| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Kinase Fusion Gene:PRKCE_SNX17 |
Kinase Fusion Protein Summary |
 Kinase Fusion gene summary Kinase Fusion gene summary |
| Kinase Fusion partner gene information | Kinase Fusion gene name: PRKCE_SNX17 | KinaseFusionDB ID: KFG4951 | FusionGDB2.0 ID: KFG4951 | Hgene | Tgene | Gene symbol | PRKCE | SNX17 | Gene ID | 5581 | 9784 | |
| Gene name | protein kinase C epsilon | sorting nexin 17 | ||||||||||
| Synonyms | PKCE|nPKC-epsilon | - | ||||||||||
| Cytomap | 2p21 | 2p23.3 | ||||||||||
| Type of gene | protein-coding | protein-coding | ||||||||||
| Description | protein kinase C epsilon type | sorting nexin-17 | ||||||||||
| Modification date | 20240403 | 20240411 | ||||||||||
| UniProtAcc | Q02156 | Q15036 | ||||||||||
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000306156, ENST00000467135, ENST00000394874, | ENST00000233575, ENST00000543024, ENST00000537606, ENST00000542478, | |||||||||
| Context (manual curation of fusion genes in KinaseFusionDB) | PubMed: PRKCE [Title/Abstract] AND SNX17 [Title/Abstract] AND fusion [Title/Abstract] | |||||||||||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | ||||||||||||
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | PRKCE | GO:0006468 | protein phosphorylation | 18556656|34593629 |
| Hgene | PRKCE | GO:0018105 | peptidyl-serine phosphorylation | 15695813 |
 Kinase Fusion gene breakpoints across PRKCE (5'-gene) Kinase Fusion gene breakpoints across PRKCE (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 Kinase Fusion gene breakpoints across SNX17 (3'-gene) Kinase Fusion gene breakpoints across SNX17 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
Top |
Kinase Fusion Gene Sample Information |
 Kinase Fusion gene information. Kinase Fusion gene information. |
 Kinase Fusion gene information from four resources (ChiTars 5.0, ChimerDB 4.0, COSMIC, and CCLE) Kinase Fusion gene information from four resources (ChiTars 5.0, ChimerDB 4.0, COSMIC, and CCLE)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Sample | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp |
| CCLE | Capan-1 | PRKCE | chr2 | 45879587 | SNX17 | chr2 | 27594136 |
| CCLE | Capan-1 | PRKCE | chr2 | 45879587 | SNX17 | chr2 | 27595490 |
| CCLE | Capan-1 | PRKCE | chr2 | 45928663 | SNX17 | chr2 | 27598373 |
Top |
Kinase Fusion ORF Analysis |
 Kinase Fusion information from ORFfinder translation from full-length transcript sequence from KinaseFusionDB. Kinase Fusion information from ORFfinder translation from full-length transcript sequence from KinaseFusionDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp | Seq length (transcript) | Seq length (amino acids) |
| ENST00000306156 | ENST00000233575 | PRKCE | chr2 | 45879587 | SNX17 | chr2 | 27594136 | 2434 | 565 |
| ENST00000306156 | ENST00000233575 | PRKCE | chr2 | 45879587 | SNX17 | chr2 | 27595490 | 2359 | 540 |
Top |
Kinase Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from KinaseFusionDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from KinaseFusionDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >Henst_Tenst_Hgene_Hchr_Hbp_Tgene_Tchr_Tbp_length(fusion AA)_AAseq >ENST00000306156_ENST00000233575_PRKCE_chr2_45879587_SNX17_chr2_27594136_length(amino acids)=565 MVVFNGLLKIKICEAVSLKPTAWSLRHAVGPRPQTFLLDPYIALNVDDSRIGQTATKQKTNSPAWHDEFVTDVCNGRKIELAVFHDAPIG YDDFVANCTIQFEELLQNGSRHFEDWAYNIHVNGVLHCRVRYSQLLGLHEQLRKEYGANVLPAFPPKKLFSLTPAEVEQRREQLEKYMQA VRQDPLLGSSETFNSFLRRAQQETQQVPTEEVSLEVLLSNGQKVLVNVLTSDQTEDVLEAVAAKLDLPDDLIGYFSLFLVREKEDGAFSF VRKLQEFELPYVSVTSLRSQEYKIVLRKSYWDSAYDDDVMENRVGLNLLYAQTVSDIERGWILVTKEQHRQLKSLQEKVSKKEFLRLAQT LRHYGYLRFDACVADFPEKDCPVVVSAGNSELSLQLRLPGQQLREGSFRVTRMRCWRVTSSVPLPSGSTSSPGRGRGEVRLELAFEYLMS KDRLQWVTITSPQAIMMSICLQSMVDELMVKKSGGSIRKMLRRRVGGTLRRSDSQQAVKSPPLLESPDATRESMVKLSSKLSAVSLRGIG -------------------------------------------------------------- >ENST00000306156_ENST00000233575_PRKCE_chr2_45879587_SNX17_chr2_27595490_length(amino acids)=540 MVVFNGLLKIKICEAVSLKPTAWSLRHAVGPRPQTFLLDPYIALNVDDSRIGQTATKQKTNSPAWHDEFVTDVCNGRKIELAVFHDAPIG YDDFVANCTIQFEELLQNGSRHFEDWLRKEYGANVLPAFPPKKLFSLTPAEVEQRREQLEKYMQAVRQDPLLGSSETFNSFLRRAQQETQ QVPTEEVSLEVLLSNGQKVLVNVLTSDQTEDVLEAVAAKLDLPDDLIGYFSLFLVREKEDGAFSFVRKLQEFELPYVSVTSLRSQEYKIV LRKSYWDSAYDDDVMENRVGLNLLYAQTVSDIERGWILVTKEQHRQLKSLQEKVSKKEFLRLAQTLRHYGYLRFDACVADFPEKDCPVVV SAGNSELSLQLRLPGQQLREGSFRVTRMRCWRVTSSVPLPSGSTSSPGRGRGEVRLELAFEYLMSKDRLQWVTITSPQAIMMSICLQSMV DELMVKKSGGSIRKMLRRRVGGTLRRSDSQQAVKSPPLLESPDATRESMVKLSSKLSAVSLRGIGSPSTDASASDVHGNFAFEGIGDEDL -------------------------------------------------------------- |
Multiple Sequence Alignment of All Fusion Protein Isoforms |
Top |
Kinase Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr2:/chr2:) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| PRKCE | SNX17 |
| FUNCTION: Calcium-independent, phospholipid- and diacylglycerol (DAG)-dependent serine/threonine-protein kinase that plays essential roles in the regulation of multiple cellular processes linked to cytoskeletal proteins, such as cell adhesion, motility, migration and cell cycle, functions in neuron growth and ion channel regulation, and is involved in immune response, cancer cell invasion and regulation of apoptosis. Mediates cell adhesion to the extracellular matrix via integrin-dependent signaling, by mediating angiotensin-2-induced activation of integrin beta-1 (ITGB1) in cardiac fibroblasts. Phosphorylates MARCKS, which phosphorylates and activates PTK2/FAK, leading to the spread of cardiomyocytes. Involved in the control of the directional transport of ITGB1 in mesenchymal cells by phosphorylating vimentin (VIM), an intermediate filament (IF) protein. In epithelial cells, associates with and phosphorylates keratin-8 (KRT8), which induces targeting of desmoplakin at desmosomes and regulates cell-cell contact. Phosphorylates IQGAP1, which binds to CDC42, mediating epithelial cell-cell detachment prior to migration. In HeLa cells, contributes to hepatocyte growth factor (HGF)-induced cell migration, and in human corneal epithelial cells, plays a critical role in wound healing after activation by HGF. During cytokinesis, forms a complex with YWHAB, which is crucial for daughter cell separation, and facilitates abscission by a mechanism which may implicate the regulation of RHOA. In cardiac myocytes, regulates myofilament function and excitation coupling at the Z-lines, where it is indirectly associated with F-actin via interaction with COPB1. During endothelin-induced cardiomyocyte hypertrophy, mediates activation of PTK2/FAK, which is critical for cardiomyocyte survival and regulation of sarcomere length. Plays a role in the pathogenesis of dilated cardiomyopathy via persistent phosphorylation of troponin I (TNNI3). Involved in nerve growth factor (NFG)-induced neurite outgrowth and neuron morphological change independently of its kinase activity, by inhibition of RHOA pathway, activation of CDC42 and cytoskeletal rearrangement. May be involved in presynaptic facilitation by mediating phorbol ester-induced synaptic potentiation. Phosphorylates gamma-aminobutyric acid receptor subunit gamma-2 (GABRG2), which reduces the response of GABA receptors to ethanol and benzodiazepines and may mediate acute tolerance to the intoxicating effects of ethanol. Upon PMA treatment, phosphorylates the capsaicin- and heat-activated cation channel TRPV1, which is required for bradykinin-induced sensitization of the heat response in nociceptive neurons. Is able to form a complex with PDLIM5 and N-type calcium channel, and may enhance channel activities and potentiates fast synaptic transmission by phosphorylating the pore-forming alpha subunit CACNA1B (CaV2.2). In prostate cancer