| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:ERG-EWSR1 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: ERG-EWSR1 | FusionPDB ID: 27340 | FusionGDB2.0 ID: 27340 | Hgene | Tgene | Gene symbol | ERG | EWSR1 | Gene ID | 2078 | 2130 |
| Gene name | ETS transcription factor ERG | EWS RNA binding protein 1 | |
| Synonyms | erg-3|p55 | EWS|EWS-FLI1|bK984G1.4 | |
| Cytomap | 21q22.2 | 22q12.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | transcriptional regulator ERGERG, ETS transcription factorFUS/ERG fusion proteinTMPRSS2/ERG fusionerythroblast transformation-specific transcription factor ERG variant 10ets-relatedtranscriptional regulator ERG (transforming protein ERG)v-ets avian | RNA-binding protein EWSEWS RNA-binding protein variant 6Ewing sarcoma breakpoint region 1Ewings sarcoma EWS-Fli1 (type 1) oncogene | |
| Modification date | 20200320 | 20200329 | |
| UniProtAcc | Q9Y282 | Q01844 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000288319, ENST00000398905, ENST00000398907, ENST00000398911, ENST00000398919, ENST00000453032, ENST00000398897, ENST00000398910, ENST00000417133, ENST00000442448, ENST00000429727, ENST00000485493, | ENST00000331029, ENST00000332035, ENST00000332050, ENST00000333395, ENST00000397938, ENST00000406548, ENST00000414183, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 19 X 16 X 5=1520 | 17 X 58 X 10=9860 |
| # samples | 31 | 48 | |
| ** MAII score | log2(31/1520*10)=-2.29373120305671 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(48/9860*10)=-4.36048133565676 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: ERG [Title/Abstract] AND EWSR1 [Title/Abstract] AND fusion [Title/Abstract] Ewing sarcoma with ERG gene rearrangements: A molecular study focusing on the prevalence of FUS-ERG and common pitfalls in detecting EWSR1-ERG fusions by FISH (pmid: 26690869) | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | EWSR1(29683123)-ERG(39764366), # samples:2 ERG(39762959)-EWSR1(29682948), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | EWSR1-ERG seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. EWSR1-ERG seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. EWSR1-ERG seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. EWSR1-ERG seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. EWSR1-ERG seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. EWSR1-ERG seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. EWSR1-ERG seems lost the major protein functional domain in Tgene partner, which is a transcription factor due to the frame-shifted ORF. ERG-EWSR1 seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. ERG-EWSR1 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. ERG-EWSR1 seems lost the major protein functional domain in Hgene partner, which is a transcription factor due to the frame-shifted ORF. ERG-EWSR1 seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. ERG-EWSR1 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ERG | GO:0045944 | positive regulation of transcription by RNA polymerase II | 18195090|23913826 |
 Fusion gene breakpoints across ERG (5'-gene) Fusion gene breakpoints across ERG (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 Fusion gene breakpoints across EWSR1 (3'-gene) Fusion gene breakpoints across EWSR1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerKB3 | . | . | ERG | chr21 | 39762917 | - | EWSR1 | chr22 | 29682912 | + |
| ChiTaRS5.0 | N/A | S82338 | ERG | chr21 | 39762959 | - | EWSR1 | chr22 | 29682948 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000398897 | ERG | chr21 | 39762917 | - | ENST00000332050 | EWSR1 | chr22 | 29682912 | + | 2288 | 754 | 971 | 1924 | 317 |
| ENST00000398897 | ERG | chr21 | 39762917 | - | ENST00000397938 | EWSR1 | chr22 | 29682912 | + | 2508 | 754 | 995 | 2143 | 382 |
| ENST00000398897 | ERG | chr21 | 39762917 | - | ENST00000406548 | EWSR1 | chr22 | 29682912 | + | 2316 | 754 | 995 | 2140 | 381 |
| ENST00000398897 | ERG | chr21 | 39762917 | - | ENST00000331029 | EWSR1 | chr22 | 29682912 | + | 2394 | 754 | 995 | 2029 | 344 |
| ENST00000398897 | ERG | chr21 | 39762917 | - | ENST00000414183 | EWSR1 | chr22 | 29682912 | + | 2323 | 754 | 995 | 2140 | 381 |
| ENST00000398897 | ERG | chr21 | 39762917 | - | ENST00000333395 | EWSR1 | chr22 | 29682912 | + | 1419 | 754 | 114 | 1055 | 313 |
| ENST00000398897 | ERG | chr21 | 39762917 | - | ENST00000332035 | EWSR1 | chr22 | 29682912 | + | 2319 | 754 | 995 | 2143 | 382 |
| ENST00000398910 | ERG | chr21 | 39762917 | - | ENST00000332050 | EWSR1 | chr22 | 29682912 | + | 2591 | 1057 | 177 | 1301 | 374 |
| ENST00000398910 | ERG | chr21 | 39762917 | - | ENST00000397938 | EWSR1 | chr22 | 29682912 | + | 2811 | 1057 | 177 | 1358 | 393 |
| ENST00000398910 | ERG | chr21 | 39762917 | - | ENST00000406548 | EWSR1 | chr22 | 29682912 | + | 2619 | 1057 | 177 | 1358 | 393 |
| ENST00000398910 | ERG | chr21 | 39762917 | - | ENST00000331029 | EWSR1 | chr22 | 29682912 | + | 2697 | 1057 | 177 | 1358 | 393 |
| ENST00000398910 | ERG | chr21 | 39762917 | - | ENST00000414183 | EWSR1 | chr22 | 29682912 | + | 2626 | 1057 | 177 | 1358 | 393 |
| ENST00000398910 | ERG | chr21 | 39762917 | - | ENST00000333395 | EWSR1 | chr22 | 29682912 | + | 1722 | 1057 | 177 | 1358 | 393 |
| ENST00000398910 | ERG | chr21 | 39762917 | - | ENST00000332035 | EWSR1 | chr22 | 29682912 | + | 2622 | 1057 | 177 | 1358 | 393 |
