| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:CAMK2D-TLL2 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: CAMK2D-TLL2 | FusionPDB ID: 12674 | FusionGDB2.0 ID: 12674 | Hgene | Tgene | Gene symbol | CAMK2D | TLL2 | Gene ID | 817 | 7093 |
| Gene name | calcium/calmodulin dependent protein kinase II delta | tolloid like 2 | |
| Synonyms | CAMKD | - | |
| Cytomap | 4q26 | 10q24.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | calcium/calmodulin-dependent protein kinase type II subunit deltaCaM kinase II delta subunitCaM-kinase II delta chainCaMK-II delta subunitcalcium/calmodulin-dependent protein kinase (CaM kinase) II deltacalcium/calmodulin-dependent protein kinase typ | tolloid-like protein 2 | |
| Modification date | 20200327 | 20200313 | |
| UniProtAcc | Q13557 | . | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000296402, ENST00000342666, ENST00000379773, ENST00000394522, ENST00000394524, ENST00000394526, ENST00000418639, ENST00000429180, ENST00000454265, ENST00000505990, ENST00000508738, ENST00000511664, ENST00000514328, ENST00000515496, ENST00000509907, | ENST00000469598, ENST00000357947, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 10 X 12 X 8=960 | 4 X 5 X 5=100 |
| # samples | 13 | 6 | |
| ** MAII score | log2(13/960*10)=-2.88452278258006 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(6/100*10)=-0.736965594166206 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: CAMK2D [Title/Abstract] AND TLL2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | CAMK2D(114421619)-TLL2(98192719), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | CAMK2D-TLL2 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. CAMK2D-TLL2 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. CAMK2D-TLL2 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. CAMK2D-TLL2 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. CAMK2D-TLL2 seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. CAMK2D-TLL2 seems lost the major protein functional domain in Hgene partner, which is a kinase due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CAMK2D | GO:0003254 | regulation of membrane depolarization | 22514276 |
| Hgene | CAMK2D | GO:0006468 | protein phosphorylation | 17179159|23283722 |
| Hgene | CAMK2D | GO:0018105 | peptidyl-serine phosphorylation | 22514276|23283722 |
| Hgene | CAMK2D | GO:0018107 | peptidyl-threonine phosphorylation | 22514276|23283722 |
| Hgene | CAMK2D | GO:0046777 | protein autophosphorylation | 22514276 |
| Hgene | CAMK2D | GO:1901897 | regulation of relaxation of cardiac muscle | 23283722 |
| Hgene | CAMK2D | GO:1902306 | negative regulation of sodium ion transmembrane transport | 22514276 |
| Hgene | CAMK2D | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity | 22514276 |
 Fusion gene breakpoints across CAMK2D (5'-gene) Fusion gene breakpoints across CAMK2D (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
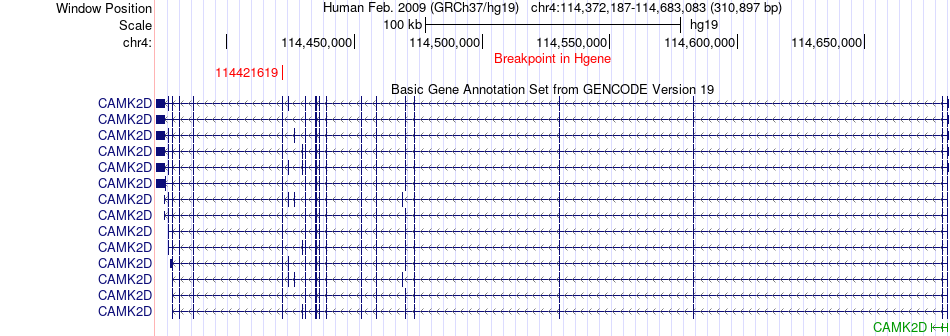 |
 Fusion gene breakpoints across TLL2 (3'-gene) Fusion gene breakpoints across TLL2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
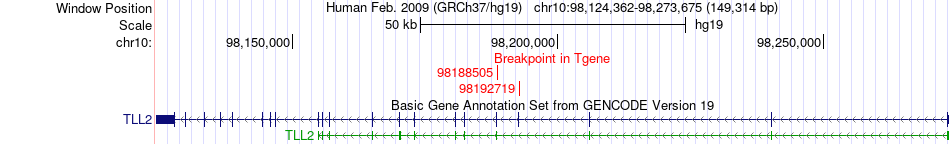 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | READ | TCGA-F5-6864-01A | CAMK2D | chr4 | 114421619 | - | TLL2 | chr10 | 98192719 | - |
| ChimerDB4 | READ | TCGA-F5-6864 | CAMK2D | chr4 | 114421619 | - | TLL2 | chr10 | 98188505 | - |
| ChimerDB4 | READ | TCGA-F5-6864 | CAMK2D | chr4 | 114421619 | - | TLL2 | chr10 | 98192719 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000394524 | CAMK2D | chr4 | 114421619 | - | ENST00000357947 | TLL2 | chr10 | 98192719 | - | 7576 | 1410 | 377 | 4093 | 1238 |
| ENST00000515496 | CAMK2D | chr4 | 114421619 | - | ENST00000357947 | TLL2 | chr10 | 98192719 | - | 7232 | 1066 | 0 | 3749 | 1249 |
| ENST00000514328 | CAMK2D | chr4 | 114421619 | - | ENST00000357947 | TLL2 | chr10 | 98192719 | - | 7196 | 1030 | 0 | 3713 | 1237 |
| ENST00000394522 | CAMK2D | chr4 | 114421619 | - | ENST00000357947 | TLL2 | chr10 | 98192719 | - | 7736 | 1570 | 495 | 4253 | 1252 |
| ENST00000505990 | CAMK2D | chr4 | 114421619 | - | ENST00000357947 | TLL2 | chr10 | 98192719 | - | 7416 | 1250 | 115 | 3933 | 1272 |
| ENST00000379773 | CAMK2D | chr4 | 114421619 | - | ENST00000357947 | TLL2 | chr10 | 98192719 | - | 7261 | 1095 | 62 | 3778 | 1238 |
| ENST00000508738 | CAMK2D | chr4 | 114421619 | - | ENST00000357947 | TLL2 | chr10 | 98192719 | - | 7232 | 1066 | 0 | 3749 | 1249 |
| ENST00000394524 | CAMK2D | chr4 | 114421619 | - | ENST00000357947 | TLL2 | chr10 | 98188505 | - | 7420 | 1410 | 377 | 3937 | 1186 |
| ENST00000515496 | CAMK2D | chr4 | 114421619 | - | ENST00000357947 | TLL2 | chr10 | 98188505 | - | 7076 | 1066 | 0 | 3593 | 1197 |
| ENST00000514328 | CAMK2D | chr4 | 114421619 | - | ENST00000357947 | TLL2 | chr10 | 98188505 | - | 7040 | 1030 | 0 | 3557 | 1185 |
| ENST00000394522 | CAMK2D | chr4 | 114421619 | - | ENST00000357947 | TLL2 | chr10 | 98188505 | - | 7580 | 1570 | 495 | 4097 | 1200 |
| ENST00000505990 | CAMK2D | chr4 | 114421619 | - | ENST00000357947 | TLL2 | chr10 | 98188505 | - | 7260 | 1250 | 115 | 3777 | 1220 |
| ENST00000379773 | CAMK2D | chr4 | 114421619 | - | ENST00000357947 | TLL2 | chr10 | 98188505 | - | 7105 | 1095 | 62 | 3622 | 1186 |
| ENST00000508738 | CAMK2D | chr4 | 114421619 | - | ENST00000357947 | TLL2 | chr10 | 98188505 | - | 7076 | 1066 | 0 | 3593 | 1197 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000394524 | ENST00000357947 | CAMK2D | chr4 | 114421619 | - | TLL2 | chr10 | 98192719 | - | 0.000435439 | 0.9995646 |
| ENST00000515496 | ENST00000357947 | CAMK2D | chr4 | 114421619 | - | TLL2 | chr10 | 98192719 | - | 0.000386968 | 0.9996131 |
