fusion gene and drug. (PubMed, 04-01-2024) |
| MeSH ID | MeSH term |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
Top |
Clinical Trials of the Found Drugs/Small Molecules |
 Statistics of the Clinical Trials of the Found Kinase Inibitors from clinicaltrials.gov (06-17-2024) Statistics of the Clinical Trials of the Found Kinase Inibitors from clinicaltrials.gov (06-17-2024) |
 Clinical Trials from clinicaltrials.gov (06-17-2024) Clinical Trials from clinicaltrials.gov (06-17-2024) |
| Fusion Gene | Kinase Inhibitor | NCT ID | Study Status | Phases | Disease | # Enrolment | Date |