| ENST00000417133 | ERG | chr21 | 39762917 | - | ENST00000332050 | EWSR1 | chr22 | 29682912 | + | 2660 | 1126 | 177 | 1370 | 397 |
| ENST00000417133 | ERG | chr21 | 39762917 | - | ENST00000397938 | EWSR1 | chr22 | 29682912 | + | 2880 | 1126 | 177 | 1427 | 416 |
| ENST00000417133 | ERG | chr21 | 39762917 | - | ENST00000406548 | EWSR1 | chr22 | 29682912 | + | 2688 | 1126 | 177 | 1427 | 416 |
| ENST00000417133 | ERG | chr21 | 39762917 | - | ENST00000331029 | EWSR1 | chr22 | 29682912 | + | 2766 | 1126 | 177 | 1427 | 416 |
| ENST00000417133 | ERG | chr21 | 39762917 | - | ENST00000414183 | EWSR1 | chr22 | 29682912 | + | 2695 | 1126 | 177 | 1427 | 416 |
| ENST00000417133 | ERG | chr21 | 39762917 | - | ENST00000333395 | EWSR1 | chr22 | 29682912 | + | 1791 | 1126 | 177 | 1427 | 416 |
| ENST00000417133 | ERG | chr21 | 39762917 | - | ENST00000332035 | EWSR1 | chr22 | 29682912 | + | 2691 | 1126 | 177 | 1427 | 416 |
| ENST00000442448 | ERG | chr21 | 39762917 | - | ENST00000332050 | EWSR1 | chr22 | 29682912 | + | 2674 | 1140 | 263 | 1384 | 373 |
| ENST00000442448 | ERG | chr21 | 39762917 | - | ENST00000397938 | EWSR1 | chr22 | 29682912 | + | 2894 | 1140 | 263 | 1441 | 392 |
| ENST00000442448 | ERG | chr21 | 39762917 | - | ENST00000406548 | EWSR1 | chr22 | 29682912 | + | 2702 | 1140 | 263 | 1441 | 392 |
| ENST00000442448 | ERG | chr21 | 39762917 | - | ENST00000331029 | EWSR1 | chr22 | 29682912 | + | 2780 | 1140 | 263 | 1441 | 392 |
| ENST00000442448 | ERG | chr21 | 39762917 | - | ENST00000414183 | EWSR1 | chr22 | 29682912 | + | 2709 | 1140 | 263 | 1441 | 392 |
| ENST00000442448 | ERG | chr21 | 39762917 | - | ENST00000333395 | EWSR1 | chr22 | 29682912 | + | 1805 | 1140 | 263 | 1441 | 392 |
| ENST00000442448 | ERG | chr21 | 39762917 | - | ENST00000332035 | EWSR1 | chr22 | 29682912 | + | 2705 | 1140 | 263 | 1441 | 392 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >27340_27340_1_ERG-EWSR1_ERG_chr21_39762917_ENST00000398897_EWSR1_chr22_29682912_ENST00000331029_length(amino acids)=344AA_BP= MGVYGQESGGFSGPGENRSMSGPDNRGRGRGGFDRGGMSRDPDEDSDNSAIYVQGLNDSVTLDDLADFFKQCGVVKMNKRTGQPMIHIYL DKETGKPKGDATVSYEDPPTAKAAVEWFDGKDFQGSKLKVSLARKKPPMNSMRGGLPPREGRGMPPPLRGGPGGPGGPGGPMGRMGGRGG DRGGFPPRGPRGSRGNPSGGGNVQHRAGDWQCPNPGCGNQNFAWRTECNQCKAPKPEGFLPPPFPPPGGDRGRGGPGGMRGGRGGLMDRG -------------------------------------------------------------- >27340_27340_2_ERG-EWSR1_ERG_chr21_39762917_ENST00000398897_EWSR1_chr22_29682912_ENST00000332035_length(amino acids)=382AA_BP= MGVYGQESGGFSGPGENRSMSGPDNRGRGRGGFDRGGMSRGGRGGGRGGMGSAGERGGFNKPGGPMDEGPDLDLGPPVDPDEDSDNSAIY VQGLNDSVTLDDLADFFKQCGVVKMNKRTGQPMIHIYLDKETGKPKGDATVSYEDPPTAKAAVEWFDGKDFQGSKLKVSLARKKPPMNSM RGGLPPREGRGMPPPLRGGPGGPGGPGGPMGRMGGRGGDRGGFPPRGPRGSRGNPSGGGNVQHRAGDWQCPNPGCGNQNFAWRTECNQCK APKPEGFLPPPFPPPGGDRGRGGPGGMRGGRGGLMDRGGPGGMFRGGRGGDRGGFRGGRGMDRGGFGGGRRGGPGGPPGPLMEQMGGRRG -------------------------------------------------------------- >27340_27340_3_ERG-EWSR1_ERG_chr21_39762917_ENST00000398897_EWSR1_chr22_29682912_ENST00000332050_length(amino acids)=317AA_BP= MDEGPDLDLGPPVDPDEDSDNSAIYVQGLNDSVTLDDLADFFKQCGVVKMNKRTGQPMIHIYLDKETGKPKGDATVSYEDPPTAKAAVEW FDGKDFQGSKLKVSLARKKPPMNSMRGGLPPREGRGMPPPLRGGPGGPGGPGGPMGRMGGRGGDRGGFPPRGPRGSRGNPSGGGNVQHRA GDWQCPNPGCGNQNFAWRTECNQCKAPKPEGFLPPPFPPPGGDRGRGGPGGMRGGRGGLMDRGGPGGMFRGGRGGDRGGFRGGRGMDRGG -------------------------------------------------------------- >27340_27340_4_ERG-EWSR1_ERG_chr21_39762917_ENST00000398897_EWSR1_chr22_29682912_ENST00000333395_length(amino acids)=313AA_BP=213 MSRTQQLISRNSPDECSVAKGGKMVGSPDTVGMNYGSYMEEKHMPPPNMTTNERRVIVPADPTLWSTDHVRQWLEWAVKEYGLPDVNILL FQNIDGKELCKMTKDDFQRLTPSYNADILLSHLHYLRETPLPHLTSDDVDKALQNSPRLMHARNTDLPYEPPRRSAWTGHGHPTPQSKAA QPSPSTVPKTEDQRPQLDPYQILGPTSSRLANPAIPLHSRLVMIRAVTLSRTPMGNRAAMDSRVAMVNKAAMGSSLPLVTHPKLDPTAKL -------------------------------------------------------------- >27340_27340_5_ERG-EWSR1_ERG_chr21_39762917_ENST00000398897_EWSR1_chr22_29682912_ENST00000397938_length(amino acids)=382AA_BP= MGVYGQESGGFSGPGENRSMSGPDNRGRGRGGFDRGGMSRGGRGGGRGGMGSAGERGGFNKPGGPMDEGPDLDLGPPVDPDEDSDNSAIY VQGLNDSVTLDDLADFFKQCGVVKMNKRTGQPMIHIYLDKETGKPKGDATVSYEDPPTAKAAVEWFDGKDFQGSKLKVSLARKKPPMNSM RGGLPPREGRGMPPPLRGGPGGPGGPGGPMGRMGGRGGDRGGFPPRGPRGSRGNPSGGGNVQHRAGDWQCPNPGCGNQNFAWRTECNQCK APKPEGFLPPPFPPPGGDRGRGGPGGMRGGRGGLMDRGGPGGMFRGGRGGDRGGFRGGRGMDRGGFGGGRRGGPGGPPGPLMEQMGGRRG -------------------------------------------------------------- >27340_27340_6_ERG-EWSR1_ERG_chr21_39762917_ENST00000398897_EWSR1_chr22_29682912_ENST00000406548_length(amino acids)=381AA_BP= MGVYGQESGGFSGPGENRSMSGPDNRGRGRGGFDRGGMSRGGRGGGRGGMGAGERGGFNKPGGPMDEGPDLDLGPPVDPDEDSDNSAIYV QGLNDSVTLDDLADFFKQCGVVKMNKRTGQPMIHIYLDKETGKPKGDATVSYEDPPTAKAAVEWFDGKDFQGSKLKVSLARKKPPMNSMR GGLPPREGRGMPPPLRGGPGGPGGPGGPMGRMGGRGGDRGGFPPRGPRGSRGNPSGGGNVQHRAGDWQCPNPGCGNQNFAWRTECNQCKA PKPEGFLPPPFPPPGGDRGRGGPGGMRGGRGGLMDRGGPGGMFRGGRGGDRGGFRGGRGMDRGGFGGGRRGGPGGPPGPLMEQMGGRRGG -------------------------------------------------------------- >27340_27340_7_ERG-EWSR1_ERG_chr21_39762917_ENST00000398897_EWSR1_chr22_29682912_ENST00000414183_length(amino acids)=381AA_BP= MGVYGQESGGFSGPGENRSMSGPDNRGRGRGGFDRGGMSRGGRGGGRGGMGAGERGGFNKPGGPMDEGPDLDLGPPVDPDEDSDNSAIYV QGLNDSVTLDDLADFFKQCGVVKMNKRTGQPMIHIYLDKETGKPKGDATVSYEDPPTAKAAVEWFDGKDFQGSKLKVSLARKKPPMNSMR GGLPPREGRGMPPPLRGGPGGPGGPGGPMGRMGGRGGDRGGFPPRGPRGSRGNPSGGGNVQHRAGDWQCPNPGCGNQNFAWRTECNQCKA PKPEGFLPPPFPPPGGDRGRGGPGGMRGGRGGLMDRGGPGGMFRGGRGGDRGGFRGGRGMDRGGFGGGRRGGPGGPPGPLMEQMGGRRGG -------------------------------------------------------------- >27340_27340_8_ERG-EWSR1_ERG_chr21_39762917_ENST00000398910_EWSR1_chr22_29682912_ENST00000331029_length(amino acids)=393AA_BP=293 MKDMIQTVPDPAAHIKEALSVVSEDQSLFECAYGTPHLAKTEMTASSSSDYGQTSKMSPRVPQQDWLSQPPARVTIKMECNPSQVNGSRN SPDECSVAKGGKMVGSPDTVGMNYGSYMEEKHMPPPNMTTNERRVIVPADPTLWSTDHVRQWLEWAVKEYGLPDVNILLFQNIDGKELCK MTKDDFQRLTPSYNADILLSHLHYLRETPLPHLTSDDVDKALQNSPRLMHARNTGGAAFIFPNTSVYPEATQRITTRPAAQPSPSTVPKT EDQRPQLDPYQILGPTSSRLANPAIPLHSRLVMIRAVTLSRTPMGNRAAMDSRVAMVNKAAMGSSLPLVTHPKLDPTAKLQVNIANRAAA -------------------------------------------------------------- >27340_27340_9_ERG-EWSR1_ERG_chr21_39762917_ENST00000398910_EWSR1_chr22_29682912_ENST00000332035_length(amino acids)=393AA_BP=293 MKDMIQTVPDPAAHIKEALSVVSEDQSLFECAYGTPHLAKTEMTASSSSDYGQTSKMSPRVPQQDWLSQPPARVTIKMECNPSQVNGSRN SPDECSVAKGGKMVGSPDTVGMNYGSYMEEKHMPPPNMTTNERRVIVPADPTLWSTDHVRQWLEWAVKEYGLPDVNILLFQNIDGKELCK MTKDDFQRLTPSYNADILLSHLHYLRETPLPHLTSDDVDKALQNSPRLMHARNTGGAAFIFPNTSVYPEATQRITTRPAAQPSPSTVPKT EDQRPQLDPYQILGPTSSRLANPAIPLHSRLVMIRAVTLSRTPMGNRAAMDSRVAMVNKAAMGSSLPLVTHPKLDPTAKLQVNIANRAAA -------------------------------------------------------------- >27340_27340_10_ERG-EWSR1_ERG_chr21_39762917_ENST00000398910_EWSR1_chr22_29682912_ENST00000332050_length(amino acids)=374AA_BP=293 MKDMIQTVPDPAAHIKEALSVVSEDQSLFECAYGTPHLAKTEMTASSSSDYGQTSKMSPRVPQQDWLSQPPARVTIKMECNPSQVNGSRN SPDECSVAKGGKMVGSPDTVGMNYGSYMEEKHMPPPNMTTNERRVIVPADPTLWSTDHVRQWLEWAVKEYGLPDVNILLFQNIDGKELCK MTKDDFQRLTPSYNADILLSHLHYLRETPLPHLTSDDVDKALQNSPRLMHARNTGGAAFIFPNTSVYPEATQRITTRPAAQPSPSTVPKT EDQRPQLDPYQILGPTSSRLANPAIPLHSRLVMIRAVTLSRTPMGNRAAMDSRVAMVNKAAMGSSLPLVTHPKLDPTAKLQVNIANRAAA -------------------------------------------------------------- >27340_27340_11_ERG-EWSR1_ERG_chr21_39762917_ENST00000398910_EWSR1_chr22_29682912_ENST00000333395_length(amino acids)=393AA_BP=293 MKDMIQTVPDPAAHIKEALSVVSEDQSLFECAYGTPHLAKTEMTASSSSDYGQTSKMSPRVPQQDWLSQPPARVTIKMECNPSQVNGSRN SPDECSVAKGGKMVGSPDTVGMNYGSYMEEKHMPPPNMTTNERRVIVPADPTLWSTDHVRQWLEWAVKEYGLPDVNILLFQNIDGKELCK MTKDDFQRLTPSYNADILLSHLHYLRETPLPHLTSDDVDKALQNSPRLMHARNTGGAAFIFPNTSVYPEATQRITTRPAAQPSPSTVPKT EDQRPQLDPYQILGPTSSRLANPAIPLHSRLVMIRAVTLSRTPMGNRAAMDSRVAMVNKAAMGSSLPLVTHPKLDPTAKLQVNIANRAAA -------------------------------------------------------------- >27340_27340_12_ERG-EWSR1_ERG_chr21_39762917_ENST00000398910_EWSR1_chr22_29682912_ENST00000397938_length(amino acids)=393AA_BP=293 MKDMIQTVPDPAAHIKEALSVVSEDQSLFECAYGTPHLAKTEMTASSSSDYGQTSKMSPRVPQQDWLSQPPARVTIKMECNPSQVNGSRN SPDECSVAKGGKMVGSPDTVGMNYGSYMEEKHMPPPNMTTNERRVIVPADPTLWSTDHVRQWLEWAVKEYGLPDVNILLFQNIDGKELCK MTKDDFQRLTPSYNADILLSHLHYLRETPLPHLTSDDVDKALQNSPRLMHARNTGGAAFIFPNTSVYPEATQRITTRPAAQPSPSTVPKT EDQRPQLDPYQILGPTSSRLANPAIPLHSRLVMIRAVTLSRTPMGNRAAMDSRVAMVNKAAMGSSLPLVTHPKLDPTAKLQVNIANRAAA -------------------------------------------------------------- >27340_27340_13_ERG-EWSR1_ERG_chr21_39762917_ENST00000398910_EWSR1_chr22_29682912_ENST00000406548_length(amino acids)=393AA_BP=293 MKDMIQTVPDPAAHIKEALSVVSEDQSLFECAYGTPHLAKTEMTASSSSDYGQTSKMSPRVPQQDWLSQPPARVTIKMECNPSQVNGSRN SPDECSVAKGGKMVGSPDTVGMNYGSYMEEKHMPPPNMTTNERRVIVPADPTLWSTDHVRQWLEWAVKEYGLPDVNILLFQNIDGKELCK MTKDDFQRLTPSYNADILLSHLHYLRETPLPHLTSDDVDKALQNSPRLMHARNTGGAAFIFPNTSVYPEATQRITTRPAAQPSPSTVPKT EDQRPQLDPYQILGPTSSRLANPAIPLHSRLVMIRAVTLSRTPMGNRAAMDSRVAMVNKAAMGSSLPLVTHPKLDPTAKLQVNIANRAAA -------------------------------------------------------------- >27340_27340_14_ERG-EWSR1_ERG_chr21_39762917_ENST00000398910_EWSR1_chr22_29682912_ENST00000414183_length(amino acids)=393AA_BP=293 MKDMIQTVPDPAAHIKEALSVVSEDQSLFECAYGTPHLAKTEMTASSSSDYGQTSKMSPRVPQQDWLSQPPARVTIKMECNPSQVNGSRN SPDECSVAKGGKMVGSPDTVGMNYGSYMEEKHMPPPNMTTNERRVIVPADPTLWSTDHVRQWLEWAVKEYGLPDVNILLFQNIDGKELCK MTKDDFQRLTPSYNADILLSHLHYLRETPLPHLTSDDVDKALQNSPRLMHARNTGGAAFIFPNTSVYPEATQRITTRPAAQPSPSTVPKT EDQRPQLDPYQILGPTSSRLANPAIPLHSRLVMIRAVTLSRTPMGNRAAMDSRVAMVNKAAMGSSLPLVTHPKLDPTAKLQVNIANRAAA -------------------------------------------------------------- >27340_27340_15_ERG-EWSR1_ERG_chr21_39762917_ENST00000417133_EWSR1_chr22_29682912_ENST00000331029_length(amino acids)=416AA_BP=316 MKDMIQTVPDPAAHIKEALSVVSEDQSLFECAYGTPHLAKTEMTASSSSDYGQTSKMSPRVPQQDWLSQPPARVTIKMECNPSQVNGSRN SPDECSVAKGGKMVGSPDTVGMNYGSYMEEKHMPPPNMTTNERRVIVPADPTLWSTDHVRQWLEWAVKEYGLPDVNILLFQNIDGKELCK MTKDDFQRLTPSYNADILLSHLHYLRETPLPHLTSDDVDKALQNSPRLMHARNTGGAAFIFPNTSVYPEATQRITTRPDLPYEPPRRSAW TGHGHPTPQSKAAQPSPSTVPKTEDQRPQLDPYQILGPTSSRLANPAIPLHSRLVMIRAVTLSRTPMGNRAAMDSRVAMVNKAAMGSSLP -------------------------------------------------------------- >27340_27340_16_ERG-EWSR1_ERG_chr21_39762917_ENST00000417133_EWSR1_chr22_29682912_ENST00000332035_length(amino acids)=416AA_BP=316 MKDMIQTVPDPAAHIKEALSVVSEDQSLFECAYGTPHLAKTEMTASSSSDYGQTSKMSPRVPQQDWLSQPPARVTIKMECNPSQVNGSRN SPDECSVAKGGKMVGSPDTVGMNYGSYMEEKHMPPPNMTTNERRVIVPADPTLWSTDHVRQWLEWAVKEYGLPDVNILLFQNIDGKELCK MTKDDFQRLTPSYNADILLSHLHYLRETPLPHLTSDDVDKALQNSPRLMHARNTGGAAFIFPNTSVYPEATQRITTRPDLPYEPPRRSAW TGHGHPTPQSKAAQPSPSTVPKTEDQRPQLDPYQILGPTSSRLANPAIPLHSRLVMIRAVTLSRTPMGNRAAMDSRVAMVNKAAMGSSLP -------------------------------------------------------------- >27340_27340_17_ERG-EWSR1_ERG_chr21_39762917_ENST00000417133_EWSR1_chr22_29682912_ENST00000332050_length(amino acids)=397AA_BP=316 MKDMIQTVPDPAAHIKEALSVVSEDQSLFECAYGTPHLAKTEMTASSSSDYGQTSKMSPRVPQQDWLSQPPARVTIKMECNPSQVNGSRN SPDECSVAKGGKMVGSPDTVGMNYGSYMEEKHMPPPNMTTNERRVIVPADPTLWSTDHVRQWLEWAVKEYGLPDVNILLFQNIDGKELCK MTKDDFQRLTPSYNADILLSHLHYLRETPLPHLTSDDVDKALQNSPRLMHARNTGGAAFIFPNTSVYPEATQRITTRPDLPYEPPRRSAW