| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:ETV6-ABL1 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: ETV6-ABL1 | FusionPDB ID: 27697 | FusionGDB2.0 ID: 27697 | Hgene | Tgene | Gene symbol | ETV6 | ABL1 | Gene ID | 2120 | 25 |
| Gene name | ETS variant transcription factor 6 | ABL proto-oncogene 1, non-receptor tyrosine kinase | |
| Synonyms | TEL|TEL/ABL|THC5 | ABL|BCR-ABL|CHDSKM|JTK7|bcr/abl|c-ABL|c-ABL1|p150|v-abl | |
| Cytomap | 12p13.2 | 9q34.12 | |
| Type of gene | protein-coding | protein-coding | |
| Description | transcription factor ETV6ETS translocation variant 6ETS variant 6ETS-related protein Tel1TEL1 oncogeneets variant gene 6 (TEL oncogene) | tyrosine-protein kinase ABL1ABL protooncogene 1 nonreceptor tyrosine kinaseAbelson tyrosine-protein kinase 1bcr/c-abl oncogene proteinc-abl oncogene 1, receptor tyrosine kinaseproto-oncogene c-Ablproto-oncogene tyrosine-protein kinase ABL1truncated | |
| Modification date | 20200313 | 20200327 | |
| UniProtAcc | P41212 | P00519 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000396373, ENST00000544715, | ENST00000318560, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 59 X 37 X 25=54575 | 21 X 149 X 8=25032 |
| # samples | 58 | 154 | |
| ** MAII score | log2(58/54575*10)=-6.55604351475058 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(154/25032*10)=-4.02277130765866 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: ETV6 [Title/Abstract] AND ABL1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | ETV6(12006495)-ABL1(133729449), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | ETV6-ABL1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ETV6-ABL1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ETV6-ABL1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ETV6-ABL1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ETV6 | GO:0000122 | negative regulation of transcription by RNA polymerase II | 10514502 |
| Tgene | ABL1 | GO:0006974 | cellular response to DNA damage stimulus | 15657060 |
| Tgene | ABL1 | GO:0006975 | DNA damage induced protein phosphorylation | 18280240 |
| Tgene | ABL1 | GO:0018108 | peptidyl-tyrosine phosphorylation | 7590236|9144171|10713049|11121037 |
| Tgene | ABL1 | GO:0038083 | peptidyl-tyrosine autophosphorylation | 10518561 |
| Tgene | ABL1 | GO:0042770 | signal transduction in response to DNA damage | 9037071|15657060 |
| Tgene | ABL1 | GO:0043065 | positive regulation of apoptotic process | 9037071 |
| Tgene | ABL1 | GO:0046777 | protein autophosphorylation | 10713049 |
| Tgene | ABL1 | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | 15657060 |
| Tgene | ABL1 | GO:0051353 | positive regulation of oxidoreductase activity | 12893824 |
| Tgene | ABL1 | GO:0051444 | negative regulation of ubiquitin-protein transferase activity | 20823226 |
| Tgene | ABL1 | GO:0070301 | cellular response to hydrogen peroxide | 10713049 |
| Tgene | ABL1 | GO:0071103 | DNA conformation change | 9558345 |
| Tgene | ABL1 | GO:0071901 | negative regulation of protein serine/threonine kinase activity | 11121037 |
| Tgene | ABL1 | GO:1990051 | activation of protein kinase C activity | 10713049 |
| Tgene | ABL1 | GO:2001020 | regulation of response to DNA damage stimulus | 9461559 |
 Fusion gene breakpoints across ETV6 (5'-gene) Fusion gene breakpoints across ETV6 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
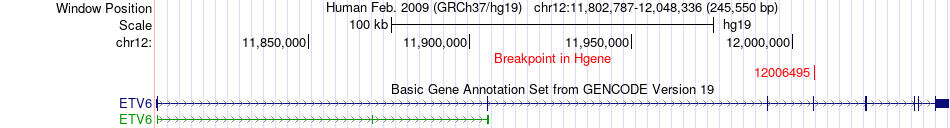 |
 Fusion gene breakpoints across ABL1 (3'-gene) Fusion gene breakpoints across ABL1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
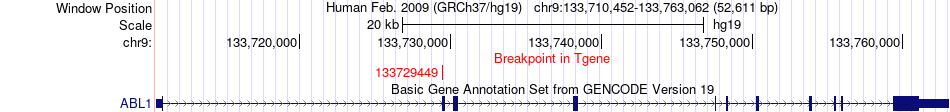 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | acute undifferentiated leukemia;acute myeloid leukemia;acute myeloblastic leukemia;acute myeloblastic leukemia with maturation;chronic myeloid leukemia;atypical chronic myeloid leukemia; acute lymphoblastic leukemia/lymphoblastic lymphoma;de novo acute no | Z35761 | ETV6 | chr12 | 12006495 | ABL1 | chr9 | 133729449 | ||
| ChimerKB3 | . | . | ETV6 | chr12 | 12022903 | + | ABL1 | chr9 | 133729450 | + |
| ChiTaRS5.0 | N/A | Z35761 | ETV6 | chr12 | 12006495 | + | ABL1 | chr9 | 133729449 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000396373 | ETV6 | chr12 | 12022903 | + | ENST00000318560 | ABL1 | chr9 | 133729450 | + | 6589 | 1283 | 274 | 4596 | 1440 |
