| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:DAB2IP-KMT2A |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: DAB2IP-KMT2A | FusionPDB ID: 21233 | FusionGDB2.0 ID: 21233 | Hgene | Tgene | Gene symbol | DAB2IP | KMT2A | Gene ID | 153090 | 4297 |
| Gene name | DAB2 interacting protein | lysine methyltransferase 2A | |
| Synonyms | AF9Q34|AIP-1|AIP1|DIP1/2 | ALL-1|CXXC7|HRX|HTRX1|MLL|MLL1|MLL1A|TRX1|WDSTS | |
| Cytomap | 9q33.2 | 11q23.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | disabled homolog 2-interacting proteinASK-interacting protein 1ASK1-interacting protein 1DAB2 interaction proteinDOC-2/DAB2 interactive proteinnGAP-like protein | histone-lysine N-methyltransferase 2ACXXC-type zinc finger protein 7lysine (K)-specific methyltransferase 2Alysine N-methyltransferase 2Amixed lineage leukemia 1myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila)trithorax-like | |
| Modification date | 20200313 | 20200319 | |
| UniProtAcc | Q5VWQ8 | Q03164 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000259371, ENST00000408936, ENST00000309989, ENST00000487716, | ENST00000420751, ENST00000354520, ENST00000389506, ENST00000534358, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 12 X 10 X 8=960 | 31 X 72 X 3=6696 |
| # samples | 12 | 79 | |
| ** MAII score | log2(12/960*10)=-3 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(79/6696*10)=-3.08337496948588 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: DAB2IP [Title/Abstract] AND KMT2A [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | |||
| Anticipated loss of major functional domain due to fusion event. | KMT2A-DAB2IP seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. KMT2A-DAB2IP seems lost the major protein functional domain in Hgene partner, which is a epigenetic factor due to the frame-shifted ORF. KMT2A-DAB2IP seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. KMT2A-DAB2IP seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. KMT2A-DAB2IP seems lost the major protein functional domain in Hgene partner, which is a transcription factor due to the frame-shifted ORF. KMT2A-DAB2IP seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. KMT2A-DAB2IP seems lost the major protein functional domain in Tgene partner, which is a tumor suppressor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | DAB2IP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 15310755 |
| Hgene | DAB2IP | GO:0000185 | activation of MAPKKK activity | 19903888 |
| Hgene | DAB2IP | GO:0007257 | activation of JUN kinase activity | 12813029 |
| Hgene | DAB2IP | GO:0008285 | negative regulation of cell proliferation | 19903888 |
| Hgene | DAB2IP | GO:0010719 | negative regulation of epithelial to mesenchymal transition | 20154697 |
| Hgene | DAB2IP | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling | 19903888 |
| Hgene | DAB2IP | GO:0016525 | negative regulation of angiogenesis | 19033661 |
| Hgene | DAB2IP | GO:0031334 | positive regulation of protein complex assembly | 18281285 |
| Hgene | DAB2IP | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway | 19948740 |
| Hgene | DAB2IP | GO:0035924 | cellular response to vascular endothelial growth factor stimulus | 19033661 |
| Hgene | DAB2IP | GO:0043065 | positive regulation of apoptotic process | 17389591|19903888 |
| Hgene | DAB2IP | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling | 15310755|17389591 |
| Hgene | DAB2IP | GO:0043254 | regulation of protein complex assembly | 19948740 |
| Hgene | DAB2IP | GO:0043407 | negative regulation of MAP kinase activity | 12813029 |
| Hgene | DAB2IP | GO:0043410 | positive regulation of MAPK cascade | 19903888 |
| Hgene | DAB2IP | GO:0043507 | positive regulation of JUN kinase activity | 15310755 |
| Hgene | DAB2IP | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity | 19903888 |
| Hgene | DAB2IP | GO:0044257 | cellular protein catabolic process | 17389591 |
| Hgene | DAB2IP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 15310755|17389591 |
| Hgene | DAB2IP | GO:0046330 | positive regulation of JNK cascade | 17389591|19903888 |
| Hgene | DAB2IP | GO:0070317 | negative regulation of G0 to G1 transition | 19903888 |
| Hgene | DAB2IP | GO:0070373 | negative regulation of ERK1 and ERK2 cascade | 19903888 |
| Hgene | DAB2IP | GO:0071158 | positive regulation of cell cycle arrest | 19903888 |
| Hgene | DAB2IP | GO:0071222 | cellular response to lipopolysaccharide | 19948740 |
| Hgene | DAB2IP | GO:0071347 | cellular response to interleukin-1 | 19948740 |
| Hgene | DAB2IP | GO:0071356 | cellular response to tumor necrosis factor | 12813029|15310755|17389591 |
| Hgene | DAB2IP | GO:0071901 | negative regulation of protein serine/threonine kinase activity | 19903888 |
| Hgene | DAB2IP | GO:0071902 | positive regulation of protein serine/threonine kinase activity | 19903888 |
| Hgene | DAB2IP | GO:1903363 | negative regulation of cellular protein catabolic process | 19903888 |
| Hgene | DAB2IP | GO:2001235 | positive regulation of apoptotic signaling pathway | 19903888 |
| Tgene | KMT2A | GO:0044648 | histone H3-K4 dimethylation | 25561738 |
| Tgene | KMT2A | GO:0045944 | positive regulation of transcription by RNA polymerase II | 20861184 |
| Tgene | KMT2A | GO:0051568 | histone H3-K4 methylation | 19556245 |
| Tgene | KMT2A | GO:0065003 | protein-containing complex assembly | 15199122 |
| Tgene | KMT2A | GO:0080182 | histone H3-K4 trimethylation | 20861184 |
| Tgene | KMT2A | GO:0097692 | histone H3-K4 monomethylation | 25561738|26324722 |
 Fusion gene breakpoints across DAB2IP (5'-gene) Fusion gene breakpoints across DAB2IP (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 Fusion gene breakpoints across KMT2A (3'-gene) Fusion gene breakpoints across KMT2A (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerKB3 | . | . | DAB2IP | chr9 | 124441060 | + | KMT2A | chr11 | 118354897 | |
| ChimerKB3 | . | . | DAB2IP | chr9 | 124441060 | + | KMT2A | chr11 | 118354897 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000259371 | DAB2IP | chr9 | 124441060 | + | ENST00000534358 | KMT2A | chr11 | 118354897 | + | 12698 | 213 | 69 | 8045 | 2658 |
| ENST00000259371 | DAB2IP | chr9 | 124441060 | + | ENST00000389506 | KMT2A | chr11 | 118354897 | + | 9782 | 213 | 69 | 8036 | 2655 |
| ENST00000259371 | DAB2IP | chr9 | 124441060 | + | ENST00000354520 | KMT2A | chr11 | 118354897 | + | 10447 | 213 | 69 | 7922 | 2617 |
| ENST00000408936 | DAB2IP | chr9 | 124441060 | + | ENST00000534358 | KMT2A | chr11 | 118354897 | + | 12895 | 410 | 182 | 8242 | 2686 |
| ENST00000408936 | DAB2IP | chr9 | 124441060 | + | ENST00000389506 | KMT2A | chr11 | 118354897 | + | 9979 | 410 | 182 | 8233 | 2683 |
| ENST00000408936 | DAB2IP | chr9 | 124441060 | + | ENST00000354520 | KMT2A | chr11 | 118354897 | + | 10644 | 410 | 182 | 8119 | 2645 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >21233_21233_1_DAB2IP-KMT2A_DAB2IP_chr9_124441060_ENST00000259371_KMT2A_chr11_118354897_ENST00000354520_length(amino acids)=2617AA_BP=48 MEPDSLLDQDDSYESPQERPGSRRSLPGSLSEKSPSMEPSAATPFRVTEKPPPVNKQENAGTLNILSTLSNGNSSKQKIPADGVHRIRVD FKFVYCQVCCEPFHKFCLEENERPLEDQLENWCCRRCKFCHVCGRQHQATKQLLECNKCRNSYHPECLGPNYPTKPTKKKKVWICTKCVR CKSCGSTTPGKGWDAQWSHDFSLCHDCAKLFAKGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSDEMYEILSNLPESVAYTCV NCTERHPAEWRLALEKELQISLKQVLTALLNSRTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDLEGVK RKMDQGNYTSVLEFSDDIVKIIQAAINSDGGQPEIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNY AQWQEREENSHTEQPPLMKKIIPAPKPKGPGEPDSPTPLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTYGDDSANDAGRLLY IGQNEWTHVNCALWSAEVFEDDDGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHRDLIK GEVVPENGFEVFRRVFVDFEGISLRRKFLNGLEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDARKRCVYTCKIV ECRPPVVEPDINSTVEHDENRTIAHSPTSFTESSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPGCRPL PSAGSPTPTTHEIVTVGDPLLSSGLRSIGSRRHSTSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLNS STSLGQNTSTSSNLQRTVVTVGNKNSHLDGSSSSEMKQSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTTSRE LNVSKIGSFAEPSSVSFSSKEALSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQKGKKS CKETFKEKHSSKSFLEPGQVTTGEEGNLKPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKELQAPRKRTVKV TLTPLKMENESQSKNALKESSPASPLQIESTSPTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQEDGSF KRRYPRRSARARSNMFFGLTPLYGVRSYGEEDIPFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHNL FREEEQCDLPKISQLDGVDDGTESDTSVTATTRKSSQIPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQ GQDSLEAQLSSLESSRRVHTSTPSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALGESPESSSSELLNLGEGLG LDSNREKDMGLFEVFSQQLPTTEPVDSSVSSSISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVTITEKSVASSES DPALLSPGVDPTPEGHMTPDHFIQGHMDADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQISNA AVQTTPPHLKPATEKLIVVNQNMQPLYVLQTLPNGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASKG LLPMSHHQHLHSFPAATQSSFPPNISNPPSGLLIGVQPPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSSTPSN IAPSDVVSNMTLINFTPSQLPNHPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAAGTSTISQDTSHLTSGSVS GLASSSSVLNVVSMQTTTTPTSSASVPGHVTLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGTSQTPSTAAITA ASSICVLPSTQTTGITAASPSGEADEHYQLQHVNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQASPTSP GGSPSSPSSGQRSASPSVPGPTKPKPKTKRFQLPLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQK ECGQPAGQVAVLPEVQVTQNPANEQESAEPKTVEEEESNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQICAESIEDAWKS LTDKVQEARSNARLKQLSFAGVNGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSARAEVHLRKSAFDMFNFLA SKHRQPPEYNPNDEEEEEVQLKSARRATSMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGNVIRSIQTDKRE KYYDSKGIGCYMFRIDDSEVVDATMHGNAARFINHSCEPNCYSRVINIDGQKHIVIFAMRKIYRGEELTYDYKFPIEDASNKLPCNCGAK -------------------------------------------------------------- >21233_21233_2_DAB2IP-KMT2A_DAB2IP_chr9_124441060_ENST00000259371_KMT2A_chr11_118354897_ENST00000389506_length(amino acids)=2655AA_BP=48 MEPDSLLDQDDSYESPQERPGSRRSLPGSLSEKSPSMEPSAATPFRVTEKPPPVNKQENAGTLNILSTLSNGNSSKQKIPADGVHRIRVD FKEDCEAENVWEMGGLGILTSVPITPRVVCFLCASSGHVEFVYCQVCCEPFHKFCLEENERPLEDQLENWCCRRCKFCHVCGRQHQATKQ LLECNKCRNSYHPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWSHDFSLCHDCAKLFAKGNFCPLCDKCYDDDDYESK MMQCGKCDRWVHSKCENLSDEMYEILSNLPESVAYTCVNCTERHPAEWRLALEKELQISLKQVLTALLNSRTTSHLLRYRQAAKPPDLNP ETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYTSVLEFSDDIVKIIQAAINSDGGQPEIKKANSMVKSFFIRQMER VFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREENSHTEQPPLMKKIIPAPKPKGPGEPDSPTPLHPPTPPILSTDRS REDSPELNPPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVNCALWSAEVFEDDDGSLKNVHMAVIRGKQLRCEFCQKPGATVG CCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVFVDFEGISLRRKFLNGLEPENIHMMIGSMTIDCLGIL NDLSDCEDKLFPIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDINSTVEHDENRTIAHSPTSFTESSSKESQNTAEIISPPSPDR PPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPTTHEIVTVGDPLLSSGLRSIGSRRHSTSSLSPQRSKLRIMSPMR TGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTSTSSNLQRTVVTVGNKNSHLDGSSSSEMKQSSASDLVSKSSSLK GEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAEPSSVSFSSKEALSFPHLHLRGQRNDRDQHTDSTQSANSSPD EDTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLEPGQVTTGEEGNLKPEFMDEVLTPEYMGQRPCNNVSS DKIGDKGLSMPGVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMENESQSKNALKESSPASPLQIESTSPTEPISASENPGDGPVAQPS PNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSARARSNMFFGLTPLYGVRSYGEEDIPFYSSSTGKKRGKRSAEGQ VDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDLPKISQLDGVDDGTESDTSVTATTRKSSQIPKRNGKENGTENLK IDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSSLESSRRVHTSTPSDKNLLDTYNTELLKSDSDNNNSDDCGNI LPSDIMDFVLKNTPSMQALGESPESSSSELLNLGEGLGLDSNREKDMGLFEVFSQQLPTTEPVDSSVSSSISAEEQFELPLELPSDLSVL TTRSPTVPSQNPSRLAVISDSGEKRVTITEKSVASSESDPALLSPGVDPTPEGHMTPDHFIQGHMDADHISSPPCGSVEQGHGNNQDLTR NSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQISNAAVQTTPPHLKPATEKLIVVNQNMQPLYVLQTLPNGVTQKIQLTSSVSSTPSV METNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQHLHSFPAATQSSFPPNISNPPSGLLIGVQPPPDPQLLVSESSQR TDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMTLINFTPSQLPNHPSLLDLGSLNTSSHRTVPNIIKRSKSSIM YFEPAPLLPQSVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQTTTTPTSSASVPGHVTLTNPRLLGTPDIGSISNLLIK ASQQSLGIQDQPVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQTTGITAASPSGEADEHYQLQHVNQLLASKTGIHSSQRDLDS ASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQASPTSPGGSPSSPSSGQRSASPSVPGPTKPKPKTKRFQLPLDKGNGKKHKVSHLRTSS SEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQVAVLPEVQVTQNPANEQESAEPKTVEEEESNFSSPLMLWLQQEQ KRKESITEKKPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARSNARLKQLSFAGVNGLRMLGILHDAVVFLIEQLSGAKHCRNYK FRFHKPEEANEPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEEEEVQLKSARRATSMDLPMPMRFRHLKKTSKEAVGVY RSPIHGRGLFCKRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCYMFRIDDSEVVDATMHGNAARFINHSCEPNCYSRVINIDGQK -------------------------------------------------------------- >21233_21233_3_DAB2IP-KMT2A_DAB2IP_chr9_124441060_ENST00000259371_KMT2A_chr11_118354897_ENST00000534358_length(amino acids)=2658AA_BP=48 MEPDSLLDQDDSYESPQERPGSRRSLPGSLSEKSPSMEPSAATPFRVTEKPPPVNKQENAGTLNILSTLSNGNSSKQKIPADGVHRIRVD FKEDCEAENVWEMGGLGILTSVPITPRVVCFLCASSGHVEFVYCQVCCEPFHKFCLEENERPLEDQLENWCCRRCKFCHVCGRQHQATKQ LLECNKCRNSYHPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWSHDFSLCHDCAKLFAKGNFCPLCDKCYDDDDYESK MMQCGKCDRWVHSKCENLSGTEDEMYEILSNLPESVAYTCVNCTERHPAEWRLALEKELQISLKQVLTALLNSRTTSHLLRYRQAAKPPD LNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYTSVLEFSDDIVKIIQAAINSDGGQPEIKKANSMVKSFFIRQ MERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREENSHTEQPPLMKKIIPAPKPKGPGEPDSPTPLHPPTPPILST DRSREDSPELNPPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVNCALWSAEVFEDDDGSLKNVHMAVIRGKQLRCEFCQKPGA TVGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVFVDFEGISLRRKFLNGLEPENIHMMIGSMTIDCL GILNDLSDCEDKLFPIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDINSTVEHDENRTIAHSPTSFTESSSKESQNTAEIISPPS PDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPTTHEIVTVGDPLLSSGLRSIGSRRHSTSSLSPQRSKLRIMS PMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTSTSSNLQRTVVTVGNKNSHLDGSSSSEMKQSSASDLVSKSS SLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAEPSSVSFSSKEALSFPHLHLRGQRNDRDQHTDSTQSANS SPDEDTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLEPGQVTTGEEGNLKPEFMDEVLTPEYMGQRPCNN VSSDKIGDKGLSMPGVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMENESQSKNALKESSPASPLQIESTSPTEPISASENPGDGPVA QPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSARARSNMFFGLTPLYGVRSYGEEDIPFYSSSTGKKRGKRSA EGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDLPKISQLDGVDDGTESDTSVTATTRKSSQIPKRNGKENGTE NLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSSLESSRRVHTSTPSDKNLLDTYNTELLKSDSDNNNSDDC GNILPSDIMDFVLKNTPSMQALGESPESSSSELLNLGEGLGLDSNREKDMGLFEVFSQQLPTTEPVDSSVSSSISAEEQFELPLELPSDL SVLTTRSPTVPSQNPSRLAVISDSGEKRVTITEKSVASSESDPALLSPGVDPTPEGHMTPDHFIQGHMDADHISSPPCGSVEQGHGNNQD LTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQISNAAVQTTPPHLKPATEKLIVVNQNMQPLYVLQTLPNGVTQKIQLTSSVSST PSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQHLHSFPAATQSSFPPNISNPPSGLLIGVQPPPDPQLLVSES SQRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMTLINFTPSQLPNHPSLLDLGSLNTSSHRTVPNIIKRSKS SIMYFEPAPLLPQSVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQTTTTPTSSASVPGHVTLTNPRLLGTPDIGSISNL LIKASQQSLGIQDQPVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQTTGITAASPSGEADEHYQLQHVNQLLASKTGIHSSQRD LDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQASPTSPGGSPSSPSSGQRSASPSVPGPTKPKPKTKRFQLPLDKGNGKKHKVSHLR TSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQVAVLPEVQVTQNPANEQESAEPKTVEEEESNFSSPLMLWLQ QEQKRKESITEKKPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARSNARLKQLSFAGVNGLRMLGILHDAVVFLIEQLSGAKHCR NYKFRFHKPEEANEPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEEEEVQLKSARRATSMDLPMPMRFRHLKKTSKEAV GVYRSPIHGRGLFCKRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCYMFRIDDSEVVDATMHGNAARFINHSCEPNCYSRVINID -------------------------------------------------------------- >21233_21233_4_DAB2IP-KMT2A_DAB2IP_chr9_124441060_ENST00000408936_KMT2A_chr11_118354897_ENST00000354520_length(amino acids)=2645AA_BP=76 MSAGGSARKSTGRSSYYYRLLRRPRLQRQRSRSRSRTRPARESPQERPGSRRSLPGSLSEKSPSMEPSAATPFRVTEKPPPVNKQENAGT LNILSTLSNGNSSKQKIPADGVHRIRVDFKFVYCQVCCEPFHKFCLEENERPLEDQLENWCCRRCKFCHVCGRQHQATKQLLECNKCRNS YHPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWSHDFSLCHDCAKLFAKGNFCPLCDKCYDDDDYESKMMQCGKCDRW VHSKCENLSDEMYEILSNLPESVAYTCVNCTERHPAEWRLALEKELQISLKQVLTALLNSRTTSHLLRYRQAAKPPDLNPETEESIPSRS SPEGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYTSVLEFSDDIVKIIQAAINSDGGQPEIKKANSMVKSFFIRQMERVFPWFSVKKS RFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREENSHTEQPPLMKKIIPAPKPKGPGEPDSPTPLHPPTPPILSTDRSREDSPELNPP PGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVNCALWSAEVFEDDDGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTSNY HFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVFVDFEGISLRRKFLNGLEPENIHMMIGSMTIDCLGILNDLSDCEDKL FPIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDINSTVEHDENRTIAHSPTSFTESSSKESQNTAEIISPPSPDRPPHSQTSGSC YYHVISKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPTTHEIVTVGDPLLSSGLRSIGSRRHSTSSLSPQRSKLRIMSPMRTGNTYSRNNV SSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTSTSSNLQRTVVTVGNKNSHLDGSSSSEMKQSSASDLVSKSSSLKGEKTKVLSSK SSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAEPSSVSFSSKEALSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTLKL SGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLEPGQVTTGEEGNLKPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSM PGVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMENESQSKNALKESSPASPLQIESTSPTEPISASENPGDGPVAQPSPNNTSCQDSQ SNNYQNLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSARARSNMFFGLTPLYGVRSYGEEDIPFYSSSTGKKRGKRSAEGQVDGADDLSTS DEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDLPKISQLDGVDDGTESDTSVTATTRKSSQIPKRNGKENGTENLKIDRPEDAGEK EHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSSLESSRRVHTSTPSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSDIMDFVL KNTPSMQALGESPESSSSELLNLGEGLGLDSNREKDMGLFEVFSQQLPTTEPVDSSVSSSISAEEQFELPLELPSDLSVLTTRSPTVPSQ NPSRLAVISDSGEKRVTITEKSVASSESDPALLSPGVDPTPEGHMTPDHFIQGHMDADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQVP VSPTVPIQNQKYVPNSTDSPGPSQISNAAVQTTPPHLKPATEKLIVVNQNMQPLYVLQTLPNGVTQKIQLTSSVSSTPSVMETNTSVLGP MGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQHLHSFPAATQSSFPPNISNPPSGLLIGVQPPPDPQLLVSESSQRTDLSTTVATP SSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMTLINFTPSQLPNHPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQ SVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQTTTTPTSSASVPGHVTLTNPRLLGTPDIGSISNLLIKASQQSLGIQD QPVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQTTGITAASPSGEADEHYQLQHVNQLLASKTGIHSSQRDLDSASGPQVSNFT QTVDAPNSMGLEQNKALSSAVQASPTSPGGSPSSPSSGQRSASPSVPGPTKPKPKTKRFQLPLDKGNGKKHKVSHLRTSSSEAHIPDQET TSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQVAVLPEVQVTQNPANEQESAEPKTVEEEESNFSSPLMLWLQQEQKRKESITEKK PKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARSNARLKQLSFAGVNGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEEAN EPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEEEEVQLKSARRATSMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLF CKRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCYMFRIDDSEVVDATMHGNAARFINHSCEPNCYSRVINIDGQKHIVIFAMRKI -------------------------------------------------------------- >21233_21233_5_DAB2IP-KMT2A_DAB2IP_chr9_124441060_ENST00000408936_KMT2A_chr11_118354897_ENST00000389506_length(amino acids)=2683AA_BP=76 MSAGGSARKSTGRSSYYYRLLRRPRLQRQRSRSRSRTRPARESPQERPGSRRSLPGSLSEKSPSMEPSAATPFRVTEKPPPVNKQENAGT LNILSTLSNGNSSKQKIPADGVHRIRVDFKEDCEAENVWEMGGLGILTSVPITPRVVCFLCASSGHVEFVYCQVCCEPFHKFCLEENERP LEDQLENWCCRRCKFCHVCGRQHQATKQLLECNKCRNSYHPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWSHDFSLC HDCAKLFAKGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSDEMYEILSNLPESVAYTCVNCTERHPAEWRLALEKELQISLKQ VLTALLNSRTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYTSVLEFSDDIVKIIQA AINSDGGQPEIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREENSHTEQPPLMKKIIPA PKPKGPGEPDSPTPLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVNCALWSAEVFEDDDG SLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVFVDFEGISL RRKFLNGLEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDINSTVEHDENRTIA HSPTSFTESSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPTTHEIVTVGDPLLSSG LRSIGSRRHSTSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTSTSSNLQRTVVTVGNK NSHLDGSSSSEMKQSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAEPSSVSFSSKEALS FPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLEPGQVTTGE EGNLKPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMENESQSKNALKESSPAS PLQIESTSPTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSARARSNMFFGLTPLYG VRSYGEEDIPFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDLPKISQLDGVDDGTES DTSVTATTRKSSQIPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSSLESSRRVHTSTPS DKNLLDTYNTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALGESPESSSSELLNLGEGLGLDSNREKDMGLFEVFSQQLPTTEP VDSSVSSSISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVTITEKSVASSESDPALLSPGVDPTPEGHMTPDHFIQ GHMDADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQISNAAVQTTPPHLKPATEKLIVVNQNMQ PLYVLQTLPNGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQHLHSFPAATQSSFPPN ISNPPSGLLIGVQPPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMTLINFTPSQLPNHP SLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQTTTTPTSSA SVPGHVTLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQTTGITAASPSGEA DEHYQLQHVNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQASPTSPGGSPSSPSSGQRSASPSVPGPTKP KPKTKRFQLPLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQVAVLPEVQVTQNPANE QESAEPKTVEEEESNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARSNARLKQLSFAGVNG LRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEEEEVQLKSA RRATSMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCYMFRIDDSEVVDAT -------------------------------------------------------------- >21233_21233_6_DAB2IP-KMT2A_DAB2IP_chr9_124441060_ENST00000408936_KMT2A_chr11_118354897_ENST00000534358_length(amino acids)=2686AA_BP=76 MSAGGSARKSTGRSSYYYRLLRRPRLQRQRSRSRSRTRPARESPQERPGSRRSLPGSLSEKSPSMEPSAATPFRVTEKPPPVNKQENAGT LNILSTLSNGNSSKQKIPADGVHRIRVDFKEDCEAENVWEMGGLGILTSVPITPRVVCFLCASSGHVEFVYCQVCCEPFHKFCLEENERP LEDQLENWCCRRCKFCHVCGRQHQATKQLLECNKCRNSYHPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWSHDFSLC HDCAKLFAKGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSGTEDEMYEILSNLPESVAYTCVNCTERHPAEWRLALEKELQIS LKQVLTALLNSRTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYTSVLEFSDDIVKI IQAAINSDGGQPEIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREENSHTEQPPLMKKI IPAPKPKGPGEPDSPTPLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVNCALWSAEVFED DDGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVFVDFEG ISLRRKFLNGLEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDINSTVEHDENR TIAHSPTSFTESSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPTTHEIVTVGDPLL SSGLRSIGSRRHSTSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTSTSSNLQRTVVTV GNKNSHLDGSSSSEMKQSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAEPSSVSFSSKE ALSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLEPGQVT TGEEGNLKPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMENESQSKNALKESS PASPLQIESTSPTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSARARSNMFFGLTP LYGVRSYGEEDIPFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDLPKISQLDGVDDG TESDTSVTATTRKSSQIPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSSLESSRRVHTS TPSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALGESPESSSSELLNLGEGLGLDSNREKDMGLFEVFSQQLPT TEPVDSSVSSSISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVTITEKSVASSESDPALLSPGVDPTPEGHMTPDH FIQGHMDADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQISNAAVQTTPPHLKPATEKLIVVNQ NMQPLYVLQTLPNGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQHLHSFPAATQSSF PPNISNPPSGLLIGVQPPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMTLINFTPSQLP NHPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQTTTTPT SSASVPGHVTLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQTTGITAASPS GEADEHYQLQHVNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQASPTSPGGSPSSPSSGQRSASPSVPGP TKPKPKTKRFQLPLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQVAVLPEVQVTQNP ANEQESAEPKTVEEEESNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARSNARLKQLSFAG VNGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEEEEVQL KSARRATSMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCYMFRIDDSEVV -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr9:/chr11:) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| DAB2IP | KMT2A |
| FUNCTION: Functions as a scaffold protein implicated in the regulation of a large spectrum of both general and specialized signaling pathways. Involved in several processes such as innate immune response, inflammation and cell growth inhibition, apoptosis, cell survival, angiogenesis, cell migration and maturation. Plays also a role in cell cycle checkpoint control; reduces G1 phase cyclin levels resulting in G0/G1 cell cycle arrest. Mediates signal transduction by receptor-mediated inflammatory signals, such as the tumor necrosis factor (TNF), interferon (IFN) or lipopolysaccharide (LPS). Modulates the balance between phosphatidylinositol 3-kinase (PI3K)-AKT-mediated cell survival and apoptosis stimulated kinase (MAP3K5)-JNK signaling pathways; sequesters both AKT1 and MAP3K5 and counterbalances the activity of each kinase by modulating their phosphorylation status in response to proinflammatory stimuli. Acts as a regulator of the endoplasmic reticulum (ER) unfolded protein response (UPR) pathway; specifically involved in transduction of the ER stress-response to the JNK cascade through ERN1. Mediates TNF-alpha-induced apoptosis activation by facilitating dissociation of inhibitor 14-3-3 from MAP3K5; recruits the PP2A phosphatase complex which dephosphorylates MAP3K5 on 'Ser-966', leading to the dissociation of 13-3-3 proteins and activation of the MAP3K5-JNK signaling pathway in endothelial cells. Mediates also