TGHGHPTPQSKAAQPSPSTVPKTEDQRPQLDPYQILGPTSSRLANPAIPLHSRLVMIRAVTLSRTPMGNRAAMDSRVAMVNKAAMGSSLP -------------------------------------------------------------- >27340_27340_18_ERG-EWSR1_ERG_chr21_39762917_ENST00000417133_EWSR1_chr22_29682912_ENST00000333395_length(amino acids)=416AA_BP=316 MKDMIQTVPDPAAHIKEALSVVSEDQSLFECAYGTPHLAKTEMTASSSSDYGQTSKMSPRVPQQDWLSQPPARVTIKMECNPSQVNGSRN SPDECSVAKGGKMVGSPDTVGMNYGSYMEEKHMPPPNMTTNERRVIVPADPTLWSTDHVRQWLEWAVKEYGLPDVNILLFQNIDGKELCK MTKDDFQRLTPSYNADILLSHLHYLRETPLPHLTSDDVDKALQNSPRLMHARNTGGAAFIFPNTSVYPEATQRITTRPDLPYEPPRRSAW TGHGHPTPQSKAAQPSPSTVPKTEDQRPQLDPYQILGPTSSRLANPAIPLHSRLVMIRAVTLSRTPMGNRAAMDSRVAMVNKAAMGSSLP -------------------------------------------------------------- >27340_27340_19_ERG-EWSR1_ERG_chr21_39762917_ENST00000417133_EWSR1_chr22_29682912_ENST00000397938_length(amino acids)=416AA_BP=316 MKDMIQTVPDPAAHIKEALSVVSEDQSLFECAYGTPHLAKTEMTASSSSDYGQTSKMSPRVPQQDWLSQPPARVTIKMECNPSQVNGSRN SPDECSVAKGGKMVGSPDTVGMNYGSYMEEKHMPPPNMTTNERRVIVPADPTLWSTDHVRQWLEWAVKEYGLPDVNILLFQNIDGKELCK MTKDDFQRLTPSYNADILLSHLHYLRETPLPHLTSDDVDKALQNSPRLMHARNTGGAAFIFPNTSVYPEATQRITTRPDLPYEPPRRSAW TGHGHPTPQSKAAQPSPSTVPKTEDQRPQLDPYQILGPTSSRLANPAIPLHSRLVMIRAVTLSRTPMGNRAAMDSRVAMVNKAAMGSSLP -------------------------------------------------------------- >27340_27340_20_ERG-EWSR1_ERG_chr21_39762917_ENST00000417133_EWSR1_chr22_29682912_ENST00000406548_length(amino acids)=416AA_BP=316 MKDMIQTVPDPAAHIKEALSVVSEDQSLFECAYGTPHLAKTEMTASSSSDYGQTSKMSPRVPQQDWLSQPPARVTIKMECNPSQVNGSRN SPDECSVAKGGKMVGSPDTVGMNYGSYMEEKHMPPPNMTTNERRVIVPADPTLWSTDHVRQWLEWAVKEYGLPDVNILLFQNIDGKELCK MTKDDFQRLTPSYNADILLSHLHYLRETPLPHLTSDDVDKALQNSPRLMHARNTGGAAFIFPNTSVYPEATQRITTRPDLPYEPPRRSAW TGHGHPTPQSKAAQPSPSTVPKTEDQRPQLDPYQILGPTSSRLANPAIPLHSRLVMIRAVTLSRTPMGNRAAMDSRVAMVNKAAMGSSLP -------------------------------------------------------------- >27340_27340_21_ERG-EWSR1_ERG_chr21_39762917_ENST00000417133_EWSR1_chr22_29682912_ENST00000414183_length(amino acids)=416AA_BP=316 MKDMIQTVPDPAAHIKEALSVVSEDQSLFECAYGTPHLAKTEMTASSSSDYGQTSKMSPRVPQQDWLSQPPARVTIKMECNPSQVNGSRN SPDECSVAKGGKMVGSPDTVGMNYGSYMEEKHMPPPNMTTNERRVIVPADPTLWSTDHVRQWLEWAVKEYGLPDVNILLFQNIDGKELCK MTKDDFQRLTPSYNADILLSHLHYLRETPLPHLTSDDVDKALQNSPRLMHARNTGGAAFIFPNTSVYPEATQRITTRPDLPYEPPRRSAW TGHGHPTPQSKAAQPSPSTVPKTEDQRPQLDPYQILGPTSSRLANPAIPLHSRLVMIRAVTLSRTPMGNRAAMDSRVAMVNKAAMGSSLP -------------------------------------------------------------- >27340_27340_22_ERG-EWSR1_ERG_chr21_39762917_ENST00000442448_EWSR1_chr22_29682912_ENST00000331029_length(amino acids)=392AA_BP=292 MKDMIQTVPDPAAHIKEALSVVSEDQSLFECAYGTPHLAKTEMTASSSSDYGQTSKMSPRVPQQDWLSQPPARVTIKMECNPSQVNGSRN SPDECSVAKGGKMVGSPDTVGMNYGSYMEEKHMPPPNMTTNERRVIVPADPTLWSTDHVRQWLEWAVKEYGLPDVNILLFQNIDGKELCK MTKDDFQRLTPSYNADILLSHLHYLRETPLPHLTSDDVDKALQNSPRLMHARNTDLPYEPPRRSAWTGHGHPTPQSKAAQPSPSTVPKTE DQRPQLDPYQILGPTSSRLANPAIPLHSRLVMIRAVTLSRTPMGNRAAMDSRVAMVNKAAMGSSLPLVTHPKLDPTAKLQVNIANRAAAT -------------------------------------------------------------- >27340_27340_23_ERG-EWSR1_ERG_chr21_39762917_ENST00000442448_EWSR1_chr22_29682912_ENST00000332035_length(amino acids)=392AA_BP=292 MKDMIQTVPDPAAHIKEALSVVSEDQSLFECAYGTPHLAKTEMTASSSSDYGQTSKMSPRVPQQDWLSQPPARVTIKMECNPSQVNGSRN SPDECSVAKGGKMVGSPDTVGMNYGSYMEEKHMPPPNMTTNERRVIVPADPTLWSTDHVRQWLEWAVKEYGLPDVNILLFQNIDGKELCK MTKDDFQRLTPSYNADILLSHLHYLRETPLPHLTSDDVDKALQNSPRLMHARNTDLPYEPPRRSAWTGHGHPTPQSKAAQPSPSTVPKTE DQRPQLDPYQILGPTSSRLANPAIPLHSRLVMIRAVTLSRTPMGNRAAMDSRVAMVNKAAMGSSLPLVTHPKLDPTAKLQVNIANRAAAT -------------------------------------------------------------- >27340_27340_24_ERG-EWSR1_ERG_chr21_39762917_ENST00000442448_EWSR1_chr22_29682912_ENST00000332050_length(amino acids)=373AA_BP=292 MKDMIQTVPDPAAHIKEALSVVSEDQSLFECAYGTPHLAKTEMTASSSSDYGQTSKMSPRVPQQDWLSQPPARVTIKMECNPSQVNGSRN SPDECSVAKGGKMVGSPDTVGMNYGSYMEEKHMPPPNMTTNERRVIVPADPTLWSTDHVRQWLEWAVKEYGLPDVNILLFQNIDGKELCK MTKDDFQRLTPSYNADILLSHLHYLRETPLPHLTSDDVDKALQNSPRLMHARNTDLPYEPPRRSAWTGHGHPTPQSKAAQPSPSTVPKTE DQRPQLDPYQILGPTSSRLANPAIPLHSRLVMIRAVTLSRTPMGNRAAMDSRVAMVNKAAMGSSLPLVTHPKLDPTAKLQVNIANRAAAT -------------------------------------------------------------- >27340_27340_25_ERG-EWSR1_ERG_chr21_39762917_ENST00000442448_EWSR1_chr22_29682912_ENST00000333395_length(amino acids)=392AA_BP=292 MKDMIQTVPDPAAHIKEALSVVSEDQSLFECAYGTPHLAKTEMTASSSSDYGQTSKMSPRVPQQDWLSQPPARVTIKMECNPSQVNGSRN SPDECSVAKGGKMVGSPDTVGMNYGSYMEEKHMPPPNMTTNERRVIVPADPTLWSTDHVRQWLEWAVKEYGLPDVNILLFQNIDGKELCK MTKDDFQRLTPSYNADILLSHLHYLRETPLPHLTSDDVDKALQNSPRLMHARNTDLPYEPPRRSAWTGHGHPTPQSKAAQPSPSTVPKTE DQRPQLDPYQILGPTSSRLANPAIPLHSRLVMIRAVTLSRTPMGNRAAMDSRVAMVNKAAMGSSLPLVTHPKLDPTAKLQVNIANRAAAT -------------------------------------------------------------- >27340_27340_26_ERG-EWSR1_ERG_chr21_39762917_ENST00000442448_EWSR1_chr22_29682912_ENST00000397938_length(amino acids)=392AA_BP=292 MKDMIQTVPDPAAHIKEALSVVSEDQSLFECAYGTPHLAKTEMTASSSSDYGQTSKMSPRVPQQDWLSQPPARVTIKMECNPSQVNGSRN SPDECSVAKGGKMVGSPDTVGMNYGSYMEEKHMPPPNMTTNERRVIVPADPTLWSTDHVRQWLEWAVKEYGLPDVNILLFQNIDGKELCK MTKDDFQRLTPSYNADILLSHLHYLRETPLPHLTSDDVDKALQNSPRLMHARNTDLPYEPPRRSAWTGHGHPTPQSKAAQPSPSTVPKTE DQRPQLDPYQILGPTSSRLANPAIPLHSRLVMIRAVTLSRTPMGNRAAMDSRVAMVNKAAMGSSLPLVTHPKLDPTAKLQVNIANRAAAT -------------------------------------------------------------- >27340_27340_27_ERG-EWSR1_ERG_chr21_39762917_ENST00000442448_EWSR1_chr22_29682912_ENST00000406548_length(amino acids)=392AA_BP=292 MKDMIQTVPDPAAHIKEALSVVSEDQSLFECAYGTPHLAKTEMTASSSSDYGQTSKMSPRVPQQDWLSQPPARVTIKMECNPSQVNGSRN SPDECSVAKGGKMVGSPDTVGMNYGSYMEEKHMPPPNMTTNERRVIVPADPTLWSTDHVRQWLEWAVKEYGLPDVNILLFQNIDGKELCK