cells, interacts with and phosphorylates STAT3, which increases DNA-binding and transcriptional activity of STAT3 and seems to be essential for prostate cancer cell invasion. Downstream of TLR4, plays an important role in the lipopolysaccharide (LPS)-induced immune response by phosphorylating and activating TICAM2/TRAM, which in turn activates the transcription factor IRF3 and subsequent cytokines production. In differentiating erythroid progenitors, is regulated by EPO and controls the protection against the TNFSF10/TRAIL-mediated apoptosis, via BCL2. May be involved in the regulation of the insulin-induced phosphorylation and activation of AKT1. Phosphorylates NLRP5/MATER and may thereby modulate AKT pathway activation in cumulus cells (PubMed:19542546). {ECO:0000269|PubMed:11884385, ECO:0000269|PubMed:1374067, ECO:0000269|PubMed:15355962, ECO:0000269|PubMed:16757566, ECO:0000269|PubMed:17603037, ECO:0000269|PubMed:17875639, ECO:0000269|PubMed:17875724, ECO:0000269|PubMed:19542546}. | FUNCTION: Critical regulator of endosomal recycling of numerous surface proteins, including integrins, signaling receptor and channels (PubMed:15121882, PubMed:15769472). Binds to NPxY sequences in the cytoplasmic tails of target cargos (PubMed:21512128). Associates with retriever and CCC complexes to prevent lysosomal degradation and promote cell surface recycling of numerous cargos such as integrins ITGB1, ITGB5 and their associated alpha subunits (PubMed:28892079, PubMed:22492727). Also required for maintenance of normal cell surface levels of APP and LRP1 (PubMed:16712798, PubMed:19005208). Interacts with membranes containing phosphatidylinositol 3-phosphate (PtdIns(3P)) (PubMed:16712798). {ECO:0000269|PubMed:15121882, ECO:0000269|PubMed:15769472, ECO:0000269|PubMed:16712798, ECO:0000269|PubMed:19005208, ECO:0000269|PubMed:21512128, ECO:0000269|PubMed:22492727, ECO:0000269|PubMed:28892079}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
 - Retained domain in the 5'-partner of fusion protein (protein functional feature from UniProt). - Retained domain in the 5'-partner of fusion protein (protein functional feature from UniProt). |
| Partner | Hgeneene | Hbp | Tgeneene | Tbp | ENST | BPexon | TotalExon | Protein feature loci | BPloci | TotalLen | Feature | Note |
| Hgene | PRKCE | 45879587 | SNX17 | 27594136 | ENST00000306156 | 1 | 15 | 1_117 | 1161 | 738 | Domain | Note=C2;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00041 |
| Hgene | PRKCE | 45879587 | SNX17 | 27595490 | ENST00000306156 | 1 | 15 | 1_117 | 1161 | 738 | Domain | Note=C2;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00041 |
 - Retained domain in the 3'-partner of fusion protein (protein functional feature from UniProt). - Retained domain in the 3'-partner of fusion protein (protein functional feature from UniProt). |
| Partner | Hgeneene | Hbp | Tgeneene | Tbp | ENST | BPexon | TotalExon | Protein feature loci | BPloci | TotalLen | Feature | Note |
Top |
Kinase Fusion Protein Structures |
 CIF files of the predicted kinase fusion proteins CIF files of the predicted kinase fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Kinase Fusion protein CIF link (fusion AA seq ID in KinaseFusionDB) | Henst | Tenst | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp | AA seq | Len(AA seq) |
| PDB file >>>201_PRKCE_SNX17 | ENST00000306156 | ENST00000233575 | PRKCE | chr2 | 45879587 | SNX17 | chr2 | 27595490 | MVVFNGLLKIKICEAVSLKPTAWSLRHAVGPRPQTFLLDPYIALNVDDSRIGQTATKQKTNSPAWHDEFVTDVCNGRKIELAVFHDAPIG YDDFVANCTIQFEELLQNGSRHFEDWLRKEYGANVLPAFPPKKLFSLTPAEVEQRREQLEKYMQAVRQDPLLGSSETFNSFLRRAQQETQ QVPTEEVSLEVLLSNGQKVLVNVLTSDQTEDVLEAVAAKLDLPDDLIGYFSLFLVREKEDGAFSFVRKLQEFELPYVSVTSLRSQEYKIV LRKSYWDSAYDDDVMENRVGLNLLYAQTVSDIERGWILVTKEQHRQLKSLQEKVSKKEFLRLAQTLRHYGYLRFDACVADFPEKDCPVVV SAGNSELSLQLRLPGQQLREGSFRVTRMRCWRVTSSVPLPSGSTSSPGRGRGEVRLELAFEYLMSKDRLQWVTITSPQAIMMSICLQSMV DELMVKKSGGSIRKMLRRRVGGTLRRSDSQQAVKSPPLLESPDATRESMVKLSSKLSAVSLRGIGSPSTDASASDVHGNFAFEGIGDEDL | 540 |
