| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Kinase Fusion Gene:RPS6KB1_SNF8 |
Kinase Fusion Protein Summary |
 Kinase Fusion gene summary Kinase Fusion gene summary |
| Kinase Fusion partner gene information | Kinase Fusion gene name: RPS6KB1_SNF8 | KinaseFusionDB ID: KFG5600 | FusionGDB2.0 ID: KFG5600 | Hgene | Tgene | Gene symbol | RPS6KB1 | SNF8 | Gene ID | 6198 | 11267 | |
| Gene name | ribosomal protein S6 kinase B1 | SNF8 subunit of ESCRT-II | ||||||||||
| Synonyms | PS6K|S6K|S6K-beta-1|S6K1|STK14A|p70 S6KA|p70(S6K)-alpha|p70-S6K|p70-alpha | DEE115|Dot3|EAP30|NEDOA|VPS22 | ||||||||||
| Cytomap | 17q23.1 | 17q21.32 | ||||||||||
| Type of gene | protein-coding | protein-coding | ||||||||||
| Description | ribosomal protein S6 kinase beta-1ribosomal protein S6 kinase Iribosomal protein S6 kinase, 70kDa, polypeptide 1serine/threonine kinase 14 alphaserine/threonine-protein kinase 14A | vacuolar-sorting protein SNF8EAP30 subunit of ELL complexELL-associated protein of 30 kDaESCRT-II complex subunit VPS22SNF8, ESCRT-II complex subunit, homolog | ||||||||||
| Modification date | 20240411 | 20240412 | ||||||||||
| UniProtAcc | P23443 | Q96H20 | ||||||||||
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000225577, ENST00000406116, ENST00000443572, ENST00000393021, ENST00000587061, | ENST00000514089, ENST00000290330, ENST00000502492, | |||||||||
| Context (manual curation of fusion genes in KinaseFusionDB) | PubMed: RPS6KB1 [Title/Abstract] AND SNF8 [Title/Abstract] AND fusion [Title/Abstract] | |||||||||||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | RPS6KB1(57970687)-SNF8(47021338), # samples:1 | |||||||||||
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | RPS6KB1 | GO:0031667 | response to nutrient levels | 29750193 |
| Hgene | RPS6KB1 | GO:0031670 | cellular response to nutrient | 22017876 |
| Hgene | RPS6KB1 | GO:0031929 | TOR signaling | 12150926 |
| Hgene | RPS6KB1 | GO:0045948 | positive regulation of translational initiation | 1922062 |
| Hgene | RPS6KB1 | GO:0071363 | cellular response to growth factor stimulus | 17936702 |
| Hgene | RPS6KB1 | GO:1904263 | positive regulation of TORC1 signaling | 22017876 |
| Tgene | SNF8 | GO:0006357 | regulation of transcription by RNA polymerase II | 10419521 |
| Tgene | SNF8 | GO:0051179 | localization | 18539118 |
 Kinase Fusion gene breakpoints across RPS6KB1 (5'-gene) Kinase Fusion gene breakpoints across RPS6KB1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
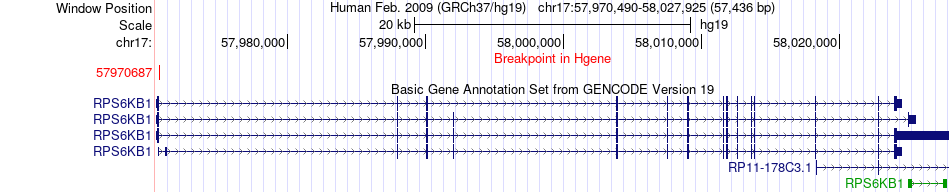 |
 Kinase Fusion gene breakpoints across SNF8 (3'-gene) Kinase Fusion gene breakpoints across SNF8 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
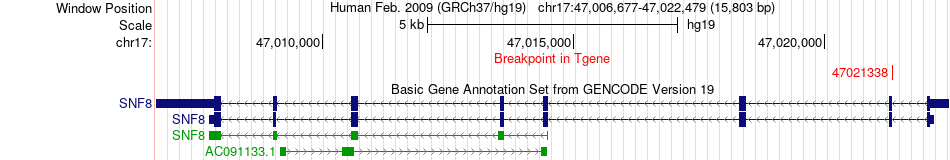 |
Top |
Kinase Fusion Gene Sample Information |
 Kinase Fusion gene information. Kinase Fusion gene information. |
 Kinase Fusion gene information from four resources (ChiTars 5.0, ChimerDB 4.0, COSMIC, and CCLE) Kinase Fusion gene information from four resources (ChiTars 5.0, ChimerDB 4.0, COSMIC, and CCLE)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Sample | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp |
| ChiTaRS5.0 | XX000001 | RPS6KB1 | chr17 | 57970687 | SNF8 | chr17 | 47021338 |
