| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Kinase Fusion Gene:USP7_PRKCB |
Kinase Fusion Protein Summary |
 Kinase Fusion gene summary Kinase Fusion gene summary |
| Kinase Fusion partner gene information | Kinase Fusion gene name: USP7_PRKCB | KinaseFusionDB ID: KFG7039 | FusionGDB2.0 ID: KFG7039 | Hgene | Tgene | Gene symbol | USP7 | PRKCB | Gene ID | 7874 | 5579 | |
| Gene name | ubiquitin specific peptidase 7 | protein kinase C beta | ||||||||||
| Synonyms | C16DELp13.2|DEL16P13.2|HAFOUS|HAUSP|TEF1 | PKC-beta|PKCB|PKCI(2)|PKCbeta|PRKCB1|PRKCB2 | ||||||||||
| Cytomap | 16p13.2 | 16p12.2-p12.1 | ||||||||||
| Type of gene | protein-coding | protein-coding | ||||||||||
| Description | ubiquitin carboxyl-terminal hydrolase 7Chromosome 16p13.2 deletion syndromeHerpes virus-associated ubiquitin-specific proteasedeubiquitinating enzyme 7ubiquitin specific peptidase 7 (herpes virus-associated)ubiquitin specific protease 7 (herpes virus | protein kinase C beta typePKC-Bprotein kinase C, beta 1 polypeptide | ||||||||||
| Modification date | 20240411 | 20240407 | ||||||||||
| UniProtAcc | Q93009 | P05771 | ||||||||||
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000344836, ENST00000381886, ENST00000535863, ENST00000566224, | ENST00000482000, ENST00000498058, ENST00000303531, ENST00000321728, | |||||||||
| Context (manual curation of fusion genes in KinaseFusionDB) | PubMed: USP7 [Title/Abstract] AND PRKCB [Title/Abstract] AND fusion [Title/Abstract] | |||||||||||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | USP7(9057063)-PRKCB(24043456), # samples:1 USP7(9057064)-PRKCB(24043457), # samples:1 | |||||||||||
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | USP7 | GO:0006307 | DNA alkylation repair | 25944111 |
| Hgene | USP7 | GO:0016579 | protein deubiquitination | 16964248|21258371|21745816|25172512 |
| Hgene | USP7 | GO:0031647 | regulation of protein stability | 27123980 |
| Hgene | USP7 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity | 11279055 |
| Hgene | USP7 | GO:0035520 | monoubiquitinated protein deubiquitination | 26280536 |
| Hgene | USP7 | GO:0042752 | regulation of circadian rhythm | 27123980 |
| Hgene | USP7 | GO:0045721 | negative regulation of gluconeogenesis | 28655758 |
| Hgene | USP7 | GO:0050821 | protein stabilization | 21258371|25172512|35216969 |
| Hgene | USP7 | GO:0051090 | regulation of DNA-binding transcription factor activity | 16964248 |
| Hgene | USP7 | GO:0075342 | symbiont-mediated disruption of host cell PML body | 20719947 |
| Hgene | USP7 | GO:1904262 | negative regulation of TORC1 signaling | 35216969 |
| Tgene | PRKCB | GO:0010827 | regulation of glucose transmembrane transport | 25982116 |
| Tgene | PRKCB | GO:0043687 | post-translational protein modification | 20228790 |
 Kinase Fusion gene breakpoints across USP7 (5'-gene) Kinase Fusion gene breakpoints across USP7 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
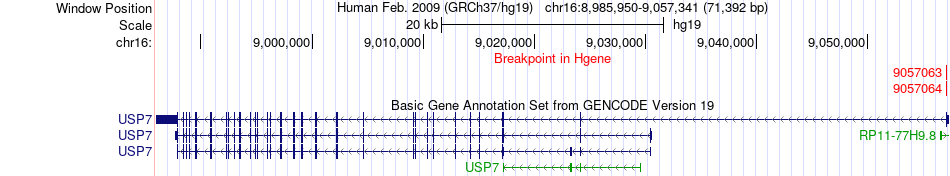 |
 Kinase Fusion gene breakpoints across PRKCB (3'-gene) Kinase Fusion gene breakpoints across PRKCB (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
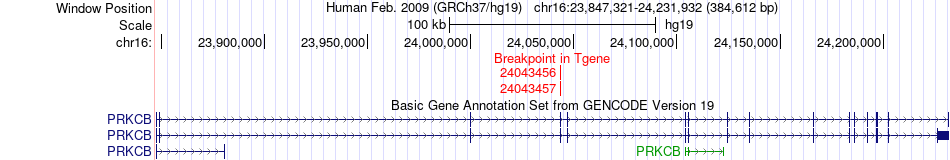 |
Top |
Kinase Fusion Gene Sample Information |
 Kinase Fusion gene information. Kinase Fusion gene information. |
 Kinase Fusion gene information from four resources (ChiTars 5.0, ChimerDB 4.0, COSMIC, and CCLE) Kinase Fusion gene information from four resources (ChiTars 5.0, ChimerDB 4.0, COSMIC, and CCLE)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Sample | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp |
| ChimerDB4 | TCGA-A6-3807-01A | USP7 | chr16 | 9057064 | PRKCB | chr16 | 24043457 |
| ChimerDB4 | TCGA-A6-3807 | USP7 | chr16 | 9057063 | PRKCB | chr16 | 24043456 |
Top |
Kinase Fusion ORF Analysis |
 Kinase Fusion information from ORFfinder translation from full-length transcript sequence from KinaseFusionDB. Kinase Fusion information from ORFfinder translation from full-length transcript sequence from KinaseFusionDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp | Seq length (transcript) | Seq length (amino acids) |
| ENST00000344836 | ENST00000321728 | USP7 | chr16 | 9057063 | PRKCB | chr16 | 24043456 | 2504 | 551 |
| ENST00000344836 | ENST00000321728 | USP7 | chr16 | 9057064 | PRKCB | chr16 | 24043457 | 2504 | 551 |
Top |
