| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Kinase Fusion Gene:STK11_TYK2 |
Kinase Fusion Protein Summary |
 Kinase Fusion gene summary Kinase Fusion gene summary |
| Kinase Fusion partner gene information | Kinase Fusion gene name: STK11_TYK2 | KinaseFusionDB ID: KFG6142 | FusionGDB2.0 ID: KFG6142 | Hgene | Tgene | Gene symbol | STK11 | TYK2 | Gene ID | 6794 | 7297 | |
| Gene name | serine/threonine kinase 11 | tyrosine kinase 2 | ||||||||||
| Synonyms | LKB1|PJS|hLKB1 | IMD35|JTK1 | ||||||||||
| Cytomap | 19p13.3 | 19p13.2 | ||||||||||
| Type of gene | protein-coding | protein-coding | ||||||||||
| Description | serine/threonine-protein kinase STK11liver kinase B1polarization-related protein LKB1renal carcinoma antigen NY-REN-19serine/threonine-protein kinase 11serine/threonine-protein kinase LKB1 | non-receptor tyrosine-protein kinase TYK2 | ||||||||||
| Modification date | 20240416 | 20240411 | ||||||||||
| UniProtAcc | Q15831 | P29597 | ||||||||||
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000326873, ENST00000585748, | ENST00000529422, ENST00000264818, ENST00000524462, ENST00000525621, ENST00000529370, | |||||||||
| Context (manual curation of fusion genes in KinaseFusionDB) | PubMed: STK11 [Title/Abstract] AND TYK2 [Title/Abstract] AND fusion [Title/Abstract] | |||||||||||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | STK11(1207202)-TYK2(10473333), # samples:3 | |||||||||||
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | STK11 | GO:0006468 | protein phosphorylation | 12805220|25329316 |
| Hgene | STK11 | GO:0018107 | peptidyl-threonine phosphorylation | 12805220 |
| Hgene | STK11 | GO:0046777 | protein autophosphorylation | 11430832 |
| Hgene | STK11 | GO:0070314 | G1 to G0 transition | 17216128 |
| Hgene | STK11 | GO:0071493 | cellular response to UV-B | 25329316 |
| Hgene | STK11 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator | 11430832 |
| Tgene | TYK2 | GO:0007259 | cell surface receptor signaling pathway via JAK-STAT | 7526154|7657660|8232552|8605876 |
| Tgene | TYK2 | GO:0032729 | positive regulation of type II interferon production | 12023369|19088061 |
| Tgene | TYK2 | GO:0032819 | positive regulation of natural killer cell proliferation | 19088061 |
| Tgene | TYK2 | GO:0035722 | interleukin-12-mediated signaling pathway | 7528775 |
| Tgene | TYK2 | GO:0042102 | positive regulation of T cell proliferation | 11114383 |
| Tgene | TYK2 | GO:0046427 | positive regulation of receptor signaling pathway via JAK-STAT | 7638186|12023369 |
| Tgene | TYK2 | GO:0051142 | positive regulation of NK T cell proliferation | 19088061 |
| Tgene | TYK2 | GO:0060333 | type II interferon-mediated signaling pathway | 8232552 |
| Tgene | TYK2 | GO:0060337 | type I interferon-mediated signaling pathway | 8232552 |
| Tgene | TYK2 | GO:1900182 | positive regulation of protein localization to nucleus | 8605876|26479788 |
 Kinase Fusion gene breakpoints across STK11 (5'-gene) Kinase Fusion gene breakpoints across STK11 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
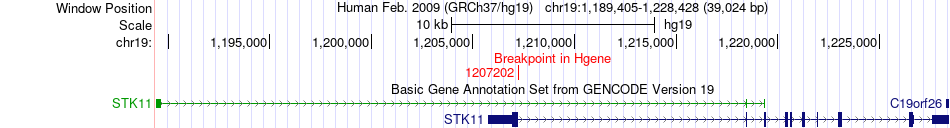 |
 Kinase Fusion gene breakpoints across TYK2 (3'-gene) Kinase Fusion gene breakpoints across TYK2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
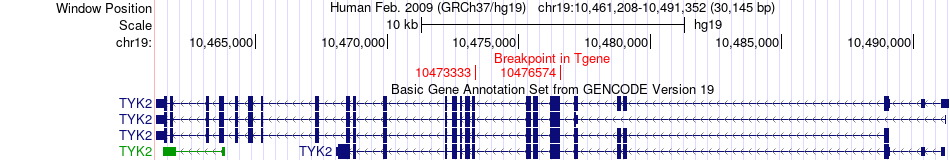 |
Top |
Kinase Fusion Gene Sample Information |
 Kinase Fusion gene information. Kinase Fusion gene information. |
 Kinase Fusion gene information from four resources (ChiTars 5.0, ChimerDB 4.0, COSMIC, and CCLE) Kinase Fusion gene information from four resources (ChiTars 5.0, ChimerDB 4.0, COSMIC, and CCLE)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Sample | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp |
| ChimerDB4 | TCGA-AQ-A04L-01B | STK11 | chr19 | 1207202 | TYK2 | chr19 | 10473333 |
| ChimerDB4 | TCGA-AQ-A04L | STK11 | chr19 | 1207202 | TYK2 | chr19 | 10476574 |
Top |
Kinase Fusion ORF Analysis |
 Kinase Fusion information from ORFfinder translation from full-length transcript sequence from KinaseFusionDB. Kinase Fusion information from ORFfinder translation from full-length transcript sequence from KinaseFusionDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp | Seq length (transcript) | Seq length (amino acids) |
| ENST00000326873 | ENST00000264818 | STK11 | chr19 | 1207202 | TYK2 | chr19 | 10473333 | 3961 | 882 |
| ENST00000326873 | ENST00000264818 | STK11 | chr19 | 1207202 | TYK2 | chr19 | 10476574 | 4699 | 1128 |
Top |
Kinase Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from KinaseFusionDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from KinaseFusionDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >Henst_Tenst_Hgene_Hchr_Hbp_Tgene_Tchr_Tbp_length(fusion AA)_AAseq >ENST00000326873_ENST00000264818_STK11_chr19_1207202_TYK2_chr19_10473333_length(amino acids)=882 MLEEGFFPSFWGFCCLFFFFFLCKILEKGSRNTRKDRSPADSGLAAGLQDPGSSMEVVDPQQLGMFTEGELMSVGMDTFIHRIDSTEVIY QPRRKRAKLIGKYLMGDLLGEGSYGKVKEVLDSETLCRRAVKILKKKKLRRIPNGEANVKKEPFVQAKLRPEDGLYLIHWSTSHPYRLIL TVAQRSQAPDGMQSLRLRKFPIEQQDGAFVLEGWGRSFPSVRELGAALQGCLLRAGDDCFSLRRCCLPQPGETSNLIIMRGARASPRTLN LSQLSFHRVDQKEITQLSHLGQGTRTNVYEGRLRVEGSGDPEEGKMDDEDPLVPGRDRGQELRVVLKVLDPSHHDIALAFYETASLMSQV SHTHLAFVHGVCVRGPENIMVTEYVEHGPLDVWLRRERGHVPMAWKMVVAQQLASALSYLENKNLVHGNVCGRNILLARLGLAEGTSPFI KLSDPGVGLGALSREERVERIPWLAPECLPGGANSLSTAMDKWGFGATLLEICFDGEAPLQSRSPSEKEHFYQRQHRLPEPSCPQLATLT SQCLTYEPTQRPSFRTILRDLTRLQPHNLADVLTVNPDSPASDPTVFHKRYLKKIRDLGEGHFGKVSLYCYDPTNDGTGEMVAVKALKAD CGPQHRSGWKQEIDILRTLYHEHIIKYKGCCEDQGEKSLQLVMEYVPLGSLRDYLPRHSIGLAQLLLFAQQICEGMAYLHAQHYIHRDLA ARNVLLDNDRLVKIGDFGLAKAVPEGHEYYRVREDGDSPVFWYAPECLKEYKFYYASDVWSFGVTLYELLTHCDSSQSPPTKFLELIGIA -------------------------------------------------------------- >ENST00000326873_ENST00000264818_STK11_chr19_1207202_TYK2_chr19_10476574_length(amino acids)=1128 MLEEGFFPSFWGFCCLFFFFFLCKILEKGSRNTRKDRSPADSGLAAGLQDPGSSMEVVDPQQLGMFTEGELMSVGMDTFIHRIDSTEVIY QPRRKRAKLIGKYLMGDLLGEGSYGKVKEVLDSETLCRRAVKILKKKKLRRIPNGEANVKNFKDCIPRSFRRHIRQHSALTRLRLRNVFR RFLRDFQPGRLSQQMVMVKYLATLERLAPRFGTERVPVCHLRLLAQAEGEPCYIRDSGVAPTDPGPESAAGPPTHEVLVTGTGGIQWWPV EEEVNKEEGSSGSSGRNPQASLFGKKAKAHKAVGQPADRPREPLWAYFCDFRDITHVVLKEHCVSIHRQDNKCLELSLPSRAAALSFVSL VDGYFRLTADSSHYLCHEVAPPRLVMSIRDGIHGPLLEPFVQAKLRPEDGLYLIHWSTSHPYRLILTVAQRSQAPDGMQSLRLRKFPIEQ QDGAFVLEGWGRSFPSVRELGAALQGCLLRAGDDCFSLRRCCLPQPGETSNLIIMRGARASPRTLNLSQLSFHRVDQKEITQLSHLGQGT RTNVYEGRLRVEGSGDPEEGKMDDEDPLVPGRDRGQELRVVLKVLDPSHHDIALAFYETASLMSQVSHTHLAFVHGVCVRGPENIMVTEY VEHGPLDVWLRRERGHVPMAWKMVVAQQLASALSYLENKNLVHGNVCGRNILLARLGLAEGTSPFIKLSDPGVGLGALSREERVERIPWL APECLPGGANSLSTAMDKWGFGATLLEICFDGEAPLQSRSPSEKEHFYQRQHRLPEPSCPQLATLTSQCLTYEPTQRPSFRTILRDLTRL QPHNLADVLTVNPDSPASDPTVFHKRYLKKIRDLGEGHFGKVSLYCYDPTNDGTGEMVAVKALKADCGPQHRSGWKQEIDILRTLYHEHI IKYKGCCEDQGEKSLQLVMEYVPLGSLRDYLPRHSIGLAQLLLFAQQICEGMAYLHAQHYIHRDLAARNVLLDNDRLVKIGDFGLAKAVP EGHEYYRVREDGDSPVFWYAPECLKEYKFYYASDVWSFGVTLYELLTHCDSSQSPPTKFLELIGIAQGQMTVLRLTELLERGERLPRPDK -------------------------------------------------------------- |
Multiple Sequence Alignment of All Fusion Protein Isoforms |
Top |
Kinase Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:1207202/chr19:10473333) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| STK11 | TYK2 |