| CCLE | BT-474 | RPS6KB1 | chr17 | 57970686 | SNF8 | chr17 | 47021337 |
Top |
Kinase Fusion ORF Analysis |
 Kinase Fusion information from ORFfinder translation from full-length transcript sequence from KinaseFusionDB. Kinase Fusion information from ORFfinder translation from full-length transcript sequence from KinaseFusionDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp | Seq length (transcript) | Seq length (amino acids) |
Top |
Kinase Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from KinaseFusionDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from KinaseFusionDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >Henst_Tenst_Hgene_Hchr_Hbp_Tgene_Tchr_Tbp_length(fusion AA)_AAseq |
Multiple Sequence Alignment of All Fusion Protein Isoforms |
Top |
Kinase Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr17:57970687/chr17:47021338) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| RPS6KB1 | SNF8 |
| FUNCTION: Serine/threonine-protein kinase that acts downstream of mTOR signaling in response to growth factors and nutrients to promote cell proliferation, cell growth and cell cycle progression (PubMed:11500364, PubMed:12801526, PubMed:14673156, PubMed:15071500, PubMed:15341740, PubMed:16286006, PubMed:17052453, PubMed:17053147, PubMed:17936702, PubMed:18952604, PubMed:19085255, PubMed:19720745, PubMed:19935711, PubMed:19995915, PubMed:23429703, PubMed:28178239, PubMed:22017876). Regulates protein synthesis through phosphorylation of EIF4B, RPS6 and EEF2K, and contributes to cell survival by repressing the pro-apoptotic function of BAD (PubMed:11500364, PubMed:12801526, PubMed:14673156, PubMed:15071500, PubMed:15341740, PubMed:16286006, PubMed:17052453, PubMed:17053147, PubMed:17936702, PubMed:18952604, PubMed:19085255, PubMed:19720745, PubMed:19935711, PubMed:19995915, PubMed:23429703, PubMed:28178239, PubMed:22017876). Under conditions of nutrient depletion, the inactive form associates with the EIF3 translation initiation complex (PubMed:16286006). Upon mitogenic stimulation, phosphorylation by the mechanistic target of rapamycin complex 1 (mTORC1) leads to dissociation from the EIF3 complex and activation (PubMed:16286006). The active form then phosphorylates and activates several substrates in the pre-initiation complex, including the EIF2B complex and the cap-binding complex component EIF4B (PubMed:16286006). Also controls translation initiation by phosphorylating a negative regulator of EIF4A, PDCD4, targeting it for ubiquitination and subsequent proteolysis (PubMed:17053147). Promotes initiation of the pioneer round of protein synthesis by phosphorylating POLDIP3/SKAR (PubMed:15341740). In response to IGF1, activates translation elongation by phosphorylating EEF2 kinase (EEF2K), which leads to its inhibition and thus activation of EEF2 (PubMed:11500364). Also plays a role in feedback regulation of mTORC2 by mTORC1 by phosphorylating RICTOR, resulting in the inhibition of mTORC2 and AKT1 signaling (PubMed:19720745, PubMed:19935711, PubMed:19995915). Also involved in feedback regulation of mTORC1 and mTORC2 by phosphorylating DEPTOR (PubMed:22017876). Mediates cell survival by phosphorylating the pro-apoptotic protein BAD and suppressing its