| 3D view using mol* of 201_PRKCE_SNX17 | ||||||||||
| PDB file >>>HKFP_289_PRKCE_SNX17 | ENST00000306156 | ENST00000233575 | PRKCE | chr2 | 45879587 | SNX17 | chr2 | 27594136 | MVVFNGLLKIKICEAVSLKPTAWSLRHAVGPRPQTFLLDPYIALNVDDSRIGQTATKQKTNSPAWHDEFVTDVCNGRKIELAVFHDAPIG YDDFVANCTIQFEELLQNGSRHFEDWAYNIHVNGVLHCRVRYSQLLGLHEQLRKEYGANVLPAFPPKKLFSLTPAEVEQRREQLEKYMQA VRQDPLLGSSETFNSFLRRAQQETQQVPTEEVSLEVLLSNGQKVLVNVLTSDQTEDVLEAVAAKLDLPDDLIGYFSLFLVREKEDGAFSF VRKLQEFELPYVSVTSLRSQEYKIVLRKSYWDSAYDDDVMENRVGLNLLYAQTVSDIERGWILVTKEQHRQLKSLQEKVSKKEFLRLAQT LRHYGYLRFDACVADFPEKDCPVVVSAGNSELSLQLRLPGQQLREGSFRVTRMRCWRVTSSVPLPSGSTSSPGRGRGEVRLELAFEYLMS KDRLQWVTITSPQAIMMSICLQSMVDELMVKKSGGSIRKMLRRRVGGTLRRSDSQQAVKSPPLLESPDATRESMVKLSSKLSAVSLRGIG | 565_PRKCE_SNX17 |
| PDB file >>>HKFP_290_PRKCE_SNX17 | ENST00000306156 | ENST00000233575 | PRKCE | chr2 | 45879587 | SNX17 | chr2 | 27595490 | MVVFNGLLKIKICEAVSLKPTAWSLRHAVGPRPQTFLLDPYIALNVDDSRIGQTATKQKTNSPAWHDEFVTDVCNGRKIELAVFHDAPIG YDDFVANCTIQFEELLQNGSRHFEDWLRKEYGANVLPAFPPKKLFSLTPAEVEQRREQLEKYMQAVRQDPLLGSSETFNSFLRRAQQETQ QVPTEEVSLEVLLSNGQKVLVNVLTSDQTEDVLEAVAAKLDLPDDLIGYFSLFLVREKEDGAFSFVRKLQEFELPYVSVTSLRSQEYKIV LRKSYWDSAYDDDVMENRVGLNLLYAQTVSDIERGWILVTKEQHRQLKSLQEKVSKKEFLRLAQTLRHYGYLRFDACVADFPEKDCPVVV SAGNSELSLQLRLPGQQLREGSFRVTRMRCWRVTSSVPLPSGSTSSPGRGRGEVRLELAFEYLMSKDRLQWVTITSPQAIMMSICLQSMV DELMVKKSGGSIRKMLRRRVGGTLRRSDSQQAVKSPPLLESPDATRESMVKLSSKLSAVSLRGIGSPSTDASASDVHGNFAFEGIGDEDL | 540_PRKCE_SNX17 |
| 3D view using mol* of HKFP_290_PRKCE_SNX17 | ||||||||||
Top |
Comparison of Fusion Protein Isoforms |
 Superimpose the 3D Structures Among All Fusion Protein Isoforms Superimpose the 3D Structures Among All Fusion Protein Isoforms * Download the pdb file and open it from the molstar online viewer. |
| 3D view using mol* of viewer/superimpose_isoforms/HKFP_289_PRKCE_SNX17_vs_HKFP_290_PRKCE_SNX17_superimposed.pdb.html |
 Comparison of the Secondary Structures of Fusion Protein Isoforms Comparison of the Secondary Structures of Fusion Protein Isoforms |
| ./secondary_str/HKFP_289_PRKCE_SNX17_vs_HKFP_290_PRKCE_SNX17.png |
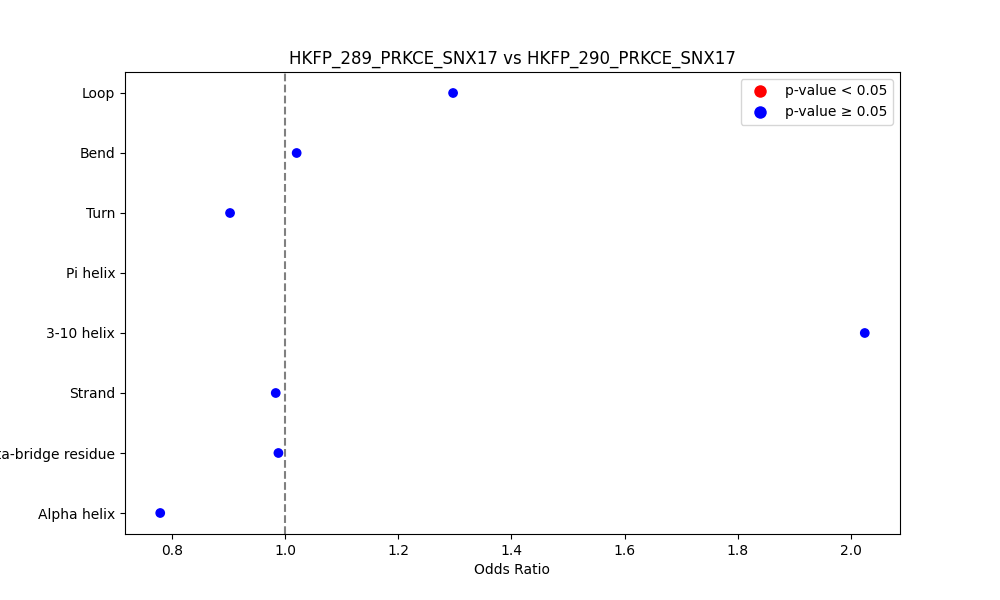 |
Top |
Comparison of Fusion Protein Sequences/Structures with Known Sequences/Structures from PDB |
Top |
pLDDT score distribution |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. * The blue color at the bottom marks the best active site residues. |
| 201_PRKCE_SNX17.png |
 |
| 201_PRKCE_SNX17.png |
 |
| HKFP_290_PRKCE_SNX17.png |
 |
Top |
Potential Active Site Information |
 The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. |
| Kinase Fusion AA seq ID in KinaseFusionDB | Site score | Size | Dscore | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
| HKFP_290_PRKCE_SNX17 | 1.054 | 365 | 1.1 | 1701.966 | 0.654 | 0.718 | 0.839 | 0.928 | 0.83 | 1.118 | 0.791 | Chain A: 2,3,4,5,8,9,10,79,102,105,106,107,108,109 ,110,112,113,116,129,132,145,146,149,150,152,153,1 66,167,170,171,172,173,174,175,177,178,180,181,182 ,183,184,206,207,208,211,234,236,244,246,247,254,2 58,261,262,263,345,395,413,414,415,416,435,437,438 ,440,477,479,480,481,483,485,486,488,496,497,498,5 00,501,502,503,504,505 |
Top |
Ramachandran Plot of Kinase Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| 201_PRKCE_SNX17_ramachandran.png |
 |
| HKFP_290_PRKCE_SNX17_ramachandran.png |
 |
Top |
Virtual Screening Results |
 Distribution of the average docking score across all approved kinase inhibitors. Distribution of the average docking score across all approved kinase inhibitors.Distribution of the number of occurrence across all approved kinase inhibitors. |
| 5'-kinase fusion protein case |
 |
| 3'-kinase fusion protein case |
Top |
 Drug information from DrugBank of the top 20 interacting small molecules. Drug information from DrugBank of the top 20 interacting small molecules.* The detailed information of individual kinase inhibitors are available in the download page. |
| Fusion gene name info | Drug | Docking score | Glide g score | Glide energy |
| HKFP_290_PRKCE_SNX17_vsw_10-DOCK_HTVS_1-001 | Encorafenib | -6.45091 | -6.884710000000001 | -48.6591 |
| HKFP_290_PRKCE_SNX17_vsw_10-DOCK_HTVS_1-001 | Encorafenib | -6.45091 | -6.884710000000001 | -48.6591 |
| HKFP_290_PRKCE_SNX17_vsw_10-DOCK_HTVS_1-001 | Lenvatinib | -6.40695 | -6.40695 | -52.788 |
| HKFP_290_PRKCE_SNX17_vsw_10-DOCK_HTVS_1-001 | Neratinib | -5.91187 | -6.09777 | -55.7999 |
| HKFP_290_PRKCE_SNX17_vsw_10-DOCK_HTVS_1-001 | Afatinib | -5.572030000000002 | -5.75433 | -49.9137 |
| HKFP_290_PRKCE_SNX17_vsw_10-DOCK_HTVS_1-001 | Afatinib | -5.572030000000002 | -5.75433 | -49.9137 |
| HKFP_290_PRKCE_SNX17_vsw_10-DOCK_HTVS_1-001 | Afatinib | -5.57063 | -5.75433 | -49.9137 |
| HKFP_290_PRKCE_SNX17_vsw_10-DOCK_HTVS_1-001 | Cobimetinib | -5.55808 | -5.56088 | -41.1163 |
| HKFP_290_PRKCE_SNX17_vsw_10-DOCK_HTVS_1-001 | Pacritinib | -5.50583 | -5.51093 | -35.4235 |
| HKFP_290_PRKCE_SNX17_vsw_10-DOCK_HTVS_1-001 | Larotrectinib | -5.41413 | -5.41413 | -36.5394 |
| HKFP_290_PRKCE_SNX17_vsw_10-DOCK_HTVS_1-001 | Encorafenib | -5.40902 | -5.84282 | -48.0211 |
| HKFP_290_PRKCE_SNX17_vsw_10-DOCK_HTVS_1-001 | Encorafenib | -5.40902 | -5.84282 | -48.0211 |
| HKFP_290_PRKCE_SNX17_vsw_10-DOCK_HTVS_1-001 | Zanubrutinib | -5.38285 | -5.38285 | -42.0712 |
| HKFP_290_PRKCE_SNX17_vsw_10-DOCK_HTVS_1-001 | Crizotinib | -5.36837 | -5.70457 | -42.6725 |
| HKFP_290_PRKCE_SNX17_vsw_10-DOCK_HTVS_1-001 | Crizotinib | -5.36837 | -5.70457 | -42.6725 |
| HKFP_290_PRKCE_SNX17_vsw_10-DOCK_HTVS_1-001 | Upadacitinib | -5.32334 | -5.32434 | -31.3896 |
| HKFP_290_PRKCE_SNX17_vsw_10-DOCK_HTVS_1-001 | Netarsudil | -5.3121 | -5.3232 | -46.1072 |
| HKFP_290_PRKCE_SNX17_vsw_10-DOCK_HTVS_1-001 | Netarsudil | -5.3121 | -5.3232 | -46.1072 |
| HKFP_290_PRKCE_SNX17_vsw_10-DOCK_HTVS_1-001 | Capmatinib | -5.2950800000000005 | -5.300680000000002 | -39.5677 |
| HKFP_290_PRKCE_SNX17_vsw_10-DOCK_HTVS_1-001 | Larotrectinib | -5.19557 | -5.19557 | -46.7222 |
| 201_PRKCE_SNX17-DOCK_HTVS_1-001 | Lapatinib | -7.58767 | -7.67647 | -63.5649 |
| 201_PRKCE_SNX17-DOCK_HTVS_1-001 | Temsirolimus | -7.10527 | -7.106669999999999 | -61.0307 |
| 201_PRKCE_SNX17-DOCK_HTVS_1-001 | Temsirolimus | -6.98728 | -6.9886800000000004 | -63.3752 |
| 201_PRKCE_SNX17-DOCK_HTVS_1-001 | Larotrectinib | -6.624860000000001 | -6.624860000000001 | -40.3367 |
| 201_PRKCE_SNX17-DOCK_HTVS_1-001 | Selpercatinib | -6.2916300000000005 | -6.3221300000000005 | -49.9729 |
| 201_PRKCE_SNX17-DOCK_HTVS_1-001 | Selpercatinib | -6.2916300000000005 | -6.3221300000000005 | -49.9729 |
| 201_PRKCE_SNX17-DOCK_HTVS_1-001 | Gilteritinib | -6.27445 | -6.30085 | -50.2262 |
| 201_PRKCE_SNX17-DOCK_HTVS_1-001 | Gilteritinib | -6.27445 | -6.30085 | -50.2262 |
| 201_PRKCE_SNX17-DOCK_HTVS_1-001 | Pralsetinib | -6.15561 | -6.24711 | -45.7595 |
| 201_PRKCE_SNX17-DOCK_HTVS_1-001 | Pralsetinib | -6.14079 | -6.23229 | -45.8777 |
| 201_PRKCE_SNX17-DOCK_HTVS_1-001 | Selpercatinib | -6.10848 | -6.13898 | -52.1676 |
| 201_PRKCE_SNX17-DOCK_HTVS_1-001 | Selpercatinib | -6.10848 | -6.13898 | -52.1676 |
| 201_PRKCE_SNX17-DOCK_HTVS_1-001 | Neratinib | -6.08814 | -6.27044 | -49.5695 |
| 201_PRKCE_SNX17-DOCK_HTVS_1-001 | Neratinib | -6.08814 | -6.27044 | -49.5695 |
| 201_PRKCE_SNX17-DOCK_HTVS_1-001 | Avapritinib | -6.082269999999999 | -6.87977 | -54.8045 |
| 201_PRKCE_SNX17-DOCK_HTVS_1-001 | Avapritinib | -6.082269999999999 | -6.87977 | -54.8045 |
| 201_PRKCE_SNX17-DOCK_HTVS_1-001 | Avapritinib | -6.082269999999999 | -6.87977 | -54.8045 |
| 201_PRKCE_SNX17-DOCK_HTVS_1-001 | Avapritinib | -6.082269999999999 | -6.87977 | -54.8045 |
| 201_PRKCE_SNX17-DOCK_HTVS_1-001 | Neratinib | -6.03656 | -6.22246 | -59.4159 |