Kinase Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from KinaseFusionDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from KinaseFusionDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >Henst_Tenst_Hgene_Hchr_Hbp_Tgene_Tchr_Tbp_length(fusion AA)_AAseq >ENST00000344836_ENST00000321728_USP7_chr16_9057063_PRKCB_chr16_24043456_length(amino acids)=551 MLYGLIHQGMKCDTCMMNVHKRCVMNVPSLCGTDHTERRGRIYIQAHIDRDVLIVLVRDAKNLVPMDPNGLSDPYVKLKLIPDPKSESKQ KTKTIKCSLNPEWNETFRFQLKESDKDRRLSVEIWDWDLTSRNDFMGSLSFGISELQKASVDGWFKLLSQEEGEYFNVPVPPEGSEANEE LRQKFERAKISQGTKVPEEKTTNTVSKFDNNGNRDRMKLTDFNFLMVLGKGSFGKVMLSERKGTDELYAVKILKKDVVIQDDDVECTMVE KRVLALPGKPPFLTQLHSCFQTMDRLYFVMEYVNGGDLMYHIQQVGRFKEPHAVFYAAEIAIGLFFLQSKGIIYRDLKLDNVMLDSEGHI KIADFGMCKENIWDGVTTKTFCGTPDYIAPEIIAYQPYGKSVDWWAFGVLLYEMLAGQAPFEGEDEDELFQSIMEHNVAYPKSMSKEAVA ICKGLMTKHPGKRLGCGPEGERDIKEHAFFRYIDWEKLERKEIQPPYKPKARDKRDTSNFDKEFTRQPVELTPTDKLFIMNLDQNEFAGF -------------------------------------------------------------- >ENST00000344836_ENST00000321728_USP7_chr16_9057064_PRKCB_chr16_24043457_length(amino acids)=551 MLYGLIHQGMKCDTCMMNVHKRCVMNVPSLCGTDHTERRGRIYIQAHIDRDVLIVLVRDAKNLVPMDPNGLSDPYVKLKLIPDPKSESKQ KTKTIKCSLNPEWNETFRFQLKESDKDRRLSVEIWDWDLTSRNDFMGSLSFGISELQKASVDGWFKLLSQEEGEYFNVPVPPEGSEANEE LRQKFERAKISQGTKVPEEKTTNTVSKFDNNGNRDRMKLTDFNFLMVLGKGSFGKVMLSERKGTDELYAVKILKKDVVIQDDDVECTMVE KRVLALPGKPPFLTQLHSCFQTMDRLYFVMEYVNGGDLMYHIQQVGRFKEPHAVFYAAEIAIGLFFLQSKGIIYRDLKLDNVMLDSEGHI KIADFGMCKENIWDGVTTKTFCGTPDYIAPEIIAYQPYGKSVDWWAFGVLLYEMLAGQAPFEGEDEDELFQSIMEHNVAYPKSMSKEAVA ICKGLMTKHPGKRLGCGPEGERDIKEHAFFRYIDWEKLERKEIQPPYKPKARDKRDTSNFDKEFTRQPVELTPTDKLFIMNLDQNEFAGF -------------------------------------------------------------- |
Multiple Sequence Alignment of All Fusion Protein Isoforms |
Top |
Kinase Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr16:9057063/chr16:24043456) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| USP7 | PRKCB |
| FUNCTION: Hydrolase that deubiquitinates target proteins such as FOXO4, DEPTOR, KAT5, p53/TP53, MDM2, ERCC6, DNMT1, UHRF1, PTEN, KMT2E/MLL5 and DAXX (PubMed:11923872, PubMed:15053880, PubMed:16964248, PubMed:18716620, PubMed:25283148, PubMed:25865756, PubMed:26678539, PubMed:28655758, PubMed:35216969). Together with DAXX, prevents MDM2 self-ubiquitination and enhances the E3 ligase activity of MDM2 towards p53/TP53, thereby promoting p53/TP53 ubiquitination and proteasomal degradation (PubMed:15053880, PubMed:16845383, PubMed:18566590, PubMed:20153724). Deubiquitinates p53/TP53, preventing degradation of p53/TP53, and enhances p53/TP53-dependent transcription regulation, cell growth repression and apoptosis (PubMed:25283148). Deubiquitinates p53/TP53 and MDM2 and strongly stabilizes p53/TP53 even in the presence of excess MDM2, and also induces p53/TP53-dependent cell growth repression and apoptosis (PubMed:11923872, PubMed:26786098). Deubiquitination of FOXO4 in presence of hydrogen peroxide is not dependent on p53/TP53 and inhibits FOXO4-induced transcriptional activity (PubMed:16964248). In association with DAXX, is involved in the deubiquitination and translocation of PTEN from the nucleus to the cytoplasm, both processes that are counteracted by PML (PubMed:18716620). Deubiquitinates KMT2E/MLL5 preventing KMT2E/MLL5 proteasomal-mediated degradation (PubMed:26678539). Involved in cell proliferation during early embryonic development. Involved in transcription-coupled nucleotide excision repair (TC-NER) in response to UV damage: recruited to DNA damage sites following interaction with KIAA1530/UVSSA and promotes deubiquitination of ERCC6, preventing UV-induced degradation of ERCC6 (PubMed:22466611, PubMed:22466612). Involved in maintenance of DNA methylation via its interaction with UHRF1 and DNMT1: acts by mediating deubiquitination of UHRF1 and DNMT1, preventing their degradation and promoting DNA methylation by DNMT1 (PubMed:21745816, PubMed:22411829). Deubiquitinates alkylation repair enzyme ALKBH3. OTUD4 recruits USP7 and USP9X to stabilize ALKBH3, thereby promoting the repair of alkylated DNA lesions (PubMed:25944111). Acts as a chromatin regulator via its association with the Polycomb group (PcG) multiprotein PRC1-like complex; may act by deubiquitinating components of the PRC1-like complex (PubMed:20601937). Able to mediate deubiquitination of histone H2B; it is however unsure whether this activity takes place in vivo (PubMed:20601937). Exhibits a preference towards 'Lys-48'-linked ubiquitin chains (PubMed:22689415). Increases regulatory T-cells (Treg) suppressive capacity by deubiquitinating and stabilizing the transcription factor FOXP3 which is crucial for Treg cell function (PubMed:23973222). Plays a role in the maintenance of the circadian clock periodicity via deubiquitination and stabilization of the CRY1 and CRY2 proteins (PubMed:27123980). Deubiquitinates REST, thereby stabilizing REST and promoting the maintenance of neural progenitor cells (PubMed:21258371). Deubiquitinates SIRT7, inhibiting SIRT7 histone deacetylase activity and regulating gluconeogenesis (PubMed:28655758). Involved in the regulation of WASH-dependent actin polymerization at the surface of endosomes and the regulation of endosomal protein recycling (PubMed:26365382). It maintains optimal WASH complex activity and precise F-actin levels via deubiquitination of TRIM27 and WASHC1 (PubMed:26365382). Mediates the deubiquitination of phosphorylated DEPTOR, promoting its stability and leading to decreased mTORC1 signaling (PubMed:35216969). {ECO:0000269|PubMed:11923872, ECO:0000269|PubMed:15053880, ECO:0000269|PubMed:16845383, ECO:0000269|PubMed:16964248, ECO:0000269|PubMed:18566590, ECO:0000269|PubMed:18716620, ECO:0000269|PubMed:20153724, ECO:0000269|PubMed:20601937, ECO:0000269|PubMed:21258371, ECO:0000269|PubMed:21745816, ECO:0000269|PubMed:22411829, ECO:0000269|PubMed:22466611, ECO:0000269|PubMed:22466612, ECO:0000269|PubMed:22689415, ECO:0000269|PubMed:23973222, ECO:0000269|PubMed:25283148, ECO:0000269|PubMed:25865756, ECO:0000269|PubMed:25944111, ECO:0000269|PubMed:26365382, ECO:0000269|PubMed:26678539, ECO:0000269|PubMed:26786098, ECO:0000269|PubMed:27123980, ECO:0000269|PubMed:28655758, ECO:0000269|PubMed:35216969}.; FUNCTION: (Microbial infection) Contributes to the overall stabilization and trans-activation capability of the herpesvirus 1 trans-acting transcriptional protein ICP0/VMW110 during HSV-1 infection. {ECO:0000269|PubMed:14506283, ECO:0000269|PubMed:16160161, ECO:0000269|PubMed:18590780}.; FUNCTION: (Microbial infection) Upon infection with Epstein-Barr virus, the interaction with viral EBNA1 increases the association of USP7 with PML proteins, which is required for the polyubiquitylation and degradation of PML. {ECO:0000269|PubMed:20719947, ECO:0000269|PubMed:24216761}. | FUNCTION: Calcium-activated, phospholipid- and diacylglycerol (DAG)-dependent serine/threonine-protein kinase involved in various cellular processes such as regulation of the B-cell receptor (BCR) signalosome, oxidative stress-induced apoptosis, androgen receptor-dependent transcription regulation, insulin signaling and endothelial cells proliferation. Plays a key role in B-cell activation by regulating BCR-induced NF-kappa-B activation. Mediates the activation of the canonical NF-kappa-B pathway (NFKB1) by direct phosphorylation of CARD11/CARMA1 at 'Ser-559', 'Ser-644' and 'Ser-652'. Phosphorylation induces CARD11/CARMA1 association with lipid rafts and recruitment of the BCL10-MALT1 complex as well as MAP3K7/TAK1, which then activates IKK complex, resulting in nuclear translocation and activation of NFKB1. Plays a direct role in the negative feedback regulation of the BCR signaling, by down-modulating BTK function via direct phosphorylation of BTK at 'Ser-180', which results in the alteration of BTK plasma membrane localization and in turn inhibition of BTK activity (PubMed:11598012). Involved in apoptosis following oxidative damage: in case of oxidative conditions, specifically phosphorylates 'Ser-36' of isoform p66Shc of SHC1, leading to mitochondrial accumulation of p66Shc, where p66Shc acts as a reactive oxygen species producer. Acts as a coactivator of androgen receptor (AR)-dependent transcription, by being recruited to AR target genes and specifically mediating phosphorylation of 'Thr-6' of histone H3 (H3T6ph), a specific tag for epigenetic transcriptional activation that prevents demethylation of histone H3 'Lys-4' (H3K4me) by LSD1/KDM1A (PubMed:20228790). In insulin signaling, may function downstream of IRS1 in muscle cells and mediate insulin-dependent DNA synthesis through the RAF1-MAPK/ERK signaling cascade. Participates in the regulation of glucose transport in adipocytes by negatively modulating the insulin-stimulated translocation of the glucose transporter SLC2A4/GLUT4. Phosphorylates SLC2A1/GLUT1, promoting glucose uptake by SLC2A1/GLUT1 (PubMed:25982116). Under high glucose in pancreatic beta-cells, is probably involved in the inhibition of the insulin gene transcription, via regulation of MYC expression. In endothelial cells, activation of PRKCB induces increased phosphorylation of RB1, increased VEGFA-induced cell proliferation, and inhibits PI3K/AKT-dependent nitric oxide synthase (NOS3/eNOS) regulation by insulin, which causes endothelial dysfunction. Also involved in triglyceride homeostasis (By similarity). Phosphorylates ATF2 which promotes cooperation between ATF2 and JUN, activating transcription (PubMed:19176525). Phosphorylates KLHL3 in response to angiotensin II signaling, decreasing the interaction between KLHL3 and WNK4 (PubMed:25313067). {ECO:0000250|UniProtKB:P68404, ECO:0000269|PubMed:11598012, ECO:0000269|PubMed:19176525, ECO:0000269|PubMed:20228790, ECO:0000269|PubMed:25313067, ECO:0000269|PubMed:25982116}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
 - Retained domain in the 5'-partner of fusion protein (protein functional feature from UniProt). - Retained domain in the 5'-partner of fusion protein (protein functional feature from UniProt). |
| Partner | Hgeneene | Hbp | Tgeneene | Tbp | ENST | BPexon | TotalExon | Protein feature loci | BPloci | TotalLen | Feature | Note |
