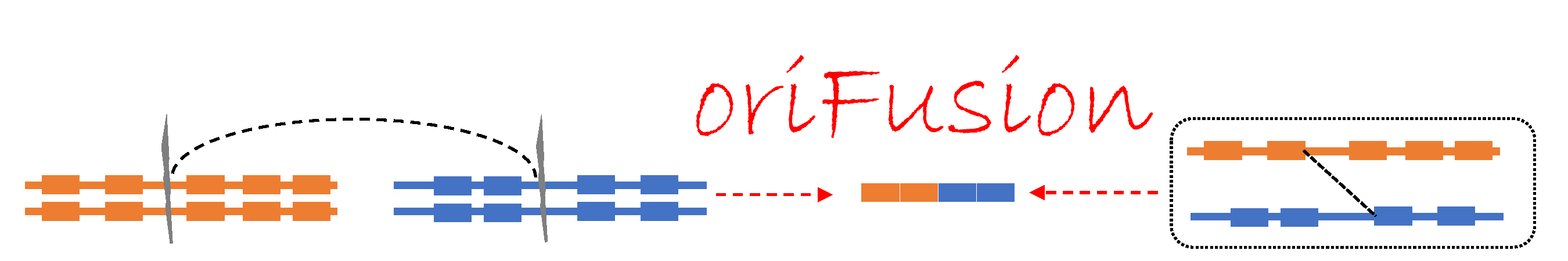
About oriFusion (orifin of Fusion genes)
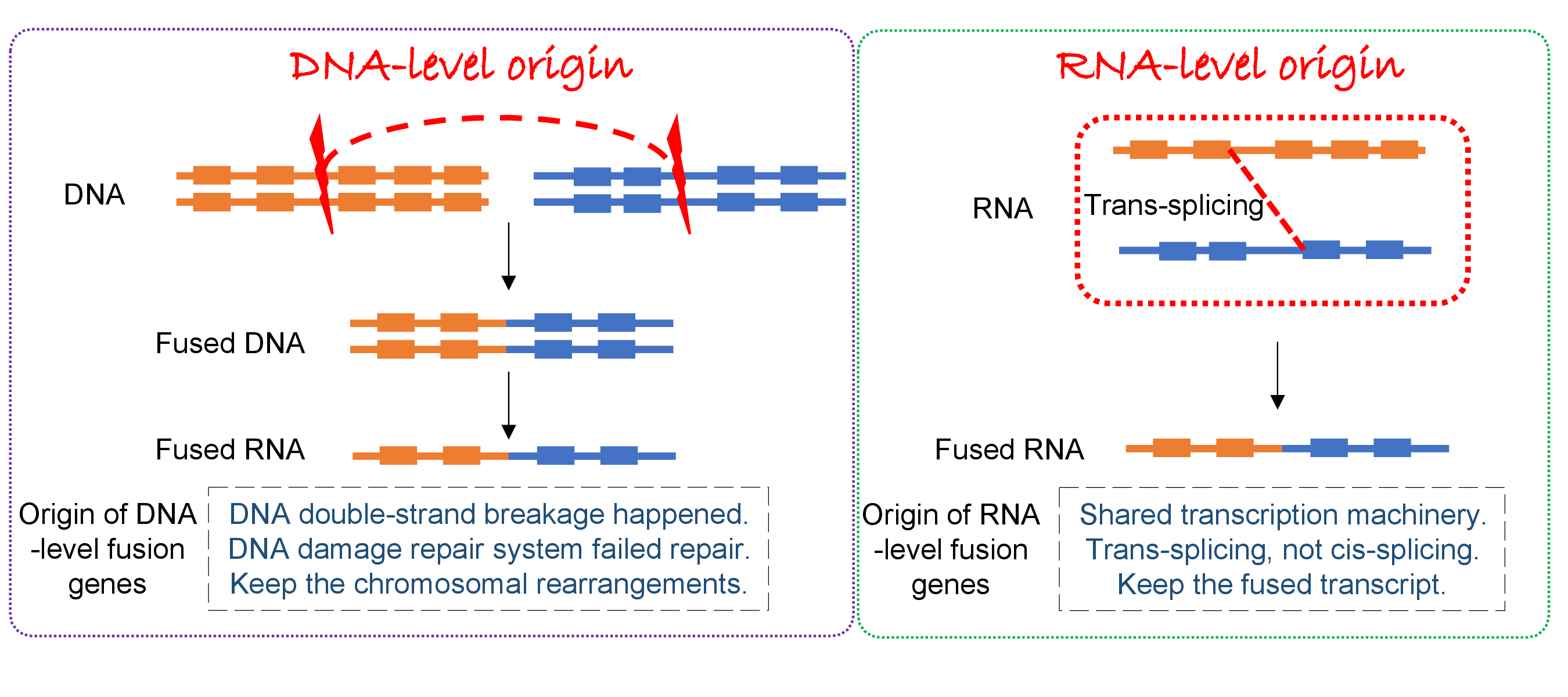
Identification of the actual genomic breakpoints of RNA-seq-identified fusion genes from the whole genome sequencing (WGS) data provides the origin of fusion gene formation, whether individual fusion genes were made by the chromosomal rearrangement at the DNA level or trans-splicing event at the RNA level. If there is a stair-shaped alignment of the split reads (unmapped reads) with the same DNA cut position, then this fusion gene was made by the chromosomal rearrangement at the DNA level. But, if not, then that fusion gene was made by the trans-splicing at the RNA level. From the knowledge of the origins of individual fusion genes, we can better understand the mechanism of DNA breakage and repair system and trans-splicing events and study to target the fusion genes in different settings. Fusion genes are usually detected from the RNA-seq data due to the cost and analysis effort. Therefore, most of the identified fusion genes’ breakpoints are at exon junction positions. We will download ~ 3600 bam files of whole genome sequencing data of TCGA tumor patients with ~ 15K fusion genes to know the actual genomic breakpoints. With our split sequencing reads analysis pipeline, we will identify the genomic breakpoints. This study will provide the origin of the fusion gene formation of ~ 15K human cancer fusion genes.
| 
 Search
Search