| FUNCTION: Tumor suppressor serine/threonine-protein kinase that controls the activity of AMP-activated protein kinase (AMPK) family members, thereby playing a role in various processes such as cell metabolism, cell polarity, apoptosis and DNA damage response. Acts by phosphorylating the T-loop of AMPK family proteins, thus promoting their activity: phosphorylates PRKAA1, PRKAA2, BRSK1, BRSK2, MARK1, MARK2, MARK3, MARK4, NUAK1, NUAK2, SIK1, SIK2, SIK3 and SNRK but not MELK. Also phosphorylates non-AMPK family proteins such as STRADA, PTEN and possibly p53/TP53. Acts as a key upstream regulator of AMPK by mediating phosphorylation and activation of AMPK catalytic subunits PRKAA1 and PRKAA2 and thereby regulates processes including: inhibition of signaling pathways that promote cell growth and proliferation when energy levels are low, glucose homeostasis in liver, activation of autophagy when cells undergo nutrient deprivation, and B-cell differentiation in the germinal center in response to DNA damage. Also acts as a regulator of cellular polarity by remodeling the actin cytoskeleton. Required for cortical neuron polarization by mediating phosphorylation and activation of BRSK1 and BRSK2, leading to axon initiation and specification. Involved in DNA damage response: interacts with p53/TP53 and recruited to the CDKN1A/WAF1 promoter to participate in transcription activation. Able to phosphorylate p53/TP53; the relevance of such result in vivo is however unclear and phosphorylation may be indirect and mediated by downstream STK11/LKB1 kinase NUAK1. Also acts as a mediator of p53/TP53-dependent apoptosis via interaction with p53/TP53: translocates to the mitochondrion during apoptosis and regulates p53/TP53-dependent apoptosis pathways. Regulates UV radiation-induced DNA damage response mediated by CDKN1A. In association with NUAK1, phosphorylates CDKN1A in response to UV radiation and contributes to its degradation which is necessary for optimal DNA repair (PubMed:25329316). {ECO:0000269|PubMed:11430832, ECO:0000269|PubMed:12805220, ECO:0000269|PubMed:14517248, ECO:0000269|PubMed:14976552, ECO:0000269|PubMed:15016379, ECO:0000269|PubMed:15733851, ECO:0000269|PubMed:15987703, ECO:0000269|PubMed:17108107, ECO:0000269|PubMed:21317932, ECO:0000269|PubMed:25329316}.; FUNCTION: [Isoform 2]: Has a role in spermiogenesis. {ECO:0000250}. | FUNCTION: Tyrosine kinase of the non-receptor type involved in numerous cytokines and interferons signaling, which regulates cell growth, development, cell migration, innate and adaptive immunity (PubMed:8232552, PubMed:7813427, PubMed:7657660, PubMed:10995743, PubMed:10542297). Plays both structural and catalytic roles in numerous interleukins and interferons (IFN-alpha/beta) signaling (PubMed:10542297). Associates with heterodimeric cytokine receptor complexes and activates STAT family members including STAT1, STAT3, STAT4 or STAT6 (PubMed:10542297, PubMed:7638186). The heterodimeric cytokine receptor complexes are composed of (1) a TYK2-associated receptor chain (IFNAR1, IL12RB1, IL10RB or IL13RA1), and (2) a second receptor chain associated either with JAK1 or JAK2 (PubMed:7813427, PubMed:10542297, PubMed:7526154, PubMed:25762719). In response to cytokine-binding to receptors, phosphorylates and activates receptors (IFNAR1, IL12RB1, IL10RB or IL13RA1), creating docking sites for STAT members (PubMed:7526154, PubMed:7657660). In turn, recruited STATs are phosphorylated by TYK2 (or JAK1/JAK2 on the second receptor chain), form homo- and heterodimers, translocate to the nucleus, and regulate cytokine/growth factor responsive genes (PubMed:7657660, PubMed:10542297, PubMed:25762719). Negatively regulates STAT3 activity by promototing phosphorylation at a specific tyrosine that differs from the site used for signaling (PubMed:29162862). {ECO:0000269|PubMed:10542297, ECO:0000269|PubMed:10995743, ECO:0000269|PubMed:25762719, ECO:0000269|PubMed:29162862, ECO:0000269|PubMed:7526154, ECO:0000269|PubMed:7638186, ECO:0000269|PubMed:7657660, ECO:0000269|PubMed:7813427, ECO:0000269|PubMed:8232552}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