MTKDDFQRLTPSYNADILLSHLHYLRETPLPHLTSDDVDKALQNSPRLMHARNTDLPYEPPRRSAWTGHGHPTPQSKAAQPSPSTVPKTE DQRPQLDPYQILGPTSSRLANPAIPLHSRLVMIRAVTLSRTPMGNRAAMDSRVAMVNKAAMGSSLPLVTHPKLDPTAKLQVNIANRAAAT -------------------------------------------------------------- >27340_27340_28_ERG-EWSR1_ERG_chr21_39762917_ENST00000442448_EWSR1_chr22_29682912_ENST00000414183_length(amino acids)=392AA_BP=292 MKDMIQTVPDPAAHIKEALSVVSEDQSLFECAYGTPHLAKTEMTASSSSDYGQTSKMSPRVPQQDWLSQPPARVTIKMECNPSQVNGSRN SPDECSVAKGGKMVGSPDTVGMNYGSYMEEKHMPPPNMTTNERRVIVPADPTLWSTDHVRQWLEWAVKEYGLPDVNILLFQNIDGKELCK MTKDDFQRLTPSYNADILLSHLHYLRETPLPHLTSDDVDKALQNSPRLMHARNTDLPYEPPRRSAWTGHGHPTPQSKAAQPSPSTVPKTE DQRPQLDPYQILGPTSSRLANPAIPLHSRLVMIRAVTLSRTPMGNRAAMDSRVAMVNKAAMGSSLPLVTHPKLDPTAKLQVNIANRAAAT -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr21:29683123/chr22:39764366) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ERG | EWSR1 |
| FUNCTION: Possible role in transport between endoplasmic reticulum and Golgi. {ECO:0000250}. | FUNCTION: Might normally function as a transcriptional repressor. EWS-fusion-proteins (EFPS) may play a role in the tumorigenic process. They may disturb gene expression by mimicking, or interfering with the normal function of CTD-POLII within the transcription initiation complex. They may also contribute to an aberrant activation of the fusion protein target genes. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
Top |
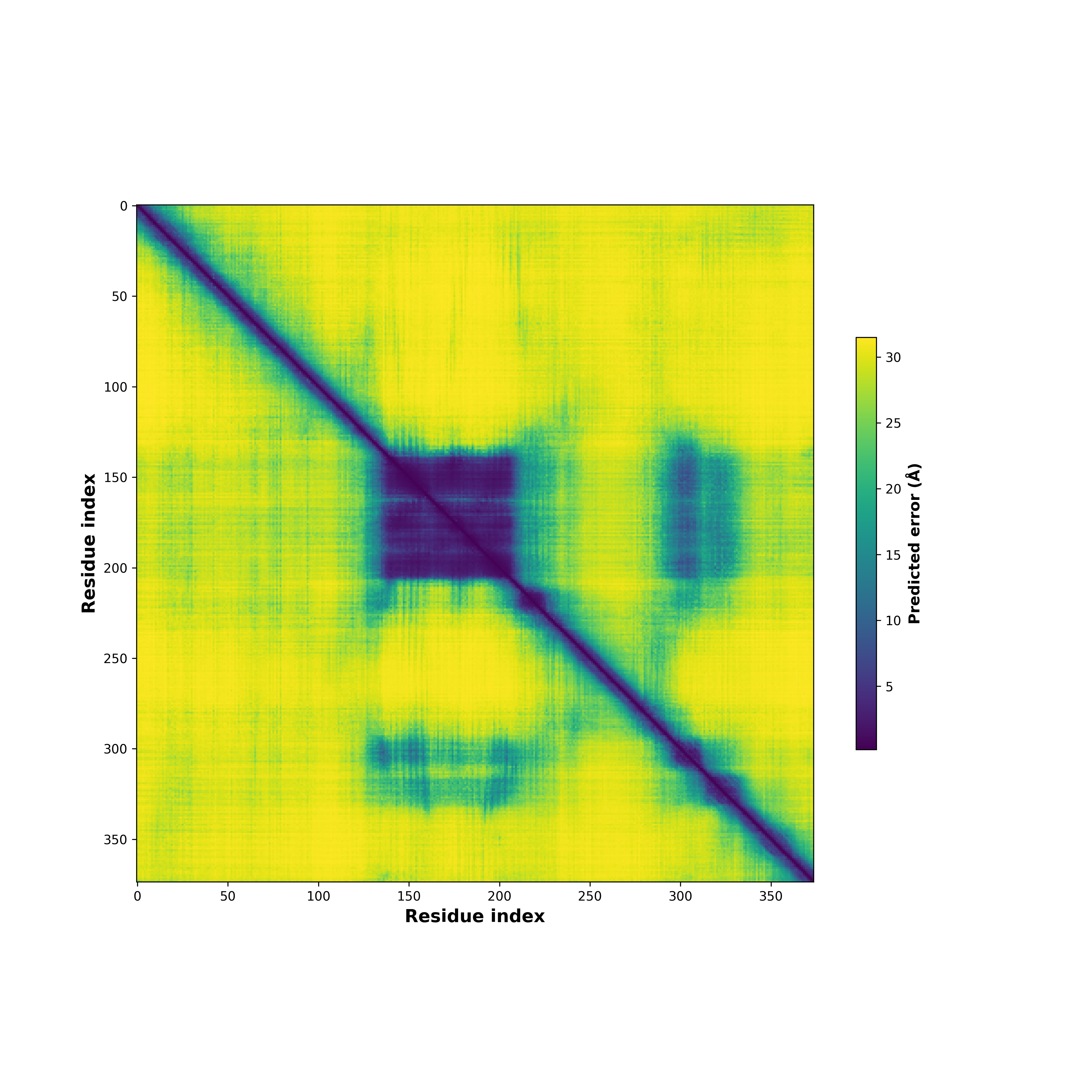
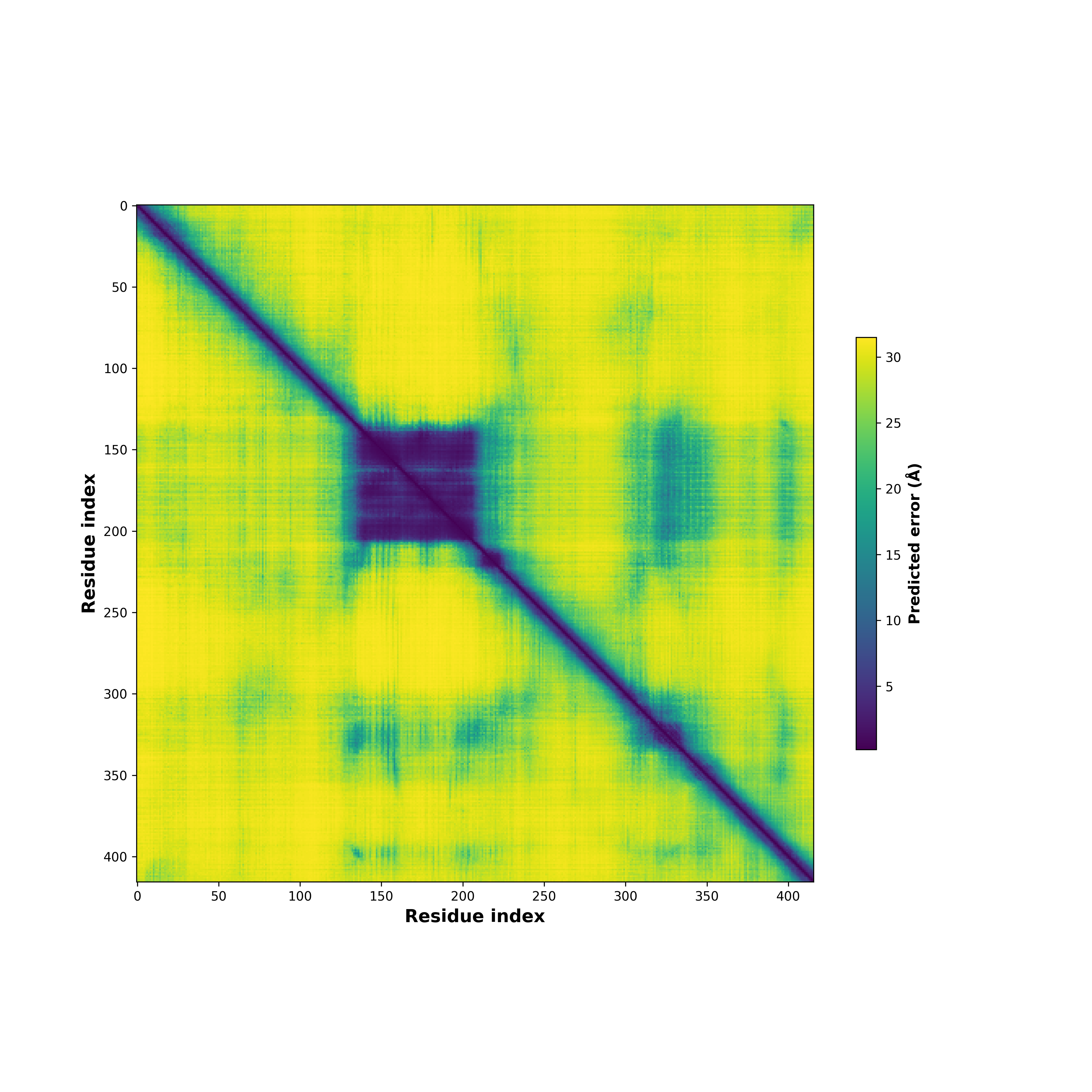
Fusion Protein Structures |
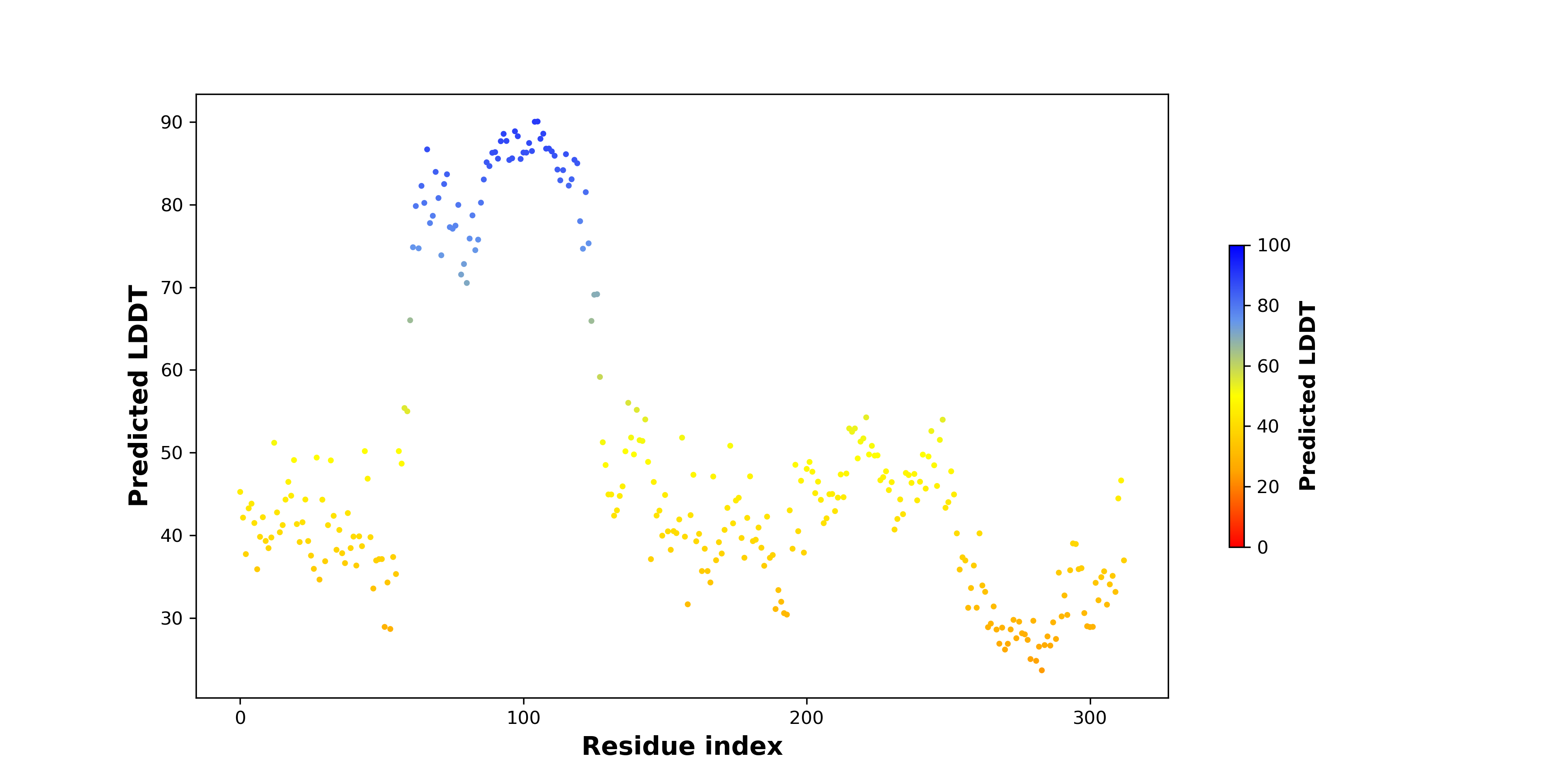
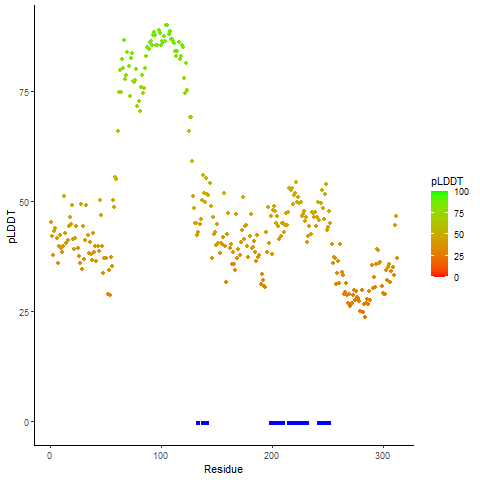
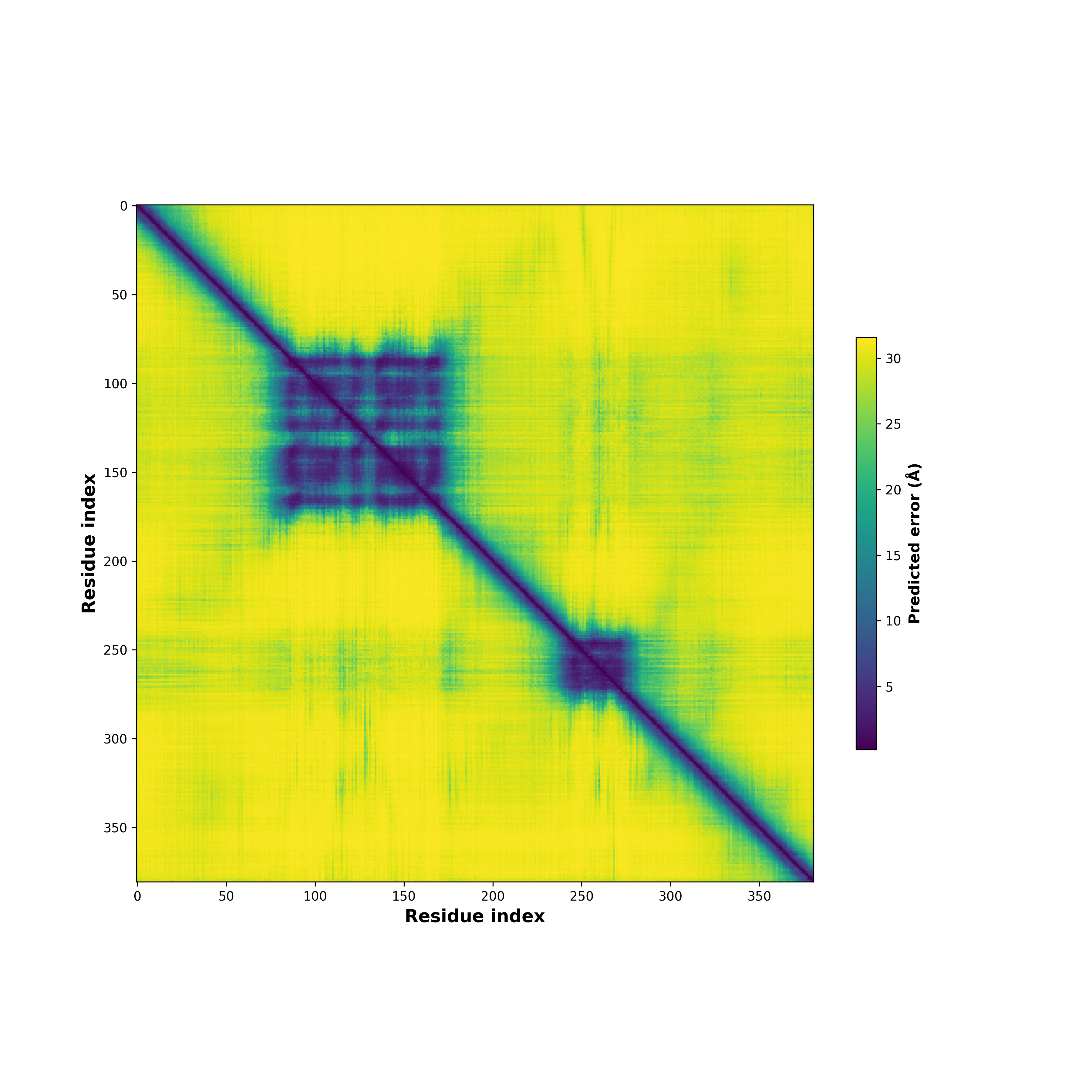
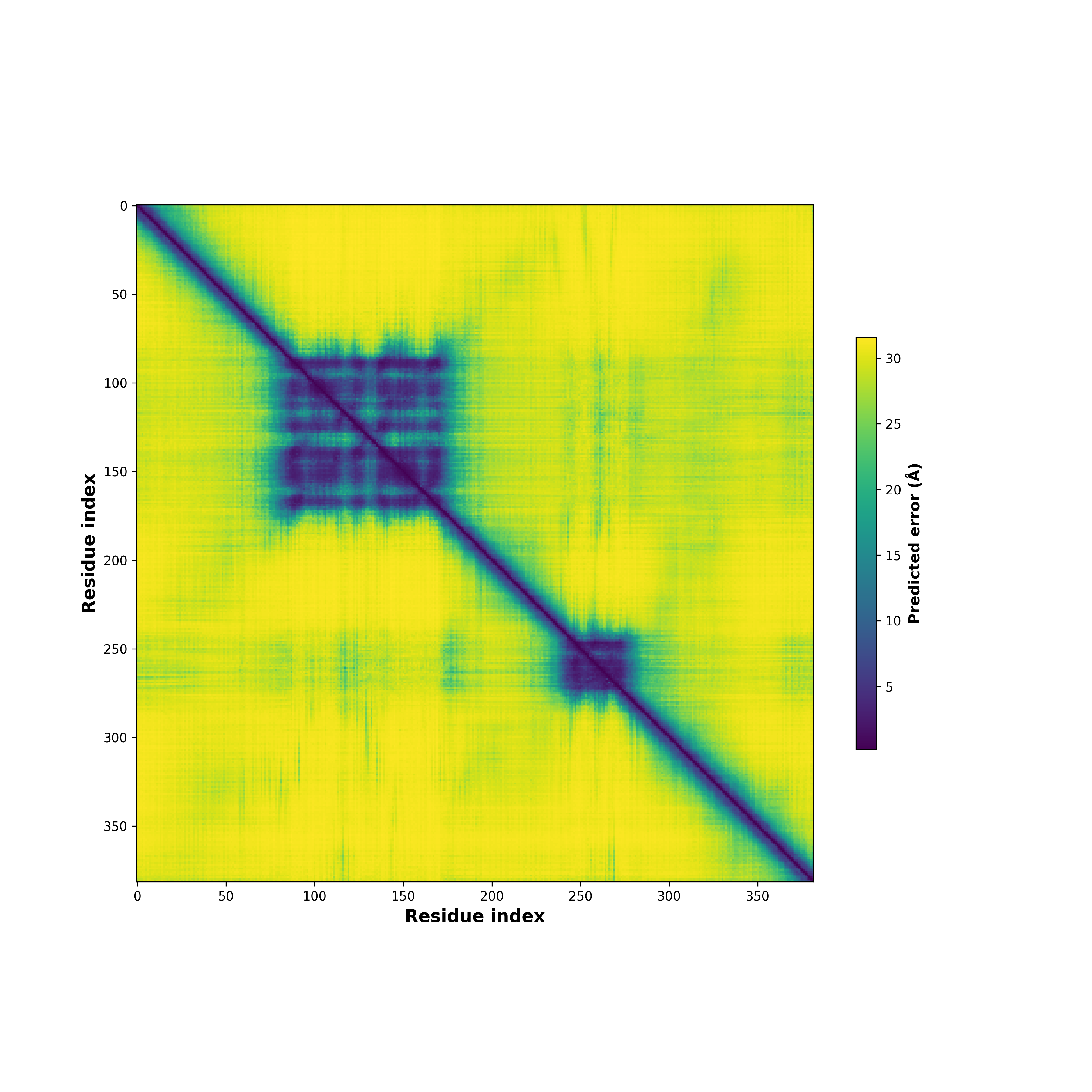
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
| PDB file (72) >>>72.pdbFusion protein BP residue: 213 CIF file (72) >>>72.cif | ERG | chr21 | 39762917 | - | EWSR1 | chr22 | 29682912 | + | MSRTQQLISRNSPDECSVAKGGKMVGSPDTVGMNYGSYMEEKHMPPPNMT TNERRVIVPADPTLWSTDHVRQWLEWAVKEYGLPDVNILLFQNIDGKELC KMTKDDFQRLTPSYNADILLSHLHYLRETPLPHLTSDDVDKALQNSPRLM HARNTDLPYEPPRRSAWTGHGHPTPQSKAAQPSPSTVPKTEDQRPQLDPY QILGPTSSRLANPAIPLHSRLVMIRAVTLSRTPMGNRAAMDSRVAMVNKA AMGSSLPLVTHPKLDPTAKLQVNIANRAAATGSRVHSDRTTPVAWVFMGR | 313 |
| 3D view using mol* of 72 (AA BP:213) | ||||||||||
 | ||||||||||
| PDB file (77) >>>77.pdbFusion protein BP residue: CIF file (77) >>>77.cif | ERG | chr21 | 39762917 | - | EWSR1 | chr22 | 29682912 | + | MDEGPDLDLGPPVDPDEDSDNSAIYVQGLNDSVTLDDLADFFKQCGVVKM NKRTGQPMIHIYLDKETGKPKGDATVSYEDPPTAKAAVEWFDGKDFQGSK LKVSLARKKPPMNSMRGGLPPREGRGMPPPLRGGPGGPGGPGGPMGRMGG RGGDRGGFPPRGPRGSRGNPSGGGNVQHRAGDWQCPNPGCGNQNFAWRTE CNQCKAPKPEGFLPPPFPPPGGDRGRGGPGGMRGGRGGLMDRGGPGGMFR GGRGGDRGGFRGGRGMDRGGFGGGRRGGPGGPPGPLMEQMGGRRGGRGGP | 317 |
| 3D view using mol* of 77 (AA BP:) | ||||||||||
 | ||||||||||
| PDB file (107) >>>107.pdbFusion protein BP residue: CIF file (107) >>>107.cif | ERG | chr21 | 39762917 | - | EWSR1 | chr22 | 29682912 | + | MGVYGQESGGFSGPGENRSMSGPDNRGRGRGGFDRGGMSRDPDEDSDNSA IYVQGLNDSVTLDDLADFFKQCGVVKMNKRTGQPMIHIYLDKETGKPKGD ATVSYEDPPTAKAAVEWFDGKDFQGSKLKVSLARKKPPMNSMRGGLPPRE GRGMPPPLRGGPGGPGGPGGPMGRMGGRGGDRGGFPPRGPRGSRGNPSGG GNVQHRAGDWQCPNPGCGNQNFAWRTECNQCKAPKPEGFLPPPFPPPGGD RGRGGPGGMRGGRGGLMDRGGPGGMFRGGRGGDRGGFRGGRGMDRGGFGG | 344 |
| 3D view using mol* of 107 (AA BP:) | ||||||||||
 | ||||||||||
| PDB file (130) >>>130.pdbFusion protein BP residue: 292 CIF file (130) >>>130.cif | ERG | chr21 | 39762917 | - | EWSR1 | chr22 | 29682912 | + | MKDMIQTVPDPAAHIKEALSVVSEDQSLFECAYGTPHLAKTEMTASSSSD YGQTSKMSPRVPQQDWLSQPPARVTIKMECNPSQVNGSRNSPDECSVAKG GKMVGSPDTVGMNYGSYMEEKHMPPPNMTTNERRVIVPADPTLWSTDHVR QWLEWAVKEYGLPDVNILLFQNIDGKELCKMTKDDFQRLTPSYNADILLS HLHYLRETPLPHLTSDDVDKALQNSPRLMHARNTDLPYEPPRRSAWTGHG HPTPQSKAAQPSPSTVPKTEDQRPQLDPYQILGPTSSRLANPAIPLHSRL VMIRAVTLSRTPMGNRAAMDSRVAMVNKAAMGSSLPLVTHPKLDPTAKLQ | 373 |
| 3D view using mol* of 130 (AA BP:292) | ||||||||||
 | ||||||||||
| PDB file (131) >>>131.pdbFusion protein BP residue: 293 CIF file (131) >>>131.cif | ERG | chr21 | 39762917 | - | EWSR1 | chr22 | 29682912 | + | MKDMIQTVPDPAAHIKEALSVVSEDQSLFECAYGTPHLAKTEMTASSSSD YGQTSKMSPRVPQQDWLSQPPARVTIKMECNPSQVNGSRNSPDECSVAKG GKMVGSPDTVGMNYGSYMEEKHMPPPNMTTNERRVIVPADPTLWSTDHVR QWLEWAVKEYGLPDVNILLFQNIDGKELCKMTKDDFQRLTPSYNADILLS HLHYLRETPLPHLTSDDVDKALQNSPRLMHARNTGGAAFIFPNTSVYPEA TQRITTRPAAQPSPSTVPKTEDQRPQLDPYQILGPTSSRLANPAIPLHSR LVMIRAVTLSRTPMGNRAAMDSRVAMVNKAAMGSSLPLVTHPKLDPTAKL | 374 |
| 3D view using mol* of 131 (AA BP:293) | ||||||||||
 | ||||||||||
| PDB file (139) >>>139.pdbFusion protein BP residue: CIF file (139) >>>139.cif | ERG | chr21 | 39762917 | - | EWSR1 | chr22 | 29682912 | + | MGVYGQESGGFSGPGENRSMSGPDNRGRGRGGFDRGGMSRGGRGGGRGGM GAGERGGFNKPGGPMDEGPDLDLGPPVDPDEDSDNSAIYVQGLNDSVTLD DLADFFKQCGVVKMNKRTGQPMIHIYLDKETGKPKGDATVSYEDPPTAKA AVEWFDGKDFQGSKLKVSLARKKPPMNSMRGGLPPREGRGMPPPLRGGPG GPGGPGGPMGRMGGRGGDRGGFPPRGPRGSRGNPSGGGNVQHRAGDWQCP NPGCGNQNFAWRTECNQCKAPKPEGFLPPPFPPPGGDRGRGGPGGMRGGR GGLMDRGGPGGMFRGGRGGDRGGFRGGRGMDRGGFGGGRRGGPGGPPGPL | 381 |
| 3D view using mol* of 139 (AA BP:) | ||||||||||
 | ||||||||||
| PDB file (140) >>>140.pdbFusion protein BP residue: CIF file (140) >>>140.cif | ERG | chr21 | 39762917 | - | EWSR1 | chr22 | 29682912 | + | MGVYGQESGGFSGPGENRSMSGPDNRGRGRGGFDRGGMSRGGRGGGRGGM GSAGERGGFNKPGGPMDEGPDLDLGPPVDPDEDSDNSAIYVQGLNDSVTL DDLADFFKQCGVVKMNKRTGQPMIHIYLDKETGKPKGDATVSYEDPPTAK AAVEWFDGKDFQGSKLKVSLARKKPPMNSMRGGLPPREGRGMPPPLRGGP GGPGGPGGPMGRMGGRGGDRGGFPPRGPRGSRGNPSGGGNVQHRAGDWQC PNPGCGNQNFAWRTECNQCKAPKPEGFLPPPFPPPGGDRGRGGPGGMRGG RGGLMDRGGPGGMFRGGRGGDRGGFRGGRGMDRGGFGGGRRGGPGGPPGP | 382 |
