| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:FGFR3-BAIAP2L1 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: FGFR3-BAIAP2L1 | FusionPDB ID: 30301 | FusionGDB2.0 ID: 30301 | Hgene | Tgene | Gene symbol | FGFR3 | BAIAP2L1 | Gene ID | 2261 | 55971 |
| Gene name | fibroblast growth factor receptor 3 | BAR/IMD domain containing adaptor protein 2 like 1 | |
| Synonyms | ACH|CD333|CEK2|HSFGFR3EX|JTK4 | IRTKS | |
| Cytomap | 4p16.3 | 7q21.3-q22.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | fibroblast growth factor receptor 3FGFR-3fibroblast growth factor receptor 3 variant 4fibroblast growth factor receptor 3-Shydroxyaryl-protein kinasetyrosine kinase JTK4 | brain-specific angiogenesis inhibitor 1-associated protein 2-like protein 1BAI1 associated protein 2 like 1BAI1-associated protein 2-like protein 1insulin receptor tyrosine kinase substrate | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | P22607 | Q9UHR4 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000260795, ENST00000340107, ENST00000352904, ENST00000412135, ENST00000440486, ENST00000481110, ENST00000474521, | ENST00000462558, ENST00000005260, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 10 X 22 X 14=3080 | 16 X 9 X 7=1008 |
| # samples | 33 | 16 | |
| ** MAII score | log2(33/3080*10)=-3.22239242133645 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(16/1008*10)=-2.65535182861255 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: FGFR3 [Title/Abstract] AND BAIAP2L1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | FGFR3(1808661)-BAIAP2L1(97991744), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | FGFR3-BAIAP2L1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. FGFR3-BAIAP2L1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. FGFR3-BAIAP2L1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. FGFR3-BAIAP2L1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. FGFR3-BAIAP2L1 seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. FGFR3-BAIAP2L1 seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. FGFR3-BAIAP2L1 seems lost the major protein functional domain in Hgene partner, which is a kinase due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | FGFR3 | GO:0008543 | fibroblast growth factor receptor signaling pathway | 8663044 |
| Hgene | FGFR3 | GO:0018108 | peptidyl-tyrosine phosphorylation | 11294897 |