pro-apoptotic function (By similarity). Phosphorylates mitochondrial URI1 leading to dissociation of a URI1-PPP1CC complex (PubMed:17936702). The free mitochondrial PPP1CC can then dephosphorylate RPS6KB1 at Thr-412, which is proposed to be a negative feedback mechanism for the RPS6KB1 anti-apoptotic function (PubMed:17936702). Mediates TNF-alpha-induced insulin resistance by phosphorylating IRS1 at multiple serine residues, resulting in accelerated degradation of IRS1 (PubMed:18952604). In cells lacking functional TSC1-2 complex, constitutively phosphorylates and inhibits GSK3B (PubMed:17052453). May be involved in cytoskeletal rearrangement through binding to neurabin (By similarity). Phosphorylates and activates the pyrimidine biosynthesis enzyme CAD, downstream of MTOR (PubMed:23429703). Following activation by mTORC1, phosphorylates EPRS and thereby plays a key role in fatty acid uptake by adipocytes and also most probably in interferon-gamma-induced translation inhibition (PubMed:28178239). {ECO:0000250|UniProtKB:P67999, ECO:0000250|UniProtKB:Q8BSK8, ECO:0000269|PubMed:11500364, ECO:0000269|PubMed:12801526, ECO:0000269|PubMed:14673156, ECO:0000269|PubMed:15071500, ECO:0000269|PubMed:15341740, ECO:0000269|PubMed:16286006, ECO:0000269|PubMed:17052453, ECO:0000269|PubMed:17053147, ECO:0000269|PubMed:17936702, ECO:0000269|PubMed:18952604, ECO:0000269|PubMed:19085255, ECO:0000269|PubMed:19720745, ECO:0000269|PubMed:19935711, ECO:0000269|PubMed:19995915, ECO:0000269|PubMed:22017876, ECO:0000269|PubMed:23429703, ECO:0000269|PubMed:28178239}. | FUNCTION: Component of the endosomal sorting complex required for transport II (ESCRT-II), which is required for multivesicular body (MVB) formation and sorting of endosomal cargo proteins into MVBs. The MVB pathway mediates delivery of transmembrane proteins into the lumen of the lysosome for degradation. The ESCRT-II complex is probably involved in the recruitment of the ESCRT-III complex. The ESCRT-II complex may also play a role in transcription regulation by participating in derepression of transcription by RNA polymerase II, possibly via its interaction with ELL. Required for degradation of both endocytosed EGF and EGFR, but not for the EGFR ligand-mediated internalization. It is also required for the degradation of CXCR4. Required for the exosomal release of SDCBP, CD63 and syndecan (PubMed:22660413). {ECO:0000269|PubMed:17714434, ECO:0000269|PubMed:17959629, ECO:0000269|PubMed:18031739, ECO:0000269|PubMed:22660413}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
 - Retained domain in the 5'-partner of fusion protein (protein functional feature from UniProt). - Retained domain in the 5'-partner of fusion protein (protein functional feature from UniProt). |
| Partner | Hgeneene | Hbp | Tgeneene | Tbp | ENST | BPexon | TotalExon | Protein feature loci | BPloci | TotalLen | Feature | Note |
 - Retained domain in the 3'-partner of fusion protein (protein functional feature from UniProt). - Retained domain in the 3'-partner of fusion protein (protein functional feature from UniProt). |
| Partner | Hgeneene | Hbp | Tgeneene | Tbp | ENST | BPexon | TotalExon | Protein feature loci | BPloci | TotalLen | Feature | Note |
Top |