| ENST00000396373 | ETV6 | chr12 | 12006495 | ENST00000318560 | ABL1 | chr9 | 133729449 | 6043 | 737 | 274 | 4050 | 1258 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000396373 | ENST00000318560 | ETV6 | chr12 | 12006495 | ABL1 | chr9 | 133729449 | 0.002629901 | 0.9973701 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >27697_27697_1_ETV6-ABL1_ETV6_chr12_12006495_ENST00000396373_ABL1_chr9_133729449_ENST00000318560_length(amino acids)=1258AA_BP=154 MSETPAQCSIKQERISYTPPESPVPSYASSTPLHVPVPRALRMEEDSIRLPAHLRLQPIYWSRDDVAQWLKWAENEFSLRPIDSNTFEMN GKALLLLTKEDFRYRSPHSGDVLYELLQHILKQRKPRILFSPFFHPGNSIHTQPEVILHQNHEEEALQRPVASDFEPQGLSEAARWNSKE NLLAGPSENDPNLFVALYDFVASGDNTLSITKGEKLRVLGYNHNGEWCEAQTKNGQGWVPSNYITPVNSLEKHSWYHGPVSRNAAEYLLS SGINGSFLVRESESSPGQRSISLRYEGRVYHYRINTASDGKLYVSSESRFNTLAELVHHHSTVADGLITTLHYPAPKRNKPTVYGVSPNY DKWEMERTDITMKHKLGGGQYGEVYEGVWKKYSLTVAVKTLKEDTMEVEEFLKEAAVMKEIKHPNLVQLLGVCTREPPFYIITEFMTYGN LLDYLRECNRQEVNAVVLLYMATQISSAMEYLEKKNFIHRDLAARNCLVGENHLVKVADFGLSRLMTGDTYTAHAGAKFPIKWTAPESLA YNKFSIKSDVWAFGVLLWEIATYGMSPYPGIDLSQVYELLEKDYRMERPEGCPEKVYELMRACWQWNPSDRPSFAEIHQAFETMFQESSI SDEVEKELGKQGVRGAVSTLLQAPELPTKTRTSRRAAEHRDTTDVPEMPHSKGQGESDPLDHEPAVSPLLPRKERGPPEGGLNEDERLLP KDKKTNLFSALIKKKKKTAPTPPKRSSSFREMDGQPERRGAGEEEGRDISNGALAFTPLDTADPAKSPKPSNGAGVPNGALRESGGSGFR SPHLWKKSSTLTSSRLATGEEEGGGSSSKRFLRSCSASCVPHGAKDTEWRSVTLPRDLQSTGRQFDSSTFGGHKSEKPALPRKRAGENRS DQVTRGTVTPPPRLVKKNEEAADEVFKDIMESSPGSSPPNLTPKPLRRQVTVAPASGLPHKEEAGKGSALGTPAAAEPVTPTSKAGSGAP GGTSKGPAEESRVRRHKHSSESPGRDKGKLSRLKPAPPPPPAASAGKAGGKPSQSPSQEAAGEAVLGAKTKATSLVDAVNSDAAKPSQPG EGLKKPVLPATPKPQSAKPSGTPISPAPVPSTLPSASSALAGDQPSSTAFIPLISTRVSLRKTRQPPERIASGAITKGVVLDSTEALCLA -------------------------------------------------------------- >27697_27697_2_ETV6-ABL1_ETV6_chr12_12022903_ENST00000396373_ABL1_chr9_133729450_ENST00000318560_length(amino acids)=1440AA_BP=336 MSETPAQCSIKQERISYTPPESPVPSYASSTPLHVPVPRALRMEEDSIRLPAHLRLQPIYWSRDDVAQWLKWAENEFSLRPIDSNTFEMN GKALLLLTKEDFRYRSPHSGDVLYELLQHILKQRKPRILFSPFFHPGNSIHTQPEVILHQNHEEDNCVQRTPRPSVDNVHHNPPTIELLH RSRSPITTNHRPSPDPEQRPLRSPLDNMIRRLSPAERAQGPRPHQENNHQESYPLSVSPMENNHCPASSESHPKPSSPRQESTRVIQLMP SPIMHPLILNPRHSVDFKQSRLSEDGLHREGKPINLSHREDLAYMNHIMVSVSPPEEHAMPIGRIAEALQRPVASDFEPQGLSEAARWNS KENLLAGPSENDPNLFVALYDFVASGDNTLSITKGEKLRVLGYNHNGEWCEAQTKNGQGWVPSNYITPVNSLEKHSWYHGPVSRNAAEYL LSSGINGSFLVRESESSPGQRSISLRYEGRVYHYRINTASDGKLYVSSESRFNTLAELVHHHSTVADGLITTLHYPAPKRNKPTVYGVSP NYDKWEMERTDITMKHKLGGGQYGEVYEGVWKKYSLTVAVKTLKEDTMEVEEFLKEAAVMKEIKHPNLVQLLGVCTREPPFYIITEFMTY GNLLDYLRECNRQEVNAVVLLYMATQISSAMEYLEKKNFIHRDLAARNCLVGENHLVKVADFGLSRLMTGDTYTAHAGAKFPIKWTAPES LAYNKFSIKSDVWAFGVLLWEIATYGMSPYPGIDLSQVYELLEKDYRMERPEGCPEKVYELMRACWQWNPSDRPSFAEIHQAFETMFQES SISDEVEKELGKQGVRGAVSTLLQAPELPTKTRTSRRAAEHRDTTDVPEMPHSKGQGESDPLDHEPAVSPLLPRKERGPPEGGLNEDERL LPKDKKTNLFSALIKKKKKTAPTPPKRSSSFREMDGQPERRGAGEEEGRDISNGALAFTPLDTADPAKSPKPSNGAGVPNGALRESGGSG FRSPHLWKKSSTLTSSRLATGEEEGGGSSSKRFLRSCSASCVPHGAKDTEWRSVTLPRDLQSTGRQFDSSTFGGHKSEKPALPRKRAGEN RSDQVTRGTVTPPPRLVKKNEEAADEVFKDIMESSPGSSPPNLTPKPLRRQVTVAPASGLPHKEEAGKGSALGTPAAAEPVTPTSKAGSG APGGTSKGPAEESRVRRHKHSSESPGRDKGKLSRLKPAPPPPPAASAGKAGGKPSQSPSQEAAGEAVLGAKTKATSLVDAVNSDAAKPSQ PGEGLKKPVLPATPKPQSAKPSGTPISPAPVPSTLPSASSALAGDQPSSTAFIPLISTRVSLRKTRQPPERIASGAITKGVVLDSTEALC LAISRNSEQMASHSAVLEAGKNLYTFCVSYVDSIQQMRNKFAFREAINKLENNLRELQICPATAGSGPAATQDFSKLLSSVKEISDIVQR -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr12:12006495/chr9:133729449) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ETV6 | ABL1 |