| 201_PRKCE_SNX17-DOCK_HTVS_1-001 | Gilteritinib | -6.028569999999999 | -6.05497 | -43.3041 |
Top |
Kinase-Substrate Information of PRKCE_SNX17 |
 Phosphorylation target of the kinase Phosphorylation target of the kinase(phosphosite, 03-17-2024) |
| Kinase | Kinase UniProt Acc | Kinase species | Substrate | Substrate UniProt Acc | Substrate phosphorylated residues | Substrate phosphorylated sites (+/-7AA) | Domain |
| PRKCE | Q02156 | human | UGT1A7 | Q9HAW7 | S432 | KAVINDKsYkENIMR | UDPGT |
| PRKCE | Q02156 | human | MAPT | P10636-8 | S352 | DFKDrVQskIGsLDN | Tubulin-binding |
| PRKCE | Q02156 | human | ADAP1 | O75689 | T276 | GFRkRWFtMDDRRLM | PH |
| PRKCE | Q02156 | human | KEAP1 | Q14145 | S602 | TRMTsGRsGVGVAVT | |
| PRKCE | Q02156 | human | ADAM17 | P78536 | T735 | kPFPAPQtPGRLQPA | |
| PRKCE | Q02156 | human | PKM | P14618 | Y105 | FAsDPILyRPVAVAL | PK |
| PRKCE | Q02156 | human | GNAI2 | P04899 | S144 | VQACFGRsREyQLND | G-alpha |
| PRKCE | Q02156 | human | NANOG | Q9H9S0 | T200 | LPMWSNQtWNNSTWS | |
| PRKCE | Q02156 | human | NANOG | Q9H9S0 | S135 | LSNILNLsYKQVKTW | Homeodomain |
| PRKCE | Q02156 | human | PRKCE | Q02156 | S316 | tPDKITNsGQRRkKL | |
| PRKCE | Q02156 | human | NANOG | Q9H9S0 | S79 | PKGKQPtsAEKSVAK | |
| PRKCE | Q02156 | human | GNAI2 | P04899 | S302 | NkYDEAAsyIQSkFE | G-alpha |
| PRKCE | Q02156 | human | PPP1R14A | Q96A00 | T38 | QkRHARVtVkYDRRE | PP1_inhibitor |
| PRKCE | Q02156 | human | MCU | Q8NE86 | S92 | VISVRLPsRRERCQF | |
| PRKCE | Q02156 | human | EGFR | P00533 | T678 | RHIVRKRtLRRLLQE | |
| PRKCE | Q02156 | human | SLC6A2 | P23975 | S259 | LWKGVKtsGKVVWIT | SNF |
| PRKCE | Q02156 | human | MIIP | Q5JXC2 | S303 | YHIHRRKsFDASDTL | MIIP |
| PRKCE | Q02156 | human | KEAP1 | Q14145 | S599 | SEVTRMTsGRsGVGV | |
| PRKCE | Q02156 | human | ADAP1 | O75689 | S87 | AARARFEsKVPSFYY | ArfGap |
| PRKCE | Q02156 | human | MAPT | P10636-8 | S324 | kVTskCGsLGNIHHk | Tubulin-binding |
| PRKCE | Q02156 | human | AURKB | Q96GD4 | S227 | GWSVHAPsLRRktMC | Pkinase |
| PRKCE | Q02156 | human | PRKCE | Q02156 | S234 | EtPDQVGsQRFSVNM | |
| PRKCE | Q02156 | human | CHAT | P28329-3 | S440 | VPTYESAsIRRFQEG | Carn_acyltransf |
| PRKCE | Q02156 | human | ITGB1 | P05556 | T789 | IyksAVttVVNPkyE | Integrin_b_cyt |
| PRKCE | Q02156 | human | STAT3 | P40763 | S727 | NtIDLPMsPrTLDSL | |
| PRKCE | Q02156 | human | TNNI3 | P19429 | S44 | KKskIsAsRKLQLKt | |
| PRKCE | Q02156 | human | NREP | Q16612 | S59 | LGSSELRsPRISYLH | Alveol-reg_P311 |
| PRKCE | Q02156 | human | PRKCE | Q02156 | K534 | GVIYRDLkLDNILLD | Pkinase |
| PRKCE | Q02156 | human | MAPT | P10636-8 | S293 | NVQskCGsKDNIkHV | Tubulin-binding |
| PRKCE | Q02156 | human | BAD | Q92934 | S99 | PFrGrsRsAPPNLWA | Bcl-2_BAD |
| PRKCE | Q02156 | human | ALDH2 | P05091 | T429 | MQILkFKtIEEVVGR | Aldedh |
| PRKCE | Q02156 | human | PRKD2 | Q9BZL6 | S706 | ARIIGEksFRRsVVG | Pkinase |
| PRKCE | Q02156 | human | PRKD2 | Q9BZL6 | S710 | GEksFRRsVVGtPAy | Pkinase |
| PRKCE | Q02156 | human | OCLN | Q16625 | T404 | HyEtDyttGGEsCDE | |
| PRKCE | Q02156 | human | KCNK3 | O14649 | T383 | TGLHSLStFRGLMKR | |
| PRKCE | Q02156 | human | FGFR1 | P11362 | S779 | PLDQysPsFPDTRSS | |
| PRKCE | Q02156 | human | TRPV1 | Q8NER1 | S801 | VPLLREAsARDRQsA | |
| PRKCE | Q02156 | human | TICAM2 | Q86XR7 | S16 | NSCPLsLsWGKRHsV | |
| PRKCE | Q02156 | human | NANOG | Q9H9S0 | T78 | sPKGKQPtsAEKSVA | |
| PRKCE | Q02156 | human | BAD | Q92934 | S75 | EIRsRHssyPAGtED | Bcl-2_BAD |
| PRKCE | Q02156 | human | MAPT | P10636-8 | S258 | PDLkNVKskIGstEN | Tubulin-binding |
| PRKCE | Q02156 | human | CREB1 | P16220 | S119 | EILsRRPsyRkILND | pKID |
| PRKCE | Q02156 | human | KIR3DL1 | P43629 | S415 | QRKITRPsQRPKtPP | |
| PRKCE | Q02156 | human | XIAP | P98170 | S87 | VGRHRKVsPNCRFIN | BIR |
| PRKCE | Q02156 | human | RPS6KB2 | Q9UBS0 | S473 | PPSGTKKsKRGRGRP | |