 - Retained domain in the 5'-partner of fusion protein (protein functional feature from UniProt). - Retained domain in the 5'-partner of fusion protein (protein functional feature from UniProt). |
| Partner | Hgeneene | Hbp | Tgeneene | Tbp | ENST | BPexon | TotalExon | Protein feature loci | BPloci | TotalLen | Feature | Note |
 - Retained domain in the 3'-partner of fusion protein (protein functional feature from UniProt). - Retained domain in the 3'-partner of fusion protein (protein functional feature from UniProt). |
| Partner | Hgeneene | Hbp | Tgeneene | Tbp | ENST | BPexon | TotalExon | Protein feature loci | BPloci | TotalLen | Feature | Note |
| Tgene | STK11 | 1207202 | TYK2 | 10473333 | ENST00000326873 | 6 | 23 | 589_875 | 455 | 1188 | Domain | Note=Protein kinase 1;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00159 |
| Tgene | STK11 | 1207202 | TYK2 | 10473333 | ENST00000326873 | 8 | 25 | 589_875 | 455 | 1188 | Domain | Note=Protein kinase 1;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00159 |
| Tgene | STK11 | 1207202 | TYK2 | 10476574 | ENST00000326873 | 3 | 23 | 589_875 | 209 | 1188 | Domain | Note=Protein kinase 1;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00159 |
| Tgene | STK11 | 1207202 | TYK2 | 10476574 | ENST00000326873 | 5 | 25 | 589_875 | 209 | 1188 | Domain | Note=Protein kinase 1;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00159 |
| Tgene | STK11 | 1207202 | TYK2 | 10473333 | ENST00000326873 | 6 | 23 | 897_1176 | 455 | 1188 | Domain | Note=Protein kinase 2;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00159 |
| Tgene | STK11 | 1207202 | TYK2 | 10473333 | ENST00000326873 | 8 | 25 | 897_1176 | 455 | 1188 | Domain | Note=Protein kinase 2;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00159 |
| Tgene | STK11 | 1207202 | TYK2 | 10476574 | ENST00000326873 | 3 | 23 | 897_1176 | 209 | 1188 | Domain | Note=Protein kinase 2;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00159 |
| Tgene | STK11 | 1207202 | TYK2 | 10476574 | ENST00000326873 | 5 | 25 | 897_1176 | 209 | 1188 | Domain | Note=Protein kinase 2;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00159 |
| Tgene | STK11 | 1207202 | TYK2 | 10476574 | ENST00000326873 | 3 | 23 | 450_529 | 209 | 1188 | Domain | Note=SH2%3B atypical |
| Tgene | STK11 | 1207202 | TYK2 | 10476574 | ENST00000326873 | 5 | 25 | 450_529 | 209 | 1188 | Domain | Note=SH2%3B atypical |
Top |
Kinase Fusion Protein Structures |
 CIF files of the predicted kinase fusion proteins CIF files of the predicted kinase fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Kinase Fusion protein CIF link (fusion AA seq ID in KinaseFusionDB) | Henst | Tenst | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp | AA seq | Len(AA seq) |
| PDB file >>>482_STK11_TYK2 | ENST00000326873 | ENST00000264818 | STK11 | chr19 | 1207202 | TYK2 | chr19 | 10476574 | MLEEGFFPSFWGFCCLFFFFFLCKILEKGSRNTRKDRSPADSGLAAGLQDPGSSMEVVDPQQLGMFTEGELMSVGMDTFIHRIDSTEVIY QPRRKRAKLIGKYLMGDLLGEGSYGKVKEVLDSETLCRRAVKILKKKKLRRIPNGEANVKNFKDCIPRSFRRHIRQHSALTRLRLRNVFR RFLRDFQPGRLSQQMVMVKYLATLERLAPRFGTERVPVCHLRLLAQAEGEPCYIRDSGVAPTDPGPESAAGPPTHEVLVTGTGGIQWWPV EEEVNKEEGSSGSSGRNPQASLFGKKAKAHKAVGQPADRPREPLWAYFCDFRDITHVVLKEHCVSIHRQDNKCLELSLPSRAAALSFVSL VDGYFRLTADSSHYLCHEVAPPRLVMSIRDGIHGPLLEPFVQAKLRPEDGLYLIHWSTSHPYRLILTVAQRSQAPDGMQSLRLRKFPIEQ QDGAFVLEGWGRSFPSVRELGAALQGCLLRAGDDCFSLRRCCLPQPGETSNLIIMRGARASPRTLNLSQLSFHRVDQKEITQLSHLGQGT RTNVYEGRLRVEGSGDPEEGKMDDEDPLVPGRDRGQELRVVLKVLDPSHHDIALAFYETASLMSQVSHTHLAFVHGVCVRGPENIMVTEY VEHGPLDVWLRRERGHVPMAWKMVVAQQLASALSYLENKNLVHGNVCGRNILLARLGLAEGTSPFIKLSDPGVGLGALSREERVERIPWL APECLPGGANSLSTAMDKWGFGATLLEICFDGEAPLQSRSPSEKEHFYQRQHRLPEPSCPQLATLTSQCLTYEPTQRPSFRTILRDLTRL QPHNLADVLTVNPDSPASDPTVFHKRYLKKIRDLGEGHFGKVSLYCYDPTNDGTGEMVAVKALKADCGPQHRSGWKQEIDILRTLYHEHI IKYKGCCEDQGEKSLQLVMEYVPLGSLRDYLPRHSIGLAQLLLFAQQICEGMAYLHAQHYIHRDLAARNVLLDNDRLVKIGDFGLAKAVP EGHEYYRVREDGDSPVFWYAPECLKEYKFYYASDVWSFGVTLYELLTHCDSSQSPPTKFLELIGIAQGQMTVLRLTELLERGERLPRPDK | 1128 |