| FUNCTION: Transcriptional repressor; binds to the DNA sequence 5'-CCGGAAGT-3'. Plays a role in hematopoiesis and malignant transformation. {ECO:0000269|PubMed:25581430}. | FUNCTION: Non-receptor tyrosine-protein kinase that plays a role in many key processes linked to cell growth and survival such as cytoskeleton remodeling in response to extracellular stimuli, cell motility and adhesion, receptor endocytosis, autophagy, DNA damage response and apoptosis. Coordinates actin remodeling through tyrosine phosphorylation of proteins controlling cytoskeleton dynamics like WASF3 (involved in branch formation); ANXA1 (involved in membrane anchoring); DBN1, DBNL, CTTN, RAPH1 and ENAH (involved in signaling); or MAPT and PXN (microtubule-binding proteins). Phosphorylation of WASF3 is critical for the stimulation of lamellipodia formation and cell migration. Involved in the regulation of cell adhesion and motility through phosphorylation of key regulators of these processes such as BCAR1, CRK, CRKL, DOK1, EFS or NEDD9. Phosphorylates multiple receptor tyrosine kinases and more particularly promotes endocytosis of EGFR, facilitates the formation of neuromuscular synapses through MUSK, inhibits PDGFRB-mediated chemotaxis and modulates the endocytosis of activated B-cell receptor complexes. Other substrates which are involved in endocytosis regulation are the caveolin (CAV1) and RIN1. Moreover, ABL1 regulates the CBL family of ubiquitin ligases that drive receptor down-regulation and actin remodeling. Phosphorylation of CBL leads to increased EGFR stability. Involved in late-stage autophagy by regulating positively the trafficking and function of lysosomal components. ABL1 targets to mitochondria in response to oxidative stress and thereby mediates mitochondrial dysfunction and cell death. In response to oxidative stress, phosphorylates serine/threonine kinase PRKD2 at 'Tyr-717' (PubMed:28428613). ABL1 is also translocated in the nucleus where it has DNA-binding activity and is involved in DNA-damage response and apoptosis. Many substrates are known mediators of DNA repair: DDB1, DDB2, ERCC3, ERCC6, RAD9A, RAD51, RAD52 or WRN. Activates the proapoptotic pathway when the DNA damage is too severe to be repaired. Phosphorylates TP73, a primary regulator for this type of damage-induced apoptosis. Phosphorylates the caspase CASP9 on 'Tyr-153' and regulates its processing in the apoptotic response to DNA damage. Phosphorylates PSMA7 that leads to an inhibition of proteasomal activity and cell cycle transition blocks. ABL1 acts also as a regulator of multiple pathological signaling cascades during infection. Several known tyrosine-phosphorylated microbial proteins have been identified as ABL1 substrates. This is the case of A36R of Vaccinia virus, Tir (translocated intimin receptor) of pathogenic E.coli and possibly Citrobacter, CagA (cytotoxin-associated gene A) of H.pylori, or AnkA (ankyrin repeat-containing protein A) of A.phagocytophilum. Pathogens can highjack ABL1 kinase signaling to reorganize the host actin cytoskeleton for multiple purposes, like facilitating intracellular movement and host cell exit. Finally, functions as its own regulator through autocatalytic activity as well as through phosphorylation of its inhibitor, ABI1. Regulates T-cell differentiation in a TBX21-dependent manner. Phosphorylates TBX21 on tyrosine residues leading to an enhancement of its transcriptional activator activity (By similarity). {ECO:0000250|UniProtKB:P00520, ECO:0000269|PubMed:10391250, ECO:0000269|PubMed:11971963, ECO:0000269|PubMed:12379650, ECO:0000269|PubMed:12531427, ECO:0000269|PubMed:12672821, ECO:0000269|PubMed:15031292, ECO:0000269|PubMed:15556646, ECO:0000269|PubMed:15657060, ECO:0000269|PubMed:15886098, ECO:0000269|PubMed:16424036, ECO:0000269|PubMed:16678104, ECO:0000269|PubMed:16943190, ECO:0000269|PubMed:17306540, ECO:0000269|PubMed:17623672, ECO:0000269|PubMed:18328268, ECO:0000269|PubMed:18945674, ECO:0000269|PubMed:19891780, ECO:0000269|PubMed:20357770, ECO:0000269|PubMed:20417104, ECO:0000269|PubMed:28428613, ECO:0000269|PubMed:9037071, ECO:0000269|PubMed:9144171, ECO:0000269|PubMed:9461559}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ETV6 | chr12:12006495 | chr9:133729449 | ENST00000396373 | + | 4 | 8 | 40_124 | 154.33333333333334 | 453.0 | Domain | PNT |
| Tgene | ABL1 | chr12:12006495 | chr9:133729449 | ENST00000318560 | 0 | 11 | 605_609 | 26.333333333333332 | 1131.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | ABL1 | chr12:12006495 | chr9:133729449 | ENST00000318560 | 0 | 11 | 782_1019 | 26.333333333333332 | 1131.0 | Compositional bias | Note=Pro-rich | |
| Tgene | ABL1 | chr12:12006495 | chr9:133729449 | ENST00000318560 | 0 | 11 | 897_903 | 26.333333333333332 | 1131.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | ABL1 | chr12:12006495 | chr9:133729449 | ENST00000318560 | 0 | 11 | 127_217 | 26.333333333333332 | 1131.0 | Domain | SH2 | |