| ENST00000514328 | ENST00000357947 | CAMK2D | chr4 | 114421619 | - | TLL2 | chr10 | 98192719 | - | 0.000372817 | 0.9996271 |
| ENST00000394522 | ENST00000357947 | CAMK2D | chr4 | 114421619 | - | TLL2 | chr10 | 98192719 | - | 0.000449711 | 0.9995503 |
| ENST00000505990 | ENST00000357947 | CAMK2D | chr4 | 114421619 | - | TLL2 | chr10 | 98192719 | - | 0.000547576 | 0.9994524 |
| ENST00000379773 | ENST00000357947 | CAMK2D | chr4 | 114421619 | - | TLL2 | chr10 | 98192719 | - | 0.000325225 | 0.9996748 |
| ENST00000508738 | ENST00000357947 | CAMK2D | chr4 | 114421619 | - | TLL2 | chr10 | 98192719 | - | 0.000386968 | 0.9996131 |
| ENST00000394524 | ENST00000357947 | CAMK2D | chr4 | 114421619 | - | TLL2 | chr10 | 98188505 | - | 0.000288424 | 0.9997116 |
| ENST00000515496 | ENST00000357947 | CAMK2D | chr4 | 114421619 | - | TLL2 | chr10 | 98188505 | - | 0.000368042 | 0.999632 |
| ENST00000514328 | ENST00000357947 | CAMK2D | chr4 | 114421619 | - | TLL2 | chr10 | 98188505 | - | 0.000167254 | 0.99983275 |
| ENST00000394522 | ENST00000357947 | CAMK2D | chr4 | 114421619 | - | TLL2 | chr10 | 98188505 | - | 0.000503682 | 0.99949634 |
| ENST00000505990 | ENST00000357947 | CAMK2D | chr4 | 114421619 | - | TLL2 | chr10 | 98188505 | - | 0.000437061 | 0.9995629 |
| ENST00000379773 | ENST00000357947 | CAMK2D | chr4 | 114421619 | - | TLL2 | chr10 | 98188505 | - | 0.000214745 | 0.99978524 |
| ENST00000508738 | ENST00000357947 | CAMK2D | chr4 | 114421619 | - | TLL2 | chr10 | 98188505 | - | 0.000368042 | 0.999632 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >12674_12674_1_CAMK2D-TLL2_CAMK2D_chr4_114421619_ENST00000379773_TLL2_chr10_98188505_ENST00000357947_length(amino acids)=1186AA_BP=342 MASTTTCTRFTDEYQLFEELGKGAFSVVRRCMKIPTGQEYAAKIINTKKLSARDHQKLEREARICRLLKHPNIVRLHDSISEEGFHYLVF DLVTGGELFEDIVAREYYSEADASHCIQQILESVNHCHLNGIVHRDLKPENLLLASKSKGAAVKLADFGLAIEVQGDQQAWFGFAGTPGY LSPEVLRKDPYGKPVDMWACGVILYILLVGYPPFWDEDQHRLYQQIKAGAYDFPSPEWDTVTPEAKDLINKMLTINPAKRITASEALKHP WICQRSTVASMMHRQETVDCLKKFNARRKLKGAILTTMLATRNFSAAKSLLKKPDGVKESTESSNTTIEDEDVKGSQRAIFKQAMRHWEK HTCVTFIERTDEESFIVFSYRTCGCCSYVGRRGGGPQAISIGKNCDKFGIVAHELGHVVGFWHEHTRPDRDQHVTIIRENIQPGQEYNFL KMEAGEVSSLGETYDFDSIMHYARNTFSRGVFLDTILPRQDDNGVRPTIGQRVRLSQGDIAQARKLYKCPACGETLQDTTGNFSAPGFPN GYPSYSHCVWRISVTPGEKIVLNFTSMDLFKSRLCWYDYVEVRDGYWRKAPLLGRFCGDKIPEPLVSTDSRLWVEFRSSSNILGKGFFAA YEATCGGDMNKDAGQIQSPNYPDDYRPSKECVWRITVSEGFHVGLTFQAFEIERHDSCAYDYLEVRDGPTEESALIGHFCGYEKPEDVKS SSNRLWMKFVSDGSINKAGFAANFFKEVDECSWPDHGGCEHRCVNTLGSYKCACDPGYELAADKKMCEVACGGFITKLNGTITSPGWPKE YPTNKNCVWQVVAPAQYRISLQFEVFELEGNDVCKYDFVEVRSGLSPDAKLHGRFCGSETPEVITSQSNNMRVEFKSDNTVSKRGFRAHF FSDKDECAKDNGGCQHECVNTFGSYLCRCRNGYWLHENGHDCKEAGCAHKISSVEGTLASPNWPDKYPSRRECTWNISSTAGHRVKLTFN EFEIEQHQECAYDHLEMYDGPDSLAPILGRFCGSKKPDPTVASGSSMFLRFYSDASVQRKGFQAVHSTECGGRLKAEVQTKELYSHAQFG DNNYPSEARCDWVIVAEDGYGVELTFRTFEVEEEADCGYDYMEAYDGYDSSAPRLGRFCGSGPLEEIYSAGDSLMIRFRTDDTINKKGFH -------------------------------------------------------------- >12674_12674_2_CAMK2D-TLL2_CAMK2D_chr4_114421619_ENST00000379773_TLL2_chr10_98192719_ENST00000357947_length(amino acids)=1238AA_BP=342 MASTTTCTRFTDEYQLFEELGKGAFSVVRRCMKIPTGQEYAAKIINTKKLSARDHQKLEREARICRLLKHPNIVRLHDSISEEGFHYLVF DLVTGGELFEDIVAREYYSEADASHCIQQILESVNHCHLNGIVHRDLKPENLLLASKSKGAAVKLADFGLAIEVQGDQQAWFGFAGTPGY LSPEVLRKDPYGKPVDMWACGVILYILLVGYPPFWDEDQHRLYQQIKAGAYDFPSPEWDTVTPEAKDLINKMLTINPAKRITASEALKHP WICQRSTVASMMHRQETVDCLKKFNARRKLKGAILTTMLATRNFSAAKSLLKKPDGVKESTESSNTTIEDEDVKDGRENTTLLHSPGTLH AAAKTFSPRVRRATTSRTERIWPGGVIPYVIGGNFTGSQRAIFKQAMRHWEKHTCVTFIERTDEESFIVFSYRTCGCCSYVGRRGGGPQA ISIGKNCDKFGIVAHELGHVVGFWHEHTRPDRDQHVTIIRENIQPGQEYNFLKMEAGEVSSLGETYDFDSIMHYARNTFSRGVFLDTILP RQDDNGVRPTIGQRVRLSQGDIAQARKLYKCPACGETLQDTTGNFSAPGFPNGYPSYSHCVWRISVTPGEKIVLNFTSMDLFKSRLCWYD YVEVRDGYWRKAPLLGRFCGDKIPEPLVSTDSRLWVEFRSSSNILGKGFFAAYEATCGGDMNKDAGQIQSPNYPDDYRPSKECVWRITVS EGFHVGLTFQAFEIERHDSCAYDYLEVRDGPTEESALIGHFCGYEKPEDVKSSSNRLWMKFVSDGSINKAGFAANFFKEVDECSWPDHGG CEHRCVNTLGSYKCACDPGYELAADKKMCEVACGGFITKLNGTITSPGWPKEYPTNKNCVWQVVAPAQYRISLQFEVFELEGNDVCKYDF VEVRSGLSPDAKLHGRFCGSETPEVITSQSNNMRVEFKSDNTVSKRGFRAHFFSDKDECAKDNGGCQHECVNTFGSYLCRCRNGYWLHEN GHDCKEAGCAHKISSVEGTLASPNWPDKYPSRRECTWNISSTAGHRVKLTFNEFEIEQHQECAYDHLEMYDGPDSLAPILGRFCGSKKPD PTVASGSSMFLRFYSDASVQRKGFQAVHSTECGGRLKAEVQTKELYSHAQFGDNNYPSEARCDWVIVAEDGYGVELTFRTFEVEEEADCG -------------------------------------------------------------- >12674_12674_3_CAMK2D-TLL2_CAMK2D_chr4_114421619_ENST00000394522_TLL2_chr10_98188505_ENST00000357947_length(amino acids)=1200AA_BP=356 MASTTTCTRFTDEYQLFEELGKGAFSVVRRCMKIPTGQEYAAKIINTKKLSARDHQKLEREARICRLLKHPNIVRLHDSISEEGFHYLVF DLVTGGELFEDIVAREYYSEADASHCIQQILESVNHCHLNGIVHRDLKPENLLLASKSKGAAVKLADFGLAIEVQGDQQAWFGFAGTPGY LSPEVLRKDPYGKPVDMWACGVILYILLVGYPPFWDEDQHRLYQQIKAGAYDFPSPEWDTVTPEAKDLINKMLTINPAKRITASEALKHP WICQRSTVASMMHRQETVDCLKKFNARRKLKGAILTTMLATRNFSAAKSLLKKPDGVKEPQTTVIHNPDGNKESTESSNTTIEDEDVKGS QRAIFKQAMRHWEKHTCVTFIERTDEESFIVFSYRTCGCCSYVGRRGGGPQAISIGKNCDKFGIVAHELGHVVGFWHEHTRPDRDQHVTI IRENIQPGQEYNFLKMEAGEVSSLGETYDFDSIMHYARNTFSRGVFLDTILPRQDDNGVRPTIGQRVRLSQGDIAQARKLYKCPACGETL QDTTGNFSAPGFPNGYPSYSHCVWRISVTPGEKIVLNFTSMDLFKSRLCWYDYVEVRDGYWRKAPLLGRFCGDKIPEPLVSTDSRLWVEF RSSSNILGKGFFAAYEATCGGDMNKDAGQIQSPNYPDDYRPSKECVWRITVSEGFHVGLTFQAFEIERHDSCAYDYLEVRDGPTEESALI GHFCGYEKPEDVKSSSNRLWMKFVSDGSINKAGFAANFFKEVDECSWPDHGGCEHRCVNTLGSYKCACDPGYELAADKKMCEVACGGFIT KLNGTITSPGWPKEYPTNKNCVWQVVAPAQYRISLQFEVFELEGNDVCKYDFVEVRSGLSPDAKLHGRFCGSETPEVITSQSNNMRVEFK SDNTVSKRGFRAHFFSDKDECAKDNGGCQHECVNTFGSYLCRCRNGYWLHENGHDCKEAGCAHKISSVEGTLASPNWPDKYPSRRECTWN ISSTAGHRVKLTFNEFEIEQHQECAYDHLEMYDGPDSLAPILGRFCGSKKPDPTVASGSSMFLRFYSDASVQRKGFQAVHSTECGGRLKA EVQTKELYSHAQFGDNNYPSEARCDWVIVAEDGYGVELTFRTFEVEEEADCGYDYMEAYDGYDSSAPRLGRFCGSGPLEEIYSAGDSLMI -------------------------------------------------------------- >12674_12674_4_CAMK2D-TLL2_CAMK2D_chr4_114421619_ENST00000394522_TLL2_chr10_98192719_ENST00000357947_length(amino acids)=1252AA_BP=356 MASTTTCTRFTDEYQLFEELGKGAFSVVRRCMKIPTGQEYAAKIINTKKLSARDHQKLEREARICRLLKHPNIVRLHDSISEEGFHYLVF DLVTGGELFEDIVAREYYSEADASHCIQQILESVNHCHLNGIVHRDLKPENLLLASKSKGAAVKLADFGLAIEVQGDQQAWFGFAGTPGY LSPEVLRKDPYGKPVDMWACGVILYILLVGYPPFWDEDQHRLYQQIKAGAYDFPSPEWDTVTPEAKDLINKMLTINPAKRITASEALKHP WICQRSTVASMMHRQETVDCLKKFNARRKLKGAILTTMLATRNFSAAKSLLKKPDGVKEPQTTVIHNPDGNKESTESSNTTIEDEDVKDG RENTTLLHSPGTLHAAAKTFSPRVRRATTSRTERIWPGGVIPYVIGGNFTGSQRAIFKQAMRHWEKHTCVTFIERTDEESFIVFSYRTCG CCSYVGRRGGGPQAISIGKNCDKFGIVAHELGHVVGFWHEHTRPDRDQHVTIIRENIQPGQEYNFLKMEAGEVSSLGETYDFDSIMHYAR NTFSRGVFLDTILPRQDDNGVRPTIGQRVRLSQGDIAQARKLYKCPACGETLQDTTGNFSAPGFPNGYPSYSHCVWRISVTPGEKIVLNF TSMDLFKSRLCWYDYVEVRDGYWRKAPLLGRFCGDKIPEPLVSTDSRLWVEFRSSSNILGKGFFAAYEATCGGDMNKDAGQIQSPNYPDD YRPSKECVWRITVSEGFHVGLTFQAFEIERHDSCAYDYLEVRDGPTEESALIGHFCGYEKPEDVKSSSNRLWMKFVSDGSINKAGFAANF FKEVDECSWPDHGGCEHRCVNTLGSYKCACDPGYELAADKKMCEVACGGFITKLNGTITSPGWPKEYPTNKNCVWQVVAPAQYRISLQFE VFELEGNDVCKYDFVEVRSGLSPDAKLHGRFCGSETPEVITSQSNNMRVEFKSDNTVSKRGFRAHFFSDKDECAKDNGGCQHECVNTFGS YLCRCRNGYWLHENGHDCKEAGCAHKISSVEGTLASPNWPDKYPSRRECTWNISSTAGHRVKLTFNEFEIEQHQECAYDHLEMYDGPDSL APILGRFCGSKKPDPTVASGSSMFLRFYSDASVQRKGFQAVHSTECGGRLKAEVQTKELYSHAQFGDNNYPSEARCDWVIVAEDGYGVEL -------------------------------------------------------------- >12674_12674_5_CAMK2D-TLL2_CAMK2D_chr4_114421619_ENST00000394524_TLL2_chr10_98188505_ENST00000357947_length(amino acids)=1186AA_BP=342 MASTTTCTRFTDEYQLFEELGKGAFSVVRRCMKIPTGQEYAAKIINTKKLSARDHQKLEREARICRLLKHPNIVRLHDSISEEGFHYLVF DLVTGGELFEDIVAREYYSEADASHCIQQILESVNHCHLNGIVHRDLKPENLLLASKSKGAAVKLADFGLAIEVQGDQQAWFGFAGTPGY LSPEVLRKDPYGKPVDMWACGVILYILLVGYPPFWDEDQHRLYQQIKAGAYDFPSPEWDTVTPEAKDLINKMLTINPAKRITASEALKHP WICQRSTVASMMHRQETVDCLKKFNARRKLKGAILTTMLATRNFSAAKSLLKKPDGVKESTESSNTTIEDEDVKGSQRAIFKQAMRHWEK HTCVTFIERTDEESFIVFSYRTCGCCSYVGRRGGGPQAISIGKNCDKFGIVAHELGHVVGFWHEHTRPDRDQHVTIIRENIQPGQEYNFL KMEAGEVSSLGETYDFDSIMHYARNTFSRGVFLDTILPRQDDNGVRPTIGQRVRLSQGDIAQARKLYKCPACGETLQDTTGNFSAPGFPN GYPSYSHCVWRISVTPGEKIVLNFTSMDLFKSRLCWYDYVEVRDGYWRKAPLLGRFCGDKIPEPLVSTDSRLWVEFRSSSNILGKGFFAA YEATCGGDMNKDAGQIQSPNYPDDYRPSKECVWRITVSEGFHVGLTFQAFEIERHDSCAYDYLEVRDGPTEESALIGHFCGYEKPEDVKS SSNRLWMKFVSDGSINKAGFAANFFKEVDECSWPDHGGCEHRCVNTLGSYKCACDPGYELAADKKMCEVACGGFITKLNGTITSPGWPKE YPTNKNCVWQVVAPAQYRISLQFEVFELEGNDVCKYDFVEVRSGLSPDAKLHGRFCGSETPEVITSQSNNMRVEFKSDNTVSKRGFRAHF FSDKDECAKDNGGCQHECVNTFGSYLCRCRNGYWLHENGHDCKEAGCAHKISSVEGTLASPNWPDKYPSRRECTWNISSTAGHRVKLTFN EFEIEQHQECAYDHLEMYDGPDSLAPILGRFCGSKKPDPTVASGSSMFLRFYSDASVQRKGFQAVHSTECGGRLKAEVQTKELYSHAQFG DNNYPSEARCDWVIVAEDGYGVELTFRTFEVEEEADCGYDYMEAYDGYDSSAPRLGRFCGSGPLEEIYSAGDSLMIRFRTDDTINKKGFH -------------------------------------------------------------- >12674_12674_6_CAMK2D-TLL2_CAMK2D_chr4_114421619_ENST00000394524_TLL2_chr10_98192719_ENST00000357947_length(amino acids)=1238AA_BP=342 MASTTTCTRFTDEYQLFEELGKGAFSVVRRCMKIPTGQEYAAKIINTKKLSARDHQKLEREARICRLLKHPNIVRLHDSISEEGFHYLVF DLVTGGELFEDIVAREYYSEADASHCIQQILESVNHCHLNGIVHRDLKPENLLLASKSKGAAVKLADFGLAIEVQGDQQAWFGFAGTPGY LSPEVLRKDPYGKPVDMWACGVILYILLVGYPPFWDEDQHRLYQQIKAGAYDFPSPEWDTVTPEAKDLINKMLTINPAKRITASEALKHP WICQRSTVASMMHRQETVDCLKKFNARRKLKGAILTTMLATRNFSAAKSLLKKPDGVKESTESSNTTIEDEDVKDGRENTTLLHSPGTLH AAAKTFSPRVRRATTSRTERIWPGGVIPYVIGGNFTGSQRAIFKQAMRHWEKHTCVTFIERTDEESFIVFSYRTCGCCSYVGRRGGGPQA ISIGKNCDKFGIVAHELGHVVGFWHEHTRPDRDQHVTIIRENIQPGQEYNFLKMEAGEVSSLGETYDFDSIMHYARNTFSRGVFLDTILP RQDDNGVRPTIGQRVRLSQGDIAQARKLYKCPACGETLQDTTGNFSAPGFPNGYPSYSHCVWRISVTPGEKIVLNFTSMDLFKSRLCWYD YVEVRDGYWRKAPLLGRFCGDKIPEPLVSTDSRLWVEFRSSSNILGKGFFAAYEATCGGDMNKDAGQIQSPNYPDDYRPSKECVWRITVS EGFHVGLTFQAFEIERHDSCAYDYLEVRDGPTEESALIGHFCGYEKPEDVKSSSNRLWMKFVSDGSINKAGFAANFFKEVDECSWPDHGG CEHRCVNTLGSYKCACDPGYELAADKKMCEVACGGFITKLNGTITSPGWPKEYPTNKNCVWQVVAPAQYRISLQFEVFELEGNDVCKYDF VEVRSGLSPDAKLHGRFCGSETPEVITSQSNNMRVEFKSDNTVSKRGFRAHFFSDKDECAKDNGGCQHECVNTFGSYLCRCRNGYWLHEN GHDCKEAGCAHKISSVEGTLASPNWPDKYPSRRECTWNISSTAGHRVKLTFNEFEIEQHQECAYDHLEMYDGPDSLAPILGRFCGSKKPD PTVASGSSMFLRFYSDASVQRKGFQAVHSTECGGRLKAEVQTKELYSHAQFGDNNYPSEARCDWVIVAEDGYGVELTFRTFEVEEEADCG -------------------------------------------------------------- >12674_12674_7_CAMK2D-TLL2_CAMK2D_chr4_114421619_ENST00000505990_TLL2_chr10_98188505_ENST00000357947_length(amino acids)=1220AA_BP=376 MASTTTCTRFTDEYQLFEELGKGAFSVVRRCMKIPTGQEYAAKIINTKKLSARDHQKLEREARICRLLKHPNIVRLHDSISEEGFHYLVF DLVTGGELFEDIVAREYYSEADASHCIQQILEAVLHCHQMGVVHRDLKPENLLLASKSKGAAVKLADFGLAIEVQGDQQAWFGFAGTPGY LSPEVLRKDPYGKPVDMWACGVILYILLVGYPPFWDEDQHRLYQQIKAGAYDFPSPEWDTVTPEAKDLINKMLTINPAKRITASEALKHP WICQRSTVASMMHRQETVDCLKKFNARRKLKGAILTTMLATRNFSAAKSLLKKPDGVKINNKANVVTSPKENIPTPALEPQTTVIHNPDG NKESTESSNTTIEDEDVKGSQRAIFKQAMRHWEKHTCVTFIERTDEESFIVFSYRTCGCCSYVGRRGGGPQAISIGKNCDKFGIVAHELG HVVGFWHEHTRPDRDQHVTIIRENIQPGQEYNFLKMEAGEVSSLGETYDFDSIMHYARNTFSRGVFLDTILPRQDDNGVRPTIGQRVRLS QGDIAQARKLYKCPACGETLQDTTGNFSAPGFPNGYPSYSHCVWRISVTPGEKIVLNFTSMDLFKSRLCWYDYVEVRDGYWRKAPLLGRF CGDKIPEPLVSTDSRLWVEFRSSSNILGKGFFAAYEATCGGDMNKDAGQIQSPNYPDDYRPSKECVWRITVSEGFHVGLTFQAFEIERHD SCAYDYLEVRDGPTEESALIGHFCGYEKPEDVKSSSNRLWMKFVSDGSINKAGFAANFFKEVDECSWPDHGGCEHRCVNTLGSYKCACDP GYELAADKKMCEVACGGFITKLNGTITSPGWPKEYPTNKNCVWQVVAPAQYRISLQFEVFELEGNDVCKYDFVEVRSGLSPDAKLHGRFC GSETPEVITSQSNNMRVEFKSDNTVSKRGFRAHFFSDKDECAKDNGGCQHECVNTFGSYLCRCRNGYWLHENGHDCKEAGCAHKISSVEG TLASPNWPDKYPSRRECTWNISSTAGHRVKLTFNEFEIEQHQECAYDHLEMYDGPDSLAPILGRFCGSKKPDPTVASGSSMFLRFYSDAS VQRKGFQAVHSTECGGRLKAEVQTKELYSHAQFGDNNYPSEARCDWVIVAEDGYGVELTFRTFEVEEEADCGYDYMEAYDGYDSSAPRLG -------------------------------------------------------------- >12674_12674_8_CAMK2D-TLL2_CAMK2D_chr4_114421619_ENST00000505990_TLL2_chr10_98192719_ENST00000357947_length(amino acids)=1272AA_BP=376 