TNF/TRAF2-induced MAP3K5-JNK activation, while it inhibits CHUK-NF-kappa-B signaling. Acts a negative regulator in the IFN-gamma-mediated JAK-STAT signaling cascade by inhibiting smooth muscle cell (VSMCs) proliferation and intimal expansion, and thus, prevents graft arteriosclerosis (GA). Acts as a GTPase-activating protein (GAP) for the ADP ribosylation factor 6 (ARF6) and Ras. Promotes hydrolysis of the ARF6-bound GTP and thus, negatively regulates phosphatidylinositol 4,5-bisphosphate (PIP2)-dependent TLR4-TIRAP-MyD88 and NF-kappa-B signaling pathways in endothelial cells in response to lipopolysaccharides (LPS). Binds specifically to phosphatidylinositol 4-phosphate (PtdIns4P) and phosphatidylinositol 3-phosphate (PtdIns3P). In response to vascular endothelial growth factor (VEGFA), acts as a negative regulator of the VEGFR2-PI3K-mediated angiogenic signaling pathway by inhibiting endothelial cell migration and tube formation. In the developing brain, promotes both the transition from the multipolar to the bipolar stage and the radial migration of cortical neurons from the ventricular zone toward the superficial layer of the neocortex in a glial-dependent locomotion process. Probable downstream effector of the Reelin signaling pathway; promotes Purkinje cell (PC) dendrites development and formation of cerebellar synapses. Functions also as a tumor suppressor protein in prostate cancer progression; prevents cell proliferation and epithelial-to-mesenchymal transition (EMT) through activation of the glycogen synthase kinase-3 beta (GSK3B)-induced beta-catenin and inhibition of PI3K-AKT and Ras-MAPK survival downstream signaling cascades, respectively. {ECO:0000269|PubMed:12813029, ECO:0000269|PubMed:17389591, ECO:0000269|PubMed:18292600, ECO:0000269|PubMed:19033661, ECO:0000269|PubMed:19903888, ECO:0000269|PubMed:19948740, ECO:0000269|PubMed:20080667, ECO:0000269|PubMed:20154697, ECO:0000269|PubMed:21700930, ECO:0000269|PubMed:22696229}. | FUNCTION: Histone methyltransferase that plays an essential role in early development and hematopoiesis (PubMed:15960975, PubMed:12453419, PubMed:15960975, PubMed:19556245, PubMed:19187761, PubMed:20677832, PubMed:21220120, PubMed:26886794). Catalytic subunit of the MLL1/MLL complex, a multiprotein complex that mediates both methylation of 'Lys-4' of histone H3 (H3K4me) complex and acetylation of 'Lys-16' of histone H4 (H4K16ac) (PubMed:15960975, PubMed:12453419, PubMed:15960975, PubMed:19556245, PubMed:24235145, PubMed:19187761, PubMed:20677832, PubMed:21220120, PubMed:26886794). Catalyzes methyl group transfer from S-adenosyl-L-methionine to the epsilon-amino group of 'Lys-4' of histone H3 (H3K4) via a non-processive mechanism. Part of chromatin remodeling machinery predominantly forms H3K4me1 and H3K4me2 methylation marks at active chromatin sites where transcription and DNA repair take place (PubMed:25561738, PubMed:15960975, PubMed:12453419, PubMed:15960975, PubMed:19556245, PubMed:19187761, PubMed:20677832, PubMed:21220120, PubMed:26886794). Has weak methyltransferase activity by itself, and requires other component of the MLL1/MLL complex to obtain full methyltransferase activity (PubMed:19187761, PubMed:26886794). Has no activity toward histone H3 phosphorylated on 'Thr-3', less activity toward H3 dimethylated on 'Arg-8' or 'Lys-9', while it has higher activity toward H3 acetylated on 'Lys-9' (PubMed:19187761). Binds to unmethylated CpG elements in the promoter of target genes and helps maintain them in the nonmethylated state (PubMed:20010842). Required for transcriptional activation of HOXA9 (PubMed:12453419, PubMed:20677832, PubMed:20010842). Promotes PPP1R15A-induced apoptosis (PubMed:10490642). Plays a critical role in the control of circadian gene expression and is essential for the transcriptional activation mediated by the CLOCK-ARNTL/BMAL1 heterodimer (By similarity). Establishes a permissive chromatin state for circadian transcription by mediating a rhythmic methylation of 'Lys-4' of histone H3 (H3K4me) and this histone modification directs the circadian acetylation at H3K9 and H3K14 allowing the recruitment of CLOCK-ARNTL/BMAL1 to chromatin (By similarity). Also has auto-methylation activity on Cys-3882 in absence of histone H3 substrate (PubMed:24235145). {ECO:0000250|UniProtKB:P55200, ECO:0000269|PubMed:10490642, ECO:0000269|PubMed:12453419, ECO:0000269|PubMed:15960975, ECO:0000269|PubMed:19187761, ECO:0000269|PubMed:19556245, ECO:0000269|PubMed:20010842, ECO:0000269|PubMed:21220120, ECO:0000269|PubMed:24235145, ECO:0000269|PubMed:26886794, ECO:0000305|PubMed:20677832}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
Top |
Fusion Protein Structures |
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
| PDB file (1159) >>>1159.pdbFusion protein BP residue: 48 CIF file (1159) >>>1159.cif | DAB2IP | chr9 | 124441060 | + | KMT2A | chr11 | 118354897 | + | MEPDSLLDQDDSYESPQERPGSRRSLPGSLSEKSPSMEPSAATPFRVTEK PPPVNKQENAGTLNILSTLSNGNSSKQKIPADGVHRIRVDFKFVYCQVCC EPFHKFCLEENERPLEDQLENWCCRRCKFCHVCGRQHQATKQLLECNKCR NSYHPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWSHD FSLCHDCAKLFAKGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENL SDEMYEILSNLPESVAYTCVNCTERHPAEWRLALEKELQISLKQVLTALL NSRTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDD QQPLDLEGVKRKMDQGNYTSVLEFSDDIVKIIQAAINSDGGQPEIKKANS MVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNY AQWQEREENSHTEQPPLMKKIIPAPKPKGPGEPDSPTPLHPPTPPILSTD RSREDSPELNPPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVN CALWSAEVFEDDDGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTS NYHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVFVDFE GISLRRKFLNGLEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCS RVYWSTTDARKRCVYTCKIVECRPPVVEPDINSTVEHDENRTIAHSPTSF TESSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSP TQRSPGCRPLPSAGSPTPTTHEIVTVGDPLLSSGLRSIGSRRHSTSSLSP QRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLNS STSLGQNTSTSSNLQRTVVTVGNKNSHLDGSSSSEMKQSSASDLVSKSSS LKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFA EPSSVSFSSKEALSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTL KLSGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLEPGQV TTGEEGNLKPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPP MQVEGSAKELQAPRKRTVKVTLTPLKMENESQSKNALKESSPASPLQIES TSPTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPD GPKPQEDGSFKRRYPRRSARARSNMFFGLTPLYGVRSYGEEDIPFYSSST GKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHNL FREEEQCDLPKISQLDGVDDGTESDTSVTATTRKSSQIPKRNGKENGTEN LKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLS SLESSRRVHTSTPSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSDIMDF VLKNTPSMQALGESPESSSSELLNLGEGLGLDSNREKDMGLFEVFSQQLP TTEPVDSSVSSSISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVI SDSGEKRVTITEKSVASSESDPALLSPGVDPTPEGHMTPDHFIQGHMDAD HISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTD SPGPSQISNAAVQTTPPHLKPATEKLIVVNQNMQPLYVLQTLPNGVTQKI QLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASKG LLPMSHHQHLHSFPAATQSSFPPNISNPPSGLLIGVQPPPDPQLLVSESS QRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNM TLINFTPSQLPNHPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLL PQSVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQTTTTP TSSASVPGHVTLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPS SGMFPQLGTSQTPSTAAITAASSICVLPSTQTTGITAASPSGEADEHYQL QHVNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALS SAVQASPTSPGGSPSSPSSGQRSASPSVPGPTKPKPKTKRFQLPLDKGNG KKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQK ECGQPAGQVAVLPEVQVTQNPANEQESAEPKTVEEEESNFSSPLMLWLQQ EQKRKESITEKKPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARS NARLKQLSFAGVNGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEE ANEPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEEEEVQ LKSARRATSMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAG EMVIEYAGNVIRSIQTDKREKYYDSKGIGCYMFRIDDSEVVDATMHGNAA RFINHSCEPNCYSRVINIDGQKHIVIFAMRKIYRGEELTYDYKFPIEDAS | 2617 |
| 3D view using mol* of 1159 (AA BP:48) | ||||||||||
 | ||||||||||