| Tgene | ABL1 | chr12:12006495 | chr9:133729449 | ENST00000318560 | 0 | 11 | 242_493 | 26.333333333333332 | 1131.0 | Domain | Protein kinase | |
| Tgene | ABL1 | chr12:12006495 | chr9:133729449 | ENST00000318560 | 0 | 11 | 61_121 | 26.333333333333332 | 1131.0 | Domain | SH3 | |
| Tgene | ABL1 | chr12:12006495 | chr9:133729449 | ENST00000318560 | 0 | 11 | 1090_1100 | 26.333333333333332 | 1131.0 | Motif | Nuclear export signal | |
| Tgene | ABL1 | chr12:12006495 | chr9:133729449 | ENST00000318560 | 0 | 11 | 381_405 | 26.333333333333332 | 1131.0 | Motif | Note=Kinase activation loop | |
| Tgene | ABL1 | chr12:12006495 | chr9:133729449 | ENST00000318560 | 0 | 11 | 709_715 | 26.333333333333332 | 1131.0 | Motif | Nuclear localization signal 2 | |
| Tgene | ABL1 | chr12:12006495 | chr9:133729449 | ENST00000318560 | 0 | 11 | 762_769 | 26.333333333333332 | 1131.0 | Motif | Nuclear localization signal 3 | |
| Tgene | ABL1 | chr12:12006495 | chr9:133729449 | ENST00000318560 | 0 | 11 | 248_256 | 26.333333333333332 | 1131.0 | Nucleotide binding | Note=ATP | |
| Tgene | ABL1 | chr12:12006495 | chr9:133729449 | ENST00000318560 | 0 | 11 | 316_322 | 26.333333333333332 | 1131.0 | Nucleotide binding | Note=ATP | |
| Tgene | ABL1 | chr12:12006495 | chr9:133729449 | ENST00000318560 | 0 | 11 | 869_968 | 26.333333333333332 | 1131.0 | Region | DNA-binding | |
| Tgene | ABL1 | chr12:12006495 | chr9:133729449 | ENST00000318560 | 0 | 11 | 953_1130 | 26.333333333333332 | 1131.0 | Region | Note=F-actin-binding |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ETV6 | chr12:12006495 | chr9:133729449 | ENST00000396373 | + | 4 | 8 | 339_420 | 154.33333333333334 | 453.0 | DNA binding | ETS |
| Tgene | ABL1 | chr12:12006495 | chr9:133729449 | ENST00000318560 | 0 | 11 | 18_22 | 26.333333333333332 | 1131.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | ABL1 | chr12:12006495 | chr9:133729449 | ENST00000318560 | 0 | 11 | 1_60 | 26.333333333333332 | 1131.0 | Region | Note=CAP |
Top |
Fusion Protein Structures |
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
| PDB file (926)CIF file (926) >>>926.cif | ETV6 | chr12 | 12022903 | + | ABL1 | chr9 | 133729450 | + | MSETPAQCSIKQERISYTPPESPVPSYASSTPLHVPVPRALRMEEDSIRL PAHLRLQPIYWSRDDVAQWLKWAENEFSLRPIDSNTFEMNGKALLLLTKE DFRYRSPHSGDVLYELLQHILKQRKPRILFSPFFHPGNSIHTQPEVILHQ NHEEDNCVQRTPRPSVDNVHHNPPTIELLHRSRSPITTNHRPSPDPEQRP LRSPLDNMIRRLSPAERAQGPRPHQENNHQESYPLSVSPMENNHCPASSE SHPKPSSPRQESTRVIQLMPSPIMHPLILNPRHSVDFKQSRLSEDGLHRE GKPINLSHREDLAYMNHIMVSVSPPEEHAMPIGRIAEALQRPVASDFEPQ GLSEAARWNSKENLLAGPSENDPNLFVALYDFVASGDNTLSITKGEKLRV LGYNHNGEWCEAQTKNGQGWVPSNYITPVNSLEKHSWYHGPVSRNAAEYL LSSGINGSFLVRESESSPGQRSISLRYEGRVYHYRINTASDGKLYVSSES RFNTLAELVHHHSTVADGLITTLHYPAPKRNKPTVYGVSPNYDKWEMERT DITMKHKLGGGQYGEVYEGVWKKYSLTVAVKTLKEDTMEVEEFLKEAAVM KEIKHPNLVQLLGVCTREPPFYIITEFMTYGNLLDYLRECNRQEVNAVVL LYMATQISSAMEYLEKKNFIHRDLAARNCLVGENHLVKVADFGLSRLMTG DTYTAHAGAKFPIKWTAPESLAYNKFSIKSDVWAFGVLLWEIATYGMSPY PGIDLSQVYELLEKDYRMERPEGCPEKVYELMRACWQWNPSDRPSFAEIH QAFETMFQESSISDEVEKELGKQGVRGAVSTLLQAPELPTKTRTSRRAAE HRDTTDVPEMPHSKGQGESDPLDHEPAVSPLLPRKERGPPEGGLNEDERL LPKDKKTNLFSALIKKKKKTAPTPPKRSSSFREMDGQPERRGAGEEEGRD ISNGALAFTPLDTADPAKSPKPSNGAGVPNGALRESGGSGFRSPHLWKKS STLTSSRLATGEEEGGGSSSKRFLRSCSASCVPHGAKDTEWRSVTLPRDL QSTGRQFDSSTFGGHKSEKPALPRKRAGENRSDQVTRGTVTPPPRLVKKN EEAADEVFKDIMESSPGSSPPNLTPKPLRRQVTVAPASGLPHKEEAGKGS ALGTPAAAEPVTPTSKAGSGAPGGTSKGPAEESRVRRHKHSSESPGRDKG KLSRLKPAPPPPPAASAGKAGGKPSQSPSQEAAGEAVLGAKTKATSLVDA VNSDAAKPSQPGEGLKKPVLPATPKPQSAKPSGTPISPAPVPSTLPSASS ALAGDQPSSTAFIPLISTRVSLRKTRQPPERIASGAITKGVVLDSTEALC LAISRNSEQMASHSAVLEAGKNLYTFCVSYVDSIQQMRNKFAFREAINKL | 1440 |
| 3D view using mol* of 926 (AA BP:) | ||||||||||
 | ||||||||||
Top |
pLDDT score distribution |
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
ETV6_pLDDT.png |
ABL1_pLDDT.png |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
ETV6_ABL1_926_pLDDT_and_active_sites.png (AA BP:) |
ETV6_ABL1_926_violinplot.png (AA BP:) |
Top |
Ramachandran Plot of Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| Fusion AA seq ID in FusionPDB and their Ramachandran plots |