 - Retained domain in the 3'-partner of fusion protein (protein functional feature from UniProt). - Retained domain in the 3'-partner of fusion protein (protein functional feature from UniProt). |
| Partner | Hgeneene | Hbp | Tgeneene | Tbp | ENST | BPexon | TotalExon | Protein feature loci | BPloci | TotalLen | Feature | Note |
| Tgene | USP7 | 9057063 | PRKCB | 24043456 | ENST00000344836 | 0 | 2 | 601_671 | 0 | 58 | Domain | Note=AGC-kinase C-terminal;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00618 |
| Tgene | USP7 | 9057063 | PRKCB | 24043456 | ENST00000344836 | 2 | 17 | 601_671 | 96 | 672 | Domain | Note=AGC-kinase C-terminal;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00618 |
| Tgene | USP7 | 9057063 | PRKCB | 24043456 | ENST00000344836 | 2 | 17 | 601_671 | 96 | 674 | Domain | Note=AGC-kinase C-terminal;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00618 |
| Tgene | USP7 | 9057064 | PRKCB | 24043457 | ENST00000344836 | 0 | 2 | 601_671 | 0 | 58 | Domain | Note=AGC-kinase C-terminal;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00618 |
| Tgene | USP7 | 9057064 | PRKCB | 24043457 | ENST00000344836 | 2 | 17 | 601_671 | 96 | 672 | Domain | Note=AGC-kinase C-terminal;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00618 |
| Tgene | USP7 | 9057064 | PRKCB | 24043457 | ENST00000344836 | 2 | 17 | 601_671 | 96 | 674 | Domain | Note=AGC-kinase C-terminal;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00618 |
| Tgene | USP7 | 9057063 | PRKCB | 24043456 | ENST00000344836 | 0 | 2 | 158_275 | 0 | 58 | Domain | Note=C2;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00041 |
| Tgene | USP7 | 9057063 | PRKCB | 24043456 | ENST00000344836 | 2 | 17 | 158_275 | 96 | 672 | Domain | Note=C2;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00041 |
| Tgene | USP7 | 9057063 | PRKCB | 24043456 | ENST00000344836 | 2 | 17 | 158_275 | 96 | 674 | Domain | Note=C2;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00041 |
| Tgene | USP7 | 9057064 | PRKCB | 24043457 | ENST00000344836 | 0 | 2 | 158_275 | 0 | 58 | Domain | Note=C2;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00041 |
| Tgene | USP7 | 9057064 | PRKCB | 24043457 | ENST00000344836 | 2 | 17 | 158_275 | 96 | 672 | Domain | Note=C2;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00041 |
| Tgene | USP7 | 9057064 | PRKCB | 24043457 | ENST00000344836 | 2 | 17 | 158_275 | 96 | 674 | Domain | Note=C2;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00041 |
| Tgene | USP7 | 9057063 | PRKCB | 24043456 | ENST00000344836 | 0 | 2 | 342_600 | 0 | 58 | Domain | Note=Protein kinase;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00159 |
| Tgene | USP7 | 9057063 | PRKCB | 24043456 | ENST00000344836 | 2 | 17 | 342_600 | 96 | 672 | Domain | Note=Protein kinase;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00159 |
| Tgene | USP7 | 9057063 | PRKCB | 24043456 | ENST00000344836 | 2 | 17 | 342_600 | 96 | 674 | Domain | Note=Protein kinase;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00159 |
| Tgene | USP7 | 9057064 | PRKCB | 24043457 | ENST00000344836 | 0 | 2 | 342_600 | 0 | 58 | Domain | Note=Protein kinase;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00159 |
| Tgene | USP7 | 9057064 | PRKCB | 24043457 | ENST00000344836 | 2 | 17 | 342_600 | 96 | 672 | Domain | Note=Protein kinase;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00159 |
| Tgene | USP7 | 9057064 | PRKCB | 24043457 | ENST00000344836 | 2 | 17 | 342_600 | 96 | 674 | Domain | Note=Protein kinase;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00159 |
Top |
Kinase Fusion Protein Structures |
 CIF files of the predicted kinase fusion proteins CIF files of the predicted kinase fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Kinase Fusion protein CIF link (fusion AA seq ID in KinaseFusionDB) | Henst | Tenst | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp | AA seq | Len(AA seq) |
| PDB file >>>546_USP7_PRKCB | ENST00000344836 | ENST00000321728 | USP7 | chr16 | 9057064 | PRKCB | chr16 | 24043457 | MLYGLIHQGMKCDTCMMNVHKRCVMNVPSLCGTDHTERRGRIYIQAHIDRDVLIVLVRDAKNLVPMDPNGLSDPYVKLKLIPDPKSESKQ KTKTIKCSLNPEWNETFRFQLKESDKDRRLSVEIWDWDLTSRNDFMGSLSFGISELQKASVDGWFKLLSQEEGEYFNVPVPPEGSEANEE LRQKFERAKISQGTKVPEEKTTNTVSKFDNNGNRDRMKLTDFNFLMVLGKGSFGKVMLSERKGTDELYAVKILKKDVVIQDDDVECTMVE KRVLALPGKPPFLTQLHSCFQTMDRLYFVMEYVNGGDLMYHIQQVGRFKEPHAVFYAAEIAIGLFFLQSKGIIYRDLKLDNVMLDSEGHI KIADFGMCKENIWDGVTTKTFCGTPDYIAPEIIAYQPYGKSVDWWAFGVLLYEMLAGQAPFEGEDEDELFQSIMEHNVAYPKSMSKEAVA ICKGLMTKHPGKRLGCGPEGERDIKEHAFFRYIDWEKLERKEIQPPYKPKARDKRDTSNFDKEFTRQPVELTPTDKLFIMNLDQNEFAGF | 551 |
| 3D view using mol* of 546_USP7_PRKCB | ||||||||||