| 3D view using mol* of 482_STK11_TYK2 | ||||||||||
| PDB file >>>TKFP_814_STK11_TYK2 | ENST00000326873 | ENST00000264818 | STK11 | chr19 | 1207202 | TYK2 | chr19 | 10473333 | MLEEGFFPSFWGFCCLFFFFFLCKILEKGSRNTRKDRSPADSGLAAGLQDPGSSMEVVDPQQLGMFTEGELMSVGMDTFIHRIDSTEVIY QPRRKRAKLIGKYLMGDLLGEGSYGKVKEVLDSETLCRRAVKILKKKKLRRIPNGEANVKKEPFVQAKLRPEDGLYLIHWSTSHPYRLIL TVAQRSQAPDGMQSLRLRKFPIEQQDGAFVLEGWGRSFPSVRELGAALQGCLLRAGDDCFSLRRCCLPQPGETSNLIIMRGARASPRTLN LSQLSFHRVDQKEITQLSHLGQGTRTNVYEGRLRVEGSGDPEEGKMDDEDPLVPGRDRGQELRVVLKVLDPSHHDIALAFYETASLMSQV SHTHLAFVHGVCVRGPENIMVTEYVEHGPLDVWLRRERGHVPMAWKMVVAQQLASALSYLENKNLVHGNVCGRNILLARLGLAEGTSPFI KLSDPGVGLGALSREERVERIPWLAPECLPGGANSLSTAMDKWGFGATLLEICFDGEAPLQSRSPSEKEHFYQRQHRLPEPSCPQLATLT SQCLTYEPTQRPSFRTILRDLTRLQPHNLADVLTVNPDSPASDPTVFHKRYLKKIRDLGEGHFGKVSLYCYDPTNDGTGEMVAVKALKAD CGPQHRSGWKQEIDILRTLYHEHIIKYKGCCEDQGEKSLQLVMEYVPLGSLRDYLPRHSIGLAQLLLFAQQICEGMAYLHAQHYIHRDLA ARNVLLDNDRLVKIGDFGLAKAVPEGHEYYRVREDGDSPVFWYAPECLKEYKFYYASDVWSFGVTLYELLTHCDSSQSPPTKFLELIGIA | 882_STK11_TYK2 |
| PDB file >>>TKFP_815_STK11_TYK2 | ENST00000326873 | ENST00000264818 | STK11 | chr19 | 1207202 | TYK2 | chr19 | 10476574 | MLEEGFFPSFWGFCCLFFFFFLCKILEKGSRNTRKDRSPADSGLAAGLQDPGSSMEVVDPQQLGMFTEGELMSVGMDTFIHRIDSTEVIY QPRRKRAKLIGKYLMGDLLGEGSYGKVKEVLDSETLCRRAVKILKKKKLRRIPNGEANVKNFKDCIPRSFRRHIRQHSALTRLRLRNVFR RFLRDFQPGRLSQQMVMVKYLATLERLAPRFGTERVPVCHLRLLAQAEGEPCYIRDSGVAPTDPGPESAAGPPTHEVLVTGTGGIQWWPV EEEVNKEEGSSGSSGRNPQASLFGKKAKAHKAVGQPADRPREPLWAYFCDFRDITHVVLKEHCVSIHRQDNKCLELSLPSRAAALSFVSL VDGYFRLTADSSHYLCHEVAPPRLVMSIRDGIHGPLLEPFVQAKLRPEDGLYLIHWSTSHPYRLILTVAQRSQAPDGMQSLRLRKFPIEQ QDGAFVLEGWGRSFPSVRELGAALQGCLLRAGDDCFSLRRCCLPQPGETSNLIIMRGARASPRTLNLSQLSFHRVDQKEITQLSHLGQGT RTNVYEGRLRVEGSGDPEEGKMDDEDPLVPGRDRGQELRVVLKVLDPSHHDIALAFYETASLMSQVSHTHLAFVHGVCVRGPENIMVTEY VEHGPLDVWLRRERGHVPMAWKMVVAQQLASALSYLENKNLVHGNVCGRNILLARLGLAEGTSPFIKLSDPGVGLGALSREERVERIPWL APECLPGGANSLSTAMDKWGFGATLLEICFDGEAPLQSRSPSEKEHFYQRQHRLPEPSCPQLATLTSQCLTYEPTQRPSFRTILRDLTRL QPHNLADVLTVNPDSPASDPTVFHKRYLKKIRDLGEGHFGKVSLYCYDPTNDGTGEMVAVKALKADCGPQHRSGWKQEIDILRTLYHEHI IKYKGCCEDQGEKSLQLVMEYVPLGSLRDYLPRHSIGLAQLLLFAQQICEGMAYLHAQHYIHRDLAARNVLLDNDRLVKIGDFGLAKAVP EGHEYYRVREDGDSPVFWYAPECLKEYKFYYASDVWSFGVTLYELLTHCDSSQSPPTKFLELIGIAQGQMTVLRLTELLERGERLPRPDK | 1128_STK11_TYK2 |
| 3D view using mol* of TKFP_815_STK11_TYK2 | ||||||||||
Top |
Comparison of Fusion Protein Isoforms |
 Superimpose the 3D Structures Among All Fusion Protein Isoforms Superimpose the 3D Structures Among All Fusion Protein Isoforms * Download the pdb file and open it from the molstar online viewer. |
 Comparison of the Secondary Structures of Fusion Protein Isoforms Comparison of the Secondary Structures of Fusion Protein Isoforms |
Top |
Comparison of Fusion Protein Sequences/Structures with Known Sequences/Structures from PDB |
Top |
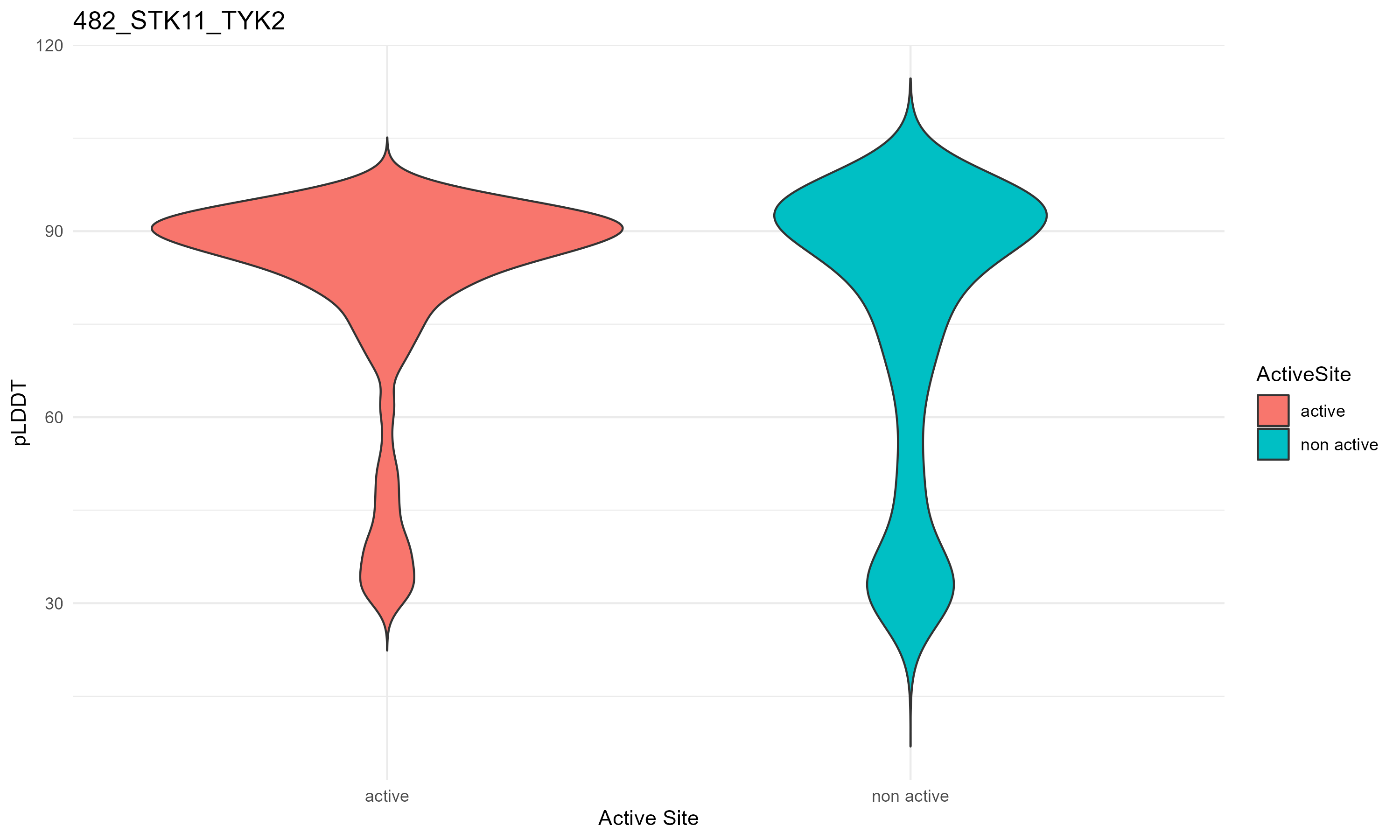
pLDDT score distribution |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. * The blue color at the bottom marks the best active site residues. |
| 482_STK11_TYK2.png |
 |
| 482_STK11_TYK2.png |
 |
| TKFP_815_STK11_TYK2.png |
 |
Top |
Potential Active Site Information |
 The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. |