Top |
Potential Active Site Information |
 The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. |
| Fusion AA seq ID in FusionPDB | Site score | Size | D score | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
| 926 | 1.061 | 90 | 1.106 | 180 | 0.516 | 0.758 | 1.053 | 1.531 | 0.737 | 2.076 | 1.027 | Chain A: 642,643,646,647,650,651,654,739,740,742,7 43,744,745,747,771,772,773,774,775,778,803,812,816 ,820 |
Top |
Potentially Interacting Small Molecules through Virtual Screening |
 The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. |
| Fusion AA seq ID in FusionPDB | ZINC ID | DrugBank ID | Drug name | Docking score | Glide gscore |
| 926 | ZINC000001530638 | DB00472 | Fluoxetine | -8.87535 | -8.87585 |
| 926 | ZINC000000000850 | DB00744 | Zileuton | -8.66624 | -8.68484 |
| 926 | ZINC000000020243 | DB00861 | Diflunisal | -8.62048 | -8.62048 |
| 926 | ZINC000011681534 | DB04861 | Nebivolol | -8.43116 | -8.45096 |
| 926 | ZINC000043206370 | DB11793 | Niraparib | -8.29133 | -8.29133 |
| 926 | ZINC000035342789 | DB00323 | Tolcapone | -6.65588 | -8.19288 |
| 926 | ZINC000004213946 | DB04861 | Nebivolol | -8.16276 | -8.18256 |
| 926 | ZINC000000001644 | DB06795 | Mafenide | -8.14513 | -8.17473 |
| 926 | ZINC000000085733 | DB00181 | Baclofen | -8.08837 | -8.08837 |
| 926 | Avapritinib | DB00181 | -6.91248 | -8.00548 | |
| 926 | Asciminib | DB00181 | -7.53678 | -8.00178 | |
| 926 | ZINC000000004321 | DB00909 | Zonisamide | -7.99504 | -7.99644 |
| 926 | ZINC000003816287 | DB06626 | Axitinib | -7.9681 | -7.9713 |
| 926 | ZINC000000897258 | DB00983 | Formoterol | -7.95725 | -7.96945 |
| 926 | ZINC000000083315 | DB00150 | Tryptophan | -7.90933 | -7.90933 |
| 926 | ZINC000005844788 | DB04861 | Nebivolol | -7.88702 | -7.90682 |
| 926 | ZINC000003872994 | DB01026 | Ketoconazole | -7.5428 | -7.8675 |
| 926 | ZINC000000537822 | DB00502 | Haloperidol | -7.82221 | -7.83561 |
| 926 | ZINC000000018635 | DB00998 | Frovatriptan | -7.83256 | -7.83306 |
| 926 | ZINC000000006481 | DB00740 | Riluzole | -7.81985 | -7.81985 |
Top |
 Drug information from DrugBank of the top 20 interacting small molecules. Drug information from DrugBank of the top 20 interacting small molecules. |
| ZINC ID | DrugBank ID | Drug name | Drug type | SMILES | Drug group |
| ZINC000001530638 | DB00472 | Fluoxetine | Small molecule | CNCCC(OC1=CC=C(C=C1)C(F)(F)F)C1=CC=CC=C1 | Approved|Vet_approved |
| ZINC000000020243 | DB00861 | Diflunisal | Small molecule | OC(=O)C1=C(O)C=CC(=C1)C1=C(F)C=C(F)C=C1 | Approved|Investigational |
| ZINC000011681534 | DB04861 | Nebivolol | Small molecule | OC(CNCC(O)C1CCC2=C(O1)C=CC(F)=C2)C1CCC2=C(O1)C=CC(F)=C2 | Approved|Investigational |
| ZINC000043206370 | DB11793 | Niraparib | Small molecule | NC(=O)C1=CC=CC2=CN(N=C12)C1=CC=C(C=C1)[C@@H]1CCCNC1 | Approved|Investigational |
| ZINC000035342789 | DB00323 | Tolcapone | Small molecule | CC1=CC=C(C=C1)C(=O)C1=CC(=C(O)C(O)=C1)[N+]([O-])=O | Approved|Withdrawn |
| ZINC000000001644 | DB06795 | Mafenide | Small molecule | NCC1=CC=C(C=C1)S(N)(=O)=O | Approved|Vet_approved |
| ZINC000000085733 | DB00181 | Baclofen | Small molecule | NCC(CC(O)=O)C1=CC=C(Cl)C=C1 | Approved |
| ZINC000000004321 | DB00909 | Zonisamide | Small molecule | NS(=O)(=O)CC1=NOC2=CC=CC=C12 | Approved|Investigational |
| ZINC000000897258 | DB00983 | Formoterol | Small molecule | COC1=CC=C(CC(C)NCC(O)C2=CC(NC=O)=C(O)C=C2)C=C1 | Approved|Investigational |
| ZINC000000083315 | DB00150 | Tryptophan | Small molecule | N[C@@H](CC1=CNC2=C1C=CC=C2)C(O)=O | Approved|Nutraceutical|Withdrawn |
| ZINC000000537822 | DB00502 | Haloperidol | Small molecule | OC1(CCN(CCCC(=O)C2=CC=C(F)C=C2)CC1)C1=CC=C(Cl)C=C1 | Approved |
| ZINC000000018635 | DB00998 | Frovatriptan | Small molecule | CN[C@@H]1CCC2=C(C1)C1=C(N2)C=CC(=C1)C(N)=O | Approved|Investigational |
| ZINC000000006481 | DB00740 | Riluzole | Small molecule | NC1=NC2=C(S1)C=C(OC(F)(F)F)C=C2 | Approved|Investigational |
Top |
Biochemical Features of Small Molecules |
 ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) |
| ZINC ID | mol_MW | dipole | SASA | FOSA | FISA | PISA | WPSA | volume | donorHB | accptHB | IP | Human Oral Absorption | Percent Human Oral Absorption | Rule Of Five | Rule Of Three |