| PRKCE | Q02156 | human | KRT18 | P05783 | S53 | IsVsrstsFrGGMGs | |
| PRKCE | Q02156 | human | GNAI2 | P04899 | S44 | LLLGAGEsGkSTIVk | G-alpha |
| PRKCE | Q02156 | human | PRKCE | Q02156 | S729 | QEEFKGFsYFGEDLM | Pkinase_C |
| PRKCE | Q02156 | human | BAD | Q92934 | S118 | GRELRRMsDEFVDsF | Bcl-2_BAD |
| PRKCE | Q02156 | human | GLS | O94925-3 | S314 | RYVGKEPsGLRFNKL | Glutaminase |
| PRKCE | Q02156 | human | IQGAP1 | P46940 | S1443 | DKMKKsksVkEDsNL | |
| PRKCE | Q02156 | human | ALDH2 | P05091 | T202 | KLGPALAtGNVVVMK | Aldedh |
| PRKCE | Q02156 | human | PRKD1 | Q15139 | S738 | ARIIGEksFRRsVVG | Pkinase |
| PRKCE | Q02156 | human | CORO1B | Q9BR76 | S2 | ______MsFRKVVRQ | |
| PRKCE | Q02156 | human | RAB11A | P62491 | S177 | TEIYRIVsQkQMSDR | |
| PRKCE | Q02156 | human | RAB5A | P20339 | T7 | _MAsrGAtRPNGPNT | |
| PRKCE | Q02156 | human | NANOG | Q9H9S0 | T280 | GLNVIQQtTRYFStP | |
| PRKCE | Q02156 | human | ITGB1 | P05556 | T788 | PIyksAVttVVNPky | Integrin_b_cyt |
| PRKCE | Q02156 | human | CHAT | P28329-3 | S476 | HKAAVPAsEKLLLLK | Carn_acyltransf |
| PRKCE | Q02156 | human | GJA1 | P17302 | S368 | QRPssRAssRAssRP | |
| PRKCE | Q02156 | human | PRKD2 | Q9BZL6 | S876 | QGLAERIsVL_____ | |
| PRKCE | Q02156 | human | IKBKB | O14920 | S177 | AkELDQGsLCtsFVG | Pkinase |
| PRKCE | Q02156 | human | ALDH2 | P05091 | S296 | KSPNIIMsDADMDWA | Aldedh |
| PRKCE | Q02156 | human | OCLN | Q16625 | T403 | DHyEtDyttGGEsCD | |
| PRKCE | Q02156 | human | PRKCE | Q02156 | S368 | NNIRKALsFDNRGEE | |
| PRKCE | Q02156 | human | ATF2 | P15336 | T52 | HkHKHEMtLKFGPAR | |
| PRKCE | Q02156 | human | PRKD1 | Q15139 | S742 | GEksFRRsVVGtPAy | Pkinase |
| PRKCE | Q02156 | human | CTNND1 | O60716 | S268 | PQVRVGGssVDLHRF | |
| PRKCE | Q02156 | human | MET | P08581 | S985 | PHLDRLVsARsVsPt |
 Biological Network Integration of This Kinase and Substrates Biological Network Integration of This Kinase and Substrates (GeneMANIA website) |
 Enriched GO biological processes of the phosphorylation target genes of the kinase Enriched GO biological processes of the phosphorylation target genes of the kinase |
| Kinase | GOID | GO term | P.adjust |
| PRKCE | ID | Description | 0.00e+00 |
| PRKCE | GO:0035767 | endothelial cell chemotaxis | 1.33e-05 |
| PRKCE | GO:0001667 | ameboidal-type cell migration | 1.33e-05 |
| PRKCE | GO:2001028 | positive regulation of endothelial cell chemotaxis | 2.30e-05 |
| PRKCE | GO:0010634 | positive regulation of epithelial cell migration | 5.39e-05 |
| PRKCE | GO:0010631 | epithelial cell migration | 5.39e-05 |
| PRKCE | GO:0090132 | epithelium migration | 5.39e-05 |
| PRKCE | GO:0090130 | tissue migration | 5.39e-05 |
| PRKCE | GO:2001026 | regulation of endothelial cell chemotaxis | 6.60e-05 |
| PRKCE | GO:0051347 | positive regulation of transferase activity | 8.62e-05 |
| PRKCE | GO:0050679 | positive regulation of epithelial cell proliferation | 1.33e-04 |
| PRKCE | GO:0033674 | positive regulation of kinase activity | 1.59e-04 |
| PRKCE | GO:0050920 | regulation of chemotaxis | 1.59e-04 |
| PRKCE | GO:0050921 | positive regulation of chemotaxis | 1.59e-04 |
| PRKCE | GO:0043542 | endothelial cell migration | 5.52e-04 |
| PRKCE | GO:0010632 | regulation of epithelial cell migration | 6.62e-04 |
| PRKCE | GO:0001938 | positive regulation of endothelial cell proliferation | 6.96e-04 |
| PRKCE | GO:0043123 | positive regulation of canonical NF-kappaB signal transduction | 6.96e-04 |
| PRKCE | GO:0002756 | MyD88-independent toll-like receptor signaling pathway | 7.21e-04 |
| PRKCE | GO:0062197 | cellular response to chemical stress | 8.37e-04 |
| PRKCE | GO:0060326 | cell chemotaxis | 9.35e-04 |
| PRKCE | GO:0010595 | positive regulation of endothelial cell migration | 1.20e-03 |
| PRKCE | GO:0042886 | amide transport | 1.48e-03 |
