| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Kinase Fusion Gene:ULK1_NOC4L |
Kinase Fusion Protein Summary |
 Kinase Fusion gene summary Kinase Fusion gene summary |
| Kinase Fusion partner gene information | Kinase Fusion gene name: ULK1_NOC4L | KinaseFusionDB ID: KFG6989 | FusionGDB2.0 ID: KFG6989 | Hgene | Tgene | Gene symbol | ULK1 | NOC4L | Gene ID | 8408 | 79050 | |
| Gene name | unc-51 like autophagy activating kinase 1 | nucleolar complex associated 4 homolog | ||||||||||
| Synonyms | ATG1|ATG1A|UNC51|Unc51.1|hATG1 | NET49|NOC4|UTP19 | ||||||||||
| Cytomap | 12q24.33 | 12q24.33 | ||||||||||
| Type of gene | protein-coding | protein-coding | ||||||||||
| Description | serine/threonine-protein kinase ULK1ATG1 autophagy related 1 homologautophagy-related protein 1 homolog | nucleolar complex protein 4 homologNOC4 protein homologNOC4-like proteinnucleolar complex-associated protein 4-like protein | ||||||||||
| Modification date | 20240407 | 20240305 | ||||||||||
| UniProtAcc | O75385 | Q9BVI4 | ||||||||||
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000321867, ENST00000540647, | ENST00000538784, ENST00000535343, ENST00000330579, | |||||||||
| Context (manual curation of fusion genes in KinaseFusionDB) | PubMed: ULK1 [Title/Abstract] AND NOC4L [Title/Abstract] AND fusion [Title/Abstract] | |||||||||||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | ULK1(132401607)-NOC4L(132635526), # samples:1 | |||||||||||
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ULK1 | GO:0000045 | autophagosome assembly | 19225151|28890335 |
| Hgene | ULK1 | GO:0006914 | autophagy | 18936157|25891078 |
| Hgene | ULK1 | GO:0008285 | negative regulation of cell population proliferation | 21795849 |
| Hgene | ULK1 | GO:0010508 | positive regulation of autophagy | 25126726|29487085|35670107 |
| Hgene | ULK1 | GO:0016241 | regulation of macroautophagy | 20921139 |
| Hgene | ULK1 | GO:0018105 | peptidyl-serine phosphorylation | 25126726|27103069 |
| Hgene | ULK1 | GO:0018107 | peptidyl-threonine phosphorylation | 27103069 |
| Hgene | ULK1 | GO:0031333 | negative regulation of protein-containing complex assembly | 25126726 |
| Hgene | ULK1 | GO:0046777 | protein autophosphorylation | 18936157 |
| Hgene | ULK1 | GO:1903059 | regulation of protein lipidation | 19225151 |
 Kinase Fusion gene breakpoints across ULK1 (5'-gene) Kinase Fusion gene breakpoints across ULK1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
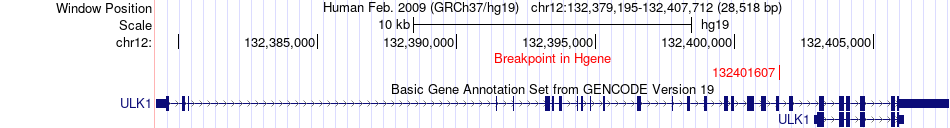 |
 Kinase Fusion gene breakpoints across NOC4L (3'-gene) Kinase Fusion gene breakpoints across NOC4L (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
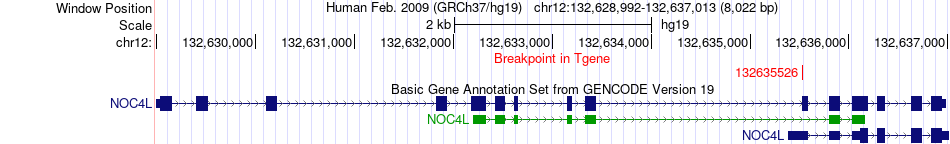 |
Top |
Kinase Fusion Gene Sample Information |
 Kinase Fusion gene information. Kinase Fusion gene information. |
 Kinase Fusion gene information from four resources (ChiTars 5.0, ChimerDB 4.0, COSMIC, and CCLE) Kinase Fusion gene information from four resources (ChiTars 5.0, ChimerDB 4.0, COSMIC, and CCLE)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Sample | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp |
| ChimerDB4 | TCGA-BC-A10U-01A | ULK1 | chr12 | 132401607 | NOC4L | chr12 | 132635526 |
Top |
Kinase Fusion ORF Analysis |
 Kinase Fusion information from ORFfinder translation from full-length transcript sequence from KinaseFusionDB. Kinase Fusion information from ORFfinder translation from full-length transcript sequence from KinaseFusionDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp | Seq length (transcript) | Seq length (amino acids) |
| ENST00000321867 | ENST00000330579 | ULK1 | chr12 | 132401607 | NOC4L | chr12 | 132635526 | 3223 | 1002 |
Top |
Kinase Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from KinaseFusionDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from KinaseFusionDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >Henst_Tenst_Hgene_Hchr_Hbp_Tgene_Tchr_Tbp_length(fusion AA)_AAseq >ENST00000321867_ENST00000330579_ULK1_chr12_132401607_NOC4L_chr12_132635526_length(amino acids)=1002 MGPGPGLPDGARGPGDAPSPGPAPPAPRPRPARPGPRLRLSPPRLGPPPPAPRPRPACAMEPGRGGTETVGKFEFSRKDLIGHGAFAVVF KGRHREKHDLEVAVKCINKKNLAKSQTLLGKEIKILKELKHENIVALYDFQEMANSVYLVMEYCNGGDLADYLHAMRTLSEDTIRLFLQQ IAGAMRLLHSKGIIHRDLKPQNILLSNPAGRRANPNSIRVKIADFGFARYLQSNMMAATLCGSPMYMAPEVIMSQHYDGKADLWSIGTIV YQCLTGKAPFQASSPQDLRLFYEKNKTLVPTIPRETSAPLRQLLLALLQRNHKDRMDFDEFFHHPFLDASPSVRKSPPVPVPSYPSSGSG SSSSSSSTSHLASPPSLGEMQQLQKTLASPADTAGFLHSSRDSGGSKDSSCDTDDFVMVPAQFPGDLVAEAPSAKPPPDSLMCSGSSLVA SAGLESHGRTPSPSPPCSSSPSPSGRAGPFSSSRCGASVPIPVPTQVQNYQRIERNLQSPTQFQTPRSSAIRRSGSTSPLGFARASPSPP AHAEHGGVLARKMSLGGGRPYTPSPQVGTIPERPGWSGTPSPQGAEMRGGRSPRPGSSAPEHSPRTSGLGCRLHSAPNLSDLHVVRPKLP KPPTDPLGAVFSPPQASPPQPSHGLQSCRNLRGSPKLPDFLQRNPLPPILGSPTKAVPSFDFPKTPSSQNLLALLARQGVVMTPPRNRTL PDLSEVGPFHGQPLGPGLRPGEDPKGPFGRSFSTSRLTDLLLKAAFGTQAPDPGSTESLQEKPMEIGGALSLLALNGLFILIHKHNLEYP DFYRKLYGLLDPSVFHVKYRARFFHLADLFLSSSHLPAYLVAAFAKRLARLALTAPPEALLMVLPFICNLLRRHPACRVLVHRPHGPELD ADPYDPGEEDPAQSRALESSLWELQALQRHYHPEVSKAASVINQALSMPEVSIAPLLELTAYEIFERDLKKKGPEPVPLEFIPAQGLLGR -------------------------------------------------------------- |
Multiple Sequence Alignment of All Fusion Protein Isoforms |
Top |
Kinase Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr12:132401607/chr12:132635526) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ULK1 | NOC4L |
| FUNCTION: Serine/threonine-protein kinase involved in autophagy in response to starvation (PubMed:18936157, PubMed:21460634, PubMed:21795849, PubMed:23524951, PubMed:25040165, PubMed:31123703, PubMed:29487085). Acts upstream of phosphatidylinositol 3-kinase PIK3C3 to regulate the formation of autophagophores, the precursors of autophagosomes (PubMed:18936157, PubMed:21460634, PubMed:21795849, PubMed:25040165). Part of regulatory feedback loops in autophagy: acts both as a downstream effector and negative regulator of mammalian target of rapamycin complex 1 (mTORC1) via interaction with RPTOR (PubMed:21795849). Activated via phosphorylation by AMPK and also acts as a regulator of AMPK by mediating phosphorylation of AMPK subunits PRKAA1, PRKAB2 and PRKAG1, leading to negatively regulate AMPK activity (PubMed:21460634). May phosphorylate ATG13/KIAA0652 and RPTOR; however