| ZINC000001530638 | 309.331 | 4.533 | 597.054 | 163.848 | 24.069 | 291.6 | 117.537 | 1015.412 | 1 | 2.25 | 9.42 | 3 | 100 | 0 | 0 |
| ZINC000000000850 | 236.288 | 5.297 | 457.642 | 85.783 | 137.035 | 198.523 | 36.3 | 755.015 | 3 | 3.7 | 8.91 | 3 | 75.351 | 0 | 0 |
| ZINC000000000850 | 236.288 | 5.297 | 450.837 | 85.532 | 135.419 | 201.893 | 27.994 | 746.839 | 3 | 3.7 | 8.57 | 3 | 76.555 | 0 | 0 |
| ZINC000000020243 | 250.201 | 8.261 | 459.081 | 0 | 144.436 | 231.005 | 83.64 | 751.879 | 1 | 1.75 | 9.879 | 3 | 86.601 | 0 | 0 |
| ZINC000011681534 | 405.441 | 2.335 | 727.789 | 280.708 | 85.7 | 267.621 | 93.76 | 1272.951 | 3 | 6.4 | 9.228 | 3 | 95.433 | 0 | 0 |
| ZINC000011681534 | 405.441 | 6.385 | 707.131 | 273.322 | 77.744 | 262.301 | 93.765 | 1256.165 | 3 | 6.4 | 9.073 | 3 | 96.543 | 0 | 0 |
| ZINC000043206370 | 320.393 | 3.754 | 607.688 | 185.794 | 143.417 | 278.476 | 0 | 1055.805 | 3 | 5 | 8.546 | 3 | 76.72 | 0 | 0 |
| ZINC000035342789 | 273.245 | 3.817 | 507.142 | 88.195 | 223.7 | 195.247 | 0 | 847.548 | 2 | 4.5 | 9.508 | 3 | 67.934 | 0 | 0 |
| ZINC000035342789 | 273.245 | 5.603 | 506.169 | 88.195 | 222.204 | 195.769 | 0 | 846.891 | 2 | 4.5 | 9.828 | 3 | 68.233 | 0 | 0 |
| ZINC000035342789 | 273.245 | 3.5 | 506.842 | 88.2 | 223.976 | 194.665 | 0 | 847.734 | 2 | 4.5 | 9.758 | 3 | 67.881 | 0 | 0 |
| ZINC000004213946 | 405.441 | 5.213 | 712.775 | 270.546 | 87.695 | 260.773 | 93.761 | 1260.01 | 3 | 6.4 | 9.251 | 3 | 94.555 | 0 | 0 |
| ZINC000004213946 | 405.441 | 3.313 | 660.255 | 261.068 | 75.583 | 229.844 | 93.76 | 1216.093 | 3 | 6.4 | 9.124 | 3 | 95.533 | 0 | 0 |
| ZINC000000001644 | 186.228 | 7.234 | 386.689 | 48.38 | 203.852 | 132.644 | 1.813 | 608.208 | 4 | 5.5 | 9.936 | 2 | 46.461 | 0 | 0 |
| ZINC000000001644 | 186.228 | 8.543 | 384.513 | 48.79 | 201.796 | 132.114 | 1.813 | 604.747 | 4 | 5.5 | 9.902 | 2 | 46.8 | 0 | 0 |
| ZINC000000085733 | 213.663 | 3.613 | 420.91 | 80.54 | 149.736 | 119.128 | 71.506 | 697.141 | 3 | 3 | 9.549 | 2 | 48.776 | 0 | 0 |
| ZINC000000004321 | 212.223 | 6.978 | 398.517 | 40.32 | 185.554 | 172.644 | 0 | 642.453 | 2 | 6 | 9.667 | 3 | 65.959 | 0 | 0 |
| ZINC000003816287 | 386.47 | 6.702 | 685.418 | 106.992 | 114.211 | 433.501 | 30.715 | 1211.931 | 2 | 4.5 | 8.542 | 3 | 100 | 0 | 1 |
| ZINC000000897258 | 344.41 | 7.083 | 674.489 | 258.736 | 187.926 | 227.826 | 0 | 1168.666 | 4 | 7.2 | 8.855 | 2 | 65.222 | 0 | 0 |
| ZINC000000897258 | 344.41 | 4.982 | 586.764 | 251.201 | 138.136 | 197.426 | 0 | 1091.391 | 4 | 7.2 | 8.677 | 3 | 73.278 | 0 | 0 |
| ZINC000000083315 | 204.228 | 3.132 | 419.183 | 46.221 | 166.648 | 206.314 | 0 | 687.589 | 4 | 3 | 8.088 | 2 | 42.506 | 0 | 1 |
| ZINC000005844788 | 405.441 | 1.102 | 724.6 | 273.703 | 87.795 | 269.339 | 93.762 | 1272.257 | 3 | 6.4 | 9.305 | 3 | 94.995 | 0 | 0 |
| ZINC000005844788 | 405.441 | 3.204 | 688.14 | 293.004 | 60.761 | 240.615 | 93.76 | 1244.959 | 3 | 6.4 | 9.151 | 3 | 100 | 0 | 0 |
| ZINC000003872994 | 531.438 | 2.643 | 843.259 | 319.79 | 80.343 | 323.603 | 119.523 | 1551.551 | 0 | 8.25 | 8.241 | 3 | 92.53 | 1 | 0 |
| ZINC000003872994 | 531.438 | 6.575 | 844.887 | 311.956 | 81.254 | 336.082 | 115.594 | 1549.966 | 0 | 8.25 | 8.233 | 3 | 92.405 | 1 | 0 |
| ZINC000003872994 | 531.438 | 5.578 | 860.223 | 333.686 | 78.629 | 331.472 | 116.437 | 1569.565 | 0 | 8.25 | 9.26 | 3 | 92.881 | 1 | 0 |
| ZINC000003872994 | 531.438 | 5.486 | 858.441 | 334.024 | 79.024 | 328.466 | 116.927 | 1566.075 | 0 | 8.25 | 9.254 | 3 | 92.765 | 1 | 0 |
| ZINC000000537822 | 375.87 | 3.194 | 684.196 | 209.603 | 68.864 | 287.369 | 118.36 | 1199.714 | 1 | 4.75 | 9.391 | 3 | 100 | 0 | 0 |
| ZINC000000537822 | 375.87 | 2.935 | 674.977 | 199.704 | 83.514 | 273.376 | 118.382 | 1195.364 | 1 | 4.75 | 9.357 | 3 | 100 | 0 | 0 |
| ZINC000000537822 | 375.87 | 4.967 | 684.834 | 209.154 | 69.652 | 287.663 | 118.365 | 1206.223 | 1 | 4.75 | 9.266 | 3 | 100 | 0 | 0 |
| ZINC000000018635 | 243.308 | 7.2 | 492.762 | 217.467 | 154.534 | 120.761 | 0 | 833.223 | 4 | 4 | 8.296 | 3 | 66.41 | 0 | 0 |
| ZINC000000006481 | 234.196 | 5.283 | 393.435 | 1.903 | 87.083 | 133.293 | 171.157 | 623.3 | 2 | 2 | 8.622 | 3 | 100 | 0 | 0 |
Top |
Drug Toxicity Information |