| Kinase Fusion AA seq ID in KinaseFusionDB | Site score | Size | Dscore | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
| TKFP_815_STK11_TYK2 | 1.059 | 1217 | 1.023 | 3709.202 | 0.44 | 0.786 | 1 | 0.476 | 1.194 | 0.398 | 0.899 | Chain A: 221,222,223,224,225,226,227,228,229,230,2 31,232,233,262,263,264,266,267,314,315,316,317,318 ,319,320,322,323,338,339,340,341,342,345,368,369,3 70,372,373,374,377,385,388,389,390,391,412,414,467 ,468,471,472,475,490,491,492,493,494,495,496,497,4 99,503,504,505,506,507,508,509,510,511,512,513,514 ,515,516,517,595,598,599,602,603,604,605,606,607,6 08,609,610,611,612,613,614,629,665,667,668,669,670 ,671,672,673,674,675,685,693,695,697,701,703,704,7 05,706,707,716,732,733,734,735,738,781,784,785,788 ,793,794,795,796,797,798,799,801,802,805,806,809,8 10,816,819,820,821,822,823,825,826,827,828,829,830 ,831,832,833,834,837,857,858,859,860,864,865,866,8 67,892,893,894,895,896,897,898,899,900,902,903,904 ,920,921,922,923,943,946,947,950,953,954,958,960,9 73,974,975,976,977,979,1107,1114,1118 |
Top |
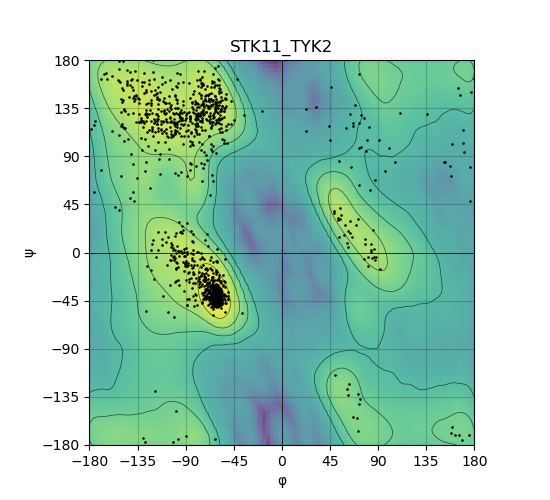
Ramachandran Plot of Kinase Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| 482_STK11_TYK2_ramachandran.png |
 |
| TKFP_815_STK11_TYK2_ramachandran.png |
 |
Top |
Virtual Screening Results |
 Distribution of the average docking score across all approved kinase inhibitors. Distribution of the average docking score across all approved kinase inhibitors.Distribution of the number of occurrence across all approved kinase inhibitors. |
| 5'-kinase fusion protein case |
| 3'-kinase fusion protein case |
Top |
 Drug information from DrugBank of the top 20 interacting small molecules. Drug information from DrugBank of the top 20 interacting small molecules.* The detailed information of individual kinase inhibitors are available in the download page. |
| Fusion gene name info | Drug | Docking score | Glide g score | Glide energy |
Top |
Kinase-Substrate Information of STK11_TYK2 |
 Phosphorylation target of the kinase Phosphorylation target of the kinase(phosphosite, 03-17-2024) |
| Kinase | Kinase UniProt Acc | Kinase species | Substrate | Substrate UniProt Acc | Substrate phosphorylated residues | Substrate phosphorylated sites (+/-7AA) | Domain |
| STK11 | Q15831 | human | PTEN | P60484 | S380 | EPDHyRYsDttDsDP | |
| STK11 | Q15831 | human | SMAD4 | Q13485 | T77 | kCVTIQRtLDGRLQV | MH1 |
| STK11 | Q15831 | human | NUAK2 | Q9H093 | T208 | HQGKFLQtFCGSPLY | Pkinase |
| STK11 | Q15831 | human | SIRT1 | Q96EB6 | S615 | GEkNERtsVAGtVRk | |
| STK11 | Q15831 | human | CRTC1 | Q6UUV9 | S151 | TSWRRtNsDsALHQs | TORC_M |
| STK11 | Q15831 | human | NUAK1 | O60285 | T211 | QKDKFLQtFCGSPLY | Pkinase |
| STK11 | Q15831 | human | MELK | Q14680 | T167 | NkDyHLQtCCGsLAY | Pkinase |
| STK11 | Q15831 | human | PRKAA1 | Q13131 | T183 | sDGEFLRtsCGsPNy | Pkinase |
| STK11 | Q15831 | human | MARK4 | Q96L34 | T214 | TLGSKLDtFCGsPPY | Pkinase |
| STK11 | Q15831 | human | BRSK2 | Q8IWQ3 | T174 | VGDSLLEtSCGSPHY | Pkinase |
| STK11 | Q15831 | human | SIRT1 | Q96EB6 | S732 | EAINEAIsVkQEVTD | |
| STK11 | Q15831 | human | TP53 | P04637 | S392 | FktEGPDsD______ | |
| STK11 | Q15831 | human | YAP1 | P46937-2 | S127 | PQHVRAHssPASLQL | |
| STK11 | Q15831 | human | SIK2 | Q9H0K1 | T175 | kSGELLAtWCGSPPY | Pkinase |
| STK11 | Q15831 | human | MARK2 | Q7KZI7 | T208 | tFGNKLDtFCGsPPY | Pkinase |
| STK11 | Q15831 | human | STK11 | Q15831 | T363 | IEDDIIYtQDFTVPG | |
| STK11 | Q15831 | human | STK11 | Q15831 | T185 | kPGNLLLtTGGtLKI | Pkinase |
| STK11 | Q15831 | human | PRMT5 | O14744 | T132 | PLNQEDNtNLARVLt | PRMT5_TIM |
| STK11 | Q15831 | human | STK11 | Q15831 | T189 | LLLtTGGtLKISDLG | Pkinase |