| 3D view using mol* of 140 (AA BP:) | ||||||||||
 | ||||||||||
| PDB file (146) | ERG | chr21 | 39762917 | - | EWSR1 | chr22 | 29682912 | + | MKDMIQTVPDPAAHIKEALSVVSEDQSLFECAYGTPHLAKTEMTASSSSD YGQTSKMSPRVPQQDWLSQPPARVTIKMECNPSQVNGSRNSPDECSVAKG GKMVGSPDTVGMNYGSYMEEKHMPPPNMTTNERRVIVPADPTLWSTDHVR QWLEWAVKEYGLPDVNILLFQNIDGKELCKMTKDDFQRLTPSYNADILLS HLHYLRETPLPHLTSDDVDKALQNSPRLMHARNTDLPYEPPRRSAWTGHG HPTPQSKAAQPSPSTVPKTEDQRPQLDPYQILGPTSSRLANPAIPLHSRL VMIRAVTLSRTPMGNRAAMDSRVAMVNKAAMGSSLPLVTHPKLDPTAKLQ | 392 |
| PDB file (149) >>>149.pdbFusion protein BP residue: 293 CIF file (149) >>>149.cif | ERG | chr21 | 39762917 | - | EWSR1 | chr22 | 29682912 | + | MKDMIQTVPDPAAHIKEALSVVSEDQSLFECAYGTPHLAKTEMTASSSSD YGQTSKMSPRVPQQDWLSQPPARVTIKMECNPSQVNGSRNSPDECSVAKG GKMVGSPDTVGMNYGSYMEEKHMPPPNMTTNERRVIVPADPTLWSTDHVR QWLEWAVKEYGLPDVNILLFQNIDGKELCKMTKDDFQRLTPSYNADILLS HLHYLRETPLPHLTSDDVDKALQNSPRLMHARNTGGAAFIFPNTSVYPEA TQRITTRPAAQPSPSTVPKTEDQRPQLDPYQILGPTSSRLANPAIPLHSR LVMIRAVTLSRTPMGNRAAMDSRVAMVNKAAMGSSLPLVTHPKLDPTAKL | 393 |
| 3D view using mol* of 149 (AA BP:293) | ||||||||||
 | ||||||||||
| PDB file (155) >>>155.pdbFusion protein BP residue: 316 CIF file (155) >>>155.cif | ERG | chr21 | 39762917 | - | EWSR1 | chr22 | 29682912 | + | MKDMIQTVPDPAAHIKEALSVVSEDQSLFECAYGTPHLAKTEMTASSSSD YGQTSKMSPRVPQQDWLSQPPARVTIKMECNPSQVNGSRNSPDECSVAKG GKMVGSPDTVGMNYGSYMEEKHMPPPNMTTNERRVIVPADPTLWSTDHVR QWLEWAVKEYGLPDVNILLFQNIDGKELCKMTKDDFQRLTPSYNADILLS HLHYLRETPLPHLTSDDVDKALQNSPRLMHARNTGGAAFIFPNTSVYPEA TQRITTRPDLPYEPPRRSAWTGHGHPTPQSKAAQPSPSTVPKTEDQRPQL DPYQILGPTSSRLANPAIPLHSRLVMIRAVTLSRTPMGNRAAMDSRVAMV | 397 |
| 3D view using mol* of 155 (AA BP:316) | ||||||||||
 | ||||||||||
| PDB file (178) >>>178.pdbFusion protein BP residue: 316 CIF file (178) >>>178.cif | ERG | chr21 | 39762917 | - | EWSR1 | chr22 | 29682912 | + | MKDMIQTVPDPAAHIKEALSVVSEDQSLFECAYGTPHLAKTEMTASSSSD YGQTSKMSPRVPQQDWLSQPPARVTIKMECNPSQVNGSRNSPDECSVAKG GKMVGSPDTVGMNYGSYMEEKHMPPPNMTTNERRVIVPADPTLWSTDHVR QWLEWAVKEYGLPDVNILLFQNIDGKELCKMTKDDFQRLTPSYNADILLS HLHYLRETPLPHLTSDDVDKALQNSPRLMHARNTGGAAFIFPNTSVYPEA TQRITTRPDLPYEPPRRSAWTGHGHPTPQSKAAQPSPSTVPKTEDQRPQL DPYQILGPTSSRLANPAIPLHSRLVMIRAVTLSRTPMGNRAAMDSRVAMV NKAAMGSSLPLVTHPKLDPTAKLQVNIANRAAATGSRVHSDRTTPVAWVF | 416 |
| 3D view using mol* of 178 (AA BP:316) | ||||||||||
 | ||||||||||
Top |
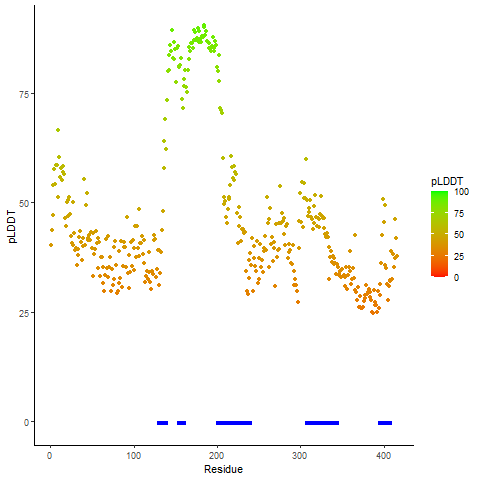
pLDDT score distribution |
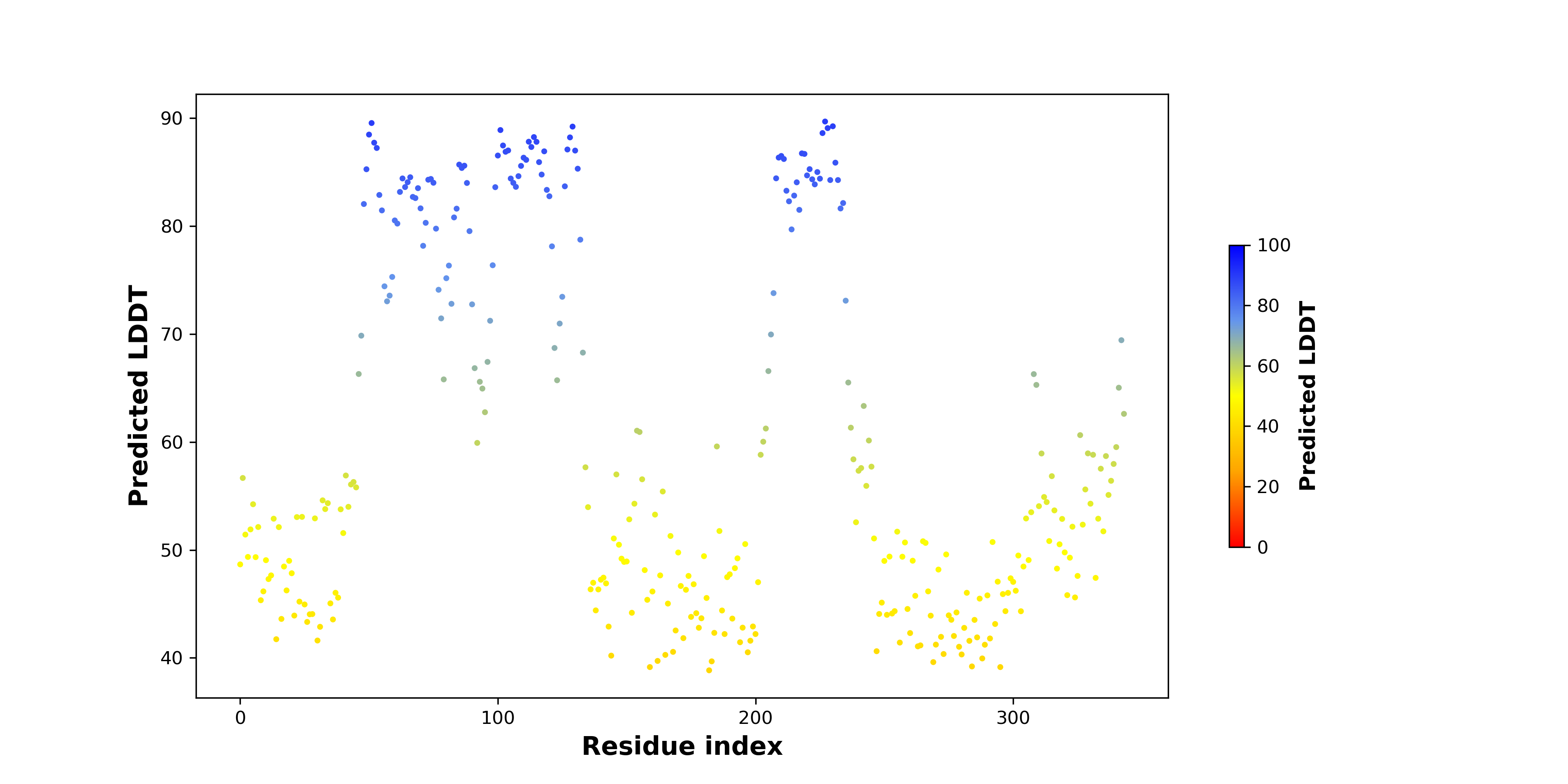
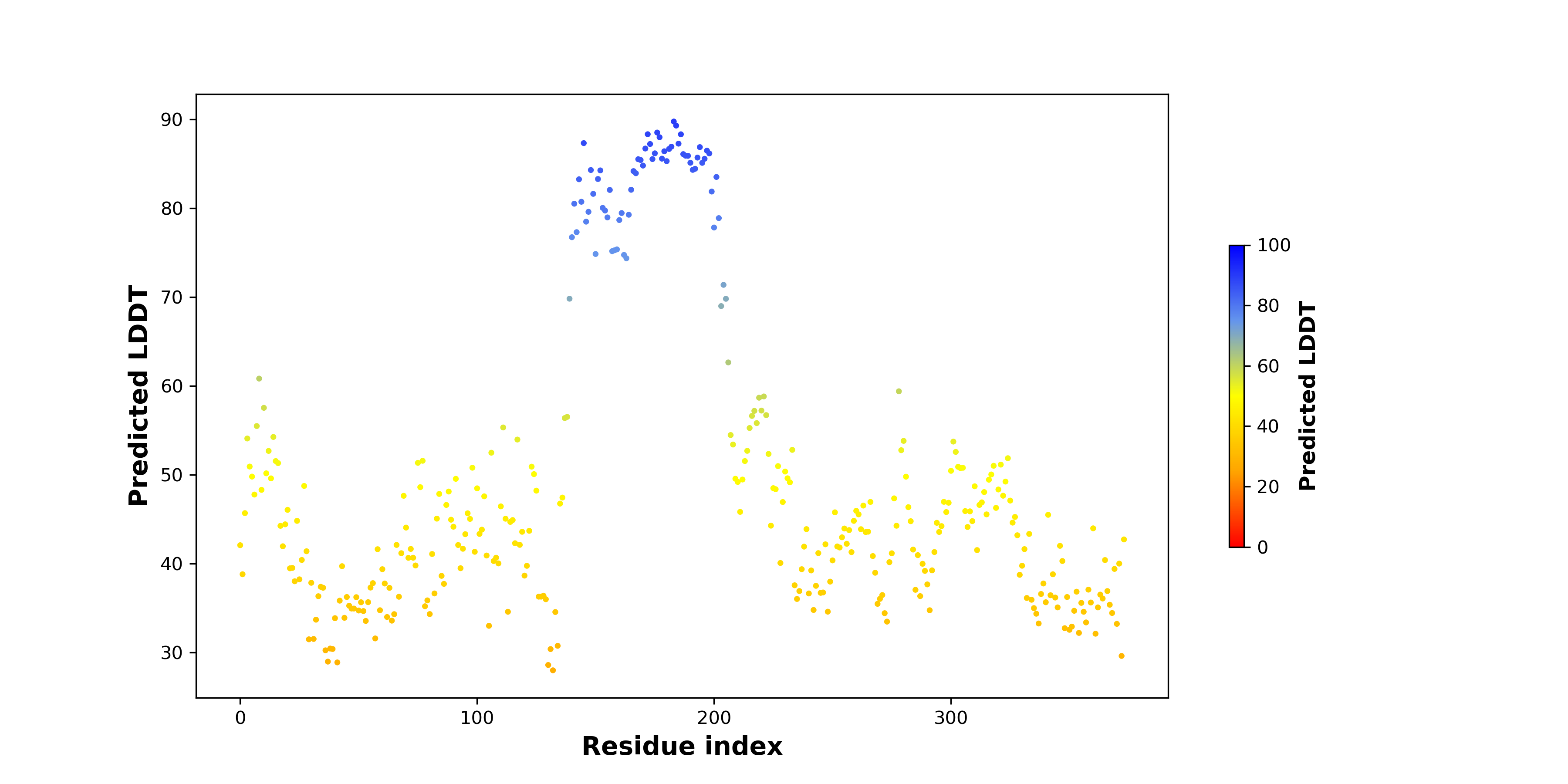
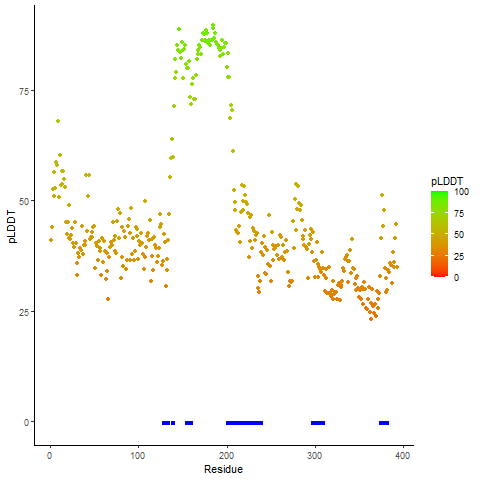
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
ERG_pLDDT.png |
EWSR1_pLDDT.png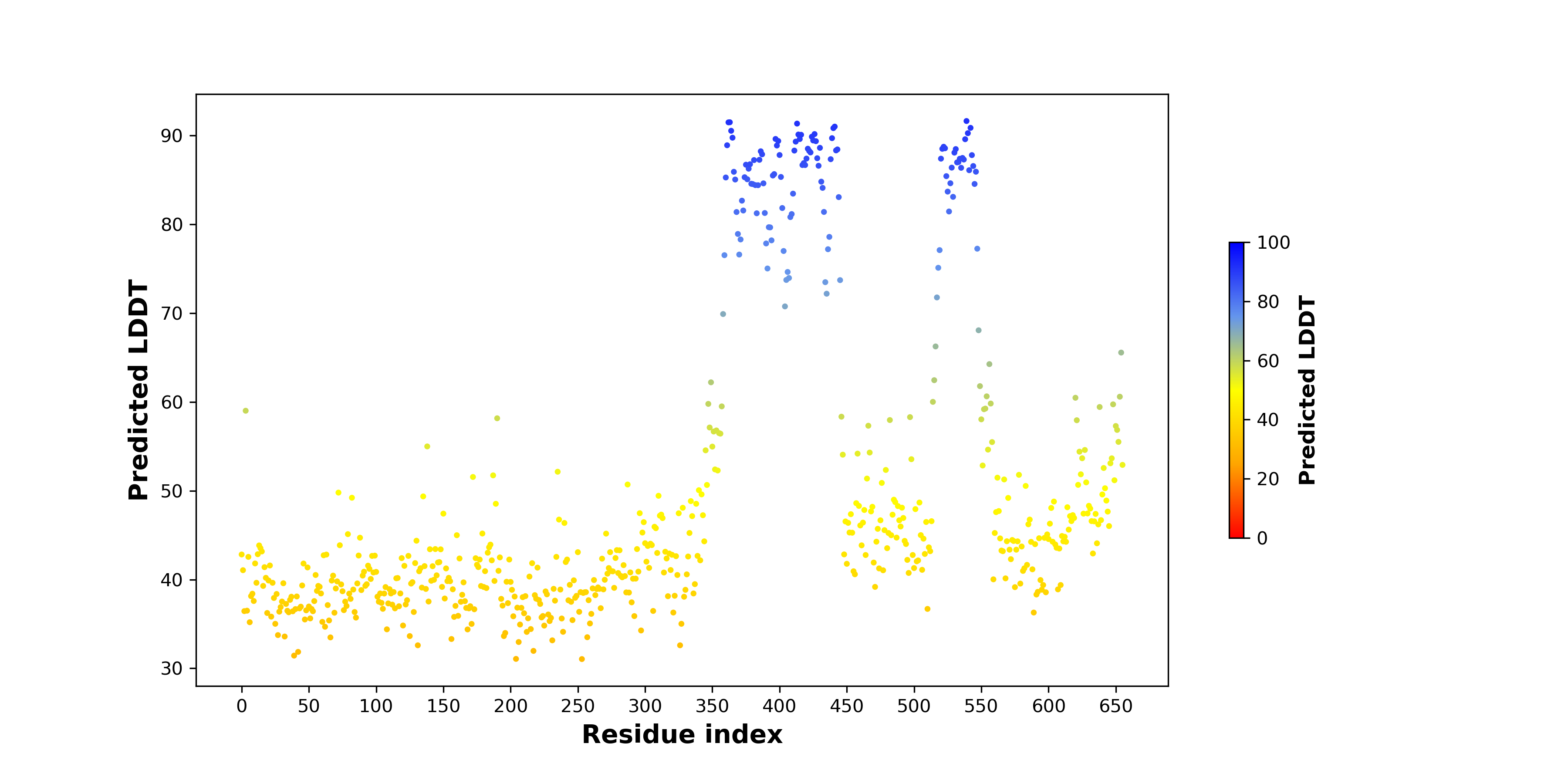 |
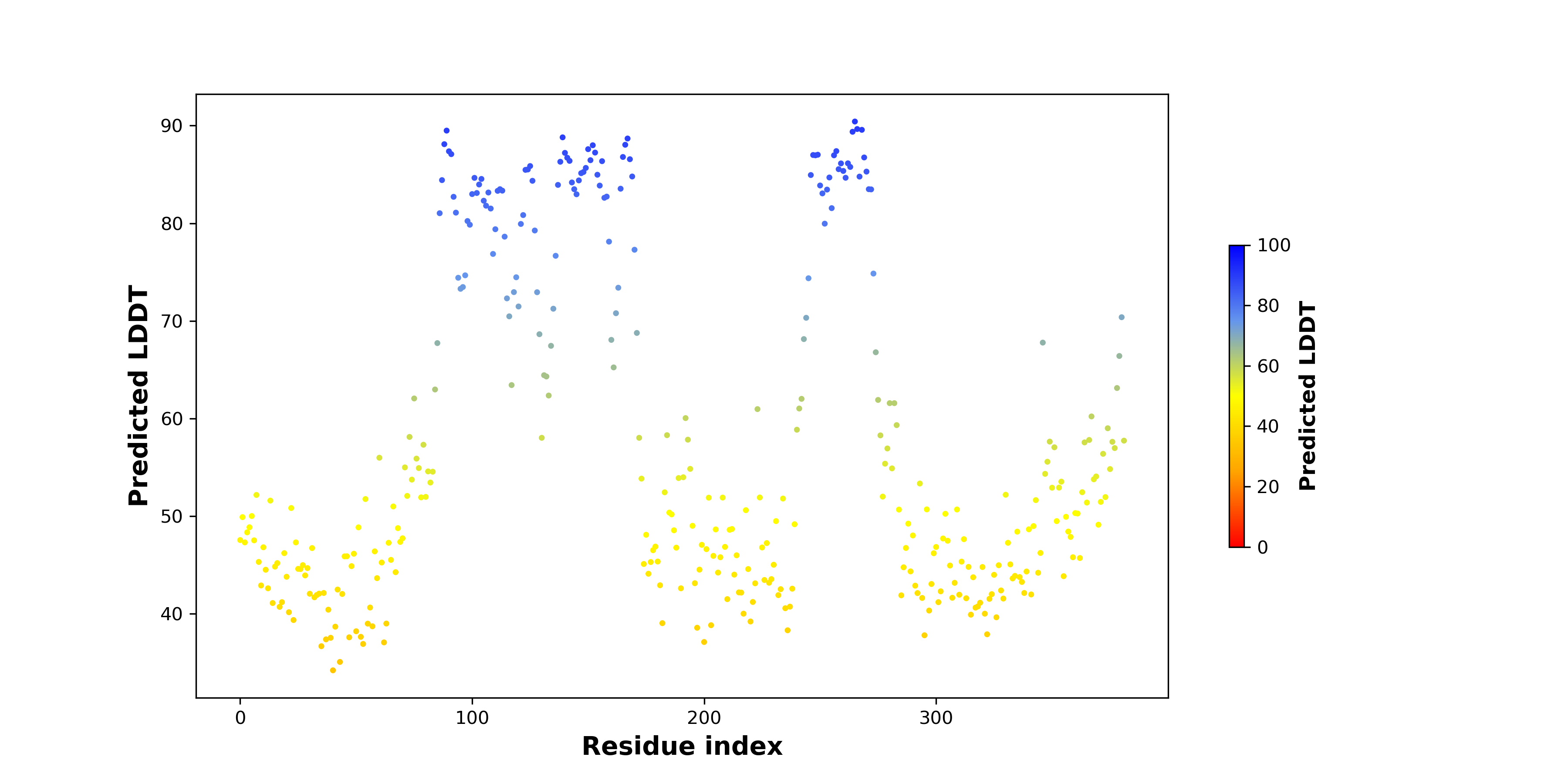
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
Top |
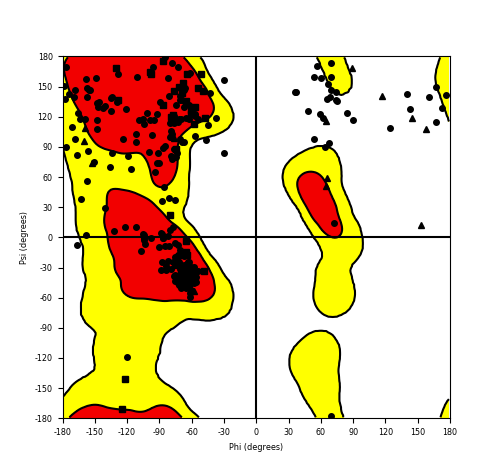
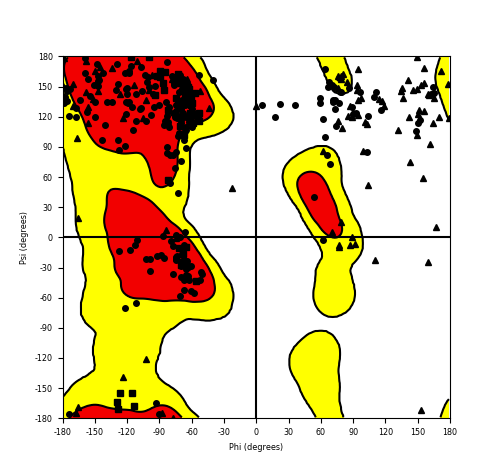
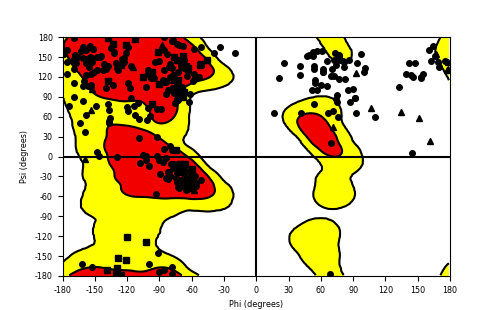
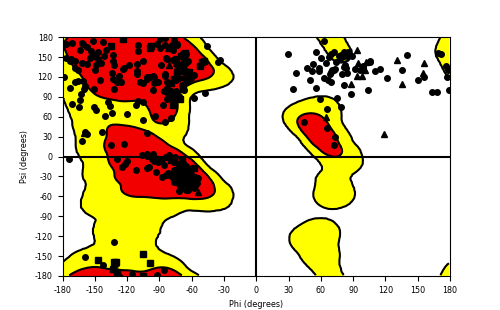
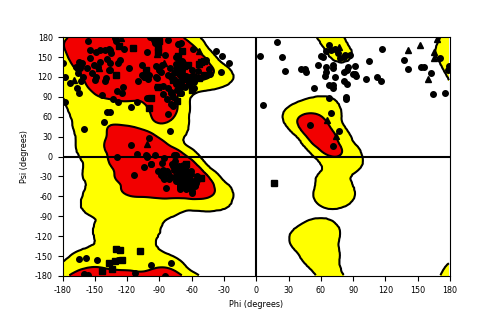
Ramachandran Plot of Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
Top |
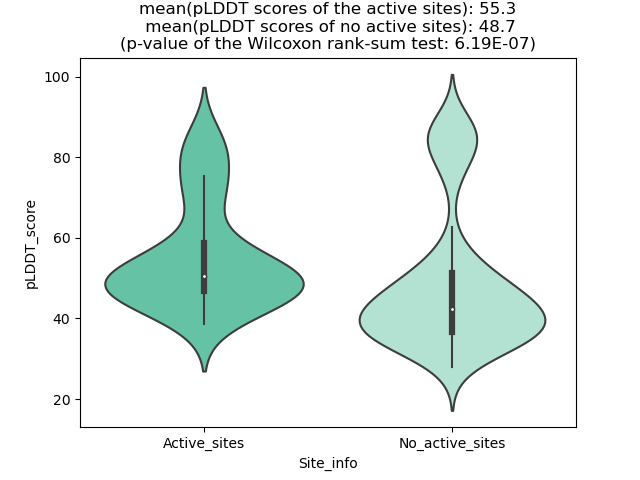
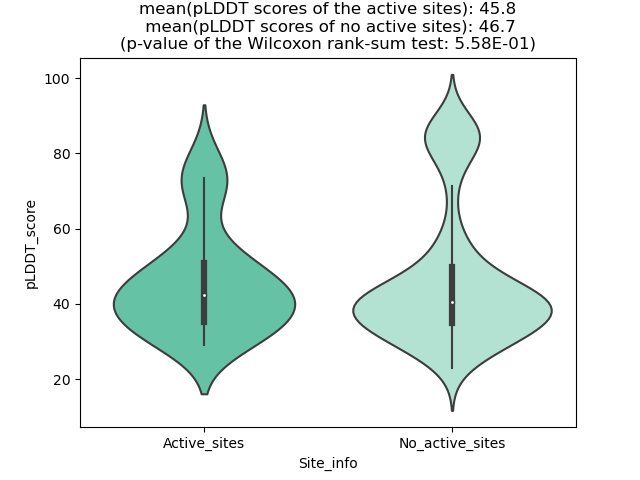
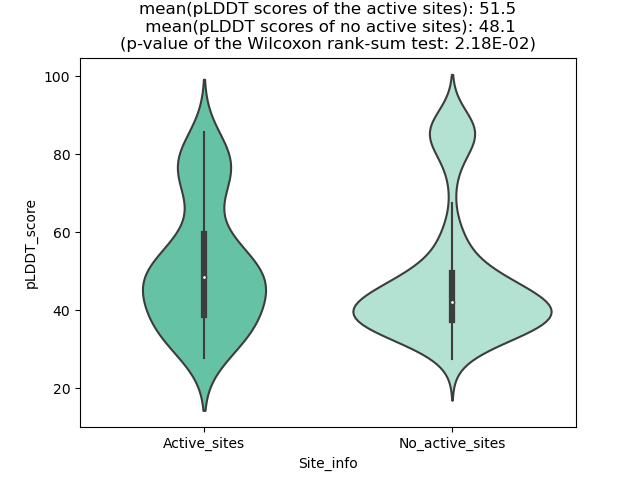
Potential Active Site Information |
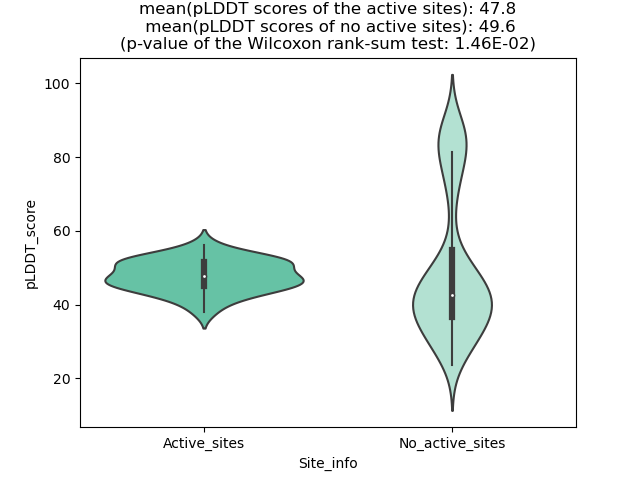
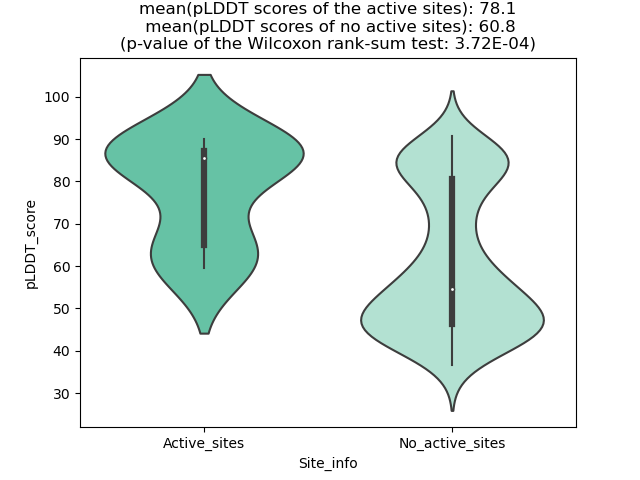
 The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. |
| Fusion AA seq ID in FusionPDB | Site score | Size | D score | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
| 72 | 1.064 | 131 | 1.144 | 461.335 | 0.612 | 0.669 | 0.816 | 1.209 | 0.605 | 1.999 | 0.949 | Chain A: 134,138,139,142,199,200,202,203,204,206,2 07,210,216,217,218,220,221,224,225,227,228,231,232 ,243,246,247,248,249,250,251,252 |
| 77 | 0.616 | 28 | 0.469 | 78.89 | 0.708 | 0.64 | 0.923 | 0.328 | 1.265 | 0.259 | 0.644 | Chain A: 176,177,178,179,182,184,192,193,194,195,2 02 |
| 107 | 0.632 | 34 | 0.512 | 107.702 | 0.707 | 0.605 | 0.829 | 0.035 | 1.225 | 0.029 | 1.113 | Chain A: 40,41,44,45,48,134,135 |
| 130 | 1.08 | 98 | 1.13 | 267.197 | 0.572 | 0.752 | 1.054 | 1.462 | 0.773 | 1.892 | 1.725 | Chain A: 160,197,200,201,203,204,207,208,304,307,3 08,309,311,312,313,314,315,316,318,319,320,322,323 ,325,326 |
| 131 | 1.055 | 323 | 1.102 | 825.258 | 0.546 | 0.717 | 0.918 | 1.047 | 0.824 | 1.271 | 1.055 | Chain A: 139,140,141,142,152,160,196,197,200,201,2 04,205,206,208,209,210,211,213,217,220,221,222,224 ,225,226,227,228,298,301,302,303,304,305,306,307,3 08,309,310,311,312,314,317,320,321,323,324,326,327 ,328,330,331,332,334 |
| 139 | 0.654 | 39 | 0.604 | 87.465 | 0.678 | 0.59 | 0.86 | 0.434 | 1.033 | 0.421 | 0.598 | Chain A: 240,241,242,243,246,247,248,256,257,258,2 59,266,267 |
| 146 | 1.043 | 295 | 1.088 | 693.546 | 0.489 | 0.706 | 0.952 | 0.922 | 0.847 | 1.089 | 0.722 | Chain A: 132,133,134,135,136,139,151,154,155,158,1 59,201,204,205,208,210,213,217,218,221,222,297,298 ,301,302,304,305,306,308,309,310,311,374,375,376,3 77,378,379,380,381,382 |
| 149 | 1.011 | 227 | 1.035 | 772.093 | 0.507 | 0.703 | 0.935 | 0.632 | 1.004 | 0.629 | 0.647 | Chain A: 129,130,131,132,133,134,139,155,158,159,1 60,201,204,205,208,210,213,217,218,221,222,225,227 ,228,230,231,233,234,236,237,238,239,298,301,302,3 05,306,309,375,376,377,378,379,380,381,382 |