 Toxicity information of individual drugs using eToxPred Toxicity information of individual drugs using eToxPred |
| ZINC ID | Smile | Surface Accessibility | Toxicity |
| ZINC000001530638 | CNCC[C@@H](Oc1ccc(C(F)(F)F)cc1)c1ccccc1 | 0.234957648 | 0.361924168 |
| ZINC000000000850 | C[C@@H](c1cc2ccccc2s1)N(O)C(N)=O | 0.136335882 | 0.223511692 |
| ZINC000000020243 | O=C(O)c1cc(-c2ccc(F)cc2F)ccc1O | 0.440409373 | 0.371648269 |
| ZINC000011681534 | O[C@H](CNC[C@@H](O)[C@@H]1CCc2cc(F)ccc2O1)[C@@H]1CCc2cc(F)ccc2O1 | 0.055550082 | 0.198725964 |
| ZINC000043206370 | NC(=O)c1cccc2cn(-c3ccc([C@@H]4CCCNC4)cc3)nc12 | 0.15810306 | 0.243905197 |
| ZINC000035342789 | Cc1ccc(C(=O)c2cc(O)c(O)c([N+](=O)[O-])c2)cc1 | 0.359735014 | 0.498165074 |
| ZINC000004213946 | O[C@@H](CNC[C@@H](O)[C@@H]1CCc2cc(F)ccc2O1)[C@@H]1CCc2cc(F)ccc2O1 | 0.055550082 | 0.198725964 |
| ZINC000000001644 | NCc1ccc(S(N)(=O)=O)cc1 | 0.497774604 | 0.389970963 |
| ZINC000000085733 | NC[C@@H](CC(=O)O)c1ccc(Cl)cc1 | 0.251203336 | 0.372196692 |
| ZINC000000004321 | NS(=O)(=O)Cc1noc2ccccc12 | 0.254569254 | 0.46155884 |
| ZINC000003816287 | CNC(=O)c1ccccc1Sc1ccc2c(/C=C/c3ccccn3)n[nH]c2c1 | 0.19874265 | 0.230949688 |
| ZINC000000897258 | COc1ccc(C[C@@H](C)NC[C@@H](O)c2ccc(O)c(NC=O)c2)cc1 | 0.133863078 | 0.246482573 |
| ZINC000000083315 | N[C@@H](Cc1c[nH]c2ccccc12)C(=O)O | 0.272359065 | 0.412747788 |
| ZINC000005844788 | O[C@@H](CNC[C@@H](O)[C@H]1CCc2cc(F)ccc2O1)[C@@H]1CCc2cc(F)ccc2O1 | 0.055550082 | 0.198725964 |
| ZINC000003872994 | CC(=O)N1CCN(c2ccc(OC[C@H]3CO[C@@](Cn4ccnc4)(c4ccc(Cl)cc4Cl)O3)cc2)CC1 | 0.087393272 | 0.387082142 |
| ZINC000000537822 | O=C(CCCN1CCC(O)(c2ccc(Cl)cc2)CC1)c1ccc(F)cc1 | 0.325389417 | 0.260800529 |
| ZINC000000018635 | CN[C@@H]1CCc2[nH]c3ccc(C(N)=O)cc3c2C1 | 0.15678731 | 0.476568578 |
| ZINC000000006481 | Nc1nc2ccc(OC(F)(F)F)cc2s1 | 0.291002026 | 0.384188974 |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| ETV6 | KAT5, GAB2, ETV6, GRB2, CRKL, ACTA1, HDAC9, FLI1, FBXL6, SKP1, L3MBTL1, IRF8, NCOR1, SIN3A, HDAC3, UBE2I, ELAVL1, PDGFRB, SUMO1, SOCS1, SOCS3, TRAF3, SOX2, HDAC6, PIN1, AP1M1, WDYHV1, LRP6, LRP5, NID2, ETV7, USP7, VPS26B, NFATC2, EGLN3, NKX2-1, TRAF2, AXIN1, ESR2, LMNA, TP53BP1, BRCA1, MDC1, KIAA1429, WWP2, ESR1, BRD4, NFE2L2, MAD2L1, B4GALT2, KLHL9, KLHL13, SUMO2, ARHGAP32, SEC16A, XPOT, PIAS2, CEP152, NUP214, NUP62, NUP98, SEC13, RBM11, ECH1, ZNF146, TRIP6, CEP85, NUP54, TAB3, ALMS1, CEP192, PPIL4, RQCD1, SDCCAG3, NUP88, TBL1Y, KIAA1671, C2orf44, GRPEL1, TNRC6C, MID1IP1, CYLD, ZMYM2, LIMD1, SPATA2, TNRC6B, TIMM13, FLOT1, LGALS3BP, FLOT2, HNRNPUL1, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| ETV6 |  |
| ABL1 | 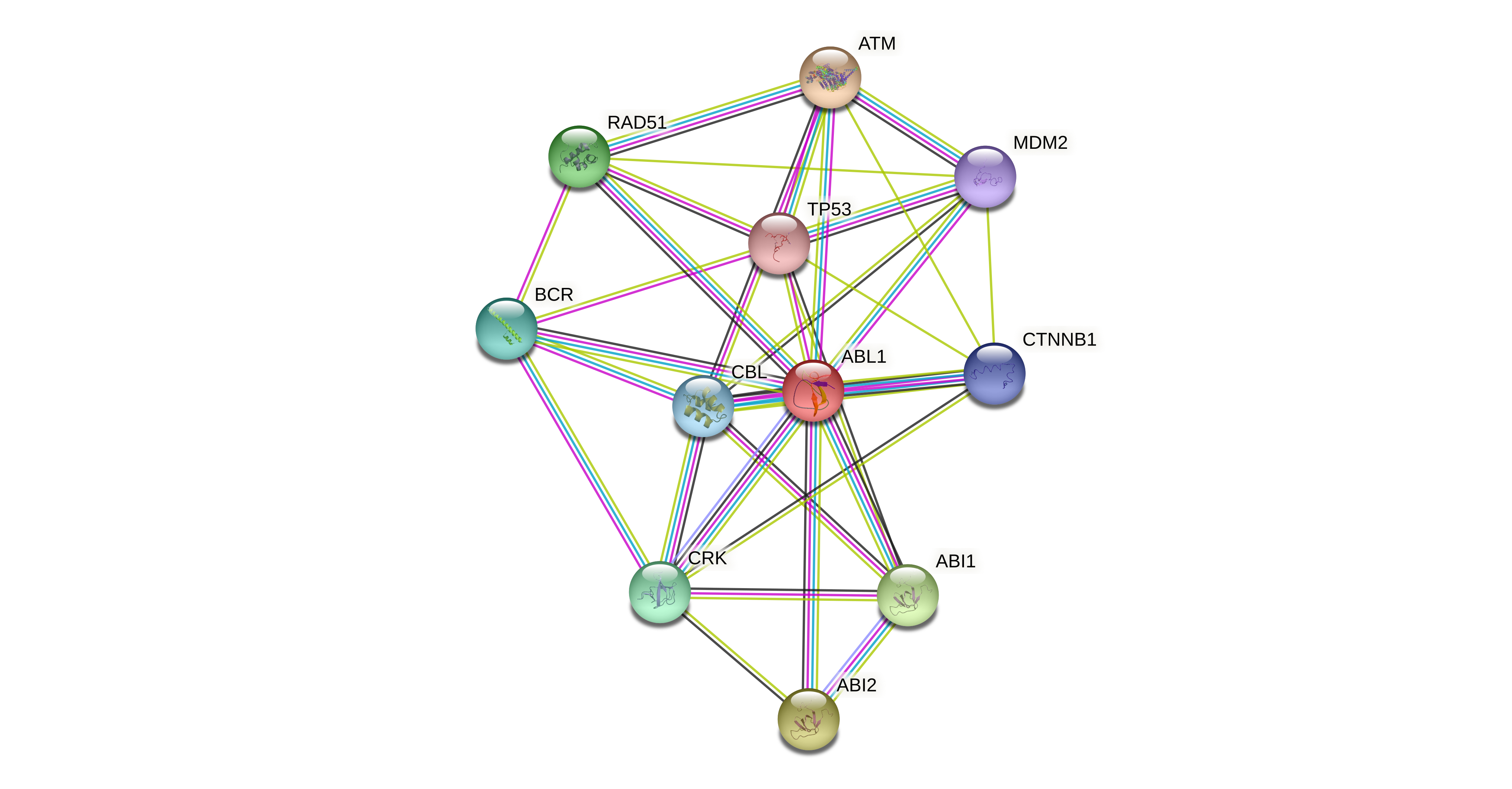 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to ETV6-ABL1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
| ETV6 | ABL1 | Nilotinib Or Dasatinib | PubMed | 32279331 |
Top |
Related Diseases to ETV6-ABL1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
| ETV6 | ABL1 | Myeloid Neoplasms | PubMed | 32279331 |
| ETV6 | ABL1 | Breast Invasive Ductal Carcinoma | MyCancerGenome | |
| ETV6 | ABL1 | Invasive Breast Carcinoma | MyCancerGenome | |
| ETV6 | ABL1 | Lung Adenocarcinoma | MyCancerGenome | |
| ETV6 | ABL1 | Appendix Adenocarcinoma | MyCancerGenome | |
| ETV6 | ABL1 | Astrocytoma | MyCancerGenome |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | ETV6 | C4015537 | THROMBOCYTOPENIA 5 | 4 | CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Hgene | ETV6 | C0023485 | Precursor B-Cell Lymphoblastic Leukemia-Lymphoma | 3 | CTD_human |
| Hgene | ETV6 | C0040034 | Thrombocytopenia | 3 | CTD_human;GENOMICS_ENGLAND |
| Hgene | ETV6 | C0023480 | Leukemia, Myelomonocytic, Chronic | 2 | ORPHANET |
| Hgene | ETV6 | C1332965 | Congenital Mesoblastic Nephroma | 2 | ORPHANET |
| Hgene | ETV6 | C0006413 | Burkitt Lymphoma | 1 | ORPHANET |
| Hgene | ETV6 | C0013146 | Drug abuse | 1 | CTD_human |
| Hgene | ETV6 | C0013170 | Drug habituation | 1 | CTD_human |
| Hgene | ETV6 | C0013222 | Drug Use Disorders | 1 | CTD_human |
| Hgene | ETV6 | C0023452 | Childhood Acute Lymphoblastic Leukemia | 1 | CTD_human |
| Hgene | ETV6 | C0023453 | L2 Acute Lymphoblastic Leukemia | 1 | CTD_human |
