| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Kinase Fusion Gene:CARS_ALK |
Kinase Fusion Protein Summary |
 Kinase Fusion gene summary Kinase Fusion gene summary |
| Kinase Fusion partner gene information | Kinase Fusion gene name: CARS_ALK | KinaseFusionDB ID: KFG917 | FusionGDB2.0 ID: KFG917 | Hgene | Tgene | Gene symbol | CARS | ALK | Gene ID | 833 | 238 | |
| Gene name | cysteinyl-tRNA synthetase 1 | ALK receptor tyrosine kinase | ||||||||||
| Synonyms | CARS|CYSRS|MCDDBH|MDBH|MGC:11246 | ALK1|CD246|NBLST3 | ||||||||||
| Cytomap | 11p15.4 | 2p23.2-p23.1 | ||||||||||
| Type of gene | protein-coding | protein-coding | ||||||||||
| Description | cysteine--tRNA ligase, cytoplasmiccysteine tRNA ligase 1, cytoplasmiccysteine translase | ALK tyrosine kinase receptorCD246 antigenanaplastic lymphoma receptor tyrosine kinasemutant anaplastic lymphoma kinase | ||||||||||
| Modification date | 20240403 | 20240411 | ||||||||||
| UniProtAcc | P49589 | Q3KRA9 | ||||||||||
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000278224, ENST00000380525, ENST00000397111, ENST00000397114, ENST00000401769, ENST00000470221, | ENST00000431873, ENST00000498037, ENST00000389048, | |||||||||
| Context (manual curation of fusion genes in KinaseFusionDB) | PubMed: CARS [Title/Abstract] AND ALK [Title/Abstract] AND fusion [Title/Abstract] | |||||||||||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | ||||||||||||
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CARS | GO:0006423 | cysteinyl-tRNA aminoacylation | 10908348|17303165 |
| Tgene | ALK | GO:0007169 | cell surface receptor protein tyrosine kinase signaling pathway | 25605972|30061385|34646012 |
| Tgene | ALK | GO:0016310 | phosphorylation | 9174053 |
| Tgene | ALK | GO:0038083 | peptidyl-tyrosine autophosphorylation | 30061385|34646012 |
| Tgene | ALK | GO:0046777 | protein autophosphorylation | 9174053 |
| Tgene | ALK | GO:0046777 | protein autophosphorylation | 9174053 |
 Kinase Fusion gene breakpoints across CARS (5'-gene) Kinase Fusion gene breakpoints across CARS (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 Kinase Fusion gene breakpoints across ALK (3'-gene) Kinase Fusion gene breakpoints across ALK (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
Top |
Kinase Fusion Gene Sample Information |
 Kinase Fusion gene information. Kinase Fusion gene information. |
 Kinase Fusion gene information from four resources (ChiTars 5.0, ChimerDB 4.0, COSMIC, and CCLE) Kinase Fusion gene information from four resources (ChiTars 5.0, ChimerDB 4.0, COSMIC, and CCLE)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Sample | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp |
| ChimerKB3 | . | CARS | chr11 | 3033424 | ALK | chr2 | 29446394 |
| ChimerKB3 | . | CARS | chr11 | 3037010 | ALK | chr2 | 29446394 |
| ChimerKB3 | . | CARS | chr11 | 3038335 | ALK | chr2 | 29445473 |
| COSMIC | 1152148 | CARS | chr11 | 3033425 | ALK | chr2 | 29446394 |
| COSMIC | 1152149 | CARS | chr11 | 3032308 | ALK | chr2 | 29446623 |
| COSMIC | 1152150 | CARS | chr11 | 3032308 | ALK | chr2 | 29446623 |
Top |
Kinase Fusion ORF Analysis |
 Kinase Fusion information from ORFfinder translation from full-length transcript sequence from KinaseFusionDB. Kinase Fusion information from ORFfinder translation from full-length transcript sequence from KinaseFusionDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp | Seq length (transcript) | Seq length (amino acids) |
| ENST00000380525 | ENST00000389048 | CARS | chr11 | 3033425 | ALK | chr2 | 29446394 | 4279 | 1252 |
| ENST00000380525 | ENST00000389048 | CARS | chr11 | 3033424 | ALK | chr2 | 29446394 | 4279 | 1252 |
| ENST00000401769 | ENST00000389048 | CARS | chr11 | 3037010 | ALK | chr2 | 29446394 | 4076 | 614 |
Top |
Kinase Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from KinaseFusionDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from KinaseFusionDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >Henst_Tenst_Hgene_Hchr_Hbp_Tgene_Tchr_Tbp_length(fusion AA)_AAseq >ENST00000380525_ENST00000389048_CARS_chr11_3033425_ALK_chr2_29446394_length(amino acids)=1252 MADSSGQQAPDYRSILSISDEAARAQALNEHLSTRSYVQGYSLSQADVDAFRQLSAPPADPQLFHVARWFRHIEALLGSPCGKGQPCRLQ ASKGRRVQPQWSPPAGTQPCRLHLYNSLTRNKEVFIPQDGKKVTWYCCGPTVYDASHMGHARSYISFDILRRVLKDYFKFDVFYCMNITD IDDKIIKRARQNHLFEQYREKRPEAAQLLEDVQAALKPFSVKLNETTDPDKKQMLERIQHAVQLATEPLEKAVQSRLTGEEVNSCVEVLL EEAKDLLSDWLDSTLGCDVTDNSIFSKLPKFWEGDFHRDMEALNVLPPDVLTRVSEYVPEIVNFVQKIVDNGYGYVSNGSVYFDTAKFAS SEKHSYGKLVPEAVGDQKALQEGEGDLSISADRLSEKRSPNDFALWKASKPGEPSWPCPWGKGRPGWHIECSAMAGTLLGASMDIHGGGF DLRFPHHDNELAQSEAYFENDCWVRYFLHTGHLTIAGCKMSKSLKNFITIKDALKKHSARQLRLAFLMHSWKDTLDYSSNTMESALQYEK FLNEFFLNVKDILRAPVDITGQFEKWGEEEAELNKNFYDKKTAIHKALCDNVDTRTVMEEMRALVSQCNLYMAARKAVRKRPNQALLENI ALYLTHMLKIFGAVEEDSSLGFPVGGPGTSLSLEATVMPYLQVLSEFREGVRKIAREQKVYRRKHQELQAMQMELQSPEYKLSKLRTSTI MTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQN IVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKI GDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPV YRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTS SGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGS -------------------------------------------------------------- >ENST00000380525_ENST00000389048_CARS_chr11_3033424_ALK_chr2_29446394_length(amino