| 155 | 1.03 | 408 | 1.091 | 1366.512 | 0.626 | 0.659 | 0.837 | 0.943 | 0.748 | 1.261 | 1.06 | Chain A: 132,133,134,135,136,137,138,139,140,141,1 43,144,148,151,152,154,155,158,159,160,197,200,201 ,203,204,205,207,208,209,210,211,212,213,215,218,2 19,221,222,225,227,303,306,307,308,309,310,311,312 ,313,314,318,319,320,321,322,323,324,325,326,327,3 28,329,330,331,332,333,334,335,337,338,339,340,342 ,343,346,347,350 |
| 178 | 1.033 | 492 | 1.069 | 1515.031 | 0.507 | 0.711 | 0.945 | 0.851 | 0.914 | 0.932 | 0.687 | Chain A: 130,132,133,134,135,139,154,155,158,159,1 60,201,204,205,210,211,213,215,217,218,220,221,222 ,224,225,227,230,231,234,235,236,237,238,239,240,3 08,309,312,313,315,318,319,321,322,325,326,328,329 ,330,331,332,333,335,336,337,340,341,344,395,397,3 98,399,400,401,402,403,404,405,406,407,408 |
Top |
Potentially Interacting Small Molecules through Virtual Screening |
 The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. |
| Fusion AA seq ID in FusionPDB | ZINC ID | DrugBank ID | Drug name | Docking score | Glide gscore |
Top |
 Drug information from DrugBank of the top 20 interacting small molecules. Drug information from DrugBank of the top 20 interacting small molecules. |
| ZINC ID | DrugBank ID | Drug name | Drug type | SMILES | Drug group |
Top |
Biochemical Features of Small Molecules |
 ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) |
| ZINC ID | mol_MW | dipole | SASA | FOSA | FISA | PISA | WPSA | volume | donorHB | accptHB | IP | Human Oral Absorption | Percent Human Oral Absorption | Rule Of Five | Rule Of Three |
Top |
Drug Toxicity Information |
 Toxicity information of individual drugs using eToxPred Toxicity information of individual drugs using eToxPred |
| ZINC ID | Smile | Surface Accessibility | Toxicity |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| ERG | JUN, ETS2, AR, HSPA4, HSPA8, NEDD4, PRKDC, XRCC5, XRCC6, PARP1, XRCC4, NME1, DDX3X, PCBP1, HNRNPA2B1, DDX5, TRIM21, TPM1, HNRNPM, EIF2S1, RPLP0, PRPF8, RPL7A, HNRNPDL, TUBB, CAD, ACTB, ATP5A1, RPS18, HNRNPU, SPTAN1, DNAJA1, DNAJA2, DNAJA4, APP, USP9X, ADAR, CDC5L, CLTC, DDX21, DDX23, DKC1, EEF1A2, ELAVL1, FBL, HNRNPA1, HNRNPAB, HNRNPC, HNRNPD, HNRNPF, HNRNPH1, HNRNPH3, HNRNPUL1, HNRNPUL2, HSPD1, ILF2, ILF3, LANCL1, MATR3, NCL, NOLC1, NONO, NPM1, NUMA1, POLR2A, PRPF40A, PUF60, RCC2, SF3B1, SF3B2, SF3B3, SFPQ, SNRNP200, SYN1, TOP1, TOP2B, XRN2, CUL3, SPOP, TRIM25, UBR5, TRIM33, RING1, UBR4, RNF2, TRIP12, HUWE1, STUB1, UHRF1, HERC2, BRD4, BRD2, BRD3, EP300, APEX1, PVT1, E7, TRIM8, MEF2D, SMARCE1, FOXP1, RERE, NFIB, FLI1, NFIA, BCL9, ARID1A, KMT2D, ZNF609, TRIM24, KDM6A, MITF, SS18L1, NCOR1, PIAS1, ATXN1L, HIVEP1, GATA4, SMARCA2, SMARCA4, ATXN1, MLLT10, LMO4, TLE1, TLE3, AES, SMARCB1, NCOA6, GSE1, SS18, NCOA2, QSER1, ARID4B, BCL7A, PRRC2B, NIPBL, TET2, ZNF746, ELMSAN1, SMARCD3, ATF7IP, BCOR, C15orf39, PAXIP1, ASXL2, C11orf30, LDB1, ZFHX4, FOXP4, RYBP, KMT2C, ARID1B, KDM2B, SMARCC2, MORC4, BCL7C, MAML1, DPF1, DPF3, DPF2, CREBBP, SMARCC1, SMARCD2, ISL2, SMARCD1, ZNF521, CIC, ARID3A, BCL7B, PYGO2, PCGF1, PAGR1, DNTTIP1, HIPK2, CPVL, ZNF703, FBRS, KIAA1551, FBRSL1, HMG20A, TCF7L2, GLTSCR1, CHD7, TCF20, TRPS1, ZNF608, PRR12, SATB2, ZNF148, ZNF281, RAD54L2, NCOR2, NCOA3, PSMB5, DBN1, NFIC, |
| EWSR1 | PRTFDC1, ZDHHC3, MSC, SLC22A24, MYO1F, KXD1, KHDRBS2, DNAJB3, FASN, NLE1, MNS1, PRUNE2, WWP2, NDUFB1, BNIP3L, NSUN4, KRR1, WWP1, RMND5B, SLC1A1, RASL11B, DFFA, WDR37, RPS15A, CPSF6, C11orf16, YY1AP1, RNF183, MTCP1, TULP2, RBPMS, KEL, MYOZ2, FAM131C, HMGA1, NPPB, HERPUD1, CD177, RPL31, VPS72, ACTL6A, RAD23A, MAGEA11, CFDP1, DMRTB1, CXADR, ZNF165, SSBP2, TPGS2, RAB37, CETP, NDUFV1, DYNLL2, NBPF3, CEACAM5, GPBP1L1, SERP2, GNPDA1, C19orf57, ELAVL3, ELAVL4, LILRA3, BAD, CCDC7, MRPS18B, CUEDC2, CNST, TSPAN3, CCDC91, TRIM37, NINL, NTNG2, CPSF7, PGLS, EPT1, MYL6, SMAD4, TMSB4Y, TRPV5, MVK, MAPK1IP1L, MDFI, MTMR9, PLSCR1, RALYL, PDHX, C10orf12, RHOXF2, MATK, SALL2, AGT, KCNMB1, SUV39H2, SMNDC1, ARHGDIA, PUF60, GSK3B, ILK, CD2BP2, BARD1, CREBBP, BTK, SF1, SNRPC, ELK1, PTK2B, CALM1, POU4F1, EZH2, IRF3, TONSL, RFX3, HLTF, SUPT4H1, ZNF184, HIST1H2BN, BLZF1, HDAC3, FXR2, HMGN4, POLR3A, ECD, ZBTB1, SCMH1, SUZ12, E2F8, TRIM5, ZNF383, DHX9, SMN1, PCM1, RAD21, NDRG1, CEBPA, ELAVL1, SIRT7, HNRNPA1, TSG101, TP53, POLR2A, YBX1, TDRD3, CUL3, CUL4A, CUL4B, CUL5, CUL2, CUL1, COPS5, COPS6, DCUN1D1, CAND1, NEDD8, KCND3, ATN1, ATXN3, ERCC5, HBP1, HSPA2, PCBP1, USP7, SRSF5, TMEM126A, SAP30BP, GORASP2, MBD3, MRPS9, HAX1, SFXN1, ITGA5, TCIRG1, RNF168, GEMIN5, HAS1, MTCH1, NDUFA5, MCAT, MRPL57, HDAC2, ESR1, FN1, VCAM1, ITGA4, CD81, PRMT1, SF3B4, EP300, PRRC2A, EWSR1, FUS, ITGB5, NONO, TRAF3, HNRNPUL1, EPAS1, CHERP, CDK12, ITCH, WBP4, rev, RPA3, RPA2, RPA1, HSPA5, RIOK2, TRAF1, TRAF2, SEC24D, TFG, SEC24A, SSBP3, PRR13, ATPAF2, PEF1, JUN, CUL7, OBSL1, CCDC8, RNF2, BMI1, EGFR, ABL1, SRPK1, ABCE1, PRMT8, RPS6KB2, ACAA2, ACAT1, EIF4H, ANXA2, HIP1R, PICALM, POR, NTRK1, SCARNA22, NPM1, KRT2, PRDX2, S100A9, YWHAZ, DDX17, PCLO, ANXA6, SEC24C, GIPC1, CSRP2, FHL1, HNRNPA3, CTTN, MARCKSL1, PARP1, SERPINB3, CCDC50, KIF2A, KRT6B, HIST1H1D, STK38, H2AFV, PACSIN2, U2AF1, RSL24D1, XRCC5, ESYT3, RPL29, SDF2L1, LRP1B, MSN, SEC23A, RPL7A, SNX18, PPIL1, RPS27A, IGHM, SUMO3, GTF2I, RUVBL1, KRT16, RBM8A, RPL8, SRSF9, ZC3HAV1L, GAPDH, ETV3, IGF2R, COL5A2, HNRNPD, ANP32B, WAC, TFAP2A, TTN, CBR3, ARGLU1, HNRNPAB, SRRT, ATP1B3, COPS7B, PRR12, ATF3, NOMO3, NOMO1, NOMO2, EYA3, C1orf198, MAZ, U2AF2, SSB, TRA2B, C1orf52, HMGB1, HMGB1P10, RPRD1A, HNRNPH3, EIF5A2, C1orf131, SEC13, MAPRE1, CSTF2T, SRSF3, LENG1, UPF1, HDGFRP2, RPS10P7, RPS10P11, RPS10, RPS10P13, RPS10P4, RPS10P22