such data need additional evidences (PubMed:18936157). Plays a role early in neuronal differentiation and is required for granule cell axon formation (PubMed:11146101). May also phosphorylate SESN2 and SQSTM1 to regulate autophagy (PubMed:25040165). Phosphorylates FLCN, promoting autophagy (PubMed:25126726). Phosphorylates AMBRA1 in response to autophagy induction, releasing AMBRA1 from the cytoskeletal docking site to induce autophagosome nucleation (PubMed:20921139). Phosphorylates ATG4B, leading to inhibit autophagy by decreasing both proteolytic activation and delipidation activities of ATG4B (PubMed:28821708). {ECO:0000269|PubMed:11146101, ECO:0000269|PubMed:18936157, ECO:0000269|PubMed:20921139, ECO:0000269|PubMed:21460634, ECO:0000269|PubMed:21795849, ECO:0000269|PubMed:23524951, ECO:0000269|PubMed:25040165, ECO:0000269|PubMed:25126726, ECO:0000269|PubMed:28821708, ECO:0000269|PubMed:29487085, ECO:0000269|PubMed:31123703}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
 - Retained domain in the 5'-partner of fusion protein (protein functional feature from UniProt). - Retained domain in the 5'-partner of fusion protein (protein functional feature from UniProt). |
| Partner | Hgeneene | Hbp | Tgeneene | Tbp | ENST | BPexon | TotalExon | Protein feature loci | BPloci | TotalLen | Feature | Note |
| Hgene | ULK1 | 132401607 | NOC4L | 132635526 | ENST00000321867 | 21 | 28 | 16_278 | 7271 | 1051 | Domain | Note=Protein kinase;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00159 |
 - Retained domain in the 3'-partner of fusion protein (protein functional feature from UniProt). - Retained domain in the 3'-partner of fusion protein (protein functional feature from UniProt). |
| Partner | Hgeneene | Hbp | Tgeneene | Tbp | ENST | BPexon | TotalExon | Protein feature loci | BPloci | TotalLen | Feature | Note |
Top |
Kinase Fusion Protein Structures |
 CIF files of the predicted kinase fusion proteins CIF files of the predicted kinase fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Kinase Fusion protein CIF link (fusion AA seq ID in KinaseFusionDB) | Henst | Tenst | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp | AA seq | Len(AA seq) |
| PDB file >>>298_ULK1_NOC4L | ENST00000321867 | ENST00000330579 | ULK1 | chr12 | 132401607 | NOC4L | chr12 | 132635526 | MGPGPGLPDGARGPGDAPSPGPAPPAPRPRPARPGPRLRLSPPRLGPPPPAPRPRPACAMEPGRGGTETVGKFEFSRKDLIGHGAFAVVF KGRHREKHDLEVAVKCINKKNLAKSQTLLGKEIKILKELKHENIVALYDFQEMANSVYLVMEYCNGGDLADYLHAMRTLSEDTIRLFLQQ IAGAMRLLHSKGIIHRDLKPQNILLSNPAGRRANPNSIRVKIADFGFARYLQSNMMAATLCGSPMYMAPEVIMSQHYDGKADLWSIGTIV YQCLTGKAPFQASSPQDLRLFYEKNKTLVPTIPRETSAPLRQLLLALLQRNHKDRMDFDEFFHHPFLDASPSVRKSPPVPVPSYPSSGSG SSSSSSSTSHLASPPSLGEMQQLQKTLASPADTAGFLHSSRDSGGSKDSSCDTDDFVMVPAQFPGDLVAEAPSAKPPPDSLMCSGSSLVA SAGLESHGRTPSPSPPCSSSPSPSGRAGPFSSSRCGASVPIPVPTQVQNYQRIERNLQSPTQFQTPRSSAIRRSGSTSPLGFARASPSPP AHAEHGGVLARKMSLGGGRPYTPSPQVGTIPERPGWSGTPSPQGAEMRGGRSPRPGSSAPEHSPRTSGLGCRLHSAPNLSDLHVVRPKLP KPPTDPLGAVFSPPQASPPQPSHGLQSCRNLRGSPKLPDFLQRNPLPPILGSPTKAVPSFDFPKTPSSQNLLALLARQGVVMTPPRNRTL PDLSEVGPFHGQPLGPGLRPGEDPKGPFGRSFSTSRLTDLLLKAAFGTQAPDPGSTESLQEKPMEIGGALSLLALNGLFILIHKHNLEYP DFYRKLYGLLDPSVFHVKYRARFFHLADLFLSSSHLPAYLVAAFAKRLARLALTAPPEALLMVLPFICNLLRRHPACRVLVHRPHGPELD ADPYDPGEEDPAQSRALESSLWELQALQRHYHPEVSKAASVINQALSMPEVSIAPLLELTAYEIFERDLKKKGPEPVPLEFIPAQGLLGR | 1002 |