| Hgene | FGFR3 | GO:0046777 | protein autophosphorylation | 11294897 |
| Tgene | BAIAP2L1 | GO:0009617 | response to bacterium | 19366662 |
| Tgene | BAIAP2L1 | GO:0030838 | positive regulation of actin filament polymerization | 21098279 |
 Fusion gene breakpoints across FGFR3 (5'-gene) Fusion gene breakpoints across FGFR3 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
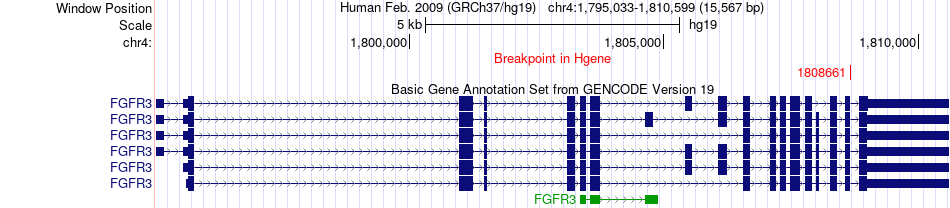 |
 Fusion gene breakpoints across BAIAP2L1 (3'-gene) Fusion gene breakpoints across BAIAP2L1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
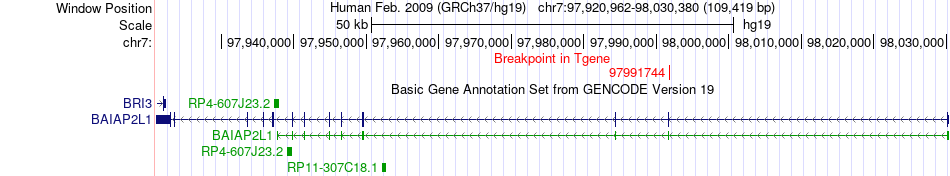 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerKB3 | . | . | FGFR3 | chr4 | 1808661 | + | BAIAP2L1 | chr7 | 97949610 | - |
| ChimerKB3 | . | . | FGFR3 | chr4 | 1808661 | + | BAIAP2L1 | chr7 | 97991744 | - |
| ChiTaRS5.0 | N/A | JE837678 | FGFR3 | chr4 | 1808661 | + | BAIAP2L1 | chr7 | 97991744 | - |
| ChiTaRS5.0 | N/A | JE837684 | FGFR3 | chr4 | 1808661 | + | BAIAP2L1 | chr7 | 97991744 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000481110 | FGFR3 | chr4 | 1808661 | + | ENST00000005260 | BAIAP2L1 | chr7 | 97949610 | - | 5659 | 2467 | 228 | 3788 | 1186 |
| ENST00000440486 | FGFR3 | chr4 | 1808661 | + | ENST00000005260 | BAIAP2L1 | chr7 | 97991744 | - | 5885 | 2530 | 223 | 4014 | 1263 |
| ENST00000412135 | FGFR3 | chr4 | 1808661 | + | ENST00000005260 | BAIAP2L1 | chr7 | 97991744 | - | 5549 | 2194 | 223 | 3678 | 1151 |
| ENST00000340107 | FGFR3 | chr4 | 1808661 | + | ENST00000005260 | BAIAP2L1 | chr7 | 97991744 | - | 5891 | 2536 | 223 | 4020 | 1265 |
| ENST00000260795 | FGFR3 | chr4 | 1808661 | + | ENST00000005260 | BAIAP2L1 | chr7 | 97991744 | - | 5731 | 2376 | 69 | 3860 | 1263 |
| ENST00000352904 | FGFR3 | chr4 | 1808661 | + | ENST00000005260 | BAIAP2L1 | chr7 | 97991744 | - | 5332 | 1977 | 6 | 3461 | 1151 |
| ENST00000440486 | FGFR3 | chr4 | 1808661 | + | ENST00000005260 | BAIAP2L1 | chr7 | 97991744 | - | 5885 | 2530 | 223 | 4014 | 1263 |
| ENST00000412135 | FGFR3 | chr4 | 1808661 | + | ENST00000005260 | BAIAP2L1 | chr7 | 97991744 | - | 5549 | 2194 | 223 | 3678 | 1151 |
| ENST00000340107 | FGFR3 | chr4 | 1808661 | + | ENST00000005260 | BAIAP2L1 | chr7 | 97991744 | - | 5891 | 2536 | 223 | 4020 | 1265 |