acids)=1252 MADSSGQQAPDYRSILSISDEAARAQALNEHLSTRSYVQGYSLSQADVDAFRQLSAPPADPQLFHVARWFRHIEALLGSPCGKGQPCRLQ ASKGRRVQPQWSPPAGTQPCRLHLYNSLTRNKEVFIPQDGKKVTWYCCGPTVYDASHMGHARSYISFDILRRVLKDYFKFDVFYCMNITD IDDKIIKRARQNHLFEQYREKRPEAAQLLEDVQAALKPFSVKLNETTDPDKKQMLERIQHAVQLATEPLEKAVQSRLTGEEVNSCVEVLL EEAKDLLSDWLDSTLGCDVTDNSIFSKLPKFWEGDFHRDMEALNVLPPDVLTRVSEYVPEIVNFVQKIVDNGYGYVSNGSVYFDTAKFAS SEKHSYGKLVPEAVGDQKALQEGEGDLSISADRLSEKRSPNDFALWKASKPGEPSWPCPWGKGRPGWHIECSAMAGTLLGASMDIHGGGF DLRFPHHDNELAQSEAYFENDCWVRYFLHTGHLTIAGCKMSKSLKNFITIKDALKKHSARQLRLAFLMHSWKDTLDYSSNTMESALQYEK FLNEFFLNVKDILRAPVDITGQFEKWGEEEAELNKNFYDKKTAIHKALCDNVDTRTVMEEMRALVSQCNLYMAARKAVRKRPNQALLENI ALYLTHMLKIFGAVEEDSSLGFPVGGPGTSLSLEATVMPYLQVLSEFREGVRKIAREQKVYRRKHQELQAMQMELQSPEYKLSKLRTSTI MTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQN IVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKI GDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPV YRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTS SGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGS -------------------------------------------------------------- >ENST00000401769_ENST00000389048_CARS_chr11_3037010_ALK_chr2_29446394_length(amino acids)=614 MEPTSGPFRTDLGYHLVGCSSGKGRRVQPQWSPPAGTQPCRLHLYNSLTRNKEVFIPQDGKKVTWYCCGPTVYDASHMGHARSYISFDIL RRVLKDYFKFDVFYCMNITDIDDKIIKRARQNHLFEQYREKRPEAAQLLEDVQAALKPFSVKLNETTDPDKKQMLERIQHAVQLATEPLE KAVQSRLTGEEVNSCVEVLLEEAKDLLSDWLDSTLGCDVTDNSIFSKLPKFWEGDFHRDMEALNVLPPDVLTRVSEYVPEIVNFVQKIVD NGYGYVSNGSVYFDTAKFASSEKHSYGKLVPEAVGDQKALQEGEGDLSISADRLSEKRSPNDFALWKASKPGEPSWPCPWGKGRPGWHIE CSAMAGTLLGASMDIHGGGFDLRFPHHDNELAQSEAYFENDCWVRYFLHTGHLTIAGCKMSKSLKNFITIKDALKKHSARQLRLAFLMHS WKDTLDYSSNTMESALQYEKFLNEFFLNVKDILRAPVDITGQFEKWGEEEAELNKNFYDKKTAIHKALCDNVDTRTVMEEMRALVSQCNL -------------------------------------------------------------- |
Multiple Sequence Alignment of All Fusion Protein Isoforms |
Top |
Kinase Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr11:/chr2:) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CARS | ALK |
| FUNCTION: Catalyzes the ATP-dependent ligation of cysteine to tRNA(Cys). {ECO:0000269|PubMed:11347887, ECO:0000269|PubMed:30824121}. | FUNCTION: Probable dioxygenase that requires molecular oxygen, alpha-ketoglutarate and iron. {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
 - Retained domain in the 5'-partner of fusion protein (protein functional feature from UniProt). - Retained domain in the 5'-partner of fusion protein (protein functional feature from UniProt). |