| PDB file (1169) >>>1169.pdbFusion protein BP residue: 76 CIF file (1169) >>>1169.cif | DAB2IP | chr9 | 124441060 | + | KMT2A | chr11 | 118354897 | + | MSAGGSARKSTGRSSYYYRLLRRPRLQRQRSRSRSRTRPARESPQERPGS RRSLPGSLSEKSPSMEPSAATPFRVTEKPPPVNKQENAGTLNILSTLSNG NSSKQKIPADGVHRIRVDFKFVYCQVCCEPFHKFCLEENERPLEDQLENW CCRRCKFCHVCGRQHQATKQLLECNKCRNSYHPECLGPNYPTKPTKKKKV WICTKCVRCKSCGSTTPGKGWDAQWSHDFSLCHDCAKLFAKGNFCPLCDK CYDDDDYESKMMQCGKCDRWVHSKCENLSDEMYEILSNLPESVAYTCVNC TERHPAEWRLALEKELQISLKQVLTALLNSRTTSHLLRYRQAAKPPDLNP ETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYTSVL EFSDDIVKIIQAAINSDGGQPEIKKANSMVKSFFIRQMERVFPWFSVKKS RFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREENSHTEQPPLMKKII PAPKPKGPGEPDSPTPLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCA LCLTYGDDSANDAGRLLYIGQNEWTHVNCALWSAEVFEDDDGSLKNVHMA VIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYC QRHRDLIKGEVVPENGFEVFRRVFVDFEGISLRRKFLNGLEPENIHMMIG SMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDARKRCVYTCKIVEC RPPVVEPDINSTVEHDENRTIAHSPTSFTESSSKESQNTAEIISPPSPDR PPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPTTHE IVTVGDPLLSSGLRSIGSRRHSTSSLSPQRSKLRIMSPMRTGNTYSRNNV SSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTSTSSNLQRTVVTVG NKNSHLDGSSSSEMKQSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVA YPGIPKLAPQVHNTTSRELNVSKIGSFAEPSSVSFSSKEALSFPHLHLRG QRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSIINEHMGSSSRD RRQKGKKSCKETFKEKHSSKSFLEPGQVTTGEEGNLKPEFMDEVLTPEYM GQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKELQAPRKRTVKVTL TPLKMENESQSKNALKESSPASPLQIESTSPTEPISASENPGDGPVAQPS PNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSARAR SNMFFGLTPLYGVRSYGEEDIPFYSSSTGKKRGKRSAEGQVDGADDLSTS DEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDLPKISQLDGVDDGT ESDTSVTATTRKSSQIPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGH KNEPKMDNCHSVSRVKTQGQDSLEAQLSSLESSRRVHTSTPSDKNLLDTY NTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALGESPESSSSEL LNLGEGLGLDSNREKDMGLFEVFSQQLPTTEPVDSSVSSSISAEEQFELP LELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVTITEKSVASSESDP ALLSPGVDPTPEGHMTPDHFIQGHMDADHISSPPCGSVEQGHGNNQDLTR NSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQISNAAVQTTPPHLKPA TEKLIVVNQNMQPLYVLQTLPNGVTQKIQLTSSVSSTPSVMETNTSVLGP MGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQHLHSFPAATQSSFP PNISNPPSGLLIGVQPPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPIS RLQTRKNKKLAPSSTPSNIAPSDVVSNMTLINFTPSQLPNHPSLLDLGSL NTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAAGTSTISQDTS HLTSGSVSGLASSSSVLNVVSMQTTTTPTSSASVPGHVTLTNPRLLGTPD IGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGTSQTPSTAAITAAS SICVLPSTQTTGITAASPSGEADEHYQLQHVNQLLASKTGIHSSQRDLDS ASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQASPTSPGGSPSSPSSGQR SASPSVPGPTKPKPKTKRFQLPLDKGNGKKHKVSHLRTSSSEAHIPDQET TSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQVAVLPEVQVTQNPA NEQESAEPKTVEEEESNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEIS SDDGFQICAESIEDAWKSLTDKVQEARSNARLKQLSFAGVNGLRMLGILH DAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSARAEVHLRKSA FDMFNFLASKHRQPPEYNPNDEEEEEVQLKSARRATSMDLPMPMRFRHLK KTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGNVIRSIQTDKREKY YDSKGIGCYMFRIDDSEVVDATMHGNAARFINHSCEPNCYSRVINIDGQK | 2645 |
| 3D view using mol* of 1169 (AA BP:76) | ||||||||||
 | ||||||||||
| PDB file (1173) >>>1173.pdbFusion protein BP residue: 48 CIF file (1173) >>>1173.cif | DAB2IP | chr9 | 124441060 | + | KMT2A | chr11 | 118354897 | + | MEPDSLLDQDDSYESPQERPGSRRSLPGSLSEKSPSMEPSAATPFRVTEK PPPVNKQENAGTLNILSTLSNGNSSKQKIPADGVHRIRVDFKEDCEAENV WEMGGLGILTSVPITPRVVCFLCASSGHVEFVYCQVCCEPFHKFCLEENE RPLEDQLENWCCRRCKFCHVCGRQHQATKQLLECNKCRNSYHPECLGPNY PTKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWSHDFSLCHDCAKLFA KGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSDEMYEILSNLP ESVAYTCVNCTERHPAEWRLALEKELQISLKQVLTALLNSRTTSHLLRYR QAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDLEGVKRK MDQGNYTSVLEFSDDIVKIIQAAINSDGGQPEIKKANSMVKSFFIRQMER VFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREENSHT EQPPLMKKIIPAPKPKGPGEPDSPTPLHPPTPPILSTDRSREDSPELNPP PGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVNCALWSAEVFEDD DGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNC VFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVFVDFEGISLRRKFLNGL EPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDARKR CVYTCKIVECRPPVVEPDINSTVEHDENRTIAHSPTSFTESSSKESQNTA EIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPGCRPLPS AGSPTPTTHEIVTVGDPLLSSGLRSIGSRRHSTSSLSPQRSKLRIMSPMR TGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTSTSS NLQRTVVTVGNKNSHLDGSSSSEMKQSSASDLVSKSSSLKGEKTKVLSSK SSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAEPSSVSFSSKEA LSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSII NEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLEPGQVTTGEEGNLKPEF MDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKELQA PRKRTVKVTLTPLKMENESQSKNALKESSPASPLQIESTSPTEPISASEN PGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQEDGSFKR RYPRRSARARSNMFFGLTPLYGVRSYGEEDIPFYSSSTGKKRGKRSAEGQ VDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDLPKI SQLDGVDDGTESDTSVTATTRKSSQIPKRNGKENGTENLKIDRPEDAGEK EHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSSLESSRRVHTST PSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALG ESPESSSSELLNLGEGLGLDSNREKDMGLFEVFSQQLPTTEPVDSSVSSS ISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVTITE KSVASSESDPALLSPGVDPTPEGHMTPDHFIQGHMDADHISSPPCGSVEQ GHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQISNAAV QTTPPHLKPATEKLIVVNQNMQPLYVLQTLPNGVTQKIQLTSSVSSTPSV METNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQHLHS FPAATQSSFPPNISNPPSGLLIGVQPPPDPQLLVSESSQRTDLSTTVATP SSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMTLINFTPSQLPN HPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAA GTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQTTTTPTSSASVPGHVTL TNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGTSQT PSTAAITAASSICVLPSTQTTGITAASPSGEADEHYQLQHVNQLLASKTG IHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQASPTSPGG SPSSPSSGQRSASPSVPGPTKPKPKTKRFQLPLDKGNGKKHKVSHLRTSS SEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQVAVL PEVQVTQNPANEQESAEPKTVEEEESNFSSPLMLWLQQEQKRKESITEKK PKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARSNARLKQLSFAGV NGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSA RAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEEEEVQLKSARRATSMDL PMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGNVIR SIQTDKREKYYDSKGIGCYMFRIDDSEVVDATMHGNAARFINHSCEPNCY SRVINIDGQKHIVIFAMRKIYRGEELTYDYKFPIEDASNKLPCNCGAKKC | 2655 |
| 3D view using mol* of 1173 (AA BP:48) | ||||||||||
 | ||||||||||
| PDB file (1175) >>>1175.pdbFusion protein BP residue: 48 CIF file (1175) >>>1175.cif | DAB2IP | chr9 | 124441060 | + | KMT2A | chr11 | 118354897 | + | MEPDSLLDQDDSYESPQERPGSRRSLPGSLSEKSPSMEPSAATPFRVTEK PPPVNKQENAGTLNILSTLSNGNSSKQKIPADGVHRIRVDFKEDCEAENV WEMGGLGILTSVPITPRVVCFLCASSGHVEFVYCQVCCEPFHKFCLEENE RPLEDQLENWCCRRCKFCHVCGRQHQATKQLLECNKCRNSYHPECLGPNY PTKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWSHDFSLCHDCAKLFA KGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSGTEDEMYEILS NLPESVAYTCVNCTERHPAEWRLALEKELQISLKQVLTALLNSRTTSHLL RYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDLEGV KRKMDQGNYTSVLEFSDDIVKIIQAAINSDGGQPEIKKANSMVKSFFIRQ MERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREEN SHTEQPPLMKKIIPAPKPKGPGEPDSPTPLHPPTPPILSTDRSREDSPEL NPPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVNCALWSAEVF EDDDGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRA KNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVFVDFEGISLRRKFL NGLEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDA RKRCVYTCKIVECRPPVVEPDINSTVEHDENRTIAHSPTSFTESSSKESQ NTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPGCRP LPSAGSPTPTTHEIVTVGDPLLSSGLRSIGSRRHSTSSLSPQRSKLRIMS PMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTS TSSNLQRTVVTVGNKNSHLDGSSSSEMKQSSASDLVSKSSSLKGEKTKVL SSKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAEPSSVSFSS KEALSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRS SIINEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLEPGQVTTGEEGNLK PEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKE LQAPRKRTVKVTLTPLKMENESQSKNALKESSPASPLQIESTSPTEPISA SENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQEDGS FKRRYPRRSARARSNMFFGLTPLYGVRSYGEEDIPFYSSSTGKKRGKRSA EGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDL PKISQLDGVDDGTESDTSVTATTRKSSQIPKRNGKENGTENLKIDRPEDA GEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSSLESSRRVH TSTPSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQ ALGESPESSSSELLNLGEGLGLDSNREKDMGLFEVFSQQLPTTEPVDSSV SSSISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVT ITEKSVASSESDPALLSPGVDPTPEGHMTPDHFIQGHMDADHISSPPCGS VEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQISN AAVQTTPPHLKPATEKLIVVNQNMQPLYVLQTLPNGVTQKIQLTSSVSST PSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQH LHSFPAATQSSFPPNISNPPSGLLIGVQPPPDPQLLVSESSQRTDLSTTV ATPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMTLINFTPSQ LPNHPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAA TAAGTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQTTTTPTSSASVPGH VTLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGT SQTPSTAAITAASSICVLPSTQTTGITAASPSGEADEHYQLQHVNQLLAS KTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQASPTS PGGSPSSPSSGQRSASPSVPGPTKPKPKTKRFQLPLDKGNGKKHKVSHLR TSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQV AVLPEVQVTQNPANEQESAEPKTVEEEESNFSSPLMLWLQQEQKRKESIT EKKPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARSNARLKQLSF AGVNGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPH GSARAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEEEEVQLKSARRATS MDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGN VIRSIQTDKREKYYDSKGIGCYMFRIDDSEVVDATMHGNAARFINHSCEP NCYSRVINIDGQKHIVIFAMRKIYRGEELTYDYKFPIEDASNKLPCNCGA | 2658 |