MASTTTCTRFTDEYQLFEELGKGAFSVVRRCMKIPTGQEYAAKIINTKKLSARDHQKLEREARICRLLKHPNIVRLHDSISEEGFHYLVF DLVTGGELFEDIVAREYYSEADASHCIQQILEAVLHCHQMGVVHRDLKPENLLLASKSKGAAVKLADFGLAIEVQGDQQAWFGFAGTPGY LSPEVLRKDPYGKPVDMWACGVILYILLVGYPPFWDEDQHRLYQQIKAGAYDFPSPEWDTVTPEAKDLINKMLTINPAKRITASEALKHP WICQRSTVASMMHRQETVDCLKKFNARRKLKGAILTTMLATRNFSAAKSLLKKPDGVKINNKANVVTSPKENIPTPALEPQTTVIHNPDG NKESTESSNTTIEDEDVKDGRENTTLLHSPGTLHAAAKTFSPRVRRATTSRTERIWPGGVIPYVIGGNFTGSQRAIFKQAMRHWEKHTCV TFIERTDEESFIVFSYRTCGCCSYVGRRGGGPQAISIGKNCDKFGIVAHELGHVVGFWHEHTRPDRDQHVTIIRENIQPGQEYNFLKMEA GEVSSLGETYDFDSIMHYARNTFSRGVFLDTILPRQDDNGVRPTIGQRVRLSQGDIAQARKLYKCPACGETLQDTTGNFSAPGFPNGYPS YSHCVWRISVTPGEKIVLNFTSMDLFKSRLCWYDYVEVRDGYWRKAPLLGRFCGDKIPEPLVSTDSRLWVEFRSSSNILGKGFFAAYEAT CGGDMNKDAGQIQSPNYPDDYRPSKECVWRITVSEGFHVGLTFQAFEIERHDSCAYDYLEVRDGPTEESALIGHFCGYEKPEDVKSSSNR LWMKFVSDGSINKAGFAANFFKEVDECSWPDHGGCEHRCVNTLGSYKCACDPGYELAADKKMCEVACGGFITKLNGTITSPGWPKEYPTN KNCVWQVVAPAQYRISLQFEVFELEGNDVCKYDFVEVRSGLSPDAKLHGRFCGSETPEVITSQSNNMRVEFKSDNTVSKRGFRAHFFSDK DECAKDNGGCQHECVNTFGSYLCRCRNGYWLHENGHDCKEAGCAHKISSVEGTLASPNWPDKYPSRRECTWNISSTAGHRVKLTFNEFEI EQHQECAYDHLEMYDGPDSLAPILGRFCGSKKPDPTVASGSSMFLRFYSDASVQRKGFQAVHSTECGGRLKAEVQTKELYSHAQFGDNNY PSEARCDWVIVAEDGYGVELTFRTFEVEEEADCGYDYMEAYDGYDSSAPRLGRFCGSGPLEEIYSAGDSLMIRFRTDDTINKKGFHARYT -------------------------------------------------------------- >12674_12674_9_CAMK2D-TLL2_CAMK2D_chr4_114421619_ENST00000508738_TLL2_chr10_98188505_ENST00000357947_length(amino acids)=1197AA_BP=353 MASTTTCTRFTDEYQLFEELGKGAFSVVRRCMKIPTGQEYAAKIINTKKLSARDHQKLEREARICRLLKHPNIVRLHDSISEEGFHYLVF DLVTGGELFEDIVAREYYSEADASHCIQQILESVNHCHLNGIVHRDLKPENLLLASKSKGAAVKLADFGLAIEVQGDQQAWFGFAGTPGY LSPEVLRKDPYGKPVDMWACGVILYILLVGYPPFWDEDQHRLYQQIKAGAYDFPSPEWDTVTPEAKDLINKMLTINPAKRITASEALKHP WICQRSTVASMMHRQETVDCLKKFNARRKLKGAILTTMLATRNFSAAKSLLKKPDGVKKRKSSSSVQMMESTESSNTTIEDEDVKGSQRA IFKQAMRHWEKHTCVTFIERTDEESFIVFSYRTCGCCSYVGRRGGGPQAISIGKNCDKFGIVAHELGHVVGFWHEHTRPDRDQHVTIIRE NIQPGQEYNFLKMEAGEVSSLGETYDFDSIMHYARNTFSRGVFLDTILPRQDDNGVRPTIGQRVRLSQGDIAQARKLYKCPACGETLQDT TGNFSAPGFPNGYPSYSHCVWRISVTPGEKIVLNFTSMDLFKSRLCWYDYVEVRDGYWRKAPLLGRFCGDKIPEPLVSTDSRLWVEFRSS SNILGKGFFAAYEATCGGDMNKDAGQIQSPNYPDDYRPSKECVWRITVSEGFHVGLTFQAFEIERHDSCAYDYLEVRDGPTEESALIGHF CGYEKPEDVKSSSNRLWMKFVSDGSINKAGFAANFFKEVDECSWPDHGGCEHRCVNTLGSYKCACDPGYELAADKKMCEVACGGFITKLN GTITSPGWPKEYPTNKNCVWQVVAPAQYRISLQFEVFELEGNDVCKYDFVEVRSGLSPDAKLHGRFCGSETPEVITSQSNNMRVEFKSDN TVSKRGFRAHFFSDKDECAKDNGGCQHECVNTFGSYLCRCRNGYWLHENGHDCKEAGCAHKISSVEGTLASPNWPDKYPSRRECTWNISS TAGHRVKLTFNEFEIEQHQECAYDHLEMYDGPDSLAPILGRFCGSKKPDPTVASGSSMFLRFYSDASVQRKGFQAVHSTECGGRLKAEVQ TKELYSHAQFGDNNYPSEARCDWVIVAEDGYGVELTFRTFEVEEEADCGYDYMEAYDGYDSSAPRLGRFCGSGPLEEIYSAGDSLMIRFR -------------------------------------------------------------- >12674_12674_10_CAMK2D-TLL2_CAMK2D_chr4_114421619_ENST00000508738_TLL2_chr10_98192719_ENST00000357947_length(amino acids)=1249AA_BP=353 MASTTTCTRFTDEYQLFEELGKGAFSVVRRCMKIPTGQEYAAKIINTKKLSARDHQKLEREARICRLLKHPNIVRLHDSISEEGFHYLVF DLVTGGELFEDIVAREYYSEADASHCIQQILESVNHCHLNGIVHRDLKPENLLLASKSKGAAVKLADFGLAIEVQGDQQAWFGFAGTPGY LSPEVLRKDPYGKPVDMWACGVILYILLVGYPPFWDEDQHRLYQQIKAGAYDFPSPEWDTVTPEAKDLINKMLTINPAKRITASEALKHP WICQRSTVASMMHRQETVDCLKKFNARRKLKGAILTTMLATRNFSAAKSLLKKPDGVKKRKSSSSVQMMESTESSNTTIEDEDVKDGREN TTLLHSPGTLHAAAKTFSPRVRRATTSRTERIWPGGVIPYVIGGNFTGSQRAIFKQAMRHWEKHTCVTFIERTDEESFIVFSYRTCGCCS YVGRRGGGPQAISIGKNCDKFGIVAHELGHVVGFWHEHTRPDRDQHVTIIRENIQPGQEYNFLKMEAGEVSSLGETYDFDSIMHYARNTF SRGVFLDTILPRQDDNGVRPTIGQRVRLSQGDIAQARKLYKCPACGETLQDTTGNFSAPGFPNGYPSYSHCVWRISVTPGEKIVLNFTSM DLFKSRLCWYDYVEVRDGYWRKAPLLGRFCGDKIPEPLVSTDSRLWVEFRSSSNILGKGFFAAYEATCGGDMNKDAGQIQSPNYPDDYRP SKECVWRITVSEGFHVGLTFQAFEIERHDSCAYDYLEVRDGPTEESALIGHFCGYEKPEDVKSSSNRLWMKFVSDGSINKAGFAANFFKE VDECSWPDHGGCEHRCVNTLGSYKCACDPGYELAADKKMCEVACGGFITKLNGTITSPGWPKEYPTNKNCVWQVVAPAQYRISLQFEVFE LEGNDVCKYDFVEVRSGLSPDAKLHGRFCGSETPEVITSQSNNMRVEFKSDNTVSKRGFRAHFFSDKDECAKDNGGCQHECVNTFGSYLC RCRNGYWLHENGHDCKEAGCAHKISSVEGTLASPNWPDKYPSRRECTWNISSTAGHRVKLTFNEFEIEQHQECAYDHLEMYDGPDSLAPI LGRFCGSKKPDPTVASGSSMFLRFYSDASVQRKGFQAVHSTECGGRLKAEVQTKELYSHAQFGDNNYPSEARCDWVIVAEDGYGVELTFR -------------------------------------------------------------- >12674_12674_11_CAMK2D-TLL2_CAMK2D_chr4_114421619_ENST00000514328_TLL2_chr10_98188505_ENST00000357947_length(amino acids)=1185AA_BP=341 MASTTTCTRFTDEYQLFEELGKGAFSVVRRCMKIPTGQEYAAKIINTKKLSARDHQKLEREARICRLLKHPNIVRLHDSISEEGFHYLVF DLVTGGELFEDIVAREYYSEADASHCIQQILESVNHCHLNGIVHRDLKPENLLLASKSKGAAVKLADFGLAIEVQGDQQAWFGFAGTPGY LSPEVLRKDPYGKPVDMWACGVILYILLVGYPPFWDEDQHRLYQQIKAGAYDFPSPEWDTVTPEAKDLINKMLTINPAKRITASEALKHP WICQRSTVASMMHRQETVDCLKKFNARRKLKGAILTTMLATRNFSAKSLLKKPDGVKESTESSNTTIEDEDVKGSQRAIFKQAMRHWEKH TCVTFIERTDEESFIVFSYRTCGCCSYVGRRGGGPQAISIGKNCDKFGIVAHELGHVVGFWHEHTRPDRDQHVTIIRENIQPGQEYNFLK MEAGEVSSLGETYDFDSIMHYARNTFSRGVFLDTILPRQDDNGVRPTIGQRVRLSQGDIAQARKLYKCPACGETLQDTTGNFSAPGFPNG YPSYSHCVWRISVTPGEKIVLNFTSMDLFKSRLCWYDYVEVRDGYWRKAPLLGRFCGDKIPEPLVSTDSRLWVEFRSSSNILGKGFFAAY EATCGGDMNKDAGQIQSPNYPDDYRPSKECVWRITVSEGFHVGLTFQAFEIERHDSCAYDYLEVRDGPTEESALIGHFCGYEKPEDVKSS SNRLWMKFVSDGSINKAGFAANFFKEVDECSWPDHGGCEHRCVNTLGSYKCACDPGYELAADKKMCEVACGGFITKLNGTITSPGWPKEY PTNKNCVWQVVAPAQYRISLQFEVFELEGNDVCKYDFVEVRSGLSPDAKLHGRFCGSETPEVITSQSNNMRVEFKSDNTVSKRGFRAHFF SDKDECAKDNGGCQHECVNTFGSYLCRCRNGYWLHENGHDCKEAGCAHKISSVEGTLASPNWPDKYPSRRECTWNISSTAGHRVKLTFNE FEIEQHQECAYDHLEMYDGPDSLAPILGRFCGSKKPDPTVASGSSMFLRFYSDASVQRKGFQAVHSTECGGRLKAEVQTKELYSHAQFGD NNYPSEARCDWVIVAEDGYGVELTFRTFEVEEEADCGYDYMEAYDGYDSSAPRLGRFCGSGPLEEIYSAGDSLMIRFRTDDTINKKGFHA -------------------------------------------------------------- >12674_12674_12_CAMK2D-TLL2_CAMK2D_chr4_114421619_ENST00000514328_TLL2_chr10_98192719_ENST00000357947_length(amino acids)=1237AA_BP=341 MASTTTCTRFTDEYQLFEELGKGAFSVVRRCMKIPTGQEYAAKIINTKKLSARDHQKLEREARICRLLKHPNIVRLHDSISEEGFHYLVF