| Partner | Hgeneene | Hbp | Tgeneene | Tbp | ENST | BPexon | TotalExon | Protein feature loci | BPloci | TotalLen | Feature | Note |
 - Retained domain in the 3'-partner of fusion protein (protein functional feature from UniProt). - Retained domain in the 3'-partner of fusion protein (protein functional feature from UniProt). |
| Partner | Hgeneene | Hbp | Tgeneene | Tbp | ENST | BPexon | TotalExon | Protein feature loci | BPloci | TotalLen | Feature | Note |
| Tgene | CARS | 3033424 | ALK | 29446394 | ENST00000380525 | 18 | 29 | 1116_1392 | 1057 | 1621 | Domain | Note=Protein kinase;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00159 |
| Tgene | CARS | 3033425 | ALK | 29446394 | ENST00000380525 | 18 | 29 | 1116_1392 | 1057 | 1621 | Domain | Note=Protein kinase;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00159 |
| Tgene | CARS | 3037010 | ALK | 29446394 | ENST00000401769 | 18 | 29 | 1116_1392 | 1057 | 1621 | Domain | Note=Protein kinase;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00159 |
Top |
Kinase Fusion Protein Structures |
 CIF files of the predicted kinase fusion proteins CIF files of the predicted kinase fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Kinase Fusion protein CIF link (fusion AA seq ID in KinaseFusionDB) | Henst | Tenst | Hgene | Hchr | Hbp | Tgene | Tchr | Tbp | AA seq | Len(AA seq) |
| PDB file >>>81_CARS_ALK | ENST00000401769 | ENST00000389048 | CARS | chr11 | 3037010 | ALK | chr2 | 29446394 | MEPTSGPFRTDLGYHLVGCSSGKGRRVQPQWSPPAGTQPCRLHLYNSLTRNKEVFIPQDGKKVTWYCCGPTVYDASHMGHARSYISFDIL RRVLKDYFKFDVFYCMNITDIDDKIIKRARQNHLFEQYREKRPEAAQLLEDVQAALKPFSVKLNETTDPDKKQMLERIQHAVQLATEPLE KAVQSRLTGEEVNSCVEVLLEEAKDLLSDWLDSTLGCDVTDNSIFSKLPKFWEGDFHRDMEALNVLPPDVLTRVSEYVPEIVNFVQKIVD NGYGYVSNGSVYFDTAKFASSEKHSYGKLVPEAVGDQKALQEGEGDLSISADRLSEKRSPNDFALWKASKPGEPSWPCPWGKGRPGWHIE CSAMAGTLLGASMDIHGGGFDLRFPHHDNELAQSEAYFENDCWVRYFLHTGHLTIAGCKMSKSLKNFITIKDALKKHSARQLRLAFLMHS WKDTLDYSSNTMESALQYEKFLNEFFLNVKDILRAPVDITGQFEKWGEEEAELNKNFYDKKTAIHKALCDNVDTRTVMEEMRALVSQCNL | 614 |
| 3D view using mol* of 81_CARS_ALK | ||||||||||
| PDB file >>>TKFP_136_CARS_ALK | ENST00000380525 | ENST00000389048 | CARS | chr11 | 3033425 | ALK | chr2 | 29446394 | MADSSGQQAPDYRSILSISDEAARAQALNEHLSTRSYVQGYSLSQADVDAFRQLSAPPADPQLFHVARWFRHIEALLGSPCGKGQPCRLQ ASKGRRVQPQWSPPAGTQPCRLHLYNSLTRNKEVFIPQDGKKVTWYCCGPTVYDASHMGHARSYISFDILRRVLKDYFKFDVFYCMNITD IDDKIIKRARQNHLFEQYREKRPEAAQLLEDVQAALKPFSVKLNETTDPDKKQMLERIQHAVQLATEPLEKAVQSRLTGEEVNSCVEVLL EEAKDLLSDWLDSTLGCDVTDNSIFSKLPKFWEGDFHRDMEALNVLPPDVLTRVSEYVPEIVNFVQKIVDNGYGYVSNGSVYFDTAKFAS SEKHSYGKLVPEAVGDQKALQEGEGDLSISADRLSEKRSPNDFALWKASKPGEPSWPCPWGKGRPGWHIECSAMAGTLLGASMDIHGGGF