| STK11 | Q15831 | human | MARK3 | P27448 | T211 | TVGGKLDtFCGsPPY | Pkinase |
| STK11 | Q15831 | human | STRADA | Q7RTN6 | T329 | GLSDSLTtSTPRPSN | Pkinase |
| STK11 | Q15831 | human | MARK3 | P27448 | S215 | KLDtFCGsPPYAAPE | Pkinase |
| STK11 | Q15831 | human | BRSK1 | Q8TDC3-2 | T205 | VGDSLLEtSCGsPHY | Pkinase |
| STK11 | Q15831 | human | STK11 | Q15831 | S428 | SSkIRRLsACkQQ__ | |
| STK11 | Q15831 | human | SIK1 | P57059 | T182 | kSGEPLStWCGSPPY | Pkinase |
| STK11 | Q15831 | human | PRMT5 | O14744 | T144 | VLtNHIHtGHHSSMF | PRMT5_TIM |
| STK11 | Q15831 | human | PAK1 | Q13153 | T109 | QWARLLQtsNITKsE | PBD |
| STK11 | Q15831 | human | STK11 | Q15831 | T402 | TEAAQLstKsRAEGR | |
| STK11 | Q15831 | human | PTBP1 | P26599-1 | T138 | sNHkELktDssPNQA | RRM_5 |
| STK11 | Q15831 | human | BRSK1 | Q8TDC3 | T189 | VGDSLLEtSCGSPHY | Pkinase |
| STK11 | Q15831 | human | CDKN1A | P38936 | T80 | LPkLYLPtGPRRGRD | |
| STK11 | Q15831 | human | SIK3 | Q9Y2K2 | T221 | TPGQLLKtWCGSPPY | Pkinase |
| STK11 | Q15831 | human | STK11 | Q15831 | T336 | KDRWRsMtVVPYLED | |
| STK11 | Q15831 | human | SIRT1 | Q96EB6 | S669 | EDDVLSSsSCGSNSD | |
| STK11 | Q15831 | human | PRMT5 | O14744 | T139 | tNLARVLtNHIHtGH | PRMT5_TIM |
| STK11 | Q15831 | human | MARK1 | Q9P0L2 | T215 | TVGNKLDtFCGsPPY | Pkinase |
| STK11 | Q15831 | human | PRKAA2 | P54646 | T172 | sDGEFLRtsCGsPNy | Pkinase |
| STK11 | Q15831 | human | STRADA | Q7RTN6 | T419 | SGIFGLVtNLEELEV | |
| STK11 | Q15831 | human | TP53 | P04637 | S15 | PsVEPPLsQEtFsDL | P53_TAD |
| STK11 | Q15831 | human | PTEN | P60484 | T383 | HyRYsDttDsDPENE | |
| STK11 | Q15831 | human | SNRK | Q9NRH2 | T173 | QPGKKLttsCGSLAY | Pkinase |
| STK11 | Q15831 | human | PTEN | P60484 | T382 | DHyRYsDttDsDPEN | |
| TYK2 | P29597 | human | TYK2 | P29597 | Y292 | QAEGEPCyIRDSGVA | Jak1_Phl |
| TYK2 | P29597 | human | TYK2 | P29597 | Y1054 | AVPEGHEyyRVREDG | PK_Tyr_Ser-Thr |
| TYK2 | P29597 | human | TYK2 | P29597 | Y1055 | VPEGHEyyRVREDGD | PK_Tyr_Ser-Thr |
| TYK2 | P29597 | human | SIVA1 | O15304 | Y53 | LFLGAQAyLDHVWDE | Siva |
| TYK2 | P29597 | human | IFNAR1 | P17181 | Y466 | VFLRCINyVFFPSLK | |
| TYK2 | P29597 | human | RACK1 | P63244 | Y194 | NHIGHtGyLNTVTVS | WD40 |
| TYK2 | P29597 | human | IFNAR1 | P17181 | Y481 | PSSSIDEyFSEQPLK | |
| TYK2 | P29597 | human | SIVA1 | O15304 | Y162 | LVDCSDMyEKVLCTS | Siva |
| TYK2 | P29597 | human | STAT3 | P40763 | Y640 | QIQsVEPytkQQLNN | SH2 |
| TYK2 | P29597 | human | STAT1 | P42224 | Y701 | DGPkGtGyIktELIs |
 Biological Network Integration of This Kinase and Substrates Biological Network Integration of This Kinase and Substrates (GeneMANIA website) |
 Enriched GO biological processes of the phosphorylation target genes of the kinase Enriched GO biological processes of the phosphorylation target genes of the kinase |
| Kinase | GOID | GO term | P.adjust |
| STK11 | ID | Description | 0.00e+00 |
| STK11 | GO:0042149 | cellular response to glucose starvation | 1.14e-07 |
| STK11 | GO:0007623 | circadian rhythm | 2.40e-07 |
| STK11 | GO:0032007 | negative regulation of TOR signaling | 4.34e-07 |
| STK11 | GO:0032006 | regulation of TOR signaling | 4.66e-07 |
| STK11 | GO:0031929 | TOR signaling | 9.10e-07 |
| STK11 | GO:0009267 | cellular response to starvation | 1.08e-06 |
| STK11 | GO:0009314 | response to radiation | 1.08e-06 |
| STK11 | GO:0048511 | rhythmic process | 1.13e-06 |
| STK11 | GO:0046777 | protein autophosphorylation | 1.59e-06 |
| STK11 | GO:0009416 | response to light stimulus | 1.59e-06 |
| STK11 | GO:0042594 | response to starvation | 2.63e-06 |
| STK11 | GO:0010212 | response to ionizing radiation | 4.73e-06 |
| STK11 | GO:0031669 | cellular response to nutrient levels | 5.85e-06 |