DLVTGGELFEDIVAREYYSEADASHCIQQILESVNHCHLNGIVHRDLKPENLLLASKSKGAAVKLADFGLAIEVQGDQQAWFGFAGTPGY LSPEVLRKDPYGKPVDMWACGVILYILLVGYPPFWDEDQHRLYQQIKAGAYDFPSPEWDTVTPEAKDLINKMLTINPAKRITASEALKHP WICQRSTVASMMHRQETVDCLKKFNARRKLKGAILTTMLATRNFSAKSLLKKPDGVKESTESSNTTIEDEDVKDGRENTTLLHSPGTLHA AAKTFSPRVRRATTSRTERIWPGGVIPYVIGGNFTGSQRAIFKQAMRHWEKHTCVTFIERTDEESFIVFSYRTCGCCSYVGRRGGGPQAI SIGKNCDKFGIVAHELGHVVGFWHEHTRPDRDQHVTIIRENIQPGQEYNFLKMEAGEVSSLGETYDFDSIMHYARNTFSRGVFLDTILPR QDDNGVRPTIGQRVRLSQGDIAQARKLYKCPACGETLQDTTGNFSAPGFPNGYPSYSHCVWRISVTPGEKIVLNFTSMDLFKSRLCWYDY VEVRDGYWRKAPLLGRFCGDKIPEPLVSTDSRLWVEFRSSSNILGKGFFAAYEATCGGDMNKDAGQIQSPNYPDDYRPSKECVWRITVSE GFHVGLTFQAFEIERHDSCAYDYLEVRDGPTEESALIGHFCGYEKPEDVKSSSNRLWMKFVSDGSINKAGFAANFFKEVDECSWPDHGGC EHRCVNTLGSYKCACDPGYELAADKKMCEVACGGFITKLNGTITSPGWPKEYPTNKNCVWQVVAPAQYRISLQFEVFELEGNDVCKYDFV EVRSGLSPDAKLHGRFCGSETPEVITSQSNNMRVEFKSDNTVSKRGFRAHFFSDKDECAKDNGGCQHECVNTFGSYLCRCRNGYWLHENG HDCKEAGCAHKISSVEGTLASPNWPDKYPSRRECTWNISSTAGHRVKLTFNEFEIEQHQECAYDHLEMYDGPDSLAPILGRFCGSKKPDP TVASGSSMFLRFYSDASVQRKGFQAVHSTECGGRLKAEVQTKELYSHAQFGDNNYPSEARCDWVIVAEDGYGVELTFRTFEVEEEADCGY -------------------------------------------------------------- >12674_12674_13_CAMK2D-TLL2_CAMK2D_chr4_114421619_ENST00000515496_TLL2_chr10_98188505_ENST00000357947_length(amino acids)=1197AA_BP=353 MASTTTCTRFTDEYQLFEELGKGAFSVVRRCMKIPTGQEYAAKIINTKKLSARDHQKLEREARICRLLKHPNIVRLHDSISEEGFHYLVF DLVTGGELFEDIVAREYYSEADASHCIQQILESVNHCHLNGIVHRDLKPENLLLASKSKGAAVKLADFGLAIEVQGDQQAWFGFAGTPGY LSPEVLRKDPYGKPVDMWACGVILYILLVGYPPFWDEDQHRLYQQIKAGAYDFPSPEWDTVTPEAKDLINKMLTINPAKRITASEALKHP WICQRSTVASMMHRQETVDCLKKFNARRKLKGAILTTMLATRNFSAAKSLLKKPDGVKKRKSSSSVQMMESTESSNTTIEDEDVKGSQRA IFKQAMRHWEKHTCVTFIERTDEESFIVFSYRTCGCCSYVGRRGGGPQAISIGKNCDKFGIVAHELGHVVGFWHEHTRPDRDQHVTIIRE NIQPGQEYNFLKMEAGEVSSLGETYDFDSIMHYARNTFSRGVFLDTILPRQDDNGVRPTIGQRVRLSQGDIAQARKLYKCPACGETLQDT TGNFSAPGFPNGYPSYSHCVWRISVTPGEKIVLNFTSMDLFKSRLCWYDYVEVRDGYWRKAPLLGRFCGDKIPEPLVSTDSRLWVEFRSS SNILGKGFFAAYEATCGGDMNKDAGQIQSPNYPDDYRPSKECVWRITVSEGFHVGLTFQAFEIERHDSCAYDYLEVRDGPTEESALIGHF CGYEKPEDVKSSSNRLWMKFVSDGSINKAGFAANFFKEVDECSWPDHGGCEHRCVNTLGSYKCACDPGYELAADKKMCEVACGGFITKLN GTITSPGWPKEYPTNKNCVWQVVAPAQYRISLQFEVFELEGNDVCKYDFVEVRSGLSPDAKLHGRFCGSETPEVITSQSNNMRVEFKSDN TVSKRGFRAHFFSDKDECAKDNGGCQHECVNTFGSYLCRCRNGYWLHENGHDCKEAGCAHKISSVEGTLASPNWPDKYPSRRECTWNISS TAGHRVKLTFNEFEIEQHQECAYDHLEMYDGPDSLAPILGRFCGSKKPDPTVASGSSMFLRFYSDASVQRKGFQAVHSTECGGRLKAEVQ TKELYSHAQFGDNNYPSEARCDWVIVAEDGYGVELTFRTFEVEEEADCGYDYMEAYDGYDSSAPRLGRFCGSGPLEEIYSAGDSLMIRFR -------------------------------------------------------------- >12674_12674_14_CAMK2D-TLL2_CAMK2D_chr4_114421619_ENST00000515496_TLL2_chr10_98192719_ENST00000357947_length(amino acids)=1249AA_BP=353 MASTTTCTRFTDEYQLFEELGKGAFSVVRRCMKIPTGQEYAAKIINTKKLSARDHQKLEREARICRLLKHPNIVRLHDSISEEGFHYLVF DLVTGGELFEDIVAREYYSEADASHCIQQILESVNHCHLNGIVHRDLKPENLLLASKSKGAAVKLADFGLAIEVQGDQQAWFGFAGTPGY LSPEVLRKDPYGKPVDMWACGVILYILLVGYPPFWDEDQHRLYQQIKAGAYDFPSPEWDTVTPEAKDLINKMLTINPAKRITASEALKHP WICQRSTVASMMHRQETVDCLKKFNARRKLKGAILTTMLATRNFSAAKSLLKKPDGVKKRKSSSSVQMMESTESSNTTIEDEDVKDGREN TTLLHSPGTLHAAAKTFSPRVRRATTSRTERIWPGGVIPYVIGGNFTGSQRAIFKQAMRHWEKHTCVTFIERTDEESFIVFSYRTCGCCS YVGRRGGGPQAISIGKNCDKFGIVAHELGHVVGFWHEHTRPDRDQHVTIIRENIQPGQEYNFLKMEAGEVSSLGETYDFDSIMHYARNTF SRGVFLDTILPRQDDNGVRPTIGQRVRLSQGDIAQARKLYKCPACGETLQDTTGNFSAPGFPNGYPSYSHCVWRISVTPGEKIVLNFTSM DLFKSRLCWYDYVEVRDGYWRKAPLLGRFCGDKIPEPLVSTDSRLWVEFRSSSNILGKGFFAAYEATCGGDMNKDAGQIQSPNYPDDYRP SKECVWRITVSEGFHVGLTFQAFEIERHDSCAYDYLEVRDGPTEESALIGHFCGYEKPEDVKSSSNRLWMKFVSDGSINKAGFAANFFKE VDECSWPDHGGCEHRCVNTLGSYKCACDPGYELAADKKMCEVACGGFITKLNGTITSPGWPKEYPTNKNCVWQVVAPAQYRISLQFEVFE LEGNDVCKYDFVEVRSGLSPDAKLHGRFCGSETPEVITSQSNNMRVEFKSDNTVSKRGFRAHFFSDKDECAKDNGGCQHECVNTFGSYLC RCRNGYWLHENGHDCKEAGCAHKISSVEGTLASPNWPDKYPSRRECTWNISSTAGHRVKLTFNEFEIEQHQECAYDHLEMYDGPDSLAPI LGRFCGSKKPDPTVASGSSMFLRFYSDASVQRKGFQAVHSTECGGRLKAEVQTKELYSHAQFGDNNYPSEARCDWVIVAEDGYGVELTFR -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr4:114421619/chr10:98192719) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CAMK2D | . |
| FUNCTION: Calcium/calmodulin-dependent protein kinase involved in the regulation of Ca(2+) homeostatis and excitation-contraction coupling (ECC) in heart by targeting ion channels, transporters and accessory proteins involved in Ca(2+) influx into the myocyte, Ca(2+) release from the sarcoplasmic reticulum (SR), SR Ca(2+) uptake and Na(+) and K(+) channel transport. Targets also transcription factors and signaling molecules to regulate heart function. In its activated form, is involved in the pathogenesis of dilated cardiomyopathy and heart failure. Contributes to cardiac decompensation and heart failure by regulating SR Ca(2+) release via direct phosphorylation of RYR2 Ca(2+) channel on 'Ser-2808'. In the nucleus, phosphorylates the MEF2 repressor HDAC4, promoting its nuclear export and binding to 14-3-3 protein, and expression of MEF2 and genes involved in the hypertrophic program. Is essential for left ventricular remodeling responses to myocardial infarction. In pathological myocardial remodeling acts downstream of the beta adrenergic receptor signaling cascade to regulate key proteins involved in ECC. Regulates Ca(2+) influx to myocytes by binding and phosphorylating the L-type Ca(2+) channel subunit beta-2 CACNB2. In addition to Ca(2+) channels, can target and regulate the cardiac sarcolemmal Na(+) channel Nav1.5/SCN5A and the K+ channel Kv4.3/KCND3, which contribute to arrhythmogenesis in heart failure. Phosphorylates phospholamban (PLN/PLB), an endogenous inhibitor of SERCA2A/ATP2A2, contributing to the enhancement of SR Ca(2+) uptake that may be important in frequency-dependent acceleration of relaxation (FDAR) and maintenance of contractile function during acidosis. May participate in the modulation of skeletal muscle function in response to exercise, by regulating SR Ca(2+) transport through phosphorylation of PLN/PLB and triadin, a ryanodine receptor-coupling factor. {ECO:0000269|PubMed:16690701, ECO:0000269|PubMed:17179159}. | FUNCTION: Might normally function as a transcriptional repressor. EWS-fusion-proteins (EFPS) may play a role in the tumorigenic process. They may disturb gene expression by mimicking, or interfering with the normal function of CTD-POLII within the transcription initiation complex. They may also contribute to an aberrant activation of the fusion protein target genes. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000296402 | - | 14 | 18 | 14_272 | 344.3333333333333 | 1391.6666666666667 | Domain | Protein kinase |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000342666 | - | 14 | 19 | 14_272 | 344.3333333333333 | 615.6666666666666 | Domain | Protein kinase |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000379773 | - | 14 | 17 | 14_272 | 344.3333333333333 | 479.0 | Domain | Protein kinase |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000394522 | - | 15 | 18 | 14_272 | 358.3333333333333 | 493.0 | Domain | Protein kinase |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000394524 | - | 14 | 18 | 14_272 | 344.3333333333333 | 479.0 | Domain | Protein kinase |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000394526 | - | 15 | 20 | 14_272 | 355.3333333333333 | 1364.6666666666667 | Domain | Protein kinase |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000418639 | - | 15 | 20 | 14_272 | 358.3333333333333 | 1367.6666666666667 | Domain | Protein kinase |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000429180 | - | 15 | 20 | 14_272 | 364.3333333333333 | 1373.6666666666667 | Domain | Protein kinase |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000454265 | - | 16 | 21 | 14_272 | 369.3333333333333 | 1378.6666666666667 | Domain | Protein kinase |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000508738 | - | 15 | 18 | 14_272 | 355.3333333333333 | 490.0 | Domain | Protein kinase |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000515496 | - | 15 | 19 | 14_272 | 355.3333333333333 | 511.0 | Domain | Protein kinase |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000296402 | - | 14 | 18 | 14_272 | 344.3333333333333 | 1391.6666666666667 | Domain | Protein kinase |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000342666 | - | 14 | 19 | 14_272 | 344.3333333333333 | 615.6666666666666 | Domain | Protein kinase |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000379773 | - | 14 | 17 | 14_272 | 344.3333333333333 | 479.0 | Domain | Protein kinase |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000394522 | - | 15 | 18 | 14_272 | 358.3333333333333 | 493.0 | Domain | Protein kinase |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000394524 | - | 14 | 18 | 14_272 | 344.3333333333333 | 479.0 | Domain | Protein kinase |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000394526 | - | 15 | 20 | 14_272 | 355.3333333333333 | 1364.6666666666667 | Domain | Protein kinase |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000418639 | - | 15 | 20 | 14_272 | 358.3333333333333 | 1367.6666666666667 | Domain | Protein kinase |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000429180 | - | 15 | 20 | 14_272 | 364.3333333333333 | 1373.6666666666667 | Domain | Protein kinase |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000454265 | - | 16 | 21 | 14_272 | 369.3333333333333 | 1378.6666666666667 | Domain | Protein kinase |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000508738 | - | 15 | 18 | 14_272 | 355.3333333333333 | 490.0 | Domain | Protein kinase |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000515496 | - | 15 | 19 | 14_272 | 355.3333333333333 | 511.0 | Domain | Protein kinase |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000296402 | - | 14 | 18 | 20_28 | 344.3333333333333 | 1391.6666666666667 | Nucleotide binding | ATP |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000342666 | - | 14 | 19 | 20_28 | 344.3333333333333 | 615.6666666666666 | Nucleotide binding | ATP |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000379773 | - | 14 | 17 | 20_28 | 344.3333333333333 | 479.0 | Nucleotide binding | ATP |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000394522 | - | 15 | 18 | 20_28 | 358.3333333333333 | 493.0 | Nucleotide binding | ATP |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000394524 | - | 14 | 18 | 20_28 | 344.3333333333333 | 479.0 | Nucleotide binding | ATP |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000394526 | - | 15 | 20 | 20_28 | 355.3333333333333 | 1364.6666666666667 | Nucleotide binding | ATP |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000418639 | - | 15 | 20 | 20_28 | 358.3333333333333 | 1367.6666666666667 | Nucleotide binding | ATP |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000429180 | - | 15 | 20 | 20_28 | 364.3333333333333 | 1373.6666666666667 | Nucleotide binding | ATP |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000454265 | - | 16 | 21 | 20_28 | 369.3333333333333 | 1378.6666666666667 | Nucleotide binding | ATP |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000508738 | - | 15 | 18 | 20_28 | 355.3333333333333 | 490.0 | Nucleotide binding | ATP |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000515496 | - | 15 | 19 | 20_28 | 355.3333333333333 | 511.0 | Nucleotide binding | ATP |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000296402 | - | 14 | 18 | 20_28 | 344.3333333333333 | 1391.6666666666667 | Nucleotide binding | ATP |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000342666 | - | 14 | 19 | 20_28 | 344.3333333333333 | 615.6666666666666 | Nucleotide binding | ATP |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000379773 | - | 14 | 17 | 20_28 | 344.3333333333333 | 479.0 | Nucleotide binding | ATP |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000394522 | - | 15 | 18 | 20_28 | 358.3333333333333 | 493.0 | Nucleotide binding | ATP |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000394524 | - | 14 | 18 | 20_28 | 344.3333333333333 | 479.0 | Nucleotide binding | ATP |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000394526 | - | 15 | 20 | 20_28 | 355.3333333333333 | 1364.6666666666667 | Nucleotide binding | ATP |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000418639 | - | 15 | 20 | 20_28 | 358.3333333333333 | 1367.6666666666667 | Nucleotide binding | ATP |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000429180 | - | 15 | 20 | 20_28 | 364.3333333333333 | 1373.6666666666667 | Nucleotide binding | ATP |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000454265 | - | 16 | 21 | 20_28 | 369.3333333333333 | 1378.6666666666667 | Nucleotide binding | ATP |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000508738 | - | 15 | 18 | 20_28 | 355.3333333333333 | 490.0 | Nucleotide binding | ATP |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000515496 | - | 15 | 19 | 20_28 | 355.3333333333333 | 511.0 | Nucleotide binding | ATP |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000296402 | - | 14 | 18 | 283_292 | 344.3333333333333 | 1391.6666666666667 | Region | Autoinhibitory domain |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000296402 | - | 14 | 18 | 291_301 | 344.3333333333333 | 1391.6666666666667 | Region | Calmodulin-binding |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000342666 | - | 14 | 19 | 283_292 | 344.3333333333333 | 615.6666666666666 | Region | Autoinhibitory domain |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000342666 | - | 14 | 19 | 291_301 | 344.3333333333333 | 615.6666666666666 | Region | Calmodulin-binding |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000379773 | - | 14 | 17 | 283_292 | 344.3333333333333 | 479.0 | Region | Autoinhibitory domain |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000379773 | - | 14 | 17 | 291_301 | 344.3333333333333 | 479.0 | Region | Calmodulin-binding |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000394522 | - | 15 | 18 | 283_292 | 358.3333333333333 | 493.0 | Region | Autoinhibitory domain |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000394522 | - | 15 | 18 | 291_301 | 358.3333333333333 | 493.0 | Region | Calmodulin-binding |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000394524 | - | 14 | 18 | 283_292 | 344.3333333333333 | 479.0 | Region | Autoinhibitory domain |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000394524 | - | 14 | 18 | 291_301 | 344.3333333333333 | 479.0 | Region | Calmodulin-binding |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000394526 | - | 15 | 20 | 283_292 | 355.3333333333333 | 1364.6666666666667 | Region | Autoinhibitory domain |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000394526 | - | 15 | 20 | 291_301 | 355.3333333333333 | 1364.6666666666667 | Region | Calmodulin-binding |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000418639 | - | 15 | 20 | 283_292 | 358.3333333333333 | 1367.6666666666667 | Region | Autoinhibitory domain |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000418639 | - | 15 | 20 | 291_301 | 358.3333333333333 | 1367.6666666666667 | Region | Calmodulin-binding |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000429180 | - | 15 | 20 | 283_292 | 364.3333333333333 | 1373.6666666666667 | Region | Autoinhibitory domain |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000429180 | - | 15 | 20 | 291_301 | 364.3333333333333 | 1373.6666666666667 | Region | Calmodulin-binding |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000454265 | - | 16 | 21 | 283_292 | 369.3333333333333 | 1378.6666666666667 | Region | Autoinhibitory domain |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000454265 | - | 16 | 21 | 291_301 | 369.3333333333333 | 1378.6666666666667 | Region | Calmodulin-binding |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000508738 | - | 15 | 18 | 283_292 | 355.3333333333333 | 490.0 | Region | Autoinhibitory domain |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000508738 | - | 15 | 18 | 291_301 | 355.3333333333333 | 490.0 | Region | Calmodulin-binding |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000515496 | - | 15 | 19 | 283_292 | 355.3333333333333 | 511.0 | Region | Autoinhibitory domain |
| Hgene | CAMK2D | chr4:114421619 | chr10:98188505 | ENST00000515496 | - | 15 | 19 | 291_301 | 355.3333333333333 | 511.0 | Region | Calmodulin-binding |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000296402 | - | 14 | 18 | 283_292 | 344.3333333333333 | 1391.6666666666667 | Region | Autoinhibitory domain |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000296402 | - | 14 | 18 | 291_301 | 344.3333333333333 | 1391.6666666666667 | Region | Calmodulin-binding |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000342666 | - | 14 | 19 | 283_292 | 344.3333333333333 | 615.6666666666666 | Region | Autoinhibitory domain |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000342666 | - | 14 | 19 | 291_301 | 344.3333333333333 | 615.6666666666666 | Region | Calmodulin-binding |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000379773 | - | 14 | 17 | 283_292 | 344.3333333333333 | 479.0 | Region | Autoinhibitory