DLRFPHHDNELAQSEAYFENDCWVRYFLHTGHLTIAGCKMSKSLKNFITIKDALKKHSARQLRLAFLMHSWKDTLDYSSNTMESALQYEK FLNEFFLNVKDILRAPVDITGQFEKWGEEEAELNKNFYDKKTAIHKALCDNVDTRTVMEEMRALVSQCNLYMAARKAVRKRPNQALLENI ALYLTHMLKIFGAVEEDSSLGFPVGGPGTSLSLEATVMPYLQVLSEFREGVRKIAREQKVYRRKHQELQAMQMELQSPEYKLSKLRTSTI MTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQN IVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKI GDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPV YRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTS SGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGS | 1252_CARS_ALK |
| PDB file >>>TKFP_137_CARS_ALK | ENST00000380525 | ENST00000389048 | CARS | chr11 | 3033424 | ALK | chr2 | 29446394 | MADSSGQQAPDYRSILSISDEAARAQALNEHLSTRSYVQGYSLSQADVDAFRQLSAPPADPQLFHVARWFRHIEALLGSPCGKGQPCRLQ ASKGRRVQPQWSPPAGTQPCRLHLYNSLTRNKEVFIPQDGKKVTWYCCGPTVYDASHMGHARSYISFDILRRVLKDYFKFDVFYCMNITD IDDKIIKRARQNHLFEQYREKRPEAAQLLEDVQAALKPFSVKLNETTDPDKKQMLERIQHAVQLATEPLEKAVQSRLTGEEVNSCVEVLL EEAKDLLSDWLDSTLGCDVTDNSIFSKLPKFWEGDFHRDMEALNVLPPDVLTRVSEYVPEIVNFVQKIVDNGYGYVSNGSVYFDTAKFAS SEKHSYGKLVPEAVGDQKALQEGEGDLSISADRLSEKRSPNDFALWKASKPGEPSWPCPWGKGRPGWHIECSAMAGTLLGASMDIHGGGF DLRFPHHDNELAQSEAYFENDCWVRYFLHTGHLTIAGCKMSKSLKNFITIKDALKKHSARQLRLAFLMHSWKDTLDYSSNTMESALQYEK FLNEFFLNVKDILRAPVDITGQFEKWGEEEAELNKNFYDKKTAIHKALCDNVDTRTVMEEMRALVSQCNLYMAARKAVRKRPNQALLENI ALYLTHMLKIFGAVEEDSSLGFPVGGPGTSLSLEATVMPYLQVLSEFREGVRKIAREQKVYRRKHQELQAMQMELQSPEYKLSKLRTSTI MTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQN IVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKI GDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPV YRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTS SGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGS | 1252_CARS_ALK |
| PDB file >>>TKFP_138_CARS_ALK | ENST00000401769 | ENST00000389048 | CARS | chr11 | 3037010 | ALK | chr2 | 29446394 | MEPTSGPFRTDLGYHLVGCSSGKGRRVQPQWSPPAGTQPCRLHLYNSLTRNKEVFIPQDGKKVTWYCCGPTVYDASHMGHARSYISFDIL RRVLKDYFKFDVFYCMNITDIDDKIIKRARQNHLFEQYREKRPEAAQLLEDVQAALKPFSVKLNETTDPDKKQMLERIQHAVQLATEPLE KAVQSRLTGEEVNSCVEVLLEEAKDLLSDWLDSTLGCDVTDNSIFSKLPKFWEGDFHRDMEALNVLPPDVLTRVSEYVPEIVNFVQKIVD NGYGYVSNGSVYFDTAKFASSEKHSYGKLVPEAVGDQKALQEGEGDLSISADRLSEKRSPNDFALWKASKPGEPSWPCPWGKGRPGWHIE CSAMAGTLLGASMDIHGGGFDLRFPHHDNELAQSEAYFENDCWVRYFLHTGHLTIAGCKMSKSLKNFITIKDALKKHSARQLRLAFLMHS WKDTLDYSSNTMESALQYEKFLNEFFLNVKDILRAPVDITGQFEKWGEEEAELNKNFYDKKTAIHKALCDNVDTRTVMEEMRALVSQCNL | 614_CARS_ALK |
| 3D view using mol* of TKFP_138_CARS_ALK | ||||||||||
Top |
Comparison of Fusion Protein Isoforms |
 Superimpose the 3D Structures Among All Fusion Protein Isoforms Superimpose the 3D Structures Among All Fusion Protein Isoforms * Download the pdb file and open it from the molstar online viewer. |