| PRKCE | GO:0046887 | positive regulation of hormone secretion | 1.62e-03 |
| PRKCE | GO:0051091 | positive regulation of DNA-binding transcription factor activity | 1.71e-03 |
| PRKCE | GO:0046883 | regulation of hormone secretion | 1.93e-03 |
| PRKCE | GO:0051092 | positive regulation of NF-kappaB transcription factor activity | 1.93e-03 |
| PRKCE | GO:0034599 | cellular response to oxidative stress | 1.95e-03 |
| PRKCE | GO:0043536 | positive regulation of blood vessel endothelial cell migration | 2.09e-03 |
| PRKCE | GO:0043122 | regulation of canonical NF-kappaB signal transduction | 2.22e-03 |
| PRKCE | GO:1905564 | positive regulation of vascular endothelial cell proliferation | 2.22e-03 |
| PRKCE | GO:0051090 | regulation of DNA-binding transcription factor activity | 2.48e-03 |
| PRKCE | GO:0050678 | regulation of epithelial cell proliferation | 2.48e-03 |
| PRKCE | GO:1903532 | positive regulation of secretion by cell | 3.08e-03 |
| PRKCE | GO:1902905 | positive regulation of supramolecular fiber organization | 3.08e-03 |
| PRKCE | GO:0072659 | protein localization to plasma membrane | 3.08e-03 |
| PRKCE | GO:0031116 | positive regulation of microtubule polymerization | 3.08e-03 |
| PRKCE | GO:0032273 | positive regulation of protein polymerization | 3.08e-03 |
| PRKCE | GO:0018105 | peptidyl-serine phosphorylation | 3.08e-03 |
| PRKCE | GO:0001936 | regulation of endothelial cell proliferation | 3.08e-03 |
| PRKCE | GO:0043534 | blood vessel endothelial cell migration | 3.08e-03 |
| PRKCE | GO:0007249 | canonical NF-kappaB signal transduction | 3.11e-03 |
| PRKCE | GO:0051495 | positive regulation of cytoskeleton organization | 3.11e-03 |
| PRKCE | GO:0045766 | positive regulation of angiogenesis | 3.11e-03 |
| PRKCE | GO:1904018 | positive regulation of vasculature development | 3.11e-03 |
| PRKCE | GO:0018209 | peptidyl-serine modification | 3.26e-03 |
| PRKCE | GO:0046879 | hormone secretion | 3.26e-03 |
| PRKCE | GO:0019932 | second-messenger-mediated signaling | 3.26e-03 |
| PRKCE | GO:0051047 | positive regulation of secretion | 3.26e-03 |
| PRKCE | GO:0031112 | positive regulation of microtubule polymerization or depolymerization | 3.27e-03 |
Top |
Related Drugs to PRKCE_SNX17 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
 Distribution of the number of studies mentioning PRKCE-SNX17 and kinase inhibitors the PubMed Abstract (04-01-2024) Distribution of the number of studies mentioning PRKCE-SNX17 and kinase inhibitors the PubMed Abstract (04-01-2024) |
| Fusion gene - drug pair 1 | Fusion gene - drug pair 2 | PMID | Publication date | DOI | Study title |
Top |
Related Diseases to PRKCE_SNX17 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Related diseases from the literature mentioned this fusion gene and drug. Related diseases from the literature mentioned this fusion gene and drug. (PubMed, 04-01-2024) |
| MeSH ID | MeSH term |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | PRKCE | C0020429 | Hyperalgesia | 3 | CTD_human |
| Hgene | PRKCE | C0458247 | Allodynia | 3 | CTD_human |
| Hgene | PRKCE | C0751211 | Hyperalgesia, Primary | 3 | CTD_human |
| Hgene | PRKCE | C0751212 | Hyperalgesia, Secondary | 3 | CTD_human |
| Hgene | PRKCE | C0751213 | Tactile Allodynia | 3 | CTD_human |
| Hgene | PRKCE | C0751214 | Hyperalgesia, Thermal | 3 | CTD_human |
| Hgene | PRKCE | C2936719 | Mechanical Allodynia | 3 | CTD_human |
Top |
Clinical Trials of the Found Drugs/Small Molecules |
 Statistics of the Clinical Trials of the Found Kinase Inibitors from clinicaltrials.gov (06-17-2024) Statistics of the Clinical Trials of the Found Kinase Inibitors from clinicaltrials.gov (06-17-2024) |
 Clinical Trials from clinicaltrials.gov (06-17-2024) Clinical Trials from clinicaltrials.gov (06-17-2024) |
| Fusion Gene | Kinase Inhibitor | NCT ID | Study Status | Phases | Disease | # Enrolment | Date |