domain |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000379773 | - | 14 | 17 | 291_301 | 344.3333333333333 | 479.0 | Region | Calmodulin-binding |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000394522 | - | 15 | 18 | 283_292 | 358.3333333333333 | 493.0 | Region | Autoinhibitory domain |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000394522 | - | 15 | 18 | 291_301 | 358.3333333333333 | 493.0 | Region | Calmodulin-binding |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000394524 | - | 14 | 18 | 283_292 | 344.3333333333333 | 479.0 | Region | Autoinhibitory domain |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000394524 | - | 14 | 18 | 291_301 | 344.3333333333333 | 479.0 | Region | Calmodulin-binding |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000394526 | - | 15 | 20 | 283_292 | 355.3333333333333 | 1364.6666666666667 | Region | Autoinhibitory domain |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000394526 | - | 15 | 20 | 291_301 | 355.3333333333333 | 1364.6666666666667 | Region | Calmodulin-binding |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000418639 | - | 15 | 20 | 283_292 | 358.3333333333333 | 1367.6666666666667 | Region | Autoinhibitory domain |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000418639 | - | 15 | 20 | 291_301 | 358.3333333333333 | 1367.6666666666667 | Region | Calmodulin-binding |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000429180 | - | 15 | 20 | 283_292 | 364.3333333333333 | 1373.6666666666667 | Region | Autoinhibitory domain |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000429180 | - | 15 | 20 | 291_301 | 364.3333333333333 | 1373.6666666666667 | Region | Calmodulin-binding |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000454265 | - | 16 | 21 | 283_292 | 369.3333333333333 | 1378.6666666666667 | Region | Autoinhibitory domain |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000454265 | - | 16 | 21 | 291_301 | 369.3333333333333 | 1378.6666666666667 | Region | Calmodulin-binding |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000508738 | - | 15 | 18 | 283_292 | 355.3333333333333 | 490.0 | Region | Autoinhibitory domain |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000508738 | - | 15 | 18 | 291_301 | 355.3333333333333 | 490.0 | Region | Calmodulin-binding |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000515496 | - | 15 | 19 | 283_292 | 355.3333333333333 | 511.0 | Region | Autoinhibitory domain |
| Hgene | CAMK2D | chr4:114421619 | chr10:98192719 | ENST00000515496 | - | 15 | 19 | 291_301 | 355.3333333333333 | 511.0 | Region | Calmodulin-binding |
| Tgene | TLL2 | chr4:114421619 | chr10:98188505 | ENST00000357947 | 3 | 21 | 351_463 | 173.33333333333334 | 1016.0 | Domain | CUB 1 | |
| Tgene | TLL2 | chr4:114421619 | chr10:98188505 | ENST00000357947 | 3 | 21 | 464_576 | 173.33333333333334 | 1016.0 | Domain | CUB 2 | |
| Tgene | TLL2 | chr4:114421619 | chr10:98188505 | ENST00000357947 | 3 | 21 | 576_617 | 173.33333333333334 | 1016.0 | Domain | EGF-like 1%3B calcium-binding | |
| Tgene | TLL2 | chr4:114421619 | chr10:98188505 | ENST00000357947 | 3 | 21 | 620_732 | 173.33333333333334 | 1016.0 | Domain | CUB 3 | |
| Tgene | TLL2 | chr4:114421619 | chr10:98188505 | ENST00000357947 | 3 | 21 | 732_772 | 173.33333333333334 | 1016.0 | Domain | EGF-like 2%3B calcium-binding | |
| Tgene | TLL2 | chr4:114421619 | chr10:98188505 | ENST00000357947 | 3 | 21 | 776_888 | 173.33333333333334 | 1016.0 | Domain | CUB 4 | |
| Tgene | TLL2 | chr4:114421619 | chr10:98188505 | ENST00000357947 | 3 | 21 | 889_1005 | 173.33333333333334 | 1016.0 | Domain | CUB 5 | |
| Tgene | TLL2 | chr4:114421619 | chr10:98192719 | ENST00000357947 | 2 | 21 | 149_349 | 121.33333333333333 | 1016.0 | Domain | Peptidase M12A | |
| Tgene | TLL2 | chr4:114421619 | chr10:98192719 | ENST00000357947 | 2 | 21 | 351_463 | 121.33333333333333 | 1016.0 | Domain | CUB 1 | |
| Tgene | TLL2 | chr4:114421619 | chr10:98192719 | ENST00000357947 | 2 | 21 | 464_576 | 121.33333333333333 | 1016.0 | Domain | CUB 2 | |
| Tgene | TLL2 | chr4:114421619 | chr10:98192719 | ENST00000357947 | 2 | 21 | 576_617 | 121.33333333333333 | 1016.0 | Domain | EGF-like 1%3B calcium-binding | |
| Tgene | TLL2 | chr4:114421619 | chr10:98192719 | ENST00000357947 | 2 | 21 | 620_732 | 121.33333333333333 | 1016.0 | Domain | CUB 3 | |
| Tgene | TLL2 | chr4:114421619 | chr10:98192719 | ENST00000357947 | 2 | 21 | 732_772 | 121.33333333333333 | 1016.0 | Domain | EGF-like 2%3B calcium-binding | |
| Tgene | TLL2 | chr4:114421619 | chr10:98192719 | ENST00000357947 | 2 | 21 | 776_888 | 121.33333333333333 | 1016.0 | Domain | CUB 4 | |
| Tgene | TLL2 | chr4:114421619 | chr10:98192719 | ENST00000357947 | 2 | 21 | 889_1005 | 121.33333333333333 | 1016.0 | Domain | CUB 5 |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | TLL2 | chr4:114421619 | chr10:98188505 | ENST00000357947 | 3 | 21 | 149_349 | 173.33333333333334 | 1016.0 | Domain | Peptidase M12A |
Top |
Fusion Protein Structures |
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
Top |
pLDDT score distribution |
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
CAMK2D_pLDDT.png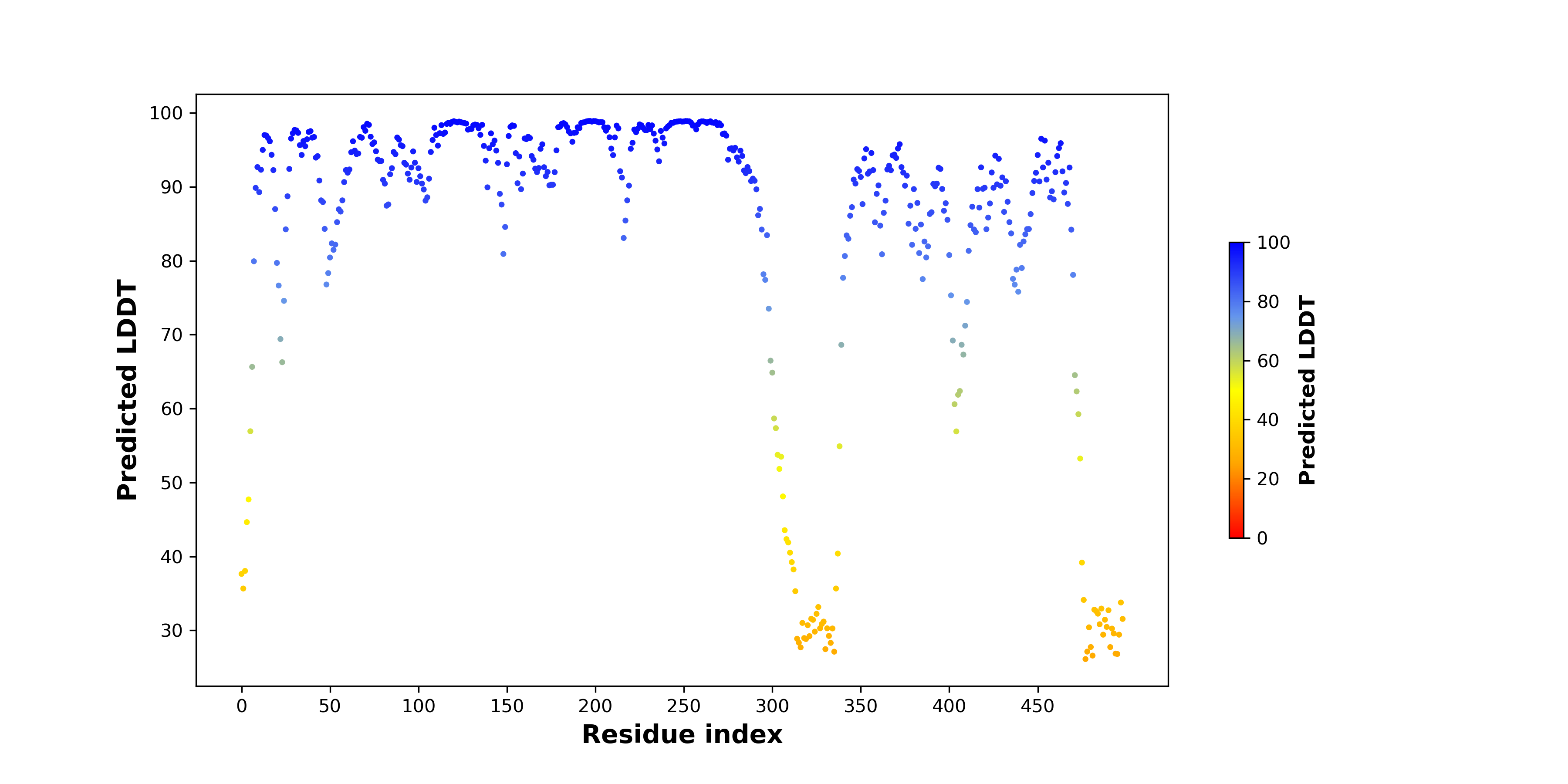 |
TLL2_pLDDT.png |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
Top |
Ramachandran Plot of Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| Fusion AA seq ID in FusionPDB and their Ramachandran plots |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| CAMK2D | |
| TLL2 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to CAMK2D-TLL2 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to CAMK2D-TLL2 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