Kinase-Substrate Information of RPS6KB1_SNF8 |
 Phosphorylation target of the kinase Phosphorylation target of the kinase(phosphosite, 03-17-2024) |
| Kinase | Kinase UniProt Acc | Kinase species | Substrate | Substrate UniProt Acc | Substrate phosphorylated residues | Substrate phosphorylated sites (+/-7AA) | Domain |
| RPS6KB1 | P23443 | human | BACH2 | Q9BYV9 | S521 | ETRTRTssSCSsYsY | |
| RPS6KB1 | P23443 | human | IRS1 | P35568 | S636 | sGDyMPMsPKsVSAP | |
| RPS6KB1 | P23443 | human | NDRG2 | Q9UN36 | S350 | RsRsRtLsQssEsGt | |
| RPS6KB1 | P23443 | human | KHSRP | Q92945 | S395 | DLLQSLrsGPPGPPG | |
| RPS6KB1 | P23443 | human | PDCD4 | Q53EL6 | S67 | kRRLRKNssRDsGrG | |
| RPS6KB1 | P23443 | human | TP63 | Q9H3D4 | S560 | LARLGCSsCLDYFTT | SAM_2 |
| RPS6KB1 | P23443 | human | MTOR | P42345 | S2448 | RsRtRtDsysAGQsV | |
| RPS6KB1 | P23443 | human | TRIB2 | Q92519 | S83 | FRAVHLHsGEELVCK | Pkinase |
| RPS6KB1 | P23443 | human | RPS6 | P62753 | S240 | RLssLRAstsKsEss | |
| RPS6KB1 | P23443 | human | RPS6KB1 | P23443 | T412 | NQVFLGFtyVAPsVL | Pkinase_C |
| RPS6KB1 | P23443 | human | SMN1 | Q16637 | S49 | AyDkAVAsFkHALkN | SMN |
| RPS6KB1 | P23443 | human | EEF2K | O00418 | S366 | sPQVRtLsGSRPPLL | |
| RPS6KB1 | P23443 | human | BACH2 | Q9BYV9 | S520 | LETRTRTssSCSsYs | |
| RPS6KB1 | P23443 | human | MXD1 | Q05195 | S145 | IERIRMDsIGSTVSS | |
| RPS6KB1 | P23443 | human | IRS1 | P35568 | S307 | TRRsRtEsItAtsPA | |
| RPS6KB1 | P23443 | human | URI1 | O94763 | S372 | AKRKRKNstGsGHsA | |
| RPS6KB1 | P23443 | human | MAPT | P10636-8 | S262 | NVKskIGstENLkHQ | Tubulin-binding |
| RPS6KB1 | P23443 | human | PDCD4 | Q53EL6 | S457 | RGRKRFVsEGDGGRL | |
| RPS6KB1 | P23443 | human | IRS1 | P35568 | S527 | RFRKRtHsAGtsPtI | |
| RPS6KB1 | P23443 | human | NCBP1 | Q09161 | S7 | _MSRRRHsDENDGGQ | |
| RPS6KB1 | P23443 | human | IRS1 | P35568 | S270 | EFRPRsKsQSssNCs | |
| RPS6KB1 | P23443 | human | H2BC3 | P33778 | S36 | RKRsRkEsysIyVyk | Histone |
| RPS6KB1 | P23443 | human | PIP5K1C | O60331 | T553 | QPRYRRRtQssGQDG | |
| RPS6KB1 | P23443 | human | POLDIP3 | Q9BY77 | S385 | PRRVNsAsSsNPPAE | |
| RPS6KB1 | P23443 | human | TARBP2 | Q15633 | S283 | ILSLRsCsLGsLGAL | |
| RPS6KB1 | P23443 | human | MTOR | P42345 | T2446 | NKRsRtRtDsysAGQ | |
| RPS6KB1 | P23443 | human | FMR1 | Q06787 | S500 | NSEASNAsETESDHR | FXMRP1_C_core |
| RPS6KB1 | P23443 | human | IRS1 | P35568 | S1101 | GCRRRHssEtFsStP | |
| RPS6KB1 | P23443 | human | RPS6 | P62753 | S244 | LRAstsKsEssQK__ | |
| RPS6KB1 | P23443 | human | GSK3B | P49841 | S9 | SGRPRttsFAEsCkP | |
| RPS6KB1 | P23443 | human | ZBTB33 | Q86T24 | T606 | yLSDRSStIPAMkDD | |
| RPS6KB1 | P23443 | human | NDRG2 | Q9UN36 | S332 | LsRsRtAsLtsAAsV | |
| RPS6KB1 | P23443 | human | MAPT | P10636-8 | S214 | GsRsRtPsLPtPPtR | |
| RPS6KB1 | P23443 | human | DEPTOR | Q8TB45 | S287 | sMssCGssGyFsssP | |
| RPS6KB1 | P23443 | human | ESR1 | P03372 | S167 | GGRERLAsTNDkGSM | Oest_recep |
| RPS6KB1 | P23443 | human | LTC4S | Q16873 | S36 | ARRAFRVsPPLtTGP | MAPEG |
| RPS6KB1 | P23443 | human | RPS6 | P62753 | S236 | AKRRRLssLRAstsK | |
| RPS6KB1 | P23443 | human | MRE11 | P49959 | T597 | sQrGrADtGLETSTr | |
| RPS6KB1 | P23443 | human | CAD | P27708 | S1859 | PPRIhRAsDPGLPAE | |
| RPS6KB1 | P23443 | human | NCBP1 | Q09161 | S22 | PHKRRKtsDANEtED | |
| RPS6KB1 | P23443 | human | DEPTOR | Q8TB45 | S286 | ssMssCGssGyFsss | |
| RPS6KB1 | P23443 | human | EGLN1 | Q9GZT9 | S125 | ADPAAAAsPCRAAAG | |
| RPS6KB1 | P23443 | human | CCT2 | P78371 | S260 | GsRVRVDstAkVAEI | Cpn60_TCP1 |
| RPS6KB1 | P23443 | human | MAPT | P10636-8 | T212 | tPGsRsRtPsLPtPP | |
| RPS6KB1 | P23443 | human | PIP5K1C | O60331 | S555 | RYRRRtQssGQDGRP | |
| RPS6KB1 | P23443 | human | GLI1 | P08151 | S84 | LTKKRALsISPLSDA | |
| RPS6KB1 | P23443 | human | MSH6 | P52701 | S309 | RMVtGNGsLKRKSSR | |
| RPS6KB1 | P23443 | human | RICTOR | Q6R327 | T1135 | NRRIRtLtEPsVDFN | RICTOR_phospho |
| RPS6KB1 | P23443 | human | SRPK2 | P78362 | S494 | HDRSRtVsAsstGDL | |
| RPS6KB1 | P23443 | human | NCBP1 | Q09161 | T21 | QPHKRRKtsDANEtE | |
| RPS6KB1 | P23443 | human | EIF4B | P23588 | S422 | RERsRtGsEssQtGt | |
| RPS6KB1 | P23443 | human | RPS6 | P62753 | S235 | IAKRRRLssLRAsts | |
| RPS6KB1 | P23443 | human | POLDIP3 | Q9BY77 | S383 | ELPRRVNsAsSsNPP | |
| RPS6KB1 | P23443 | human | DEPTOR | Q8TB45 | S291 | CGssGyFsssPtLss | |
| RPS6KB1 | P23443 | human | EPRS1 | P07814 | S999 | NQGGGLsssGAGEGQ | |
| RPS6KB1 | P23443 | human | CDK1 | P06493 | S39 | MkkIRLEsEEEGVPs | Pkinase |
| RPS6KB1 | P23443-2 | human | GSK3B | P49841 | S9 | SGRPRttsFAEsCkP | |
| RPS6KB1 | P23443-2 | human | EPRS1 | P07814 | S999 | NQGGGLsssGAGEGQ |
 Biological Network Integration of This Kinase and Substrates Biological Network Integration of This Kinase and Substrates (GeneMANIA website) |
 Enriched GO biological processes of the phosphorylation target genes of the kinase Enriched GO biological processes of the phosphorylation target genes of the kinase |
| Kinase | GOID | GO term | P.adjust |
| RPS6KB1 | ID | Description | 0.00e+00 |
| RPS6KB1 | GO:0045927 | positive regulation of growth | 4.45e-04 |
| RPS6KB1 | GO:0006446 | regulation of translational initiation | 4.45e-04 |
| RPS6KB1 | GO:0032868 | response to insulin | 4.45e-04 |
| RPS6KB1 | GO:0032869 | cellular response to insulin stimulus | 9.65e-04 |
| RPS6KB1 | GO:0043086 | negative regulation of catalytic activity | 9.65e-04 |
| RPS6KB1 | GO:1900034 | regulation of cellular response to heat | 9.65e-04 |
| RPS6KB1 | GO:0006413 | translational initiation | 9.65e-04 |
| RPS6KB1 | GO:0051348 | negative regulation of transferase activity | 9.69e-04 |
| RPS6KB1 | GO:0045936 | negative regulation of phosphate metabolic process | 1.25e-03 |
| RPS6KB1 | GO:0010563 | negative regulation of phosphorus metabolic process | 1.25e-03 |
| RPS6KB1 | GO:0043434 | response to peptide hormone | 2.49e-03 |
| RPS6KB1 | GO:0031929 | TOR signaling | 2.49e-03 |
| RPS6KB1 | GO:0018105 | peptidyl-serine phosphorylation | 2.79e-03 |
| RPS6KB1 | GO:0018209 | peptidyl-serine modification | 3.19e-03 |
| RPS6KB1 | GO:0071375 | cellular response to peptide hormone stimulus | 3.19e-03 |
| RPS6KB1 | GO:0033673 | negative regulation of kinase activity | 3.47e-03 |
| RPS6KB1 | GO:0006403 | RNA localization | 3.47e-03 |
| RPS6KB1 | GO:0006913 | nucleocytoplasmic transport | 3.47e-03 |
| RPS6KB1 | GO:0051169 | nuclear transport | 3.47e-03 |
| RPS6KB1 | GO:0046777 | protein autophosphorylation | 3.47e-03 |