, DACH1, ANO1, NCOR1, MLX, SUMO1P3, SUMO1, WIZ, PFDN6, ARFIP2, ZHX3, EEF1B2, MBIP, BAG6, DENND2A, PRCC, SRSF1, EIF4ENIF1, SPTAN1, CDCA8, PLS3, API5P1, API5, PSMA4, DNTTIP1, AKAP8, NCOA3, SMTN, FBRS, SMARCE1, ERC1, WBP11, SPTBN1, NFRKB, OLA1, ZNF207, R3HDM1, TRIM33, SAFB, UBFD1, SRSF7, SRSF2, GATA6, VDAC1P1, VDAC1, IL16, GMNN, ILF2, MED4, QKI, VCL, MFAP1, SNAP29, PADI1, BCL9, BCL9L, PKM, GPATCH11, CASC3, PSMC6, CACYBP, RPL12P6, RPL12P32, RPL12P14, RPL12, RPL12P2, RPL12P35, RPL12P19, TPI1, TPI1P1, CHAF1A, MIA3, CIC, SDCBP, CA2, FKBP3, ACE, NKX2-5, CSTF2, PFDN2, UBTF, FAM207A, LOC729774, BRD8, C12orf45, C1orf35, TCF20, SOD1, SPAG7, MED8, ETS2, ALDOC, FKBP4, INCENP, CEP85, CECR2, TFE3, SUPT16HP1, SUPT16H, MAPT, HTATSF1, RPS18P12, RPS18P5, KPNA2, TMX1, CKAP4, HSPE1, COPRS, PTGES3, LAMP2, ERLIN2, CTNNBL1, TOMM22, NRBF2, C9orf78, NCOA6, MED26, RANBP1, LOC389842, LOC727803, HMGB3, CANX, PUS7, RPSAP19, RPSA, RPSAP18, RPSAP58, RPSAP15, RPSAP8, RPSAP9, RPSAP12, RPSAP29, RPSAP61, ARHGAP17, USF1, PSIP1, SNRPEP2, SNRPE, NUDT5, PPM1G, OTUB1, AHCY, COPS3, NSMCE2, SAE1, PROSER1, GRWD1, CREB5, TAF9B, RBM33, EDF1, PGK1, FAM114A2, SRRM1, RAD23B, CIAPIN1, CIAPIN1P, LRRC59, PABPN1, KMT2A, RPRD1B, GPATCH8, CCDC43, DGCR14, PPP1R2, ERICH1, EIF5A, EIF5AL1, BAG3, PCNA, SOX7, PNISR, FAM168A, MED15, SRSF11, SIRT1, RSF1, MAML1, HPRT1, SPDL1, CRTC3, CEP55, CDV3, ALYREF, RNF40, STOML2, DGCR8, NUCKS1, UBN2, PSMD7, WNT10A, HMBS, KHDRBS1, VBP1, NCSTN, CDCA2, SFSWAP, ZRANB2, DDB1, RBBP6, ZEB1, SRSF6, LOC644422, EIF2S1, RFX5, RPS19, RPS19P3, TALDO1, CWC15, CDCA5, LOC645086, C11orf58, TXN, STX12, PHRF1, BSG, TAF4, SH3GL1, LIN37, HRNR, FAM192A, RRBP1, KIAA0907, GOLGB1, PAX9, P4HB, CHMP5, LDHB, CALR, SUMO2P1, SUMO2, PDIA6, AHSA1, EN2, CCDC124, RPLP0P6, RPLP0P2, RPLP0P3, RPLP0, NIPBL, PDLIM4, PRKCSH, C15orf39, HNRNPDL, PDIA4, NUP210, RPLP2P3, RPLP2, PRDX4, DAZAP1, UBE2T, PHAX, AMOT, MARCKS, LOC284685, SMARCC1, BCORL1, RFC4, GLRX3, ANP32E, HYOU1, NPM3, ATF7IP, SARNP, TRA2A, HDGF, STIP1, PELP1, KCTD12, GLO1, PCF11, CLIC1, DNAJC8, RNF114, SLC4A1AP, FAM50A, GTF2A1, PRPF40A, CDC37, PPIAP22, PPIA, SMARCC2, MEGF11, KIAA1143, DENR, LAMP1, MYBL2, PITX1, UBE2MP1, UBE2M, CHTF8, OTX1, NACA, FNBP4, GTF2F2, GLTSCR1, GTF2E1, PQBP1, EMD, RNF113A, GPALPP1, SNRPA, RRP15, RPS25, RPS25P8, GMEB2, LNPEP, DNAJB1, IGBP1P1, IGBP1, HINT1, ARID1A, PPIB, ANXA11, MATR3, Sgol2, PPARGC1A, MCM2, Ksr1, UBASH3B, SFPQ, CAPN13, HEY1, BRCA1, MTCH2, PPIE, TBX3, BMP4, CTNNB1, GSK3A, HNF1B, TCF7L2, TRIP4, YAF2, ZNF217, AAR2, PIH1D1, EFTUD2, TNIP2, CHD3, CHD4, HEXIM1, MEPCE, LARP7, RUNX1, AGR2, RECQL4, REST, CDK9, SMARCA4, DDIT3, FLI1, TP53BP1, MDC1, METTL3, METTL14, KIAA1429, RC3H1, RC3H2, ATG16L1, PHB, DISC1, NR2C2, UBQLN2, ZFYVE21, XRCC6, AGRN, USP19, HIST1H4A, APEX1, DDX5, SNRNP70, SNRPB, SNRPD1, SNRPD2, SNRPD3, RNU1-1, RBMX, HNRNPM, HNRNPA2B1, TAF15, DDX3X, TARDBP, CLINT1, HNRNPL, NUMA1, ZFR, SNRNP200, ZNF326, HNRNPK, SF3B1, TOE1, HSPA8, SNRPB2, DDX20, GOT2, ILF3, PRPF6, ZNF638, HNRNPF, HNRNPH1, HNRNPR, VCP, CAD, CCAR2, DDX23, GEMIN4, HSPA1A, PCMTD1, POTEF, PRMT5, RBM45, SAFB2, SF3B2, SF3B3, SNRPF, SRSF4, THRAP3, TIA1, TTC7A, ZCCHC8, ITFG1, ARAF, BRD7, SOX2, ARIH2, PLEKHA4, NGB, OPTN, ZC3H18, CELF1, MKI67, INS, Apc2, FBP1, N, ZNF768, SYNCRIP, KDM5C, DDX58, OGT, SPOP, UFL1, DDRGK1, WDR5, TPX2, MALL, SOX21, POU3F3, PTP4A3, TRIM8, RCHY1, nsp14, SOX5, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| ERG | 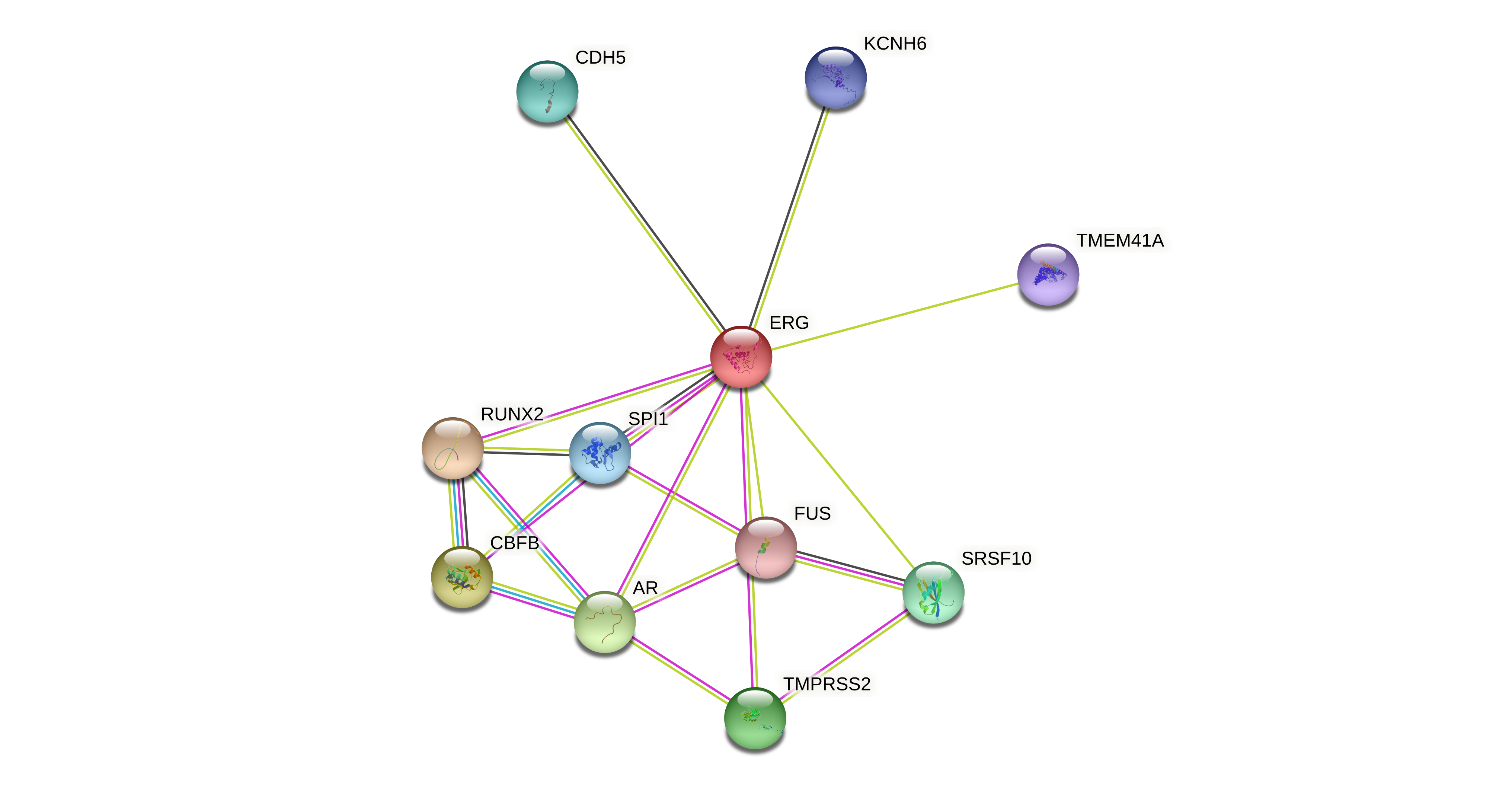 |
| EWSR1 | 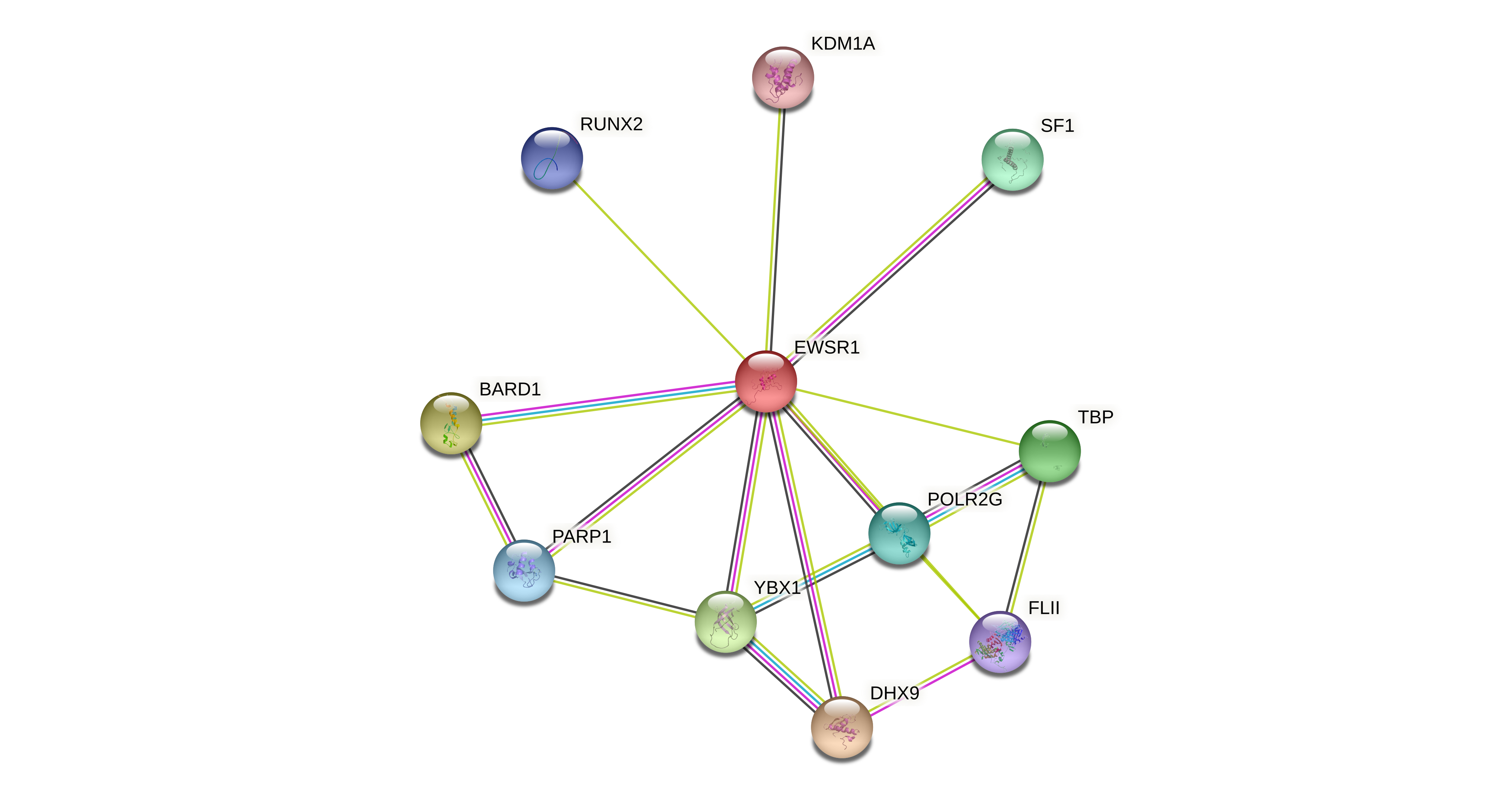 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to ERG-EWSR1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to ERG-EWSR1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
| ERG | EWSR1 | Ewing Sarcoma Undifferentiated Pleomorphic Sarcoma | MyCancerGenome |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | ERG | C0033578 | Prostatic Neoplasms | 7 | CTD_human |
| Hgene | ERG | C0376358 | Malignant neoplasm of prostate | 7 | CTD_human |
| Hgene | ERG | C0023418 | leukemia | 1 | CTD_human |
| Hgene | ERG | C0023452 | Childhood Acute Lymphoblastic Leukemia | 1 | CTD_human |
| Hgene | ERG | C0023453 | L2 Acute Lymphoblastic Leukemia | 1 | CTD_human |
| Hgene | ERG | C0023467 | Leukemia, Myelocytic, Acute | 1 | CTD_human |
| Hgene | ERG | C0026998 | Acute Myeloid Leukemia, M1 | 1 | CTD_human |
| Hgene | ERG | C0279980 | Extra-osseous Ewing's sarcoma | 1 | ORPHANET |
| Hgene | ERG | C1879321 | Acute Myeloid Leukemia (AML-M2) | 1 | CTD_human |
| Hgene | ERG | C1961102 | Precursor Cell Lymphoblastic Leukemia Lymphoma | 1 | CTD_human |
| Tgene | EWSR1 | C0553580 | Ewings sarcoma | 3 | CTD_human;ORPHANET |
| Tgene | EWSR1 | C0002736 | Amyotrophic Lateral Sclerosis | 1 | GENOMICS_ENGLAND |
| Tgene | EWSR1 | C0033578 | Prostatic Neoplasms | 1 | CTD_human |
| Tgene | EWSR1 | C0206651 | Clear Cell Sarcoma of Soft Tissue | 1 | ORPHANET |
| Tgene | EWSR1 | C0206663 | Neuroectodermal Tumor, Primitive | 1 | CTD_human |
| Tgene | EWSR1 | C0279980 | Extra-osseous Ewing's sarcoma | 1 | ORPHANET |
| Tgene | EWSR1 | C0334584 | Spongioblastoma | 1 | CTD_human |
| Tgene | EWSR1 | C0334596 | Medulloepithelioma | 1 | CTD_human |
| Tgene | EWSR1 | C0376358 | Malignant neoplasm of prostate | 1 | CTD_human |
| Tgene | EWSR1 | C0700367 | Ependymoblastoma | 1 | CTD_human |
| Tgene | EWSR1 | C0751675 | Cerebral Primitive Neuroectodermal Tumor | 1 | CTD_human |
| Tgene | EWSR1 | C1275278 | Extraskeletal Myxoid Chondrosarcoma | 1 | ORPHANET |