| 3D view using mol* of 298_ULK1_NOC4L | ||||||||||
| PDB file >>>HKFP_435_ULK1_NOC4L | ENST00000321867 | ENST00000330579 | ULK1 | chr12 | 132401607 | NOC4L | chr12 | 132635526 | MGPGPGLPDGARGPGDAPSPGPAPPAPRPRPARPGPRLRLSPPRLGPPPPAPRPRPACAMEPGRGGTETVGKFEFSRKDLIGHGAFAVVF KGRHREKHDLEVAVKCINKKNLAKSQTLLGKEIKILKELKHENIVALYDFQEMANSVYLVMEYCNGGDLADYLHAMRTLSEDTIRLFLQQ IAGAMRLLHSKGIIHRDLKPQNILLSNPAGRRANPNSIRVKIADFGFARYLQSNMMAATLCGSPMYMAPEVIMSQHYDGKADLWSIGTIV YQCLTGKAPFQASSPQDLRLFYEKNKTLVPTIPRETSAPLRQLLLALLQRNHKDRMDFDEFFHHPFLDASPSVRKSPPVPVPSYPSSGSG SSSSSSSTSHLASPPSLGEMQQLQKTLASPADTAGFLHSSRDSGGSKDSSCDTDDFVMVPAQFPGDLVAEAPSAKPPPDSLMCSGSSLVA SAGLESHGRTPSPSPPCSSSPSPSGRAGPFSSSRCGASVPIPVPTQVQNYQRIERNLQSPTQFQTPRSSAIRRSGSTSPLGFARASPSPP AHAEHGGVLARKMSLGGGRPYTPSPQVGTIPERPGWSGTPSPQGAEMRGGRSPRPGSSAPEHSPRTSGLGCRLHSAPNLSDLHVVRPKLP KPPTDPLGAVFSPPQASPPQPSHGLQSCRNLRGSPKLPDFLQRNPLPPILGSPTKAVPSFDFPKTPSSQNLLALLARQGVVMTPPRNRTL PDLSEVGPFHGQPLGPGLRPGEDPKGPFGRSFSTSRLTDLLLKAAFGTQAPDPGSTESLQEKPMEIGGALSLLALNGLFILIHKHNLEYP DFYRKLYGLLDPSVFHVKYRARFFHLADLFLSSSHLPAYLVAAFAKRLARLALTAPPEALLMVLPFICNLLRRHPACRVLVHRPHGPELD ADPYDPGEEDPAQSRALESSLWELQALQRHYHPEVSKAASVINQALSMPEVSIAPLLELTAYEIFERDLKKKGPEPVPLEFIPAQGLLGR | 1002_ULK1_NOC4L |
Top |
Comparison of Fusion Protein Isoforms |
 Superimpose the 3D Structures Among All Fusion Protein Isoforms Superimpose the 3D Structures Among All Fusion Protein Isoforms * Download the pdb file and open it from the molstar online viewer. |
 Comparison of the Secondary Structures of Fusion Protein Isoforms Comparison of the Secondary Structures of Fusion Protein Isoforms |
Top |
Comparison of Fusion Protein Sequences/Structures with Known Sequences/Structures from PDB |
Top |
pLDDT score distribution |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. * The blue color at the bottom marks the best active site residues. |
| 298_ULK1_NOC4L.png |
 |
| 298_ULK1_NOC4L.png |
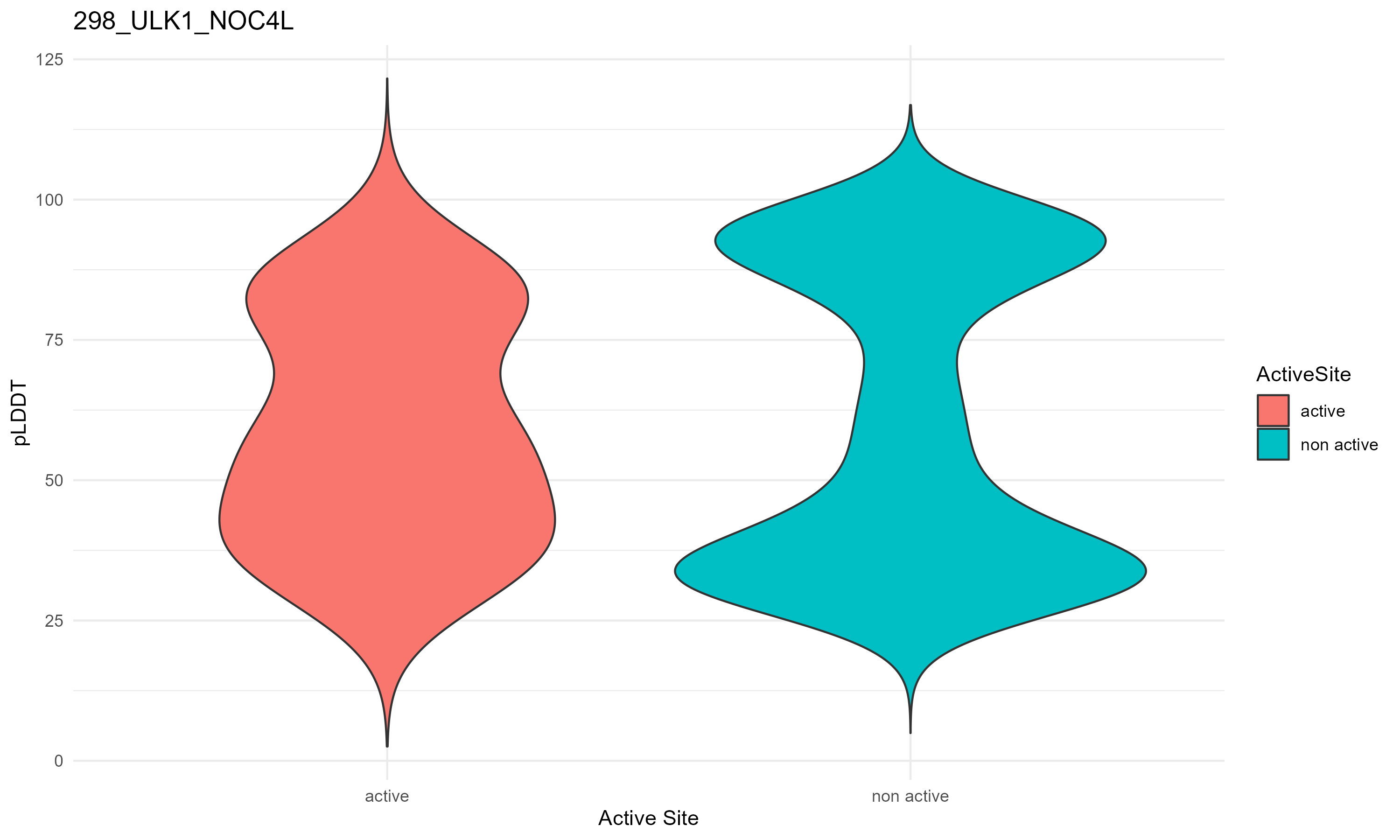 |
Top |
Potential Active Site Information |
 The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. |
| Kinase Fusion AA seq ID in KinaseFusionDB | Site score | Size | Dscore | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
Top |
Ramachandran Plot of Kinase Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| 298_ULK1_NOC4L_ramachandran.png |
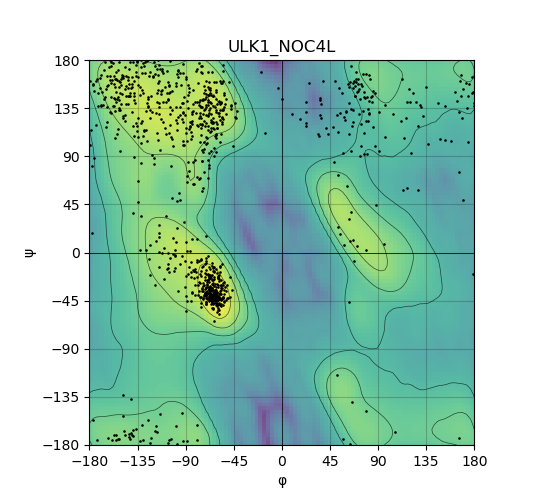 |
Top |
Virtual Screening Results |
 Distribution of the average docking score across all approved kinase inhibitors. Distribution of the average docking score across all approved kinase inhibitors.Distribution of the number of occurrence across all approved kinase inhibitors. |
| 5'-kinase fusion protein case |
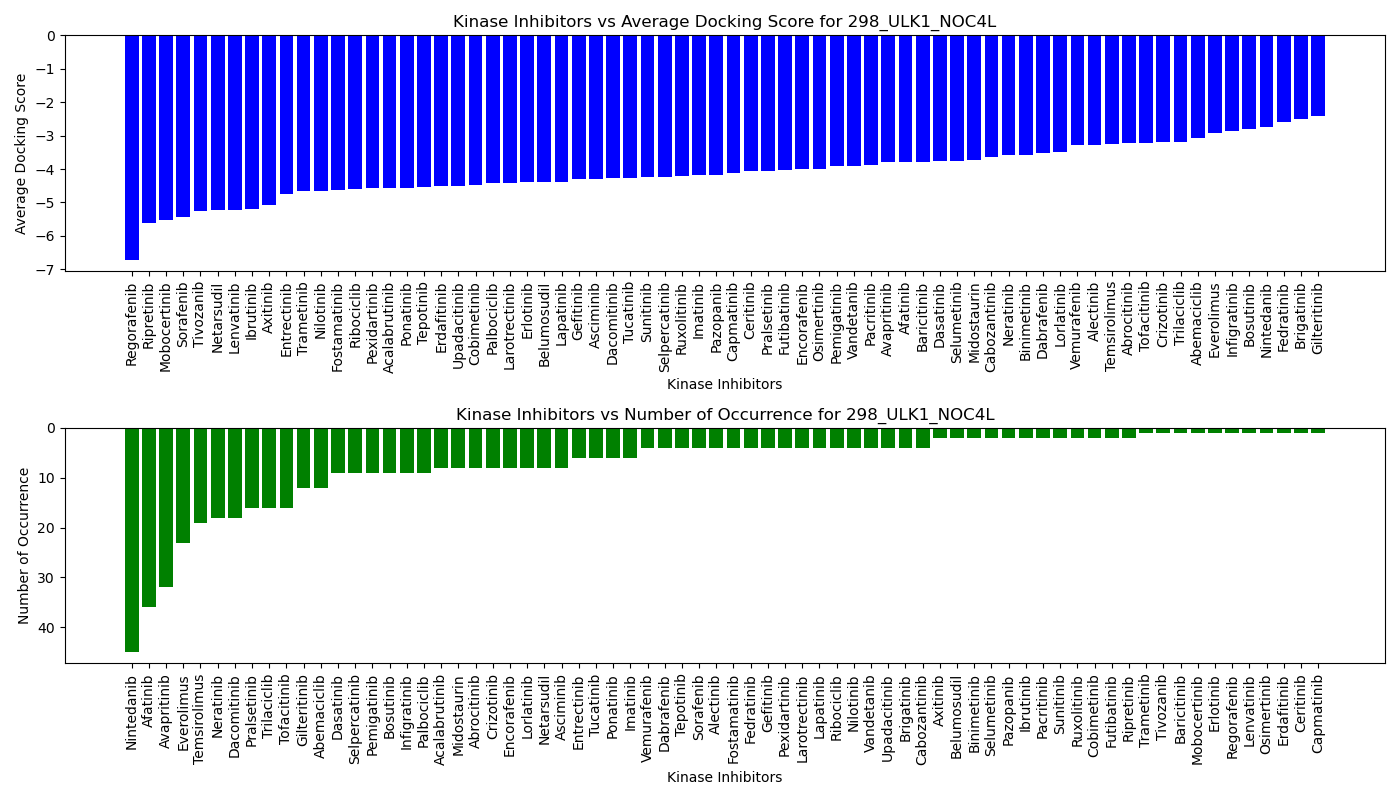 |
| 3'-kinase fusion protein case |
Top |
 Drug information from DrugBank of the top 20 interacting small molecules. Drug information from DrugBank of the top 20 interacting small molecules.* The detailed information of individual kinase inhibitors are available in the download page. |
| Fusion gene name info | Drug | Docking score | Glide g score | Glide energy |
| 298_ULK1_NOC4L-DOCK_HTVS_1-001 | Sorafenib | -6.85849 | -6.870589999999999 | -46.9871 |
| 298_ULK1_NOC4L-DOCK_HTVS_1-001 | Sorafenib | -6.85849 | -6.870589999999999 | -46.9871 |
| 298_ULK1_NOC4L-DOCK_HTVS_1-001 | Regorafenib | -6.715669999999999 | -6.715669999999999 | -46.6774 |
| 298_ULK1_NOC4L-DOCK_HTVS_1-001 | Netarsudil | -6.67315 | -6.68425 | -52.6775 |
| 298_ULK1_NOC4L-DOCK_HTVS_1-001 | Netarsudil | -6.67315 | -6.68425 | -52.6775 |
| 298_ULK1_NOC4L-DOCK_HTVS_1-001 | Netarsudil | -6.25082 | -6.26192 | -50.6674 |
| 298_ULK1_NOC4L-DOCK_HTVS_1-001 | Netarsudil | -6.25082 | -6.26192 | -50.6674 |
| 298_ULK1_NOC4L-DOCK_HTVS_1-001 | Acalabrutinib | -5.80595 | -5.82005 | -47.3507 |
| 298_ULK1_NOC4L-DOCK_HTVS_1-001 | Acalabrutinib | -5.80595 | -5.82005 | -47.3507 |
| 298_ULK1_NOC4L-DOCK_HTVS_1-001 | Pralsetinib | -5.708080000000001 | -5.799580000000001 | -49.1268 |
| 298_ULK1_NOC4L-DOCK_HTVS_1-001 | Encorafenib | -5.68456 | -6.07306 | -54.4653 |
| 298_ULK1_NOC4L-DOCK_HTVS_1-001 | Palbociclib | -5.63255 | -6.03945 | -45.5088 |
| 298_ULK1_NOC4L-DOCK_HTVS_1-001 | Palbociclib | -5.63255 | -6.03945 | -45.5088 |
| 298_ULK1_NOC4L-DOCK_HTVS_1-001 | Ripretinib | -5.617999999999999 | -5.625 | -46.1294 |
| 298_ULK1_NOC4L-DOCK_HTVS_1-001 | Ripretinib | -5.617999999999999 | -5.625 | -46.1294 |
| 298_ULK1_NOC4L-DOCK_HTVS_1-001 | Palbociclib | -5.59316 | -6.0000599999999995 | -45.0931 |
| 298_ULK1_NOC4L-DOCK_HTVS_1-001 | Mobocertinib | -5.52531 | -5.53301 | -50.5531 |
| 298_ULK1_NOC4L-DOCK_HTVS_1-001 | Nilotinib | -5.46626 | -5.60586 | -45.5429 |
| 298_ULK1_NOC4L-DOCK_HTVS_1-001 | Nilotinib | -5.46626 | -5.60586 | -45.5429 |
| 298_ULK1_NOC4L-DOCK_HTVS_1-001 | Ponatinib | -5.46138 | -5.66798 | -46.5059 |
Top |
Kinase-Substrate Information of ULK1_NOC4L |
 Phosphorylation target of the kinase Phosphorylation target of the kinase(phosphosite, 03-17-2024) |
| Kinase | Kinase UniProt Acc | Kinase species | Substrate | Substrate UniProt Acc | Substrate phosphorylated residues | Substrate phosphorylated sites (+/-7AA) | Domain |
| ULK1 | O75385 | human | CDC37 | Q16543 | S339 | HMQRCIDsGLWVPNs | CDC37_C |
| ULK1 | O75385 | human | ATG9A | Q7Z3C6 | S761 | QsIPRsAsYPCAAPR | |
| ULK1 | O75385 | human | FBP1 | P09467 | S88 | LVMNMLKsSFATCVL | FBPase |
| ULK1 | O75385 | human | RIPK1 | Q13546 | T337 | ssRsNsAtEQPGSLH | |
| ULK1 | O75385 | human | NR3C2 | P08235 | S843 | NEEKMHQsAMYELCQ | Hormone_recep |
| ULK1 | O75385 | human | RIPK1 | Q13546 | S296 | YLsQLEEsVEEDVks | |
| ULK1 | O75385 | human | SMCR8 | Q8TEV9 | S400 | PSQDRPPsSsLEECP | |
| ULK1 | O75385 | human | STING1 | Q86WV6 | S366 | QEPELLIsGMEkPLP | |
| ULK1 | O75385 | human | BNIP3L | O60238 | S35 | PPAGLNssWVELPMN | BNIP3 |
| ULK1 | O75385 | human | ATG13 | O75143 | S355 | DTETVSNssEGRAsP | |
| ULK1 | O75385 | human | YAP1 | P46937 | S227 | VQQNMMNsASGPLPD | |
| ULK1 | O75385 | human | ATG16L1 | Q676U5 | S278 | PAGGLLDsITNIFGR | |
| ULK1 | O75385 | human | SQSTM1 | Q13501 | S349 | sskEVDPstGELQsL | |
| ULK1 | O75385 | human | ATG9A | Q7Z3C6 | S14 | EyQRLEAsysDsPPG | |
| ULK1 | O75385 | human | RIPK1 | Q13546 | S335 | VPssRsNsAtEQPGS | |
| ULK1 | O75385 | human | PFKM | P08237 | S762 | yEIDLDTsDHAHLEH | |
| ULK1 | O75385 | human | BECN1 | Q14457 | S30 | QPLkLDTsFkILDRV | |
| ULK1 | O75385 | human | RPTOR | Q8N122 | S792 | DKMRRAssYSsLNSL | |
| ULK1 | O75385 | human | RPTOR | Q8N122 | S877 | HIHQAGGsPPAssts | |
| ULK1 | O75385 | human | BECN1 | Q14457 | S337 | LVPyGNHsYLESLTD | APG6 |
| ULK1 | O75385 | human | ENO1 | P06733 | S282 | QLADLyksFIkDyPV | Enolase_C |
| ULK1 | O75385 | human | BECN1 | Q14457 | S96 | MstEsANsFTLIGEA | |
| ULK1 | O75385 | human | RB1CC1 | Q8TDY2 | S986 | ELQsLEQsHLkELED | |
| ULK1 | O75385 | human | RB1CC1 | Q8TDY2 | S943 | EMENIMHsQNCEIKE | |
| ULK1 | O75385 | human | SMCR8 | Q8TEV9 | S562 | IRFQASIsPPELGET | |
| ULK1 | O75385 | human | SDCBP | O00560 | S60 | LsQyMGLsLNEEEIR | Peptidase_M50 |
| ULK1 | O75385 | human | ATG4B | Q9Y4P1 | S316 | SIAELDPsIAVGFFC | Peptidase_C54 |
| ULK1 | O75385 | human | SESN2 | P58004 | S254 | NNsGGFEsARDVEAL | PA26 |
| ULK1 | O75385 | human | ULK1 | O75385 | S317 | SHLASPPsLGEMQQL | |
| ULK1 | O75385 | human | RIPK1 | Q13546 | S291 | kFRPFYLsQLEEsVE | |
| ULK1 | O75385 | human | SDCBP | O00560 | S6 | __MsLyPsLEDLkVD | |
| ULK1 | O75385 | human | HK1 | P19367 | S124 | NIVHGSGsQLFDHVA | Hexokinase_1 |
| ULK1 | O75385 | human | RIPK1 | Q13546 | S345 | EQPGSLHssQGLGMG | |
| ULK1 | O75385 | human | BNIP3 | Q12983 | S17 | QEESLQGsWVELHFs | BNIP3 |
| ULK1 | O75385 | human | SESN2 | P58004 | S73 | GLEALMSsGRVDNLA | PA26 |
| ULK1 | O75385 | human | AMBRA1 | Q9C0C7 | S635 | ssSRLELsSSAsPQE | |
| ULK1 | O75385 | human | SMCR8 | Q8TEV9 | S492 | SKsDsQAsLtVPLsP | |
| ULK1 | O75385 | human | DENND3 | A2RUS2 | S490 | ELAPRNSsLRLtDTA | |
| ULK1 | O75385 | human | FLCN | Q8NFG4 | S406 | CVRIIPYsSQYEEAY | Folliculin_C |
| ULK1 | O75385 | human | AMBRA1 | Q9C0C7 | S465 | SQASVYTsATEGRGF | |
| ULK1 | O75385 | human | RPTOR | Q8N122 | S855 | QRVLDtssLtQsAPA | |
| ULK1 | O75385 | human | RPTOR | Q8N122 | S859 | DtssLtQsAPAsPtN | |
| ULK1 | O75385 | human | SQSTM1 | Q13501 | S403 | ESLSQMLsMGFsDEG | UBA_5 |
| ULK1 | O75385 | human | SEC23A | Q15436 | S207 | LQEMLGLskVPLTQA | Sec23_trunk |
| ULK1 | O75385 | human | ATG13 | O75143 | S389 | INQVTLTsLDIPFAM | |
| ULK1 | O75385 | human | FBP1 | P09467 | S63 | HLyGIAGsTNVTGDQ | FBPase |
| ULK1 | O75385 | human | SMCR8 | Q8TEV9 | T666 | ASRDISktsLDNYSD | |
| ULK1 | O75385 | human | PFKM | P08237 | S74 | EATWESVsMMLQLGG | PFK |
| ULK1 | O75385 | human | RIPK1 | Q13546 | S357 | GMGPVEEsWFAPSLE | |
| ULK1 | O75385 | human | PRKN | O60260 | S108 | sLTRVDLsssVLPGD | |
| ULK1 | O75385 | human | ATG101 | Q9BSB4 | S203 | ITDALGTsVTTTMRR | |
| ULK1 | O75385 | human | RIPK1 | Q13546 | S389 | ANyHLyGsRMDRQTK | |
| ULK1 | O75385 | human | SEC23B | Q15437 | S186 | SCEGISksYVFRGTk | Sec23_trunk |
| ULK1 | O75385 | human | ENO1 | P06733 | S115 | ANAILGVsLAVCkAG | Enolase_N |
| ULK1 | O75385 | human | RPTOR | Q8N122 | S863 | LtQsAPAsPtNkGVH | |
| ULK1 | O75385 | human | FLCN | Q8NFG4 | S537 | EDTQkLLsILGAsEE | Folliculin_C |
| ULK1 | O75385 | human | FUNDC1 | Q8IVP5 | S17 | DyEsDDDsyEVLDLT | |
| ULK1 | O75385 | human | FLCN | Q8NFG4 | S542 | LLsILGAsEEDNVKL | Folliculin_C |
| ULK1 | O75385 | human | HK1 | P19367 | S364 | TRLGVEPsDDDCVSV | Hexokinase_2 |
| ULK1 | O75385 | human | RIPK1 | Q13546 | S320 | AVVkRMQsLQLdCVA | |
| ULK1 | O75385 | human | RB1CC1 | Q8TDY2 | S1323 | EMQNVRTsLIAEQQT | |
| ULK1 | O75385 | human | RIPK1 | Q13546 | S333 | VAVPssRsNsAtEQP | |
| ULK1 | O75385 | human | RIPK1 | Q13546 | S346 | QPGSLHssQGLGMGP | |
| ULK1 | O75385 | human | MAD1L1 | Q9Y6D9 | S546 | RTKVLHMsLNPTsVA | MAD |
| ULK1 | O75385 | human | BECN1 | Q14457 | S15 | NNSTMQVsFVCQRCS | |
| ULK1 | O75385 | human | SEC16A | O15027 | S846 | LAQPINFsVSLSNSH | |
| ULK1 | O75385 | human | ATG14 | Q6ZNE5 | S29 | LARDLVDsVDDAEGL | |
| ULK1 | O75385 | human | SEC23A | Q15436 | T405 | FKMGFGGtLEIKTSR | Sec23_BS |
| ULK1 | O75385 | human | ATG101 | Q9BSB4 | S11 | RSEVLEVsVEGRQVE | ATG101 |
| ULK1 | O75385 | human | PIK3C3 | Q8NEB9 | S249 | ESsPILTsFELVKVP | |
| ULK1 | O75385 | human | DENND3 | A2RUS2 | S472 | THRRMVVsMPNLQDI | |
| ULK1 | O75385 | human | RIPK1 | Q13546 | S262 | YCPREIIsLMKLCWE | PK_Tyr_Ser-Thr |
| ULK1 | O75385 | human | BECN1 | Q14457 | S279 | ATFHIWHsGQFGTIN | APG6 |
| ULK1 | O75385 | human | FLII | Q13045 | S64 | KLEHLSVsHNNLTTL | LRR_8 |
 Biological Network Integration of This Kinase and Substrates Biological Network Integration of This Kinase and Substrates (GeneMANIA website) |
 Enriched GO biological processes of the phosphorylation target genes of the kinase Enriched GO biological processes of the phosphorylation target genes of the kinase |
| Kinase | GOID | GO term | P.adjust |
| ULK1 | ID | Description | 0.00e+00 |
| ULK1 | GO:0016236 | macroautophagy | 2.30e-21 |
| ULK1 | GO:0061912 | selective autophagy | 4.62e-21 |
| ULK1 | GO:0000422 | autophagy of mitochondrion | 4.62e-21 |
| ULK1 | GO:0061726 | mitochondrion disassembly | 4.62e-21 |
| ULK1 | GO:1903008 | organelle disassembly | 1.34e-20 |
| ULK1 | GO:0010508 | positive regulation of autophagy | 1.70e-20 |
| ULK1 | GO:0010506 | regulation of autophagy | 7.32e-20 |
| ULK1 | GO:0000045 | autophagosome assembly | 2.46e-18 |
| ULK1 | GO:1905037 | autophagosome organization | 4.91e-18 |
| ULK1 | GO:0022411 | cellular component disassembly | 2.18e-17 |
| ULK1 | GO:0007033 | vacuole organization | 4.47e-16 |
| ULK1 | GO:0031331 | positive regulation of cellular catabolic process | 5.89e-16 |
| ULK1 | GO:0000423 | mitophagy | 4.68e-13 |
| ULK1 | GO:0098780 | response to mitochondrial depolarisation | 5.46e-13 |
| ULK1 | GO:0016241 | regulation of macroautophagy | 2.58e-11 |
| ULK1 | GO:0016239 | positive regulation of macroautophagy | 9.94e-11 |
| ULK1 | GO:0016237 | lysosomal microautophagy | 1.92e-09 |
| ULK1 | GO:0034497 | protein localization to phagophore assembly site | 6.13e-09 |
| ULK1 | GO:1903059 | regulation of protein lipidation | 2.60e-07 |
| ULK1 | GO:0042594 | response to starvation | 4.33e-07 |
| ULK1 | GO:0006605 | protein targeting | 6.95e-07 |