| PDB file >>>TKFP_935_USP7_PRKCB | ENST00000344836 | ENST00000321728 | USP7 | chr16 | 9057063 | PRKCB | chr16 | 24043456 | MLYGLIHQGMKCDTCMMNVHKRCVMNVPSLCGTDHTERRGRIYIQAHIDRDVLIVLVRDAKNLVPMDPNGLSDPYVKLKLIPDPKSESKQ KTKTIKCSLNPEWNETFRFQLKESDKDRRLSVEIWDWDLTSRNDFMGSLSFGISELQKASVDGWFKLLSQEEGEYFNVPVPPEGSEANEE LRQKFERAKISQGTKVPEEKTTNTVSKFDNNGNRDRMKLTDFNFLMVLGKGSFGKVMLSERKGTDELYAVKILKKDVVIQDDDVECTMVE KRVLALPGKPPFLTQLHSCFQTMDRLYFVMEYVNGGDLMYHIQQVGRFKEPHAVFYAAEIAIGLFFLQSKGIIYRDLKLDNVMLDSEGHI KIADFGMCKENIWDGVTTKTFCGTPDYIAPEIIAYQPYGKSVDWWAFGVLLYEMLAGQAPFEGEDEDELFQSIMEHNVAYPKSMSKEAVA ICKGLMTKHPGKRLGCGPEGERDIKEHAFFRYIDWEKLERKEIQPPYKPKARDKRDTSNFDKEFTRQPVELTPTDKLFIMNLDQNEFAGF | 551_USP7_PRKCB |
| PDB file >>>TKFP_936_USP7_PRKCB | ENST00000344836 | ENST00000321728 | USP7 | chr16 | 9057064 | PRKCB | chr16 | 24043457 | MLYGLIHQGMKCDTCMMNVHKRCVMNVPSLCGTDHTERRGRIYIQAHIDRDVLIVLVRDAKNLVPMDPNGLSDPYVKLKLIPDPKSESKQ KTKTIKCSLNPEWNETFRFQLKESDKDRRLSVEIWDWDLTSRNDFMGSLSFGISELQKASVDGWFKLLSQEEGEYFNVPVPPEGSEANEE LRQKFERAKISQGTKVPEEKTTNTVSKFDNNGNRDRMKLTDFNFLMVLGKGSFGKVMLSERKGTDELYAVKILKKDVVIQDDDVECTMVE KRVLALPGKPPFLTQLHSCFQTMDRLYFVMEYVNGGDLMYHIQQVGRFKEPHAVFYAAEIAIGLFFLQSKGIIYRDLKLDNVMLDSEGHI KIADFGMCKENIWDGVTTKTFCGTPDYIAPEIIAYQPYGKSVDWWAFGVLLYEMLAGQAPFEGEDEDELFQSIMEHNVAYPKSMSKEAVA ICKGLMTKHPGKRLGCGPEGERDIKEHAFFRYIDWEKLERKEIQPPYKPKARDKRDTSNFDKEFTRQPVELTPTDKLFIMNLDQNEFAGF | 551_USP7_PRKCB |
Top |
Comparison of Fusion Protein Isoforms |
 Superimpose the 3D Structures Among All Fusion Protein Isoforms Superimpose the 3D Structures Among All Fusion Protein Isoforms * Download the pdb file and open it from the molstar online viewer. |
 Comparison of the Secondary Structures of Fusion Protein Isoforms Comparison of the Secondary Structures of Fusion Protein Isoforms |
Top |
Comparison of Fusion Protein Sequences/Structures with Known Sequences/Structures from PDB |
Top |
pLDDT score distribution |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. * The blue color at the bottom marks the best active site residues. |
| 546_USP7_PRKCB.png |
 |
| 546_USP7_PRKCB.png |
 |
Top |
Potential Active Site Information |
 The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. |
| Kinase Fusion AA seq ID in KinaseFusionDB | Site score | Size | Dscore | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
Top |
Ramachandran Plot of Kinase Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| 546_USP7_PRKCB_ramachandran.png |
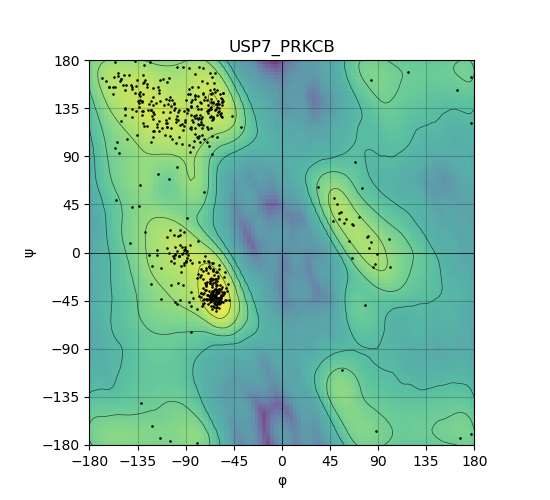 |
Top |
Virtual Screening Results |
 Distribution of the average docking score across all approved kinase inhibitors. Distribution of the average docking score across all approved kinase inhibitors.Distribution of the number of occurrence across all approved kinase inhibitors. |
| 5'-kinase fusion protein case |
| 3'-kinase fusion protein case |
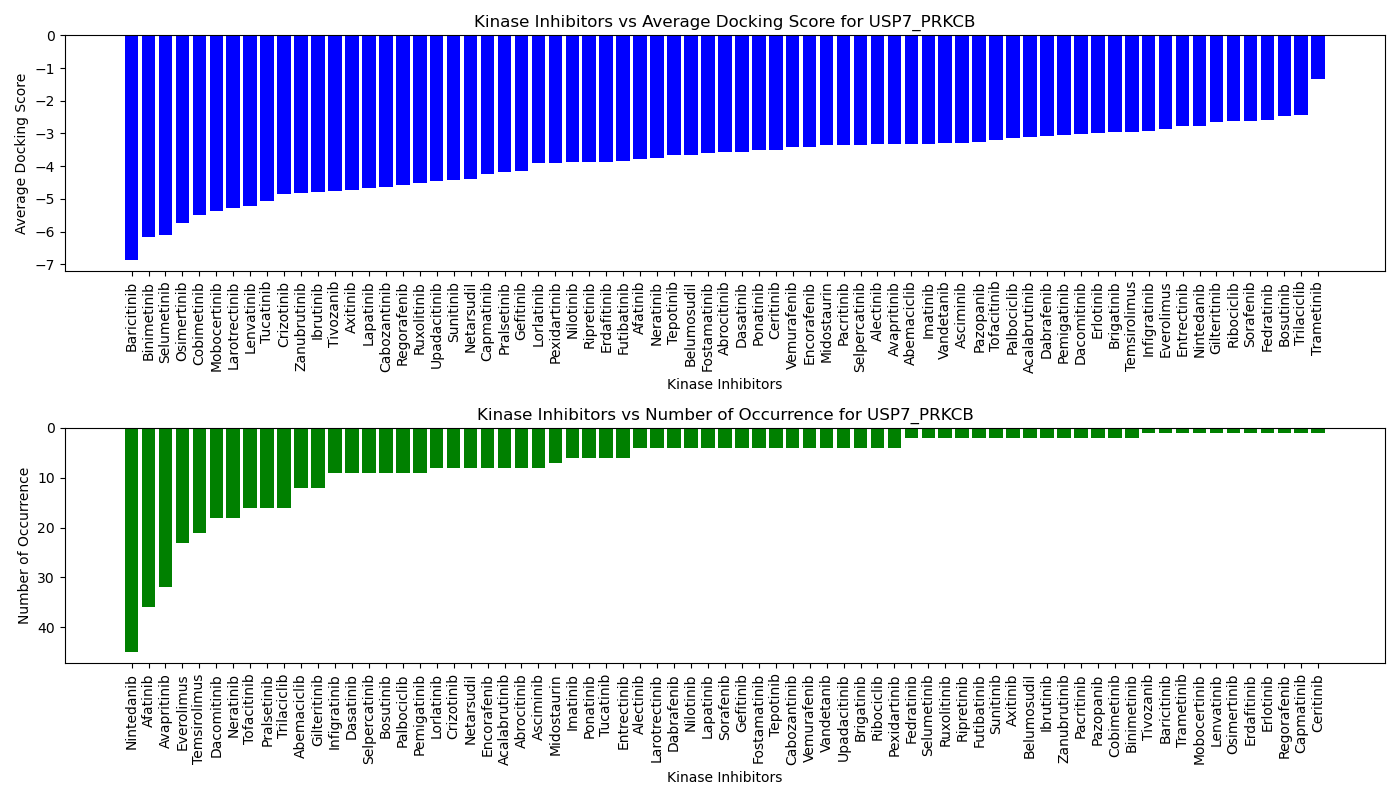 |
Top |
 Drug information from DrugBank of the top 20 interacting small molecules. Drug information from DrugBank of the top 20 interacting small molecules.* The detailed information of individual kinase inhibitors are available in the download page. |
| Fusion gene name info | Drug | Docking score | Glide g score | Glide energy |
| 546_USP7_PRKCB-DOCK_HTVS_1-001 | Baricitinib | -6.85982 | -6.85982 | -41.7277 |
| 546_USP7_PRKCB-DOCK_HTVS_1-001 | Afatinib | -6.414569999999999 | -6.59687 | -50.5344 |
| 546_USP7_PRKCB-DOCK_HTVS_1-001 | Afatinib | -6.414569999999999 | -6.59687 | -50.5344 |
| 546_USP7_PRKCB-DOCK_HTVS_1-001 | Afatinib | -6.41317 | -6.59687 | -50.5344 |
| 546_USP7_PRKCB-DOCK_HTVS_1-001 | Lapatinib | -6.2001800000000005 | -6.2889800000000005 | -58.3708 |
| 546_USP7_PRKCB-DOCK_HTVS_1-001 | Binimetinib | -6.15665 | -6.16535 | -53.1595 |
| 546_USP7_PRKCB-DOCK_HTVS_1-001 | Binimetinib | -6.15665 | -6.16535 | -53.1595 |
| 546_USP7_PRKCB-DOCK_HTVS_1-001 | Selumetinib | -6.10584 | -6.11454 | -55.4153 |
| 546_USP7_PRKCB-DOCK_HTVS_1-001 | Selumetinib | -6.10584 | -6.11454 | -55.4153 |
| 546_USP7_PRKCB-DOCK_HTVS_1-001 | Neratinib | -5.939 | -6.1249 | -56.6159 |
| 546_USP7_PRKCB-DOCK_HTVS_1-001 | Cabozantinib | -5.914969999999999 | -5.959969999999999 | -47.3991 |
| 546_USP7_PRKCB-DOCK_HTVS_1-001 | Cabozantinib | -5.914969999999999 | -5.959969999999999 | -47.3991 |
| 546_USP7_PRKCB-DOCK_HTVS_1-001 | Larotrectinib | -5.89876 | -5.89876 | -44.8851 |
| 546_USP7_PRKCB-DOCK_HTVS_1-001 | Osimertinib | -5.75093 | -5.75863 | -54.1605 |
| 546_USP7_PRKCB-DOCK_HTVS_1-001 | Afatinib | -5.7121699999999995 | -5.89447 | -52.1349 |
| 546_USP7_PRKCB-DOCK_HTVS_1-001 | Afatinib | -5.7121699999999995 | -5.89447 | -52.1349 |
| 546_USP7_PRKCB-DOCK_HTVS_1-001 | Afatinib | -5.71077 | -5.89447 | -52.1349 |
| 546_USP7_PRKCB-DOCK_HTVS_1-001 | Pralsetinib | -5.69971 | -5.7912099999999995 | -59.2513 |
| 546_USP7_PRKCB-DOCK_HTVS_1-001 | Tucatinib | -5.6771199999999995 | -6.05232 | -49.0189 |
| 546_USP7_PRKCB-DOCK_HTVS_1-001 | Tucatinib | -5.6771199999999995 | -6.05232 | -49.0189 |
Top |
Kinase-Substrate Information of USP7_PRKCB |
 Phosphorylation target of the kinase Phosphorylation target of the kinase(phosphosite, 03-17-2024) |