| ENST00000260795 | FGFR3 | chr4 | 1808661 | + | ENST00000005260 | BAIAP2L1 | chr7 | 97991744 | - | 5731 | 2376 | 69 | 3860 | 1263 |
| ENST00000352904 | FGFR3 | chr4 | 1808661 | + | ENST00000005260 | BAIAP2L1 | chr7 | 97991744 | - | 5332 | 1977 | 6 | 3461 | 1151 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000440486 | ENST00000005260 | FGFR3 | chr4 | 1808661 | + | BAIAP2L1 | chr7 | 97991744 | - | 0.001116543 | 0.9988834 |
| ENST00000412135 | ENST00000005260 | FGFR3 | chr4 | 1808661 | + | BAIAP2L1 | chr7 | 97991744 | - | 0.001734099 | 0.9982659 |
| ENST00000340107 | ENST00000005260 | FGFR3 | chr4 | 1808661 | + | BAIAP2L1 | chr7 | 97991744 | - | 0.001075541 | 0.99892443 |
| ENST00000260795 | ENST00000005260 | FGFR3 | chr4 | 1808661 | + | BAIAP2L1 | chr7 | 97991744 | - | 0.00091506 | 0.99908495 |
| ENST00000352904 | ENST00000005260 | FGFR3 | chr4 | 1808661 | + | BAIAP2L1 | chr7 | 97991744 | - | 0.001339923 | 0.9986601 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >30301_30301_1_FGFR3-BAIAP2L1_FGFR3_chr4_1808661_ENST00000260795_BAIAP2L1_chr7_97991744_ENST00000005260_length(amino acids)=1263AA_BP=769 MPEDAAAPAPAMGAPACALALCVAVAIVAGASSESLGTEQRVVGRAAEVPGPEPGQQEQLVFGSGDAVELSCPPPGGGPMGPTVWVKDGT GLVPSERVLVGPQRLQVLNASHEDSGAYSCRQRLTQRVLCHFSVRVTDAPSSGDDEDGEDEAEDTGVDTGAPYWTRPERMDKKLLAVPAA NTVRFRCPAAGNPTPSISWLKNGREFRGEHRIGGIKLRHQQWSLVMESVVPSDRGNYTCVVENKFGSIRQTYTLDVLERSPHRPILQAGL PANQTAVLGSDVEFHCKVYSDAQPHIQWLKHVEVNGSKVGPDGTPYVTVLKTAGANTTDKELEVLSLHNVTFEDAGEYTCLAGNSIGFSH HSAWLVVLPAEEELVEADEAGSVYAGILSYGVGFFLFILVVAAVTLCRLRSPPKKGLGSPTVHKISRFPLKRQVSLESNASMSSNTPLVR IARLSSGEGPTLANVSELELPADPKWELSRARLTLGKPLGEGCFGQVVMAEAIGIDKDRAAKPVTVAVKMLKDDATDKDLSDLVSEMEMM KMIGKHKNIINLLGACTQGGPLYVLVEYAAKGNLREFLRARRPPGLDYSFDTCKPPEEQLTFKDLVSCAYQVARGMEYLASQKCIHRDLA ARNVLVTEDNVMKIADFGLARDVHNLDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLLWEIFTLGGSPYPGIPVEELFKLLKE GHRMDKPANCTHDLYMIMRECWHAAPSQRPTFKQLVEDLDRVLTVTSTDNVMEQFNPGLRNLINLGKNYEKAVNAMILAGKAYYDGVAKI GEIATGSPVSTELGHVLIEISSTHKKLNESLDENFKKFHKEIIHELEKKIELDVKYMNATLKRYQTEHKNKLESLEKSQAELKKIRRKSQ GSRNALKYEHKEIEYVETVTSRQSEIQKFIADGCKEALLEEKRRFCFLVDKHCGFANHIHYYHLQSAELLNSKLPRWQETCVDAIKVPEK IMNMIEEIKTPASTPVSGTPQASPMIERSNVVRKDYDTLSKCSPKMPPAPSGRAYTSPLIDMFNNPATAAPNSQRVNNSTGTSEDPSLQR SVSVATGLNMMKKQKVKTIFPHTAGSNKTLLSFAQGDVITLLIPEEKDGWLYGEHDVSKARGWFPSSYTKLLEENETEAVTVPTPSPTPV RSISTVNLSENSSVVIPPPDYLECLSMGAAADRRADSARTTSTFKAPASKPETAAPNDANGTAKPPFLSGENPFATVKLRPTVTNDRSAP -------------------------------------------------------------- >30301_30301_2_FGFR3-BAIAP2L1_FGFR3_chr4_1808661_ENST00000340107_BAIAP2L1_chr7_97991744_ENST00000005260_length(amino acids)=1265AA_BP=771 MPEDAAAPAPAMGAPACALALCVAVAIVAGASSESLGTEQRVVGRAAEVPGPEPGQQEQLVFGSGDAVELSCPPPGGGPMGPTVWVKDGT GLVPSERVLVGPQRLQVLNASHEDSGAYSCRQRLTQRVLCHFSVRVTDAPSSGDDEDGEDEAEDTGVDTGAPYWTRPERMDKKLLAVPAA NTVRFRCPAAGNPTPSISWLKNGREFRGEHRIGGIKLRHQQWSLVMESVVPSDRGNYTCVVENKFGSIRQTYTLDVLERSPHRPILQAGL PANQTAVLGSDVEFHCKVYSDAQPHIQWLKHVEVNGSKVGPDGTPYVTVLKSWISESVEADVRLRLANVSERDGGEYLCRATNFIGVAEK AFWLSVHGPRAAEEELVEADEAGSVYAGILSYGVGFFLFILVVAAVTLCRLRSPPKKGLGSPTVHKISRFPLKRQVSLESNASMSSNTPL VRIARLSSGEGPTLANVSELELPADPKWELSRARLTLGKPLGEGCFGQVVMAEAIGIDKDRAAKPVTVAVKMLKDDATDKDLSDLVSEME MMKMIGKHKNIINLLGACTQGGPLYVLVEYAAKGNLREFLRARRPPGLDYSFDTCKPPEEQLTFKDLVSCAYQVARGMEYLASQKCIHRD LAARNVLVTEDNVMKIADFGLARDVHNLDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLLWEIFTLGGSPYPGIPVEELFKLL KEGHRMDKPANCTHDLYMIMRECWHAAPSQRPTFKQLVEDLDRVLTVTSTDNVMEQFNPGLRNLINLGKNYEKAVNAMILAGKAYYDGVA KIGEIATGSPVSTELGHVLIEISSTHKKLNESLDENFKKFHKEIIHELEKKIELDVKYMNATLKRYQTEHKNKLESLEKSQAELKKIRRK SQGSRNALKYEHKEIEYVETVTSRQSEIQKFIADGCKEALLEEKRRFCFLVDKHCGFANHIHYYHLQSAELLNSKLPRWQETCVDAIKVP EKIMNMIEEIKTPASTPVSGTPQASPMIERSNVVRKDYDTLSKCSPKMPPAPSGRAYTSPLIDMFNNPATAAPNSQRVNNSTGTSEDPSL QRSVSVATGLNMMKKQKVKTIFPHTAGSNKTLLSFAQGDVITLLIPEEKDGWLYGEHDVSKARGWFPSSYTKLLEENETEAVTVPTPSPT PVRSISTVNLSENSSVVIPPPDYLECLSMGAAADRRADSARTTSTFKAPASKPETAAPNDANGTAKPPFLSGENPFATVKLRPTVTNDRS -------------------------------------------------------------- >30301_30301_3_FGFR3-BAIAP2L1_FGFR3_chr4_1808661_ENST00000352904_BAIAP2L1_chr7_97991744_ENST00000005260_length(amino acids)=1151AA_BP=657 MPEDAAAPAPAMGAPACALALCVAVAIVAGASSESLGTEQRVVGRAAEVPGPEPGQQEQLVFGSGDAVELSCPPPGGGPMGPTVWVKDGT GLVPSERVLVGPQRLQVLNASHEDSGAYSCRQRLTQRVLCHFSVRVTDAPSSGDDEDGEDEAEDTGVDTGAPYWTRPERMDKKLLAVPAA NTVRFRCPAAGNPTPSISWLKNGREFRGEHRIGGIKLRHQQWSLVMESVVPSDRGNYTCVVENKFGSIRQTYTLDVLERSPHRPILQAGL PANQTAVLGSDVEFHCKVYSDAQPHIQWLKHVEVNGSKVGPDGTPYVTVLKVSLESNASMSSNTPLVRIARLSSGEGPTLANVSELELPA DPKWELSRARLTLGKPLGEGCFGQVVMAEAIGIDKDRAAKPVTVAVKMLKDDATDKDLSDLVSEMEMMKMIGKHKNIINLLGACTQGGPL YVLVEYAAKGNLREFLRARRPPGLDYSFDTCKPPEEQLTFKDLVSCAYQVARGMEYLASQKCIHRDLAARNVLVTEDNVMKIADFGLARD VHNLDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLLWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPANCTHDLYMIMRECW HAAPSQRPTFKQLVEDLDRVLTVTSTDNVMEQFNPGLRNLINLGKNYEKAVNAMILAGKAYYDGVAKIGEIATGSPVSTELGHVLIEISS THKKLNESLDENFKKFHKEIIHELEKKIELDVKYMNATLKRYQTEHKNKLESLEKSQAELKKIRRKSQGSRNALKYEHKEIEYVETVTSR QSEIQKFIADGCKEALLEEKRRFCFLVDKHCGFANHIHYYHLQSAELLNSKLPRWQETCVDAIKVPEKIMNMIEEIKTPASTPVSGTPQA SPMIERSNVVRKDYDTLSKCSPKMPPAPSGRAYTSPLIDMFNNPATAAPNSQRVNNSTGTSEDPSLQRSVSVATGLNMMKKQKVKTIFPH TAGSNKTLLSFAQGDVITLLIPEEKDGWLYGEHDVSKARGWFPSSYTKLLEENETEAVTVPTPSPTPVRSISTVNLSENSSVVIPPPDYL -------------------------------------------------------------- >30301_30301_4_FGFR3-BAIAP2L1_FGFR3_chr4_1808661_ENST00000412135_BAIAP2L1_chr7_97991744_ENST00000005260_length(amino acids)=1151AA_BP=657 MPEDAAAPAPAMGAPACALALCVAVAIVAGASSESLGTEQRVVGRAAEVPGPEPGQQEQLVFGSGDAVELSCPPPGGGPMGPTVWVKDGT GLVPSERVLVGPQRLQVLNASHEDSGAYSCRQRLTQRVLCHFSVRVTDAPSSGDDEDGEDEAEDTGVDTGAPYWTRPERMDKKLLAVPAA NTVRFRCPAAGNPTPSISWLKNGREFRGEHRIGGIKLRHQQWSLVMESVVPSDRGNYTCVVENKFGSIRQTYTLDVLERSPHRPILQAGL PANQTAVLGSDVEFHCKVYSDAQPHIQWLKHVEVNGSKVGPDGTPYVTVLKVSLESNASMSSNTPLVRIARLSSGEGPTLANVSELELPA DPKWELSRARLTLGKPLGEGCFGQVVMAEAIGIDKDRAAKPVTVAVKMLKDDATDKDLSDLVSEMEMMKMIGKHKNIINLLGACTQGGPL YVLVEYAAKGNLREFLRARRPPGLDYSFDTCKPPEEQLTFKDLVSCAYQVARGMEYLASQKCIHRDLAARNVLVTEDNVMKIADFGLARD VHNLDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLLWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPANCTHDLYMIMRECW HAAPSQRPTFKQLVEDLDRVLTVTSTDNVMEQFNPGLRNLINLGKNYEKAVNAMILAGKAYYDGVAKIGEIATGSPVSTELGHVLIEISS THKKLNESLDENFKKFHKEIIHELEKKIELDVKYMNATLKRYQTEHKNKLESLEKSQAELKKIRRKSQGSRNALKYEHKEIEYVETVTSR QSEIQKFIADGCKEALLEEKRRFCFLVDKHCGFANHIHYYHLQSAELLNSKLPRWQETCVDAIKVPEKIMNMIEEIKTPASTPVSGTPQA SPMIERSNVVRKDYDTLSKCSPKMPPAPSGRAYTSPLIDMFNNPATAAPNSQRVNNSTGTSEDPSLQRSVSVATGLNMMKKQKVKTIFPH TAGSNKTLLSFAQGDVITLLIPEEKDGWLYGEHDVSKARGWFPSSYTKLLEENETEAVTVPTPSPTPVRSISTVNLSENSSVVIPPPDYL -------------------------------------------------------------- >30301_30301_5_FGFR3-BAIAP2L1_FGFR3_chr4_1808661_ENST00000440486_BAIAP2L1_chr7_97991744_ENST00000005260_length(amino acids)=1263AA_BP=769 MPEDAAAPAPAMGAPACALALCVAVAIVAGASSESLGTEQRVVGRAAEVPGPEPGQQEQLVFGSGDAVELSCPPPGGGPMGPTVWVKDGT GLVPSERVLVGPQRLQVLNASHEDSGAYSCRQRLTQRVLCHFSVRVTDAPSSGDDEDGEDEAEDTGVDTGAPYWTRPERMDKKLLAVPAA NTVRFRCPAAGNPTPSISWLKNGREFRGEHRIGGIKLRHQQWSLVMESVVPSDRGNYTCVVENKFGSIRQTYTLDVLERSPHRPILQAGL PANQTAVLGSDVEFHