|
Home |
Download |
Statistics |
Examples |
Help |
Contact |
Terms of Use |
Navigation
0. Overview of KinaseFusionDB pipeline.1. Fusion Gene Data Collection.
2. Open Reading Frame (ORF) Analysis
3. Creation of Fusion Transcript and Amino Acid Sequences.
4. Protean Features Retention Analysis.
5. 3D structure prediction of fusion proteins.
6. Preparation of protein and active site identification.
7. Receptor grid generation and Virtual screening.
8. ADME studies.
9. Toxicity and accessibility
10. Understanding of KinaseFusionDB's Annotation Category.
- FusionGene Search Result Page.
- FusionGene Annotation Result Page.
-- 1) Fusion Gene Summary.
-- 2) Fusion Gene Sample Information.
-- 3) Fusion Gene ORF Analysis.
-- 4) Fusion Amino Acid Sequences.
-- 5) Fusion Protein Features.
-- 6) Fusion Protein Structures.
-- 7) pLDDT scores.
-- 8) Ramachandra plot of Fusion Protein Structures.
-- 9) Potential Active Site Information.
-- 10) Potentially Interacting Small Molecules through Virtual Screening.
-- 11) Biochemical Features of Small Molecules.
-- 12) Drug Toxicity Information.
-- 13) Fusion Protein-Protein Interaction.
-- 13) Related Drugs.
-- 14) Related Diseases.
12. Download Data and Contact Us.
0. Overview of KinaseFusionDB pipeline.
We set up a pipeline of screening interacting small molecules of human fusion proteins, named as FusionScreen. We analyze the open reading frame of the curated human fusion genes. For the in-frame fusion transcripts, we make the full-length fusion transcript nucleotide sequence. Then, we make a fusion protein amino acid sequence. Next, we input this fusion protein amino acid sequence into AlphaFold2 to predict its 3D protein structure. After having the 3D structures of fusion proteins, we check the pLDDT scores around the best actie sites of individual fusion protein structures. Then we choose the fusion proteins tha we perform virtual screening of potential interacting/inhibiting molecules against the FDA-approved drugs. Further, we investigate the pharmacokinetics of absorption, distribution, metabolism, and excretion (ADME) and toxicology. Finally, we release this knowledge into KinaseFusionDB.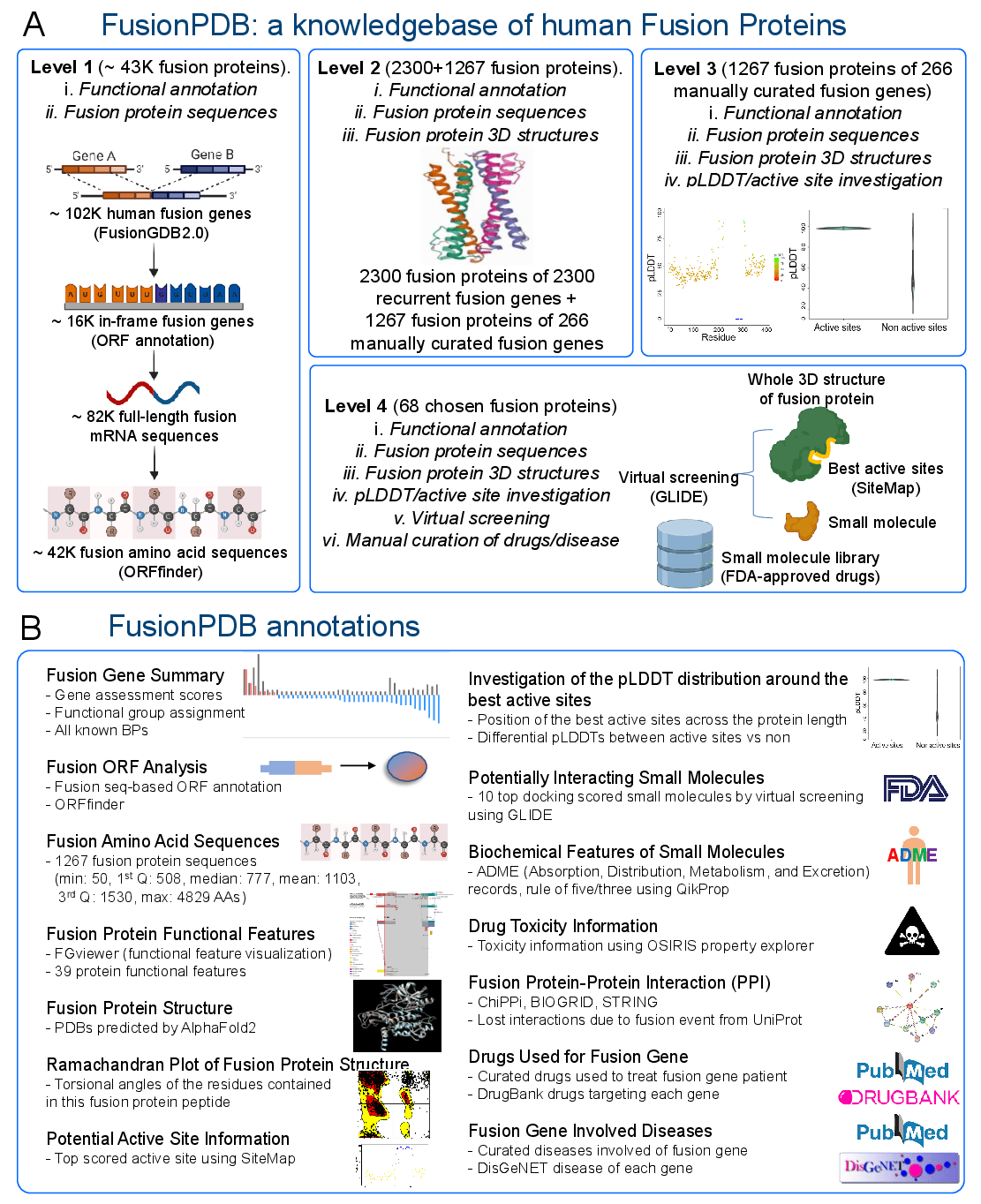
1. Fusion Gene Data Collection
We downloaded the fusion gene information from FusionGDB2.0. Detailed information is on the statistics page.2. Creation of Fusion Transcript and Amino Acid Sequences
Two different genes can form fusion genes with multiple breakpoints based on multiple gene isoforms. Therefore, we considered all gene isoforms at each breakpoint. To help with the identification and validation of fusion genes, we focused on the in-frame fusion genes. For more reliable fusion genes, we checked the distance between the two breakpoints in case of intra-chromosomal rearrangements and created fusion sequences when those genes are apart more than 100kb. We also selected fusion genes when both of their breakpoints are aligned at the exon junction. To call each exon sequence of the given breakpoint, transcription start/end sites, and CDS start/end sites, we used the nibFrag utility from UCSC Genome Browser based on ENCODE hg19 genome structure. By adding these exon sequences, we made the full-length fusion transcript sequences of the in-frame fusion genes. For these fusion transcript sequences, we input to the ORFfinder and chose the longest ORF as the potential fusion protein sequence.3. Open Reading Frame (ORF) Analysis
To check the coding potential, we analyzed the ORF of the fusion transcript sequences. First, we investigated the ORF whether in-frame or frame-shift if both breakpoints are located in the coding sequence (CDS) area. If not, we reported the location of individual breakpoint is in 5'-UTR, CDS, or 3'-UTR. Second, to have the potential amino acid sequence, we ran ORFfinder by NCBI. Third, we ran the in-house classifier (to be available soon) between the coding genes mapped by Ribo-seq reads with high reliability and non-coding genes not mapped by any Ribo-seq reads.4. Protein Features Retention Analysis
We searched the retention of 39 protein features of UniProt (six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features) at the fusion amino acid sequence level. Through this process, we also checked the retention of protein-protein interaction (PPI) at the fusion protein. Detailed information about all of the protein features is on the UniProt page.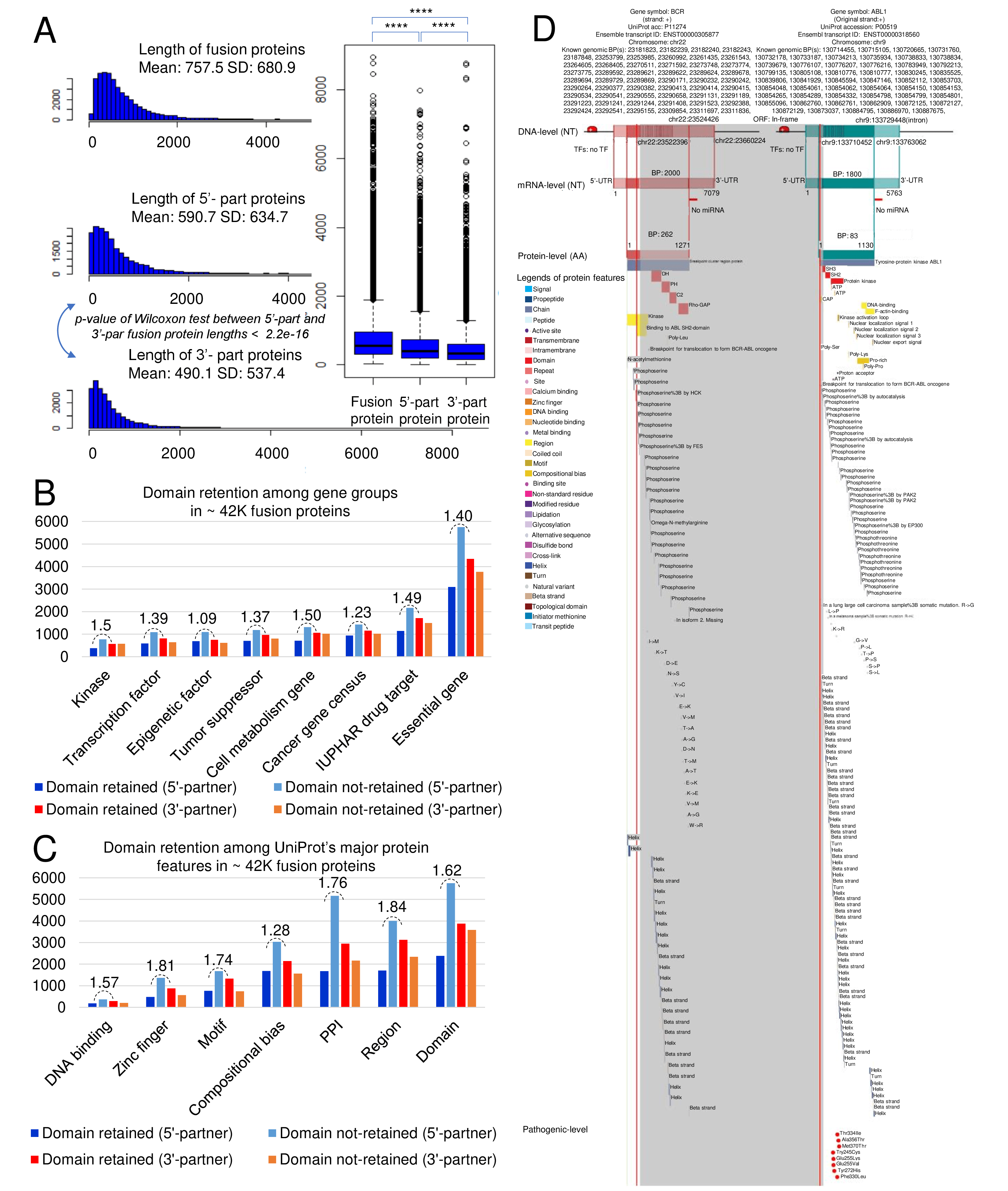
FGviewer provides functional feature annotations at four different levels: DNA-, RNA-, protein-, and pathogenic levels. The same breakpoint line across four tiers will classify between FG involving or non-involving zone with multiple types of functional features.
5. Predict 3D structure of fusion proteins.
The biological mechanism and function of any protein are based on its 3D structure. In 2020, AlphaFold2, an artificial intelligence-based protein structure prediction tool has been developed by DeepMind which uses amino acid sequence as an input. The DeepMind and EMBL-EBI have developed the publicly available database of predicted protein 3D structures. In our study, we are predicting the 3D structure of ~3300 fusion proteins, for the first time, with the help of AlphaFold2. We have used GPU-based Linux version AlphaFold v2.0. and followed the step-by-step procedures as described in GitHub repository (https://github.com/deepmind/alphafold). We have created the fasta files for ~ 3300 fusion protein’s amino acids (of 2300 recurrent fusion genes and 266 manually curated fusion genes) as input for AlphaFold. For alignment, we have downloaded the genetic databases (size: 2.2 TB) such as uniref90, uniport, uniclust30, small_bfd, pdb_seqres, pdb_mmcif, pdb70, params, mgnify, and bfd. The model parameters were also made available by Alphafold to download. The output folder provides the predicted PDB structures on the basis of model confidence and ranked as ranked_0.pdb to ranked_4.pdb.The figure below shows the superimpose result between the known protein structures of the ABL1 kinase domain of BCR-ABL1 fusion protein and our predicted 3D structures. Overall, the comparison between the known 3D structures of BCR-ABL1 and our predicted ones was in good alignment as less than 1 Å RMSD.

The figure below shows the superimposed structures of predicted BCR-ABL1 protein by AlphaFold2 and known structure of ABL1 from PDB (5MO4) due to lack of 3D structure of fusion proteins (A). The blue color is the predicted structure by AlphaFold2, and grey indicates the reference PDB. This fusion protein was predicted based on the most frequent breakpoint (BCR:chr22: 23632600-ABL1:chr9: 133729451) between different transcript isoforms BCR:ENST00000305877-ABL1:ENST00000318560 and BCR:ENST00000359540-ABL1:ENST00000318560. Both fusion transcript isoforms were translated into the same fusion protein amino acid sequence with a length of 1,530 AA. RMSD between the predicted structure and PDB reference (5MO4) is 0.642. Predicted whole 3D structures of five NTRK fusion proteins and the best predicted active sites based on the whole 3D structures (B).

6. Preparation of protein and active site identification
The PDB structures of predicted proteins may contain missing atoms, missing connectivity information, water molecules, etc., which need to be removed or preprocessed before using the 3D structure for screening. All ~3300 predicted protein structures were preprocessed by the protein preparation module of Maestro from Schrödinger (Release 2022-2, LLC, New York, NY, 2021), by removing the water molecules, assigning the bond orders, and adding the hydrogen bonds to the crystal structure of receptor molecule. Restrained minimization was done by the constraint of the RMSD and OPLS force field. Since most of the fusion protein structures are unknown, experimentally verified active sites are less reporting. Various computational approaches have been applied to predict the active sites of unknown proteins. In this study we used, SiteMap, Schrödinger Release 2021-4, Schrödinger, LLC, New York, NY, 2021 to identify active sites of all ~3300 predicted proteins. SiteMap provides the site score for the predicted active site on the basis of multiple factors such as the size of the site, exposure to the solvent, hydrophobicity and hydrophilicity, hydrogen bond donor and acceptor, etc.To provide the reliability of the fusion protein 3D structure prediction, we checked the significance of the higher pLDDT scores in our whole 3D structure-based predicted active sites versus non-active sites. In this way, the users can have some evidence that at least the active sites are having better pLDDT scores and their pLDDT score ranges. If they satisfy with this distribution, then they can check the virtual screening result. We also provide the distribution of pLDDT and predicted aligned error (PAE) scores across the fusion protein length.


As shown below, the active sites (1 at the X-axis) predicted based on the whole 3D structure of fusion proteins showed higher interaction with the approved-drugs compared to the known domains (2 at the X-axis).

7. Receptor grid generation and Virtual screening.
The grid size represents the volume of a receptor’s active sites where the ligand can search for binding while docking. The grid around the receptor was generated using a module available with Glide, Schrödinger, LLC, New York, NY, 2021;. The dimensions of the grid were selected by considering the active sites predicted by SiteMap. Virtual screening is a computational technique used in the drug discovery process to get small molecules that are most likely to bind to receptor or target molecule). In this work, we considered all FDA-approved ligand libraries of the ZINC database accessed on 10th December 2021. These selected libraries were processed by LigPrep module and made available for virtual screening. In silico screening and docking analysis of the 68 chosen fusion protein against the FDA approved ligands were carried out using Schrödinger Release 2021-4 (Glide, Schrödinger, LLC, New York, NY, 2021.).8. ADME studies.
It has been reported that nearly 40% of proposed drug candidates are not able to qualify the clinical trials because of poor ADME properties of the drug molecules. It is recommended that before going for expensive experimental procedures, one should check the ADME property computationally. In this study we used QikProp, Schrödinger Release 2022-2: QikProp, Schrödinger, LLC, New York, NY, 2021 to calculate the ADME (i.e. Absorption, Distribution, Metabolism, and Excretion) features of our ligand library. The predicted ADME properties of the screened ligands includes hydrophobic component of the SASA, dipole moment, solvent accessible surface area, total polar surface area, hydrophilic component of the SASA, molecular weight (MW), predicted polarizability, predicted octanol/water partition coefficient, predicted octanol/gas partition coefficient, predicted hexadecane/gas partition coefficient, predicted water/gas partition coefficient, etc.9. Toxicity and accessibility
In addition to drug screening, ADME, and toxicity prediction are significantly important steps of computer-aided drug designing process. Toxicity prediction aims to identify the undesirable effects of putative drugs on humans, animals, plants, and the environment. We used eToxPred (https://github.com/pulimeng/etoxpred), a machine learning-based technique for the estimation of toxicity and accessibility of ligand library.10. Understanding KinaseFusionDB's Annotation Categories
Search page, example: ERG
Input query
- Official HUGO gene symbol or Entrez gene ID.

Browse the fusion genes per Level and gene group
FusionGene Search Result Page

Select your fusion gene from the gene list.
We provide a classification in the first query search result into three sections according to our three Levels of fusion protein categories.
- Level 4 provides fusion protein sequence, fusion protein 3D structure, pLDDT score distribution checking around the best active sites, and virtual screening results (68 chosen fusion proteins from 266 manually curated in-frame fusion genes).
- Level 3 provides fusion protein sequence, fusion protein 3D structure, and pLDDT score distribution checking around the best active sites (1267 fusion proteins from 266 manually curated in-frame fusion genes).
- Level 2 provides the functional annotation, fusion protein sequences, and whole 3D structure of fusion proteins (1267 fusion proteins from 266 manually curated in-frame fusion genes + 2300 fusion proteins from 2300 recurrent fusion genes).
- Level 1 provides the functional annotation, fusion protein sequences of ~ 43K fusion proteins from ~ 16K in-frame fusion genes.
Users can expect what kinds of fusion protein analysis information are available for their searched fusion protein.
FusionGene Annotation Result Page

These are FusionGeneDB's annotation categories for your query with links to their corresponding annotation parts.
1) Fusion Gene Summary.
This category shows the information of the fusion gene. Firstly, it shows each partner gene's overall information from basic information such as symbol, alias, and locations and ENST accessions involved in fusion gene. Specifically, the DoF score provides all possible combinations of each gene in pan-cancer fusion genes. From the # samples, the user can Words in blue are linked to their respective databases. FusionGeneSummary table also shows the tissue and cancer type information including manually curated PubMed article information.






2) Fusion Gene Sample Information.
This category shows fusion gene breakpoint, cancer type, and sample information.
2) Fusion Gene ORF analysis.
This category shows the coding potential study results from three approaches. First, we investigated the ORF whether in-frame or frame-shift if both breakpoints are located in the coding sequence (CDS) area. If not, we reported the location of individual breakpoints is in 5'-UTR, CDS, or 3'-UTR. Second, to have the potential amino acid sequence, we ran ORFfinder by NCBI. Third, we ran the in-house classifier (to be available soon) between the coding genes mapped by Ribo-seq reads with high reliability and non-coding genes not mapped by any Ribo-seq reads.

3) Fusion Amino Acid Sequences.

4) Fusion Protein Features.
This category provides the retention information of 39 protein features of fusion proteins based on their multiple isoform gene structures and multiple breakpoints. By focusing on the type of protein features, the user can understand the overall function of fusion genes and make a story in pathogenesis study. In this updated version, we also added the link for our FGviewer, a tool for visualizing functional features of the human fusion genes. FGviewer provides functional feature annotations at four different levels: DNA-, RNA-, protein-, and pathogenic levels. The same breakpoint line across four tiers will classify between FG involving or non-involving zone with multiple types of functional features.
This image shows the introduction of FGviewer and link to have the functional feature visualization and analysis of fusion genes.

This image shows the overview of the FGviewer result page of the TMPRSS2-ERG query search.
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt. (Six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features)
Retention analysis result of each fusion partner protein across 39 protein features of UniProt. (Six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features)Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation results can be downloaded at
download page.
* Minus value of BPloci means that the break point is located before the CDS.- In-frame and retained protein feature among the 13 regional features.
After fusion protein formed, these protein domains or features were still intact.

- In-frame and not-retained protein feature among the 13 regional features.
After fusion protein formed, these protein domains or features were still intact.

5) Fusion Protein Structures.
This category visualize the 3D strucure of the predicted fusion proteins.


6) pLDDT scores.
This category visualize the reliability related information of the predicted fusion protein structures such as pLDDT score distribution across individual fusion partner wild type proteins and fusion protein sequences, protein domain visualization, PAE heatmap, and visualization of the difference of the pLDDT scores between active sites and non-active sites.



7) Ramachandra plot of Fusion Protein Structures
This category shows the distribution of torsion angles in the fusion protein structures.
8) Potential Active Site Information
This table provides the detailed information of individual active sites of fusion protein.
9) Potentially Interacting Small Molecules through Virtual Screening
This table provides the top 10 FDA-approved drugs that potentially interact with fusion proteins.
This table provides the cross reference of the top 10 poentially interacting FDA-approved drugs with DrugBank.

This table provides the fusion protein-top 10 drug network.

10) Biochemical Features of Small Molecules
This table provides the detailed biochemical features of the top 10 screened FDA-approved small molecules.
11) Drug Toxicity Information
This table provides disease information related to each fusion partner.
12) Fusion Protein-Protein Interaction
This category provides diverse annotation information on the PPIs in fusion proteins.


13) Related Drugs
This table provides information of drugs that were used to treat fusion gene patient from manual curation and multiple resources.
14) Related Diseases
This table provides information of diseases that were expressed fusion gene from manual curation and multiple resources.