| 3D view using mol* of viewer/superimpose_isoforms/TKFP_136_CARS_ALK_vs_TKFP_138_CARS_ALK_superimposed.pdb.html |
 Comparison of the Secondary Structures of Fusion Protein Isoforms Comparison of the Secondary Structures of Fusion Protein Isoforms |
| ./secondary_str/TKFP_136_CARS_ALK_vs_TKFP_138_CARS_ALK.png |
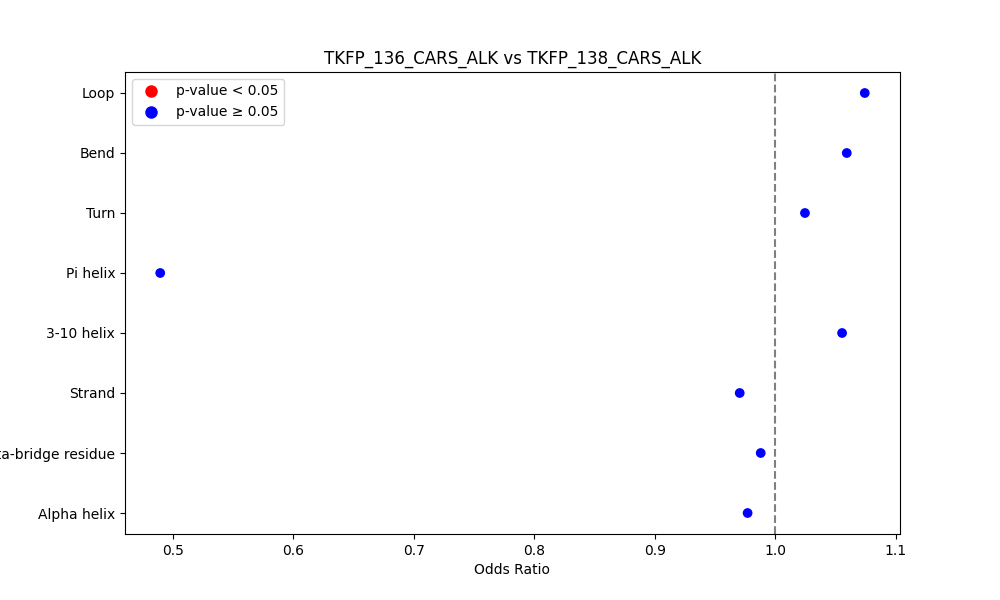 |
Top |
Comparison of Fusion Protein Sequences/Structures with Known Sequences/Structures from PDB |
Top |
pLDDT score distribution |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. * The blue color at the bottom marks the best active site residues. |
| 81_CARS_ALK.png |
 |
| 81_CARS_ALK.png |
 |
| TKFP_138_CARS_ALK.png |
 |
Top |
Potential Active Site Information |
 The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. |
| Kinase Fusion AA seq ID in KinaseFusionDB | Site score | Size | Dscore | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
| TKFP_138_CARS_ALK | 1.065 | 295 | 1.056 | 1066.73 | 0.503 | 0.795 | 0.964 | 0.599 | 1.107 | 0.541 | 0.718 | Chain A: 68,69,70,71,72,73,74,77,79,80,82,83,84,87 ,107,109,112,114,117,118,120,121,122,145,148,149,1 52,164,165,168,204,205,208,209,210,212,213,228,232 ,312,313,314,315,316,317,337,357,360,361,378,379,3 80,381,382,385,386,410,411,412,413,419,420,421,422 ,423,425 |
Top |
Ramachandran Plot of Kinase Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| 81_CARS_ALK_ramachandran.png |
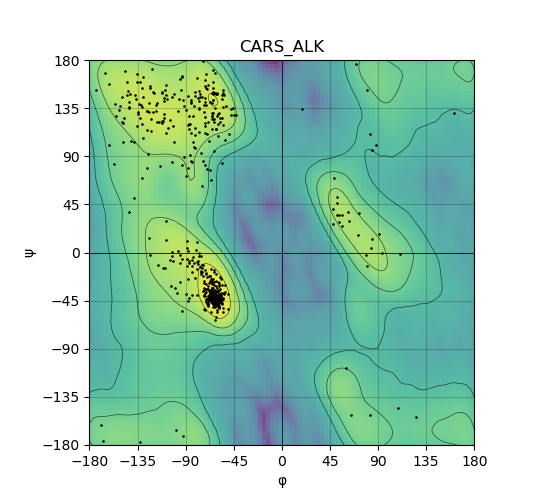 |
| TKFP_138_CARS_ALK_ramachandran.png |
 |
Top |
Virtual Screening Results |
 Distribution of the average docking score across all approved kinase inhibitors. Distribution of the average docking score across all approved kinase inhibitors.Distribution of the number of occurrence across all approved kinase inhibitors. |
| 5'-kinase fusion protein case |
| 3'-kinase fusion protein case |
 |
Top |
 Drug information from DrugBank of the top 20 interacting small molecules. Drug information from DrugBank of the top 20 interacting small molecules.* The detailed information of individual kinase inhibitors are available in the download page. |
| Fusion gene name info | Drug | Docking score | Glide g score | Glide energy |
| 81_CARS_ALK-DOCK_HTVS_1-001 | Pralsetinib | -7.05163 | -7.14313 | -60.4963 |
| 81_CARS_ALK-DOCK_HTVS_1-001 | Larotrectinib | -6.8300399999999994 | -6.8300399999999994 | -57.9763 |
| 81_CARS_ALK-DOCK_HTVS_1-001 | Belumosudil | -6.45809 | -6.46579 | -54.1226 |
| 81_CARS_ALK-DOCK_HTVS_1-001 | Belumosudil | -6.431019999999999 | -6.43872 | -58.0764 |