| 3D view using mol* of 1175 (AA BP:48) | ||||||||||
 | ||||||||||
| PDB file (1185) | DAB2IP | chr9 | 124441060 | + | KMT2A | chr11 | 118354897 | + | MSAGGSARKSTGRSSYYYRLLRRPRLQRQRSRSRSRTRPARESPQERPGS RRSLPGSLSEKSPSMEPSAATPFRVTEKPPPVNKQENAGTLNILSTLSNG NSSKQKIPADGVHRIRVDFKEDCEAENVWEMGGLGILTSVPITPRVVCFL CASSGHVEFVYCQVCCEPFHKFCLEENERPLEDQLENWCCRRCKFCHVCG RQHQATKQLLECNKCRNSYHPECLGPNYPTKPTKKKKVWICTKCVRCKSC GSTTPGKGWDAQWSHDFSLCHDCAKLFAKGNFCPLCDKCYDDDDYESKMM QCGKCDRWVHSKCENLSDEMYEILSNLPESVAYTCVNCTERHPAEWRLAL EKELQISLKQVLTALLNSRTTSHLLRYRQAAKPPDLNPETEESIPSRSSP EGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYTSVLEFSDDIVKIIQA AINSDGGQPEIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNS GMLPNAVLPPSLDHNYAQWQEREENSHTEQPPLMKKIIPAPKPKGPGEPD SPTPLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTYGDDSAND AGRLLYIGQNEWTHVNCALWSAEVFEDDDGSLKNVHMAVIRGKQLRCEFC QKPGATVGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVV PENGFEVFRRVFVDFEGISLRRKFLNGLEPENIHMMIGSMTIDCLGILND LSDCEDKLFPIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDINST VEHDENRTIAHSPTSFTESSSKESQNTAEIISPPSPDRPPHSQTSGSCYY HVISKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPTTHEIVTVGDPLLSSG LRSIGSRRHSTSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDL ESSAKVVDHVLGPLNSSTSLGQNTSTSSNLQRTVVTVGNKNSHLDGSSSS EMKQSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVH NTTSRELNVSKIGSFAEPSSVSFSSKEALSFPHLHLRGQRNDRDQHTDST QSANSSPDEDTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQKGKKSCKET FKEKHSSKSFLEPGQVTTGEEGNLKPEFMDEVLTPEYMGQRPCNNVSSDK IGDKGLSMPGVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMENESQSK NALKESSPASPLQIESTSPTEPISASENPGDGPVAQPSPNNTSCQDSQSN NYQNLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSARARSNMFFGLTPLYG VRSYGEEDIPFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTR TVISSGGEERLASHNLFREEEQCDLPKISQLDGVDDGTESDTSVTATTRK SSQIPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSV SRVKTQGQDSLEAQLSSLESSRRVHTSTPSDKNLLDTYNTELLKSDSDNN NSDDCGNILPSDIMDFVLKNTPSMQALGESPESSSSELLNLGEGLGLDSN REKDMGLFEVFSQQLPTTEPVDSSVSSSISAEEQFELPLELPSDLSVLTT RSPTVPSQNPSRLAVISDSGEKRVTITEKSVASSESDPALLSPGVDPTPE GHMTPDHFIQGHMDADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVS PTVPIQNQKYVPNSTDSPGPSQISNAAVQTTPPHLKPATEKLIVVNQNMQ PLYVLQTLPNGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLN PSLPTSQSLFPSASKGLLPMSHHQHLHSFPAATQSSFPPNISNPPSGLLI GVQPPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAP SSTPSNIAPSDVVSNMTLINFTPSQLPNHPSLLDLGSLNTSSHRTVPNII KRSKSSIMYFEPAPLLPQSVGGTAATAAGTSTISQDTSHLTSGSVSGLAS SSSVLNVVSMQTTTTPTSSASVPGHVTLTNPRLLGTPDIGSISNLLIKAS QQSLGIQDQPVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQTTG ITAASPSGEADEHYQLQHVNQLLASKTGIHSSQRDLDSASGPQVSNFTQT VDAPNSMGLEQNKALSSAVQASPTSPGGSPSSPSSGQRSASPSVPGPTKP KPKTKRFQLPLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGA EAEQQDTASVEQSSQKECGQPAGQVAVLPEVQVTQNPANEQESAEPKTVE EEESNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQICAESI EDAWKSLTDKVQEARSNARLKQLSFAGVNGLRMLGILHDAVVFLIEQLSG AKHCRNYKFRFHKPEEANEPPLNPHGSARAEVHLRKSAFDMFNFLASKHR QPPEYNPNDEEEEEVQLKSARRATSMDLPMPMRFRHLKKTSKEAVGVYRS PIHGRGLFCKRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCYMFR IDDSEVVDATMHGNAARFINHSCEPNCYSRVINIDGQKHIVIFAMRKIYR | 2683 |
| PDB file (1186) | DAB2IP | chr9 | 124441060 | + | KMT2A | chr11 | 118354897 | + | MSAGGSARKSTGRSSYYYRLLRRPRLQRQRSRSRSRTRPARESPQERPGS RRSLPGSLSEKSPSMEPSAATPFRVTEKPPPVNKQENAGTLNILSTLSNG NSSKQKIPADGVHRIRVDFKEDCEAENVWEMGGLGILTSVPITPRVVCFL CASSGHVEFVYCQVCCEPFHKFCLEENERPLEDQLENWCCRRCKFCHVCG RQHQATKQLLECNKCRNSYHPECLGPNYPTKPTKKKKVWICTKCVRCKSC GSTTPGKGWDAQWSHDFSLCHDCAKLFAKGNFCPLCDKCYDDDDYESKMM QCGKCDRWVHSKCENLSGTEDEMYEILSNLPESVAYTCVNCTERHPAEWR LALEKELQISLKQVLTALLNSRTTSHLLRYRQAAKPPDLNPETEESIPSR SSPEGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYTSVLEFSDDIVKI IQAAINSDGGQPEIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVS SNSGMLPNAVLPPSLDHNYAQWQEREENSHTEQPPLMKKIIPAPKPKGPG EPDSPTPLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTYGDDS ANDAGRLLYIGQNEWTHVNCALWSAEVFEDDDGSLKNVHMAVIRGKQLRC EFCQKPGATVGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHRDLIKG EVVPENGFEVFRRVFVDFEGISLRRKFLNGLEPENIHMMIGSMTIDCLGI LNDLSDCEDKLFPIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDI NSTVEHDENRTIAHSPTSFTESSSKESQNTAEIISPPSPDRPPHSQTSGS CYYHVISKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPTTHEIVTVGDPLL SSGLRSIGSRRHSTSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTA TDLESSAKVVDHVLGPLNSSTSLGQNTSTSSNLQRTVVTVGNKNSHLDGS SSSEMKQSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAP QVHNTTSRELNVSKIGSFAEPSSVSFSSKEALSFPHLHLRGQRNDRDQHT DSTQSANSSPDEDTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQKGKKSC KETFKEKHSSKSFLEPGQVTTGEEGNLKPEFMDEVLTPEYMGQRPCNNVS SDKIGDKGLSMPGVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMENES QSKNALKESSPASPLQIESTSPTEPISASENPGDGPVAQPSPNNTSCQDS QSNNYQNLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSARARSNMFFGLTP LYGVRSYGEEDIPFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYN FTRTVISSGGEERLASHNLFREEEQCDLPKISQLDGVDDGTESDTSVTAT TRKSSQIPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNC HSVSRVKTQGQDSLEAQLSSLESSRRVHTSTPSDKNLLDTYNTELLKSDS DNNNSDDCGNILPSDIMDFVLKNTPSMQALGESPESSSSELLNLGEGLGL DSNREKDMGLFEVFSQQLPTTEPVDSSVSSSISAEEQFELPLELPSDLSV LTTRSPTVPSQNPSRLAVISDSGEKRVTITEKSVASSESDPALLSPGVDP TPEGHMTPDHFIQGHMDADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQV PVSPTVPIQNQKYVPNSTDSPGPSQISNAAVQTTPPHLKPATEKLIVVNQ NMQPLYVLQTLPNGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTT GLNPSLPTSQSLFPSASKGLLPMSHHQHLHSFPAATQSSFPPNISNPPSG LLIGVQPPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKK LAPSSTPSNIAPSDVVSNMTLINFTPSQLPNHPSLLDLGSLNTSSHRTVP NIIKRSKSSIMYFEPAPLLPQSVGGTAATAAGTSTISQDTSHLTSGSVSG LASSSSVLNVVSMQTTTTPTSSASVPGHVTLTNPRLLGTPDIGSISNLLI KASQQSLGIQDQPVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQ TTGITAASPSGEADEHYQLQHVNQLLASKTGIHSSQRDLDSASGPQVSNF TQTVDAPNSMGLEQNKALSSAVQASPTSPGGSPSSPSSGQRSASPSVPGP TKPKPKTKRFQLPLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGT PGAEAEQQDTASVEQSSQKECGQPAGQVAVLPEVQVTQNPANEQESAEPK TVEEEESNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQICA ESIEDAWKSLTDKVQEARSNARLKQLSFAGVNGLRMLGILHDAVVFLIEQ LSGAKHCRNYKFRFHKPEEANEPPLNPHGSARAEVHLRKSAFDMFNFLAS KHRQPPEYNPNDEEEEEVQLKSARRATSMDLPMPMRFRHLKKTSKEAVGV YRSPIHGRGLFCKRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCY MFRIDDSEVVDATMHGNAARFINHSCEPNCYSRVINIDGQKHIVIFAMRK | 2686 |
Top |
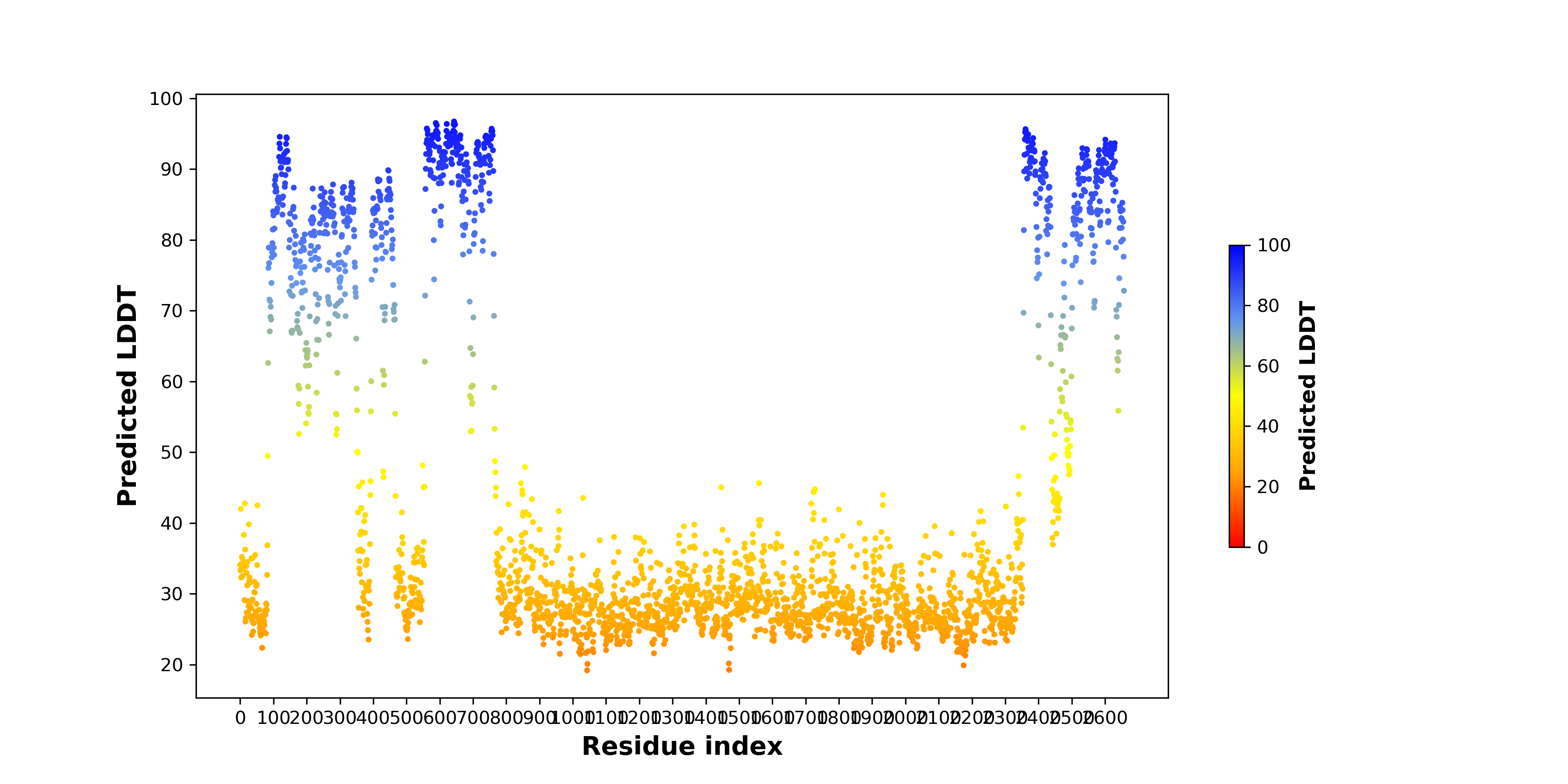
pLDDT score distribution |
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
DAB2IP_pLDDT.png |
KMT2A_pLDDT.png |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
Top |
Ramachandran Plot of Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| Fusion AA seq ID in FusionPDB and their Ramachandran plots |
| DAB2IP_KMT2A_1173.png |
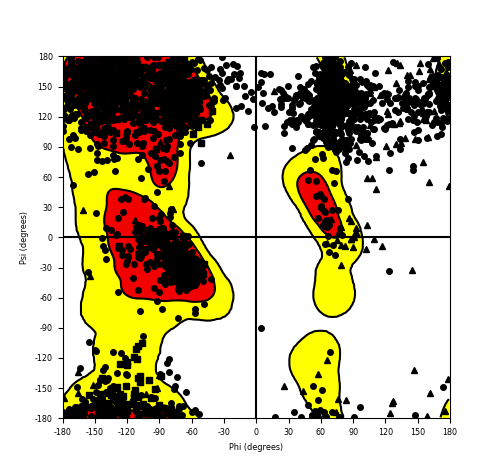 |
Top |
Potential Active Site Information |
 The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. |
| Fusion AA seq ID in FusionPDB | Site score | Size | D score | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
| 1173 | 1.115 | 152 | 1.084 | 237.013 | 0.339 | 0.87 | 1.182 | 0.871 | 1.159 | 0.752 | 0.913 | Chain A: 89,90,91,92,93,106,107,108,109,123,124,12 5,557,576,577,578,580,581,584,608,684,685,687,688, 690,691,692,696,707,2412,2415,2416 |
Top |
Potentially Interacting Small Molecules through Virtual Screening |
 The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. |
| Fusion AA seq ID in FusionPDB | ZINC ID | DrugBank ID | Drug name | Docking score | Glide gscore |
| 1173 | ZINC000043206370 | DB11793 | Niraparib | -9.26297 | -9.26297 |
| 1173 | ZINC000001611274 | DB01030 | Topotecan | -8.89063 | -9.15863 |
| 1173 | ZINC000085537017 | DB06441 | Cangrelor | -6.80318 | -9.02948 |
| 1173 | ZINC000003938482 | DB01263 | Posaconazole | -6.53352 | -8.62212 |
| 1173 | ZINC000000968328 | DB00412 | Rosiglitazone | -8.07864 | -8.60284 |
| 1173 | ZINC000001999441 | DB04861 | Nebivolol | -8.57589 | -8.59569 |
| 1173 | ZINC000018324776 | DB00862 | Vardenafil | -7.81841 | -8.58941 |
| 1173 | ZINC000004474682 | DB00287 | Travoprost | -8.53846 | -8.53846 |
| 1173 | ZINC000053084692 | DB06654 | Safinamide | -7.75157 | -8.52967 |
| 1173 | ZINC000000004319 | DB00598 | Labetalol | -6.54243 | -8.48023 |
| 1173 | ZINC000100052685 | DB01088 | Iloprost | -8.44841 | -8.44841 |