| RPS6KB1 | GO:1901653 | cellular response to peptide | 6.48e-03 |
| RPS6KB1 | GO:0051028 | mRNA transport | 8.35e-03 |
| RPS6KB1 | GO:0033002 | muscle cell proliferation | 8.39e-03 |
| RPS6KB1 | GO:0030850 | prostate gland development | 8.98e-03 |
| RPS6KB1 | GO:0001558 | regulation of cell growth | 9.68e-03 |
| RPS6KB1 | GO:0043467 | regulation of generation of precursor metabolites and energy | 9.68e-03 |
| RPS6KB1 | GO:0045727 | positive regulation of translation | 9.68e-03 |
| RPS6KB1 | GO:0060525 | prostate glandular acinus development | 9.68e-03 |
| RPS6KB1 | GO:1905214 | regulation of RNA binding | 9.68e-03 |
| RPS6KB1 | GO:1900180 | regulation of protein localization to nucleus | 9.68e-03 |
| RPS6KB1 | GO:0031400 | negative regulation of protein modification process | 1.03e-02 |
| RPS6KB1 | GO:0010921 | regulation of phosphatase activity | 1.20e-02 |
| RPS6KB1 | GO:0002181 | cytoplasmic translation | 1.20e-02 |
| RPS6KB1 | GO:0050657 | nucleic acid transport | 1.20e-02 |
| RPS6KB1 | GO:0050658 | RNA transport | 1.20e-02 |
| RPS6KB1 | GO:0031100 | animal organ regeneration | 1.20e-02 |
| RPS6KB1 | GO:0048524 | positive regulation of viral process | 1.20e-02 |
| RPS6KB1 | GO:0051236 | establishment of RNA localization | 1.20e-02 |
| RPS6KB1 | GO:0030307 | positive regulation of cell growth | 1.23e-02 |
| RPS6KB1 | GO:0034605 | cellular response to heat | 1.23e-02 |
| RPS6KB1 | GO:0001655 | urogenital system development | 1.23e-02 |
| RPS6KB1 | GO:0048639 | positive regulation of developmental growth | 1.23e-02 |
| RPS6KB1 | GO:0051168 | nuclear export | 1.23e-02 |
| RPS6KB1 | GO:0034250 | positive regulation of amide metabolic process | 1.23e-02 |
| RPS6KB1 | GO:0038203 | TORC2 signaling | 1.23e-02 |
| RPS6KB1 | GO:1900452 | regulation of long-term synaptic depression | 1.23e-02 |
| RPS6KB1 | GO:0006406 | mRNA export from nucleus | 1.25e-02 |
| RPS6KB1 | GO:0016049 | cell growth | 1.28e-02 |
| RPS6KB1 | GO:0043201 | response to leucine | 1.28e-02 |
Top |
Related Drugs to RPS6KB1_SNF8 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
 Distribution of the number of studies mentioning RPS6KB1-SNF8 and kinase inhibitors the PubMed Abstract (04-01-2024) Distribution of the number of studies mentioning RPS6KB1-SNF8 and kinase inhibitors the PubMed Abstract (04-01-2024) |
| Fusion gene - drug pair 1 | Fusion gene - drug pair 2 | PMID | Publication date | DOI | Study title |
Top |
Related Diseases to RPS6KB1_SNF8 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Related diseases from the literature mentioned this fusion gene and drug. Related diseases from the literature mentioned this fusion gene and drug. (PubMed, 04-01-2024) |
| MeSH ID | MeSH term |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
Top |
Clinical Trials of the Found Drugs/Small Molecules |
 Statistics of the Clinical Trials of the Found Kinase Inibitors from clinicaltrials.gov (06-17-2024) Statistics of the Clinical Trials of the Found Kinase Inibitors from clinicaltrials.gov (06-17-2024) |
 Clinical Trials from clinicaltrials.gov (06-17-2024) Clinical Trials from clinicaltrials.gov (06-17-2024) |
| Fusion Gene | Kinase Inhibitor | NCT ID | Study Status | Phases | Disease | # Enrolment | Date |