| 81_CARS_ALK-DOCK_HTVS_1-001 | Tivozanib | -6.28022 | -6.28022 | -51.1618 |
| 81_CARS_ALK-DOCK_HTVS_1-001 | Lenvatinib | -6.04542 | -6.04542 | -52.0039 |
| 81_CARS_ALK-DOCK_HTVS_1-001 | Tofacitinib | -5.9429300000000005 | -5.95443 | -40.6479 |
| 81_CARS_ALK-DOCK_HTVS_1-001 | Tofacitinib | -5.9429300000000005 | -5.95443 | -40.6479 |
| 81_CARS_ALK-DOCK_HTVS_1-001 | Midostaurin | -5.89931 | -5.89931 | -54.4718 |
| 81_CARS_ALK-DOCK_HTVS_1-001 | Tofacitinib | -5.88566 | -5.8971599999999995 | -41.1829 |
| 81_CARS_ALK-DOCK_HTVS_1-001 | Tofacitinib | -5.88566 | -5.8971599999999995 | -41.1829 |
| 81_CARS_ALK-DOCK_HTVS_1-001 | Pralsetinib | -5.8168 | -5.9083 | -60.3563 |
| 81_CARS_ALK-DOCK_HTVS_1-001 | Nilotinib | -5.5254 | -6.4502 | -59.7425 |
| 81_CARS_ALK-DOCK_HTVS_1-001 | Nilotinib | -5.5254 | -6.4502 | -59.7425 |
| 81_CARS_ALK-DOCK_HTVS_1-001 | Tofacitinib | -5.5176099999999995 | -5.52911 | -39.8861 |
| 81_CARS_ALK-DOCK_HTVS_1-001 | Tofacitinib | -5.5176099999999995 | -5.52911 | -39.8861 |
| 81_CARS_ALK-DOCK_HTVS_1-001 | Pralsetinib | -5.50852 | -5.60002 | -57.1214 |
| 81_CARS_ALK-DOCK_HTVS_1-001 | Pexidartinib | -5.490559999999999 | -6.1913599999999995 | -44.619 |
| 81_CARS_ALK-DOCK_HTVS_1-001 | Pexidartinib | -5.490559999999999 | -6.1913599999999995 | -44.619 |
| 81_CARS_ALK-DOCK_HTVS_1-001 | Baricitinib | -5.4888900000000005 | -5.4888900000000005 | -46.0658 |
Top |
Kinase-Substrate Information of CARS_ALK |
 Phosphorylation target of the kinase Phosphorylation target of the kinase(phosphosite, 03-17-2024) |
| Kinase | Kinase UniProt Acc | Kinase species | Substrate | Substrate UniProt Acc | Substrate phosphorylated residues | Substrate phosphorylated sites (+/-7AA) | Domain |
| ALK | Q9UM73 | human | ALK | Q9UM73 | Y1604 | tAPGAGHyEDTILkS | |
| ALK | Q9UM73 | human | ELP3 | Q9H9T3 | Y202 | GHtSNNIyEAVkySE | Radical_SAM |
| ALK | Q9UM73 | human | ALK | Q9UM73 | Y1278 | FGMARDIyRAsyyRK | PK_Tyr_Ser-Thr |
| ALK | Q9UM73 | human | CDK9 | P50750 | Y19 | FCDEVSkyEkLAkIG | |
| ALK | Q9UM73 | human | SFPQ | P23246 | Y293 | RRPGEktyTQRCRLF | |
| ALK | Q9UM73 | human | ATIC | P31939 | Y104 | RVVACNLyPFVkTVA | MGS |
| ALK | Q9UM73 | human | SRCIN1 | Q9C0H9 | Y396 | LVKGEGLyADPyGLL | |
| ALK | Q9UM73 | human | STAT3 | P40763 | Y705 | DPGsAAPyLktKFIC | |
| ALK | Q9UM73 | human | PTPN11 | Q06124 | Y580 | REDsARVyENVGLMQ | |
| ALK | Q9UM73 | human | PBK | Q96KB5 | Y74 | NPICNDHyRsVyQkR | Pkinase |
| ALK | Q9UM73 | human | VASP | P50552 | Y39 | AFSRVQIyHNPtANs | WH1 |
| ALK | Q9UM73 | human | PTPN11 | Q06124 | Y542 | sKRkGHEytNIKysL | |
| ALK | Q9UM73 | human | GRB2 | P62993 | Y160 | QVPQQPtyVQALFDF |
 Biological Network Integration of This Kinase and Substrates Biological Network Integration of This Kinase and Substrates (GeneMANIA website) |
 Enriched GO biological processes of the phosphorylation target genes of the kinase Enriched GO biological processes of the phosphorylation target genes of the kinase |
| Kinase | GOID | GO term | P.adjust |
Top |
Related Drugs to CARS_ALK |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
 Distribution of the number of studies mentioning CARS-ALK and kinase inhibitors the PubMed Abstract (04-01-2024) Distribution of the number of studies mentioning CARS-ALK and kinase inhibitors the PubMed Abstract (04-01-2024) |
| Fusion gene - drug pair 1 | Fusion gene - drug pair 2 | PMID | Publication date | DOI | Study title |
Top |
Related Diseases to CARS_ALK |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
| CARS | ALK | Lung Adenocarcinoma | MyCancerGenome | |
| CARS | ALK | Acute Myeloid Leukemia | MyCancerGenome | |
| CARS | ALK | Cancer Of Unknown Primary | MyCancerGenome | |
| CARS | ALK | Diffuse Large B-Cell Lymphoma | MyCancerGenome |
 Related diseases from the literature mentioned this fusion gene and drug. Related diseases from the literature mentioned this fusion gene and drug. (PubMed, 04-01-2024) |
| MeSH ID | MeSH term |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Tgene | ALK | C0007131 | Non-Small Cell Lung Carcinoma | 28 | CGI;CTD_human |
| Tgene | ALK | C0027819 | Neuroblastoma | 13 | CGI;CTD_human;ORPHANET |
| Tgene | ALK | C0152013 | Adenocarcinoma of lung (disorder) | 8 | CGI;CTD_human |
| Tgene | ALK | C2751681 | NEUROBLASTOMA, SUSCEPTIBILITY TO, 3 | 8 | CLINGEN;UNIPROT |
| Tgene | ALK | C0206180 | Ki-1+ Anaplastic Large Cell Lymphoma | 6 | CGI;CTD_human |
| Tgene | ALK | C0334121 | Inflammatory Myofibroblastic Tumor | 4 | CGI;CTD_human;ORPHANET |
| Tgene | ALK | C0018199 | Granuloma, Plasma Cell | 3 | CTD_human |
| Tgene | ALK | C0007621 | Neoplastic Cell Transformation | 2 | CTD_human |
| Tgene | ALK | C0027627 | Neoplasm Metastasis | 2 | CTD_human |
| Tgene | ALK | C0238463 | Papillary thyroid carcinoma | 2 | ORPHANET |
Top |
Clinical Trials of the Found Drugs/Small Molecules |
 Statistics of the Clinical Trials of the Found Kinase Inibitors from clinicaltrials.gov (06-17-2024) Statistics of the Clinical Trials of the Found Kinase Inibitors from clinicaltrials.gov (06-17-2024) |
 Clinical Trials from clinicaltrials.gov (06-17-2024) Clinical Trials from clinicaltrials.gov (06-17-2024) |
| Fusion Gene | Kinase Inhibitor | NCT ID | Study Status | Phases | Disease | # Enrolment | Date |