| 1173 | ZINC000058581064 | DB08930 | Dolutegravir | -8.06844 | -8.35564 |
| 1173 | ZINC000000601305 | DB00808 | Indapamide | -8.24757 | -8.24997 |
| 1173 | ZINC000001530948 | DB01041 | Thalidomide | -8.19085 | -8.19085 |
| 1173 | ZINC000030691754 | DB12364 | Betrixaban | -8.1799 | -8.1803 |
| 1173 | ZINC000001550477 | DB01259 | Lapatinib | -8.07014 | -8.15894 |
| 1173 | ZINC000000025958 | DB12332 | Rucaparib | -8.10151 | -8.11651 |
| 1173 | ZINC000012468792 | DB00654 | Latanoprost | -8.10321 | -8.10321 |
| 1173 | ZINC000003812913 | DB00929 | Misoprostol | -8.06702 | -8.06702 |
| 1173 | ZINC000000000856 | DB00983 | Formoterol | -8.01027 | -8.02247 |
Top |
 Drug information from DrugBank of the top 20 interacting small molecules. Drug information from DrugBank of the top 20 interacting small molecules. |
| ZINC ID | DrugBank ID | Drug name | Drug type | SMILES | Drug group |
| ZINC000043206370 | DB11793 | Niraparib | Small molecule | NC(=O)C1=CC=CC2=CN(N=C12)C1=CC=C(C=C1)[C@@H]1CCCNC1 | Approved|Investigational |
| ZINC000001611274 | DB01030 | Topotecan | Small molecule | CC[C@@]1(O)C(=O)OCC2=C1C=C1N(CC3=CC4=C(C=CC(O)=C4CN(C)C)N=C13)C2=O | Approved|Investigational |
| ZINC000085537017 | DB06441 | Cangrelor | Small molecule | CSCCNC1=C2N=CN([C@@H]3O[C@H](COP(O)(=O)OP(O)(=O)C(Cl)(Cl)P(O)(O)=O)[C@@H](O)[C@H]3O)C2=NC(SCCC(F)(F)F)=N1 | Approved |
| ZINC000003938482 | DB01263 | Posaconazole | Small molecule | [H][C@@](C)(O)[C@]([H])(CC)N1N=CN(C1=O)C1=CC=C(C=C1)N1CCN(CC1)C1=CC=C(OC[C@]2([H])CO[C@](CN3C=NC=N3)(C2)C2=C(F)C=C(F)C=C2)C=C1 | Approved|Investigational|Vet_approved |
| ZINC000018324776 | DB00862 | Vardenafil | Small molecule | CCCC1=NC(C)=C2N1NC(=NC2=O)C1=C(OCC)C=CC(=C1)S(=O)(=O)N1CCN(CC)CC1 | Approved |
| ZINC000004474682 | DB00287 | Travoprost | Small molecule | CC(C)OC(=O)CCCC=C/C[C@H]1[C@@H](O)C[C@@H](O)[C@@H]1C=C[C@@H](O)COC1=CC=CC(=C1)C(F)(F)F | Approved |
| ZINC000053084692 | DB06654 | Safinamide | Small molecule | C[C@H](NCC1=CC=C(OCC2=CC(F)=CC=C2)C=C1)C(N)=O | Approved|Investigational |
| ZINC000058581064 | DB08930 | Dolutegravir | Small molecule | [H][C@]12CN3C=C(C(=O)NCC4=CC=C(F)C=C4F)C(=O)C(O)=C3C(=O)N1[C@H](C)CCO2 | Approved |
| ZINC000000601305 | DB00808 | Indapamide | Small molecule | CC1CC2=CC=CC=C2N1NC(=O)C1=CC(=C(Cl)C=C1)S(N)(=O)=O | Approved |
| ZINC000030691754 | DB12364 | Betrixaban | Small molecule | COC1=CC=C(NC(=O)C2=CC=C(C=C2)C(=N)N(C)C)C(=C1)C(=O)NC1=CC=C(Cl)C=N1 | Approved|Investigational |
| ZINC000001550477 | DB01259 | Lapatinib | Small molecule | CS(=O)(=O)CCNCC1=CC=C(O1)C1=CC2=C(C=C1)N=CN=C2NC1=CC(Cl)=C(OCC2=CC(F)=CC=C2)C=C1 | Approved|Investigational |
| ZINC000000025958 | DB12332 | Rucaparib | Small molecule | CNCC1=CC=C(C=C1)C1=C2CCNC(=O)C3=C2C(N1)=CC(F)=C3 | Approved|Investigational |
| ZINC000012468792 | DB00654 | Latanoprost | Small molecule | CC(C)OC(=O)CCCC=C/C[C@H]1[C@@H](O)C[C@@H](O)[C@@H]1CC[C@@H](O)CCC1=CC=CC=C1 | Approved|Investigational |
Top |
Biochemical Features of Small Molecules |
 ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) |
| ZINC ID | mol_MW | dipole | SASA | FOSA | FISA | PISA | WPSA | volume | donorHB | accptHB | IP | Human Oral Absorption | Percent Human Oral Absorption | Rule Of Five | Rule Of Three |
| ZINC000043206370 | 320.393 | 3.754 | 607.688 | 185.794 | 143.417 | 278.476 | 0 | 1055.805 | 3 | 5 | 8.546 | 3 | 76.72 | 0 | 0 |
| ZINC000001611274 | 421.452 | 10.894 | 681.175 | 353.281 | 180.162 | 147.732 | 0 | 1247.085 | 2 | 10.5 | 8.905 | 2 | 63.438 | 0 | 0 |
| ZINC000001611274 | 421.452 | 9.949 | 676.628 | 345.411 | 185.261 | 145.956 | 0 | 1238.283 | 2 | 10.5 | 8.936 | 2 | 62.055 | 0 | 0 |
| ZINC000001611274 | 421.452 | 8.244 | 688.002 | 362.818 | 173.087 | 152.098 | 0 | 1254.803 | 2 | 8.75 | 7.843 | 2 | 69.25 | 0 | 1 |
| ZINC000085537017 | 776.353 | 7.43 | 927.054 | 287.655 | 393.585 | 54.42 | 191.394 | 1783.369 | 3 | 20.6 | 8.462 | 1 | 0 | 3 | 1 |
| ZINC000085537017 | 776.353 | 5.036 | 918.307 | 262.294 | 312.532 | 51.82 | 291.661 | 1775.566 | 3 | 20.6 | 8.603 | 1 | 0 | 3 | 1 |
| ZINC000003938482 | 700.787 | 6.593 | 1133.73 | 453.341 | 130.579 | 474.963 | 74.848 | 2110.183 | 1 | 11.2 | 8.262 | 1 | 82.417 | 3 | 1 |
| ZINC000003938482 | 700.787 | 8.545 | 1114.625 | 456.955 | 127.996 | 456.482 | 73.192 | 2101.798 | 1 | 11.2 | 8.612 | 1 | 82.52 | 3 | 1 |
| ZINC000000968328 | 357.426 | 1.444 | 634.074 | 175.676 | 137.918 | 287.735 | 32.745 | 1113.731 | 1 | 5.25 | 8.373 | 3 | 96.012 | 0 | 0 |
| ZINC000000968328 | 357.426 | 2.563 | 632.369 | 175.708 | 136.258 | 287.719 | 32.684 | 1111.853 | 1 | 5.25 | 8.348 | 3 | 96.294 | 0 | 0 |
| ZINC000000968328 | 357.426 | 1.098 | 615.115 | 188.498 | 120.189 | 271.15 | 35.279 | 1101.068 | 1 | 5.25 | 8.417 | 3 | 100 | 0 | 0 |
| ZINC000000968328 | 357.426 | 4.059 | 637.846 | 177.05 | 121.495 | 296.234 | 43.067 | 1111.876 | 1 | 5.25 | 8.521 | 3 | 100 | 0 | 0 |
| ZINC000001999441 | 405.441 | 4.722 | 712.226 | 273.488 | 84.211 | 260.774 | 93.753 | 1256.147 | 3 | 6.4 | 9.229 | 3 | 95.143 | 0 | 0 |
| ZINC000001999441 | 405.441 | 0.513 | 723.645 | 269.655 | 91.189 | 269.032 | 93.768 | 1265.844 | 3 | 6.4 | 9.27 | 3 | 94.039 | 0 | 0 |
| ZINC000018324776 | 488.604 | 6.83 | 851.417 | 623.618 | 134.756 | 93.044 | 0 | 1545.426 | 1 | 14.25 | 9.351 | 3 | 73.754 | 0 | 0 |
| ZINC000018324776 | 488.604 | 9.849 | 848.688 | 580.954 | 164.708 | 103.026 | 0 | 1542.734 | 1 | 12.25 | 9.078 | 3 | 72.527 | 0 | 0 |
| ZINC000018324776 | 488.604 | 8.683 | 837.703 | 563.885 | 162.912 | 110.906 | 0 | 1520.003 | 1 | 12.25 | 9.043 | 3 | 72.14 | 0 | 0 |
| ZINC000018324776 | 488.604 | 9.048 | 794.363 | 553.059 | 145.645 | 95.659 | 0 | 1503.896 | 1 | 14.25 | 9.407 | 3 | 70.208 | 0 | 0 |
| ZINC000018324776 | 488.604 | 8.871 | 816.03 | 566.343 | 150.772 | 98.915 | 0 | 1508.433 | 1 | 12.25 | 9.14 | 3 | 74.269 | 0 | 0 |
| ZINC000018324776 | 488.604 | 8.857 | 831.394 | 614.938 | 123.142 | 93.314 | 0 | 1524.919 | 1 | 12.25 | 8.996 | 3 | 80.597 | 0 | 0 |
| ZINC000018324776 | 488.604 | 9.702 | 848.752 | 580.696 | 165.188 | 102.869 | 0 | 1543.973 | 1 | 12.25 | 9.077 | 3 | 72.472 | 0 | 0 |
| ZINC000004474682 | 500.554 | 7.814 | 798.264 | 377.787 | 163.81 | 139.044 | 117.622 | 1521.375 | 3 | 7.85 | 9.535 | 3 | 85.406 | 1 | 2 |
| ZINC000053084692 | 302.348 | 6.36 | 612.581 | 156.033 | 105.388 | 304.286 | 46.874 | 1036.218 | 3 | 4.75 | 9.083 | 3 | 76.961 | 0 | 0 |
| ZINC000053084692 | 302.348 | 3.053 | 603.233 | 143.432 | 120.534 | 292.37 | 46.897 | 1022.597 | 3 | 4.75 | 9.137 | 3 | 72.683 | 0 | 0 |
| ZINC000000004319 | 328.41 | 5.655 | 664.549 | 169.051 | 199.646 | 295.852 | 0 | 1140.615 | 4 | 5.45 | 9.067 | 2 | 70.589 | 0 | 0 |
| ZINC000000004319 | 328.41 | 3.937 | 652.909 | 170.739 | 198.631 | 283.539 | 0 | 1134.882 | 4 | 5.45 | 9.364 | 2 | 70.662 | 0 | 0 |
| ZINC000100052685 | 478.627 | 5.289 | 955.739 | 563.979 | 167.746 | 224.014 | 0 | 1701.681 | 2 | 7.4 | 9.749 | 1 | 90.88 | 1 | 2 |
| ZINC000058581064 | 419.384 | 10.752 | 671.499 | 281.937 | 165.698 | 150.485 | 73.379 | 1193.652 | 0 | 8.45 | 8.755 | 3 | 83.672 | 0 | 0 |
| ZINC000058581064 | 419.384 | 10.808 | 670.317 | 276.157 | 165.522 | 158.31 | 70.328 | 1193.761 | 0 | 8.45 | 8.94 | 3 | 83.698 | 0 | 0 |
| ZINC000000601305 | 365.834 | 2.673 | 651.585 | 129.631 | 190.345 | 277.736 | 53.872 | 1108.18 | 3 | 8.5 | 8.619 | 3 | 76.119 | 0 | 0 |
| ZINC000001530948 | 258.233 | 6.265 | 444.715 | 83.913 | 189.185 | 171.618 | 0 | 765.624 | 1 | 6 | 10.446 | 3 | 68.301 | 0 | 0 |
| ZINC000030691754 | 451.911 | 10.54 | 790.066 | 238.941 | 130.476 | 348.605 | 72.044 | 1393.504 | 2 | 7.25 | 8.365 | 1 | 100 | 0 | 1 |
| ZINC000001550477 | 581.06 | 8.763 | 984.429 | 219.392 | 136.563 | 520.447 | 108.027 | 1738.439 | 1 | 8.25 | 8.353 | 1 | 75.112 | 2 | 1 |
| ZINC000001550477 | 581.06 | 8.228 | 914.704 | 239.853 | 89.107 | 476.75 | 108.994 | 1676.884 | 1 | 8.25 | 8.025 | 1 | 82.627 | 2 | 1 |
| ZINC000000025958 | 323.369 | 9.363 | 601.398 | 214.232 | 112.926 | 227.819 | 46.421 | 1032.273 | 3 | 4 | 8.52 | 3 | 85.064 | 0 | 0 |
| ZINC000000025958 | 323.369 | 9.466 | 597.964 | 212.386 | 112.007 | 227.152 | 46.419 | 1026.229 | 3 | 4 | 8.513 | 3 | 85.039 | 0 | 0 |
| ZINC000012468792 | 432.599 | 2.028 | 800.281 | 456.672 | 137.125 | 206.485 | 0 | 1494.735 | 3 | 7.1 | 9.572 | 1 | 100 | 0 | 1 |
| ZINC000003812913 | 382.539 | 3.507 | 771.329 | 596.521 | 174.657 | 0.151 | 0 | 1405.474 | 2 | 6.45 | 10.343 | 3 | 92.472 | 0 | 1 |
| ZINC000000000856 | 344.41 | 6.104 | 642.08 | 271.184 | 183.213 | 187.682 | 0 | 1147.526 | 4 | 7.2 | 8.665 | 2 | 65.328 | 0 | 0 |
| ZINC000000000856 | 344.41 | 4.876 | 597.045 | 248.042 | 184.569 | 164.434 | 0 | 1098.954 | 4 | 7.2 | 8.741 | 2 | 63.468 | 0 | 0 |
Top |
Drug Toxicity Information |
 Toxicity information of individual drugs using eToxPred Toxicity information of individual drugs using eToxPred |