| STK11 | GO:0032886 | regulation of microtubule-based process | 6.56e-06 |
| STK11 | GO:0009411 | response to UV | 6.74e-06 |
| STK11 | GO:0050770 | regulation of axonogenesis | 7.66e-06 |
| STK11 | GO:0031668 | cellular response to extracellular stimulus | 9.68e-06 |
| STK11 | GO:0018105 | peptidyl-serine phosphorylation | 1.23e-05 |
| STK11 | GO:0072331 | signal transduction by p53 class mediator | 1.23e-05 |
| STK11 | GO:0018209 | peptidyl-serine modification | 1.54e-05 |
| STK11 | GO:0006109 | regulation of carbohydrate metabolic process | 1.70e-05 |
| STK11 | GO:1901796 | regulation of signal transduction by p53 class mediator | 2.81e-05 |
| STK11 | GO:0071496 | cellular response to external stimulus | 2.93e-05 |
| STK11 | GO:0090398 | cellular senescence | 2.96e-05 |
| STK11 | GO:0042752 | regulation of circadian rhythm | 3.40e-05 |
| STK11 | GO:0010332 | response to gamma radiation | 4.84e-05 |
| STK11 | GO:2000772 | regulation of cellular senescence | 5.88e-05 |
| STK11 | GO:0042593 | glucose homeostasis | 6.84e-05 |
| STK11 | GO:0033500 | carbohydrate homeostasis | 6.84e-05 |
| STK11 | GO:0051347 | positive regulation of transferase activity | 7.40e-05 |
| STK11 | GO:0097091 | synaptic vesicle clustering | 7.40e-05 |
| STK11 | GO:0055089 | fatty acid homeostasis | 8.69e-05 |
| STK11 | GO:0007409 | axonogenesis | 1.09e-04 |
| STK11 | GO:0045860 | positive regulation of protein kinase activity | 1.09e-04 |
| STK11 | GO:0044839 | cell cycle G2/M phase transition | 1.09e-04 |
| STK11 | GO:0070314 | G1 to G0 transition | 1.09e-04 |
| STK11 | GO:0010975 | regulation of neuron projection development | 1.09e-04 |
| STK11 | GO:0001678 | intracellular glucose homeostasis | 1.09e-04 |
| STK11 | GO:0016055 | Wnt signaling pathway | 1.12e-04 |
| STK11 | GO:0071479 | cellular response to ionizing radiation | 1.12e-04 |
| STK11 | GO:0198738 | cell-cell signaling by wnt | 1.12e-04 |
| STK11 | GO:0034504 | protein localization to nucleus | 1.57e-04 |
| STK11 | GO:0097009 | energy homeostasis | 1.63e-04 |
| STK11 | GO:0031667 | response to nutrient levels | 1.66e-04 |
| STK11 | GO:0097193 | intrinsic apoptotic signaling pathway | 1.72e-04 |
| STK11 | GO:0045913 | positive regulation of carbohydrate metabolic process | 1.76e-04 |
| STK11 | GO:0071478 | cellular response to radiation | 1.76e-04 |
| STK11 | GO:0043153 | entrainment of circadian clock by photoperiod | 1.92e-04 |
| STK11 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator | 1.92e-04 |
Top |
Related Drugs to STK11_TYK2 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
 Distribution of the number of studies mentioning STK11-TYK2 and kinase inhibitors the PubMed Abstract (04-01-2024) Distribution of the number of studies mentioning STK11-TYK2 and kinase inhibitors the PubMed Abstract (04-01-2024) |
| Fusion gene - drug pair 1 | Fusion gene - drug pair 2 | PMID | Publication date | DOI | Study title |
Top |
Related Diseases to STK11_TYK2 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Related diseases from the literature mentioned this fusion gene and drug. Related diseases from the literature mentioned this fusion gene and drug. (PubMed, 04-01-2024) |
| MeSH ID | MeSH term |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
Top |
Clinical Trials of the Found Drugs/Small Molecules |
 Statistics of the Clinical Trials of the Found Kinase Inibitors from clinicaltrials.gov (06-17-2024) Statistics of the Clinical Trials of the Found Kinase Inibitors from clinicaltrials.gov (06-17-2024) |
 Clinical Trials from clinicaltrials.gov (06-17-2024) Clinical Trials from clinicaltrials.gov (06-17-2024) |
| Fusion Gene | Kinase Inhibitor | NCT ID | Study Status | Phases | Disease | # Enrolment | Date |