| Hgene | ETV6 | C0023467 | Leukemia, Myelocytic, Acute | 1 | CGI;CTD_human;GENOMICS_ENGLAND |
| Hgene | ETV6 | C0029231 | Organic Mental Disorders, Substance-Induced | 1 | CTD_human |
| Hgene | ETV6 | C0038580 | Substance Dependence | 1 | CTD_human |
| Hgene | ETV6 | C0038586 | Substance Use Disorders | 1 | CTD_human |
| Hgene | ETV6 | C0087031 | Juvenile-Onset Still Disease | 1 | CTD_human |
| Hgene | ETV6 | C0236969 | Substance-Related Disorders | 1 | CTD_human |
| Hgene | ETV6 | C0238463 | Papillary thyroid carcinoma | 1 | ORPHANET |
| Hgene | ETV6 | C0376544 | Hematopoietic Neoplasms | 1 | CTD_human |
| Hgene | ETV6 | C0376545 | Hematologic Neoplasms | 1 | CTD_human |
| Hgene | ETV6 | C0740858 | Substance abuse problem | 1 | CTD_human |
| Hgene | ETV6 | C1292769 | Precursor B-cell lymphoblastic leukemia | 1 | ORPHANET |
| Hgene | ETV6 | C1510472 | Drug Dependence | 1 | CTD_human |
| Hgene | ETV6 | C1832388 | Platelet Disorder, Familial, with Associated Myeloid Malignancy | 1 | ORPHANET |
| Hgene | ETV6 | C1838656 | Macrocytosis, Familial | 1 | CTD_human |
| Hgene | ETV6 | C1961102 | Precursor Cell Lymphoblastic Leukemia Lymphoma | 1 | CTD_human |
| Hgene | ETV6 | C3495559 | Juvenile arthritis | 1 | CTD_human |
| Hgene | ETV6 | C3714758 | Juvenile psoriatic arthritis | 1 | CTD_human |
| Hgene | ETV6 | C4316881 | Prescription Drug Abuse | 1 | CTD_human |
| Hgene | ETV6 | C4552091 | Polyarthritis, Juvenile, Rheumatoid Factor Negative | 1 | CTD_human |
| Hgene | ETV6 | C4704862 | Polyarthritis, Juvenile, Rheumatoid Factor Positive | 1 | CTD_human |
| Tgene | ABL1 | C0023473 | Myeloid Leukemia, Chronic | 3 | CGI;CTD_human;ORPHANET |
| Tgene | ABL1 | C0023452 | Childhood Acute Lymphoblastic Leukemia | 2 | CTD_human |
| Tgene | ABL1 | C0023453 | L2 Acute Lymphoblastic Leukemia | 2 | CTD_human |
| Tgene | ABL1 | C1961102 | Precursor Cell Lymphoblastic Leukemia Lymphoma | 2 | CGI;CTD_human |
| Tgene | ABL1 | C0001418 | Adenocarcinoma | 1 | CTD_human |
| Tgene | ABL1 | C0003706 | Arachnodactyly | 1 | GENOMICS_ENGLAND |
| Tgene | ABL1 | C0005941 | Bone Diseases, Developmental | 1 | CTD_human |
| Tgene | ABL1 | C0006142 | Malignant neoplasm of breast | 1 | CTD_human |
| Tgene | ABL1 | C0006413 | Burkitt Lymphoma | 1 | ORPHANET |
| Tgene | ABL1 | C0014859 | Esophageal Neoplasms | 1 | CTD_human |
| Tgene | ABL1 | C0015544 | Failure to Thrive | 1 | CTD_human |
| Tgene | ABL1 | C0018798 | Congenital Heart Defects | 1 | CTD_human |
| Tgene | ABL1 | C0023903 | Liver neoplasms | 1 | CTD_human |
| Tgene | ABL1 | C0027659 | Neoplasms, Experimental | 1 | CTD_human |
| Tgene | ABL1 | C0032927 | Precancerous Conditions | 1 | CTD_human |
| Tgene | ABL1 | C0039075 | Syndactyly | 1 | GENOMICS_ENGLAND |
| Tgene | ABL1 | C0151491 | Congenital musculoskeletal anomalies | 1 | CTD_human |
| Tgene | ABL1 | C0205641 | Adenocarcinoma, Basal Cell | 1 | CTD_human |
| Tgene | ABL1 | C0205642 | Adenocarcinoma, Oxyphilic | 1 | CTD_human |
| Tgene | ABL1 | C0205643 | Carcinoma, Cribriform | 1 | CTD_human |
| Tgene | ABL1 | C0205644 | Carcinoma, Granular Cell | 1 | CTD_human |
| Tgene | ABL1 | C0205645 | Adenocarcinoma, Tubular | 1 | CTD_human |
| Tgene | ABL1 | C0265610 | Clinodactyly of fingers | 1 | GENOMICS_ENGLAND |
| Tgene | ABL1 | C0282313 | Condition, Preneoplastic | 1 | CTD_human |
| Tgene | ABL1 | C0345904 | Malignant neoplasm of liver | 1 | CTD_human |
| Tgene | ABL1 | C0546837 | Malignant neoplasm of esophagus | 1 | CTD_human |
| Tgene | ABL1 | C0596263 | Carcinogenesis | 1 | CTD_human |
| Tgene | ABL1 | C0678222 | Breast Carcinoma | 1 | CTD_human |
| Tgene | ABL1 | C1257931 | Mammary Neoplasms, Human | 1 | CTD_human |
| Tgene | ABL1 | C1292769 | Precursor B-cell lymphoblastic leukemia | 1 | ORPHANET |
| Tgene | ABL1 | C1458155 | Mammary Neoplasms | 1 | CTD_human |
| Tgene | ABL1 | C1961099 | Precursor T-Cell Lymphoblastic Leukemia-Lymphoma | 1 | CGI;ORPHANET |
| Tgene | ABL1 | C4539857 | CONGENITAL HEART DEFECTS AND SKELETAL MALFORMATIONS SYNDROME | 1 | GENOMICS_ENGLAND;UNIPROT |
| Tgene | ABL1 | C4551485 | Clinodactyly | 1 | GENOMICS_ENGLAND |
| Tgene | ABL1 | C4704874 | Mammary Carcinoma, Human | 1 | CTD_human |