| ULK1 | GO:0071496 | cellular response to external stimulus | 7.92e-07 |
| ULK1 | GO:0006096 | glycolytic process | 9.50e-07 |
| ULK1 | GO:0006497 | protein lipidation | 1.01e-06 |
| ULK1 | GO:0031669 | cellular response to nutrient levels | 1.01e-06 |
| ULK1 | GO:0031667 | response to nutrient levels | 1.01e-06 |
| ULK1 | GO:0016052 | carbohydrate catabolic process | 1.07e-06 |
| ULK1 | GO:0050746 | regulation of lipoprotein metabolic process | 1.26e-06 |
| ULK1 | GO:0042158 | lipoprotein biosynthetic process | 1.27e-06 |
| ULK1 | GO:0009267 | cellular response to starvation | 1.84e-06 |
| ULK1 | GO:0044804 | nucleophagy | 1.85e-06 |
| ULK1 | GO:0031668 | cellular response to extracellular stimulus | 1.95e-06 |
| ULK1 | GO:0006090 | pyruvate metabolic process | 3.61e-06 |
| ULK1 | GO:0062197 | cellular response to chemical stress | 4.65e-06 |
| ULK1 | GO:0042157 | lipoprotein metabolic process | 7.58e-06 |
| ULK1 | GO:0010639 | negative regulation of organelle organization | 1.24e-05 |
| ULK1 | GO:0010821 | regulation of mitochondrion organization | 1.45e-05 |
| ULK1 | GO:0007034 | vacuolar transport | 2.57e-05 |
| ULK1 | GO:0030242 | autophagy of peroxisome | 2.57e-05 |
| ULK1 | GO:0034727 | piecemeal microautophagy of the nucleus | 2.57e-05 |
| ULK1 | GO:0001558 | regulation of cell growth | 3.09e-05 |
| ULK1 | GO:1903146 | regulation of autophagy of mitochondrion | 3.13e-05 |
| ULK1 | GO:0006002 | fructose 6-phosphate metabolic process | 3.28e-05 |
| ULK1 | GO:0006006 | glucose metabolic process | 4.92e-05 |
| ULK1 | GO:0072594 | establishment of protein localization to organelle | 5.77e-05 |
| ULK1 | GO:0006623 | protein targeting to vacuole | 6.02e-05 |
| ULK1 | GO:0042149 | cellular response to glucose starvation | 6.96e-05 |
| ULK1 | GO:1903599 | positive regulation of autophagy of mitochondrion | 8.06e-05 |
| ULK1 | GO:0016049 | cell growth | 9.57e-05 |
Top |
Related Drugs to ULK1_NOC4L |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
 Distribution of the number of studies mentioning ULK1-NOC4L and kinase inhibitors the PubMed Abstract (04-01-2024) Distribution of the number of studies mentioning ULK1-NOC4L and kinase inhibitors the PubMed Abstract (04-01-2024) |
| Fusion gene - drug pair 1 | Fusion gene - drug pair 2 | PMID | Publication date | DOI | Study title |
Top |
Related Diseases to ULK1_NOC4L |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Related diseases from the literature mentioned this fusion gene and drug. Related diseases from the literature mentioned this fusion gene and drug. (PubMed, 04-01-2024) |
| MeSH ID | MeSH term |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
Top |
Clinical Trials of the Found Drugs/Small Molecules |
 Statistics of the Clinical Trials of the Found Kinase Inibitors from clinicaltrials.gov (06-17-2024) Statistics of the Clinical Trials of the Found Kinase Inibitors from clinicaltrials.gov (06-17-2024) |
 Clinical Trials from clinicaltrials.gov (06-17-2024) Clinical Trials from clinicaltrials.gov (06-17-2024) |
| Fusion Gene | Kinase Inhibitor | NCT ID | Study Status | Phases | Disease | # Enrolment | Date |