| Kinase | Kinase UniProt Acc | Kinase species | Substrate | Substrate UniProt Acc | Substrate phosphorylated residues | Substrate phosphorylated sites (+/-7AA) | Domain |
| PRKCB | P05771-2 | human | MAPT | P10636-8 | S352 | DFKDrVQskIGsLDN | Tubulin-binding |
| PRKCB | P05771-2 | human | ITGB2 | P05107 | S745 | FEkEKLksQWNNDNP | Integrin_b_cyt |
| PRKCB | P05771-2 | human | TRPV1 | Q8NER1 | T705 | WKLQRAItILDTEKS | |
| PRKCB | P05771-2 | human | DAB2 | P98082 | S24 | QAAPKAPsKKEKKKG | |
| PRKCB | P05771-2 | human | PRKCB | P05771-2 | S660 | QsEFEGFsFVNsEFL | Pkinase_C |
| PRKCB | P05771-2 | human | MAPT | P10636-8 | S258 | PDLkNVKskIGstEN | Tubulin-binding |
| PRKCB | P05771-2 | human | PPARA | Q07869 | S230 | VKARVILsGKASNNP | |
| PRKCB | P05771-2 | human | ATF2 | P15336 | S121 | LAtPIIRsKIEEPSV | |
| PRKCB | P05771-2 | human | KIR3DL1 | P43629 | S415 | QRKITRPsQRPKtPP | |
| PRKCB | P05771-2 | human | EIF6 | P56537 | S235 | QPstIAtsMRDsLID | |
| PRKCB | P05771-2 | human | PTPN11 | Q06124 | S591 | GLMQQQksFR_____ | |
| PRKCB | P05771-2 | human | GSK3B | P49841 | S9 | SGRPRttsFAEsCkP | |
| PRKCB | P05771-2 | human | MYH9 | P35579 | S1916 | AMNREVssLkNkLRr | Myosin_tail_1 |
| PRKCB | P05771-2 | human | RPS6KB2 | Q9UBS0 | S473 | PPSGTKKsKRGRGRP | |
| PRKCB | P05771-2 | human | ITGB2 | P05107 | T758 | NPLFksAtttVMNPk | Integrin_b_cyt |
| PRKCB | P05771-2 | human | TRPV6 | Q9H1D0 | S184 | LARRAsVsARAtGTA | Ank_2 |
| PRKCB | P05771-2 | human | C5AR1 | P21730 | S334 | sVVREsKsFTRsTVD | |
| PRKCB | P05771-2 | human | NCF1 | P14598 | S320 | QRsRKRLsQDAYRRN | NECFESHC |
| PRKCB | P05771-2 | human | MFN1 | Q8IWA4 | S284 | QNRIFFVsAkEVLsA | |
| PRKCB | P05771-2 | human | EIF3A | Q14152 | S1364 | RAEKDREsLRRtKNE | |
| PRKCB | P05771-2 | human | LMNB1 | P20700 | S405 | VtVsRAsssRsVRtt | |
| PRKCB | P05771-2 | human | SYT6 | Q5T7P8 | T418 | RLKKKKttIKKNTLN | C2 |
| PRKCB | P05771-2 | human | MCU | Q8NE86 | S92 | VISVRLPsRRERCQF | |
| PRKCB | P05771-2 | human | EIF4G1 | Q04637-8 | S1093 | FAPGGRLsWGKGSSG | |
| PRKCB | P05771-2 | human | MFN1 | Q8IWA4 | S290 | VsAkEVLsARKQKAQ | |
| PRKCB | P05771-2 | human | PTPN11 | Q06124 | S576 | CAEMREDsARVyENV | |
| PRKCB | P05771-2 | human | NCF1 | P14598 | S315 | AHsIHQRsRKRLsQD | NECFESHC |
| PRKCB | P05771-2 | human | TRPV6 | Q9H1D0 | T728 | MPSVSRstSRssANW | |
| PRKCB | P05771-2 | human | RAB11A | P62491 | S177 | TEIYRIVsQkQMSDR | |
| PRKCB | P05771-2 | human | PPARA | Q07869 | S179 | RFGRMPRsEKAKLkA | |
| PRKCB | P05771-2 | human | NCF1 | P14598 | S328 | QDAYRRNsVRFLQQR | NECFESHC |
| PRKCB | P05771-2 | human | MAPT | P10636-8 | S324 | kVTskCGsLGNIHHk | Tubulin-binding |
| PRKCB | P05771-2 | human | TP73 | O15350 | S388 | VPQPLVDsYRQQQQL | |
| PRKCB | P05771-2 | human | CHAT | P28329-3 | S476 | HKAAVPAsEKLLLLK | Carn_acyltransf |
| PRKCB | P05771-2 | human | LMNB1 | P20700 | S395 | LkLsPsPssRVtVsR | |
| PRKCB | P05771-2 | human | CHAT | P28329-3 | S346 | LLKHVTQssRKLIRA | Carn_acyltransf |
| PRKCB | P05771-2 | human | CHAT | P28329-3 | S440 | VPTYESAsIRRFQEG | Carn_acyltransf |
| PRKCB | P05771-2 | human | FUS | P35637 | S257 | GrGGMGGsDrGGFNk | |
| PRKCB | P05771-2 | human | SYT6 | Q5T7P8 | T417 | RRLKKKKttIKKNTL | C2 |
| PRKCB | P05771-2 | human | CHAT | P28329-3 | S347 | LKHVTQssRKLIRAD | Carn_acyltransf |
| PRKCB | P05771-2 | human | CHAT | P28329-3 | T255 | TVLVKDStNRDSLDM | Carn_acyltransf |
| PRKCB | P05771-2 | human | NCF1 | P14598 | S370 | PAVPPRPsADLILNR | p47_phox_C |
| PRKCB | P05771-2 | human | NCF1 | P14598 | S303 | RGAPPRRssIRNAHs | NECFESHC |
| PRKCB | P05771-2 | human | GHRL | Q9UBU3 | S41 | RVQQRKEsKKPPAKL | Motilin_ghrelin |
| PRKCB | P05771-2 | human | MAPT | P10636-8 | S293 | NVQskCGsKDNIkHV | Tubulin-binding |
| PRKCB | P05771-2 | human | KCNE1 | P15382 | S102 | VQARVLEsYRSCYVV | ISK_Channel |
| PRKCB | P05771-2 | human | NCF1 | P14598 | S359 | EERQtQRsKPQPAVP | p47_phox_C |
| PRKCB | P05771-2 | human | NCF1 | P14598 | S304 | GAPPRRssIRNAHsI | NECFESHC |
| PRKCB | P05771-2 | human | MFN1 | Q8IWA4 | S86 | AFFGRTSsGKSSVIN | Dynamin_N |
| PRKCB | P05771-2 | human | GSK3A | P49840 | S21 | sGrARtssFAEPGGG | |
| PRKCB | P05771-2 | human | NCF1 | P14598 | S379 | DLILNRCsEstKRKL | p47_phox_C |
| PRKCB | P05771-2 | human | SYT6 | Q5T7P8 | T283 | DRKCKLQtRVHRKTL | C2 |
| PRKCB | P05771 | human | ANXA1 | P04083 | S28 | yVQtVksskGGPGsA | |
| PRKCB | P05771 | human | MAPT | P10636-8 | S352 | DFKDrVQskIGsLDN | Tubulin-binding |
| PRKCB | P05771 | human | ITGB2 | P05107 | S745 | FEkEKLksQWNNDNP | Integrin_b_cyt |
| PRKCB | P05771 | human | PRKCB | P05771-2 | S660 | QsEFEGFsFVNsEFL | Pkinase_C |
| PRKCB | P05771 | human | PTPN11 | Q06124 | S591 | GLMQQQksFR_____ | |
| PRKCB | P05771 | human | MYH9 | P35579 | S1916 | AMNREVssLkNkLRr | Myosin_tail_1 |
| PRKCB | P05771 | human | ANXA2 | P07355 | S26 | tPPsAyGsVkAytNF | |