| ZINC ID | Smile | Surface Accessibility | Toxicity |
| ZINC000043206370 | NC(=O)c1cccc2cn(-c3ccc([C@@H]4CCCNC4)cc3)nc12 | 0.15810306 | 0.243905197 |
| ZINC000001611274 | CC[C@@]1(O)C(=O)OCc2c1cc1n(c2=O)Cc2cc3c(CN(C)C)c(O)ccc3nc2-1 | 0.083656413 | 0.374781684 |
| ZINC000085537017 | CSCCNc1nc(SCCC(F)(F)F)nc2c1ncn2[C@@H]1O[C@H](CO[P@@](=O)(O)O[P@](=O)(O)C(Cl)(Cl)P(=O)(O)O)[C@@H](O)[C@H]1O | 0.017889541 | 0.259362736 |
| ZINC000003938482 | CC[C@@H]([C@H](C)O)n1ncn(-c2ccc(N3CCN(c4ccc(OC[C@@H]5CO[C@@](Cn6cncn6)(c6ccc(F)cc6F)C5)cc4)CC3)cc2)c1=O | 0.031724381 | 0.429306683 |
| ZINC000000968328 | CN(CCOc1ccc(C[C@@H]2SC(=O)NC2=O)cc1)c1ccccn1 | 0.155326871 | 0.273380926 |
| ZINC000001999441 | O[C@@H](CNC[C@@H](O)[C@@H]1CCc2cc(F)ccc2O1)[C@H]1CCc2cc(F)ccc2O1 | 0.055550082 | 0.198725964 |
| ZINC000018324776 | CCCc1nc(C)c2c(=O)[nH]c(-c3cc(S(=O)(=O)N4CCN(CC)CC4)ccc3OCC)nn12 | 0.150680014 | 0.356552314 |
| ZINC000004474682 | CC(C)OC(=O)CCC/C=CC[C@H]1[C@@H](O)C[C@@H](O)[C@@H]1/C=C/[C@@H](O)COc1cccc(C(F)(F)F)c1 | 0.043771914 | 0.166030074 |
| ZINC000053084692 | C[C@H](NCc1ccc(OCc2cccc(F)c2)cc1)C(N)=O | 0.279955479 | 0.265373208 |
| ZINC000000004319 | C[C@H](CCc1ccccc1)NC[C@H](O)c1ccc(O)c(C(N)=O)c1 | 0.163888793 | 0.349082082 |
| ZINC000100052685 | CC#CC[C@H](C)[C@@H](O)/C=C/[C@@H]1[C@@H](O)C[C@@H]2C/C(=C/CCCC(=O)OCC(=O)c3ccccc3)C[C@H]21 | 0.031863798 | 0.193548025 |
| ZINC000058581064 | C[C@@H]1CCO[C@H]2Cn3cc(C(=O)NCc4ccc(F)cc4F)c(=O)c(O)c3C(=O)N21 | 0.07265705 | 0.3133548 |
| ZINC000000601305 | C[C@H]1Cc2ccccc2N1NC(=O)c1ccc(Cl)c(S(N)(=O)=O)c1 | 0.166136479 | 0.17734677 |
| ZINC000001530948 | O=C1CC[C@@H](N2C(=O)c3ccccc3C2=O)C(=O)N1 | 0.202402232 | 0.556220101 |
| ZINC000030691754 | COc1ccc(NC(=O)c2ccc(C(=N)N(C)C)cc2)c(C(=O)Nc2ccc(Cl)cn2)c1 | 0.274619295 | 0.219857351 |
| ZINC000001550477 | CS(=O)(=O)CCNCc1ccc(-c2ccc3ncnc(Nc4ccc(OCc5cccc(F)c5)c(Cl)c4)c3c2)o1 | 0.18759569 | 0.281219747 |
| ZINC000000025958 | CNCc1ccc(-c2[nH]c3cc(F)cc4c3c2CCNC4=O)cc1 | 0.204259799 | 0.179994505 |
| ZINC000012468792 | CC(C)OC(=O)CCC/C=CC[C@H]1[C@@H](O)C[C@@H](O)[C@@H]1CC[C@@H](O)CCc1ccccc1 | 0.057567479 | 0.159818744 |
| ZINC000003812913 | CCCC[C@](C)(O)C/C=C/[C@H]1[C@H](O)CC(=O)[C@@H]1CCCCCCC(=O)OC | 0.055724408 | 0.308697552 |
| ZINC000000000856 | COc1ccc(C[C@H](C)NC[C@@H](O)c2ccc(O)c(NC=O)c2)cc1 | 0.133863078 | 0.246482573 |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| DAB2IP | DAB2, TP53, TP63, TP73, YWHAQ, TRAF2, RIPK1, MAP3K5, TNFRSF1A, TRADD, PIK3R1, AKT1, HRAS, CUL1, CUL4A, SKP1, RBX1, FBXW7, RFPL3, NTRK1, XPO1, EXOSC9, RAB4B, IWS1, UGGT1, RNASEH2B, Cdk1, Shoc2, Mis12, Cep152, CDH1, CTDSPL, ANKS1A, CLIP1, MCC, CD83, CGN, TMEM206, SNX17, CBY1, TRIM25, NRAS, KIAA1429, E6, YWHAE, nsp16, C11orf52, KRAS, LYN, OCLN, RAB35, RHOB, SMARCA2, CLEC4A, TMEM9, RASAL2, ANKS1B, EFNB1, YWHAB, YWHAG, YWHAH, SLC31A1, ASXL2, DDX3Y, |
| KMT2A | PPIE, PPP1R15A, KMT2A, ASH2L, HCFC1, HCFC2, MEN1, RBBP5, WDR5, AVP, INS, OXT, MAP3K5, HDAC1, CTBP1, CBX4, BMI1, CREBBP, SMARCB1, CXXC1, MYB, CTNNB1, SNW1, E2F2, E2F4, E2F6, PSIP1, MLLT4, POLR2A, KAT8, RNF2, TP53, tat, SBF1, MTM1, SET, HIST1H3A, HIST1H4A, KAT6A, ELL, AFF1, AFF4, CDK9, CCNT1, CTR9, LEO1, PAF1, CDC73, WDR61, MLLT3, DOT1L, SKP2, HIST3H3, SVIL, HIST2H3C, SIN3A, MLLT1, RUNX1, CBFB, H3F3A, SIRT7, ASB2, TCEB1, TCEB2, CBX8, TOP1, TAF6, NCL, HECW2, LGR4, CSNK2A2, SENP3, SYMPK, PKN1, PIH1D1, KRAS, TAF1, CHD3, SMARCA2, SMARCC2, SMARCC1, HDAC2, RBBP4, RBBP7, TBP, MBD3, SAP30, RAN, TAF9, TASP1, HIST1H2BG, EWSR1, DYNLT1, KIF11, ING4, Cbx1, Set, ZNF131, ASB7, AR, CSNK2A1, OGT, BRD4, KDM5B, KDM5D, KDM6B, CKS1B, CKS2, CHD4, ESR2, AGR2, EZH2, FBXW7, SOX2, MED17, WDR82, ACTR8, GEMIN5, RUVBL1, MCM2, RTF1, MED15, MED8, MRGBP, MED14, HIST1H2BB, HIST1H2AB, KIAA1429, SP1, MYC, NR2C2, IRF2, PLEKHA4, DPY30, PTEN, NPM1, RPL13, CCT6A, TMCO1, ITGB1, ESR1, MYCN, SUPT5H, CIT, Pik3r2, FASN, LDLR, CDC27, KMT2B, SETD1A, KDM6A, KANSL2, BUB3, SETD1B, KANSL1, RERE, MCRS1, KMT2C, ZZZ3, MBIP, YEATS2, PAXIP1, KANSL3, KMT2D, BOD1L1, CSRP2BP, ZNF608, ATN1, PHF20L1, PHF20, NCOA6, TADA3, APEX1, ASF1A, CBX3, CD3EAP, CENPA, NUP50, POLR1E, TERF2IP, ZNF330, NAA40, PRKRA, SRSF4, KRR1, RPL13A, ABT1, CSNK2B, OASL, SRSF5, SRSF6, RPS9, FGFBP1, RPLP0, RPL36, KAL1, RPL3, PML, ELF1, ELF2, FEV, NHLH1, EN1, KLF12, KLF3, NFIX, NFYC, YY1, BRD3, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| DAB2IP |  |
| KMT2A |  |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to DAB2IP-KMT2A |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to DAB2IP-KMT2A |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
| DAB2IP | KMT2A | Anal Squamous Cell Carcinoma | MyCancerGenome | |
| DAB2IP | KMT2A | Cancer Of Unknown Primary | MyCancerGenome | |
| DAB2IP | KMT2A | Nos | MyCancerGenome | |
| DAB2IP | KMT2A | Invasive Breast Carcinoma | MyCancerGenome | |
| DAB2IP | KMT2A | Lung Adenocarcinoma | MyCancerGenome | |
| DAB2IP | KMT2A | Poorly Differentiated Non-Small Cell Lung Cancer | MyCancerGenome |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | DAB2IP | C0024121 | Lung Neoplasms | 1 | CTD_human |
| Hgene | DAB2IP | C0027051 | Myocardial Infarction | 1 | CTD_human |
| Hgene | DAB2IP | C0033578 | Prostatic Neoplasms | 1 | CTD_human |
| Hgene | DAB2IP | C0034065 | Pulmonary Embolism | 1 | CTD_human |
| Hgene | DAB2IP | C0085096 | Peripheral Vascular Diseases | 1 | CTD_human |
| Hgene | DAB2IP | C0162871 | Aortic Aneurysm, Abdominal | 1 | CTD_human |
| Hgene | DAB2IP | C0242379 | Malignant neoplasm of lung | 1 | CTD_human |
| Hgene | DAB2IP | C0376358 | Malignant neoplasm of prostate | 1 | CTD_human |
| Hgene | DAB2IP | C0524702 | Pulmonary Thromboembolisms | 1 | CTD_human |
| Tgene | KMT2A | C2826025 | Mixed phenotype acute leukemia | 3 | ORPHANET |
| Tgene | KMT2A | C0023418 | leukemia | 2 | CTD_human |
| Tgene | KMT2A | C0023452 | Childhood Acute Lymphoblastic Leukemia | 2 | CTD_human |
| Tgene | KMT2A | C0023453 | L2 Acute Lymphoblastic Leukemia | 2 | CTD_human |
| Tgene | KMT2A | C0023466 | Leukemia, Monocytic, Chronic | 2 | CTD_human |
| Tgene | KMT2A | C0023467 | Leukemia, Myelocytic, Acute | 2 | CTD_human |
| Tgene | KMT2A | C0023470 | Myeloid Leukemia | 2 | CTD_human |
| Tgene | KMT2A | C0026998 | Acute Myeloid Leukemia, M1 | 2 | CTD_human |
| Tgene | KMT2A | C1854630 | Growth Deficiency and Mental Retardation with Facial Dysmorphism | 2 | CTD_human;GENOMICS_ENGLAND;ORPHANET |
| Tgene | KMT2A | C1879321 | Acute Myeloid Leukemia (AML-M2) | 2 | CTD_human |
| Tgene | KMT2A | C1961102 | Precursor Cell Lymphoblastic Leukemia Lymphoma | 2 | CTD_human |
| Tgene | KMT2A | C0001418 | Adenocarcinoma | 1 | CTD_human |
| Tgene | KMT2A | C0004403 | Autosome Abnormalities | 1 | CTD_human |
| Tgene | KMT2A | C0005684 | Malignant neoplasm of urinary bladder | 1 | CTD_human |
| Tgene | KMT2A | C0005695 | Bladder Neoplasm | 1 | CTD_human |
| Tgene | KMT2A | C0007138 | Carcinoma, Transitional Cell | 1 | CTD_human |
| Tgene | KMT2A | C0008625 | Chromosome Aberrations | 1 | CTD_human |
| Tgene | KMT2A | C0023448 | Lymphoid leukemia | 1 | CTD_human |
| Tgene | KMT2A | C0023465 | Acute monocytic leukemia | 1 | CTD_human |
| Tgene | KMT2A | C0023479 | Acute myelomonocytic leukemia | 1 | CTD_human |
| Tgene | KMT2A | C0024623 | Malignant neoplasm of stomach | 1 | CTD_human |
| Tgene | KMT2A | C0033578 | Prostatic Neoplasms | 1 | CTD_human |
| Tgene | KMT2A | C0036341 | Schizophrenia | 1 | PSYGENET |
| Tgene | KMT2A | C0038356 | Stomach Neoplasms | 1 | CTD_human |
| Tgene | KMT2A | C0149925 | Small cell carcinoma of lung | 1 | CTD_human |
| Tgene | KMT2A | C0205641 | Adenocarcinoma, Basal Cell | 1 | CTD_human |
| Tgene | KMT2A | C0205642 | Adenocarcinoma, Oxyphilic | 1 | CTD_human |
| Tgene | KMT2A | C0205643 | Carcinoma, Cribriform | 1 | CTD_human |
| Tgene | KMT2A | C0205644 | Carcinoma, Granular Cell | 1 | CTD_human |
| Tgene | KMT2A | C0205645 | Adenocarcinoma, Tubular | 1 | CTD_human |
| Tgene | KMT2A | C0270972 | Cornelia De Lange Syndrome | 1 | ORPHANET |
| Tgene | KMT2A | C0280141 | Acute Undifferentiated Leukemia | 1 | ORPHANET |
| Tgene | KMT2A | C0376358 | Malignant neoplasm of prostate | 1 | CTD_human |
| Tgene | KMT2A | C0856823 | Undifferentiated type acute leukemia | 1 | ORPHANET |
| Tgene | KMT2A | C1535926 | Neurodevelopmental Disorders | 1 | CTD_human |
| Tgene | KMT2A | C1708349 | Hereditary Diffuse Gastric Cancer | 1 | CTD_human |
| Tgene | KMT2A | C2239176 | Liver carcinoma | 1 | CTD_human |
| Tgene | KMT2A | C2930974 | Acute erythroleukemia | 1 | CTD_human |
| Tgene | KMT2A | C2930975 | Acute erythroleukemia - M6a subtype | 1 | CTD_human |
| Tgene | KMT2A | C2930976 | Acute myeloid leukemia FAB-M6 | 1 | CTD_human |
| Tgene | KMT2A | C2930977 | Acute erythroleukemia - M6b subtype | 1 | CTD_human |