| PRKCB | P05771 | human | LIN28B | Q6ZN17 | S243 | EQSKkGPsVQKRKKT | |
| PRKCB | P05771 | human | ORAI1 | Q96D31 | S27 | GGSTTsGsRRsRRRs | |
| PRKCB | P05771 | human | KCNC4 | Q03721 | S9 | ISSVCVssYRGRKsG | Potassium_chann |
| PRKCB | P05771 | human | ITGB2 | P05107 | T758 | NPLFksAtttVMNPk | Integrin_b_cyt |
| PRKCB | P05771 | human | PIK3CG | P48736 | S582 | LWHFRYEsLKHPKAY | PI3Ka |
| PRKCB | P05771 | human | AKT1 | P31749 | S473 | RPHFPQFsysAsGtA | Pkinase_C |
| PRKCB | P05771 | human | NCF1 | P14598 | S320 | QRsRKRLsQDAYRRN | NECFESHC |
| PRKCB | P05771 | human | IBTK | Q9P2D0 | S1200 | ASSLHsVsSksFRDF | |
| PRKCB | P05771 | human | EPHA2 | P29317 | S892 | ADFDPRVsIRLPsts | |
| PRKCB | P05771 | human | KCNC4 | Q03721 | S21 | KsGNKPPsKTCLKEE | Potassium_chann |
| PRKCB | P05771 | human | PRKAA1 | Q13131 | S496 | AtPQRsGsVsNyRSC | AdenylateSensor |
| PRKCB | P05771 | human | PTPN11 | Q06124 | S576 | CAEMREDsARVyENV | |
| PRKCB | P05771 | human | NOX5 | Q96PH1-4 | T494 | KRLsRSVtMRKsQRS | FAD_binding_8 |
| PRKCB | P05771 | human | EPB41 | P11171-4 | S312 | QAQTRQAsALIDRPA | FA |
| PRKCB | P05771 | human | STMN1 | P16949 | S25 | QAFELILsPrskEsV | Stathmin |
| PRKCB | P05771 | human | TYR | P14679 | S527 | DYHsLYQsHL_____ | |
| PRKCB | P05771 | human | MAPT | P10636-8 | S324 | kVTskCGsLGNIHHk | Tubulin-binding |
| PRKCB | P05771 | human | MARCKS | P29966 | S163 | KRFsFkKsFkLsGFs | MARCKS |
| PRKCB | P05771 | human | CHAT | P28329-3 | S440 | VPTYESAsIRRFQEG | Carn_acyltransf |
| PRKCB | P05771 | human | STMN1 | P16949 | S38 | sVPEFPLsPPkKkDL | Stathmin |
| PRKCB | P05771 | human | MSX2 | P35548 | T135 | HMsPTTCtLRKHKtN | |
| PRKCB | P05771 | human | CHAT | P28329-3 | S347 | LKHVTQssRKLIRAD | Carn_acyltransf |
| PRKCB | P05771 | human | CHAT | P28329-3 | T255 | TVLVKDStNRDSLDM | Carn_acyltransf |
| PRKCB | P05771 | human | TYR | P14679 | S523 | MEKEDYHsLYQsHL_ | |
| PRKCB | P05771 | human | MAPT | P10636-8 | S293 | NVQskCGsKDNIkHV | Tubulin-binding |
| PRKCB | P05771 | human | NCF1 | P14598 | S304 | GAPPRRssIRNAHsI | NECFESHC |
| PRKCB | P05771 | human | SHC1 | P29353 | T206 | VPGAkGAtRRRKPCs | PID |
| PRKCB | P05771 | human | GSK3A | P49840 | S21 | sGrARtssFAEPGGG | |
| PRKCB | P05771 | human | IBTK | Q9P2D0 | S1203 | LHsVsSksFRDFLLE | |
| PRKCB | P05771 | human | TRPM8 | Q7Z2W7 | S1040 | CKEKNMEssVCCFKN | |
| PRKCB | P05771 | human | MARCKS | P29966 | S159 | kkKKKRFsFkKsFkL | MARCKS |
| PRKCB | P05771 | human | NOX5 | Q96PH1-4 | S498 | RSVtMRKsQRSsKGS | FAD_binding_8 |
| PRKCB | P05771 | human | GRN | P28799 | S81 | IFTVSGTsSCCPFPE | Granulin |
| PRKCB | P05771 | human | SHC1 | P29353 | S213 | tRRRKPCsRPLSSIL | PID |
| PRKCB | P05771 | human | MAPT | P10636-8 | S258 | PDLkNVKskIGstEN | Tubulin-binding |
| PRKCB | P05771 | human | KCNC4 | Q03721 | S8 | MISSVCVssYRGRKs | Potassium_chann |
| PRKCB | P05771 | human | ATF2 | P15336 | S121 | LAtPIIRsKIEEPSV | |
| PRKCB | P05771 | human | GSK3B | P49841 | S9 | SGRPRttsFAEsCkP | |
| PRKCB | P05771 | human | OCLN | Q16625 | S490 | RLKQVkGsADYKSKK | Occludin_ELL |
| PRKCB | P05771 | human | RPS6KB2 | Q9UBS0 | S473 | PPSGTKKsKRGRGRP | |
| PRKCB | P05771 | human | ORAI1 | Q96D31 | S30 | TTsGsRRsRRRsGDG | |
| PRKCB | P05771 | human | MSX2 | P35548 | T141 | CtLRKHKtNRKPRTP | |
| PRKCB | P05771 | human | CFLAR | O15519-2 | S193 | LQAAIQKsLKDPSNN | |
| PRKCB | P05771 | human | C5AR1 | P21730 | S334 | sVVREsKsFTRsTVD | |
| PRKCB | P05771 | human | KCNC4 | Q03721 | S15 | ssYRGRKsGNKPPsK | Potassium_chann |
| PRKCB | P05771 | human | CLDN1 | O95832 | T191 | CSCPRkTtsYPtPRP | |
| PRKCB | P05771 | human | KLHL3 | Q9UH77 | S433 | PMNTRRSsVGVGVVE | Kelch_1 |
| PRKCB | P05771 | human | IRS1 | P35568 | S323 | MVGGKPGsFRVRAss | |
| PRKCB | P05771 | human | SHC1 | P29353 | S139 | EEWTRHGsFVNkPtR | |
| PRKCB | P05771 | human | NCF1 | P14598 | S315 | AHsIHQRsRKRLsQD | NECFESHC |
| PRKCB | P05771 | human | CFLAR | O15519 | S193 | LQAAIQksLkDPSNN | |
| PRKCB | P05771 | human | TRPM8 | Q7Z2W7 | S1041 | KEKNMEssVCCFKNE | |
| PRKCB | P05771 | human | NCF1 | P14598 | S328 | QDAYRRNsVRFLQQR | NECFESHC |
| PRKCB | P05771 | human | CHAT | P28329-3 | S476 | HKAAVPAsEKLLLLK | Carn_acyltransf |
| PRKCB | P05771 | human | CHAT | P28329-3 | S346 | LLKHVTQssRKLIRA | Carn_acyltransf |
| PRKCB | P05771 | human | ILF3 | Q12906 | S647 | rGrGRGGsIRGRGRG | |
| PRKCB | P05771 | human | H3C1 | P68431 | T6 | __ArtkQtArkstGG | Histone |
| PRKCB | P05771 | human | FCER1G | P30273 | S69 | DGVytGLstRNQEty | ITAM |
| PRKCB | P05771 | human | IRS1 | P35568 | S441 | SPCDFRSsFRsVtPD | |
| PRKCB | P05771 | human | ACSL4 | O60488 | T328 | LAHVLELtAEISCFT | AMP-binding |
| PRKCB | P05771 | human | AKT1 | P31749 | T308 | kDGAtMKtFCGtPEy | Pkinase |
 Biological Network Integration of This Kinase and Substrates Biological Network Integration of This Kinase and Substrates (GeneMANIA website) |
 Enriched GO biological processes of the phosphorylation target genes of the kinase Enriched GO biological processes of the phosphorylation target genes of the kinase |
| Kinase | GOID | GO term | P.adjust |
| PRKCB | ID | Description | 0.00e+00 |
| PRKCB | GO:0043434 | response to peptide hormone | 6.72e-08 |
| PRKCB | GO:1903829 | positive regulation of protein localization | 1.29e-07 |
| PRKCB | GO:0032868 | response to insulin | 3.59e-07 |
| PRKCB | GO:0008286 | insulin receptor signaling pathway | 2.13e-06 |
| PRKCB | GO:0097284 | hepatocyte apoptotic process | 2.13e-06 |
| PRKCB | GO:0032869 | cellular response to insulin stimulus | 4.55e-06 |
| PRKCB | GO:0046879 | hormone secretion | 8.73e-06 |
| PRKCB | GO:0071375 | cellular response to peptide hormone stimulus | 8.73e-06 |
| PRKCB | GO:0009914 | hormone transport | 1.02e-05 |
| PRKCB | GO:0002790 | peptide secretion | 1.27e-05 |
| PRKCB | GO:0046883 | regulation of hormone secretion | 1.37e-05 |
| PRKCB | GO:0015833 | peptide transport | 1.81e-05 |
| PRKCB | GO:0010827 | regulation of glucose transmembrane transport | 2.50e-05 |
| PRKCB | GO:0030073 | insulin secretion | 2.60e-05 |
| PRKCB | GO:1901653 | cellular response to peptide | 2.70e-05 |
| PRKCB | GO:0023061 | signal release | 2.82e-05 |
| PRKCB | GO:1903532 | positive regulation of secretion by cell | 2.82e-05 |
| PRKCB | GO:0043467 | regulation of generation of precursor metabolites and energy | 2.82e-05 |
| PRKCB | GO:0046887 | positive regulation of hormone secretion | 2.95e-05 |
| PRKCB | GO:0045834 | positive regulation of lipid metabolic process | 3.39e-05 |
| PRKCB | GO:1903828 | negative regulation of protein localization | 3.49e-05 |
| PRKCB | GO:0051222 | positive regulation of protein transport | 3.89e-05 |
| PRKCB | GO:0051047 | positive regulation of secretion | 3.90e-05 |
| PRKCB | GO:0001774 | microglial cell activation | 3.90e-05 |
| PRKCB | GO:0042060 | wound healing | 4.85e-05 |
| PRKCB | GO:0002269 | leukocyte activation involved in inflammatory response | 4.85e-05 |
| PRKCB | GO:1904951 | positive regulation of establishment of protein localization | 4.85e-05 |
| PRKCB | GO:0002274 | myeloid leukocyte activation | 5.41e-05 |
| PRKCB | GO:0030072 | peptide hormone secretion | 5.41e-05 |
| PRKCB | GO:0050796 | regulation of insulin secretion | 5.43e-05 |
| PRKCB | GO:0032386 | regulation of intracellular transport | 5.72e-05 |
| PRKCB | GO:0006816 | calcium ion transport | 6.04e-05 |
| PRKCB | GO:0061900 | glial cell activation | 6.04e-05 |
| PRKCB | GO:0019216 | regulation of lipid metabolic process | 6.04e-05 |
| PRKCB | GO:0006887 | exocytosis | 6.94e-05 |
| PRKCB | GO:0035265 | organ growth | 7.07e-05 |
| PRKCB | GO:0046324 | regulation of glucose import | 7.07e-05 |
| PRKCB | GO:0042886 | amide transport | 7.20e-05 |
| PRKCB | GO:0050994 | regulation of lipid catabolic process | 7.28e-05 |
| PRKCB | GO:0051098 | regulation of binding | 7.36e-05 |
| PRKCB | GO:1904659 | glucose transmembrane transport | 8.83e-05 |
| PRKCB | GO:0006109 | regulation of carbohydrate metabolic process | 9.31e-05 |
| PRKCB | GO:0008645 | hexose transmembrane transport | 9.76e-05 |
| PRKCB | GO:0015749 | monosaccharide transmembrane transport | 1.10e-04 |
| PRKCB | GO:0090276 | regulation of peptide hormone secretion | 1.11e-04 |
| PRKCB | GO:0046626 | regulation of insulin receptor signaling pathway | 1.15e-04 |
| PRKCB | GO:0002791 | regulation of peptide secretion | 1.17e-04 |
| PRKCB | GO:0005979 | regulation of glycogen biosynthetic process | 1.17e-04 |
| PRKCB | GO:0010962 | regulation of glucan biosynthetic process | 1.17e-04 |
Top |
Related Drugs to USP7_PRKCB |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
 Distribution of the number of studies mentioning USP7-PRKCB and kinase inhibitors the PubMed Abstract (04-01-2024) Distribution of the number of studies mentioning USP7-PRKCB and kinase inhibitors the PubMed Abstract (04-01-2024) |
| Fusion gene - drug pair 1 | Fusion gene - drug pair 2 | PMID | Publication date | DOI | Study title |
Top |
Related Diseases to USP7_PRKCB |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Related diseases from the literature mentioned this fusion gene and drug. Related diseases from the literature mentioned this fusion gene and drug. (PubMed, 04-01-2024) |
| MeSH ID | MeSH term |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
Top |
Clinical Trials of the Found Drugs/Small Molecules |
 Statistics of the Clinical Trials of the Found Kinase Inibitors from clinicaltrials.gov (06-17-2024) Statistics of the Clinical Trials of the Found Kinase Inibitors from clinicaltrials.gov (06-17-2024) |
 Clinical Trials from clinicaltrials.gov (06-17-2024) Clinical Trials from clinicaltrials.gov (06-17-2024) |
| Fusion Gene | Kinase Inhibitor | NCT ID | Study Status | Phases | Disease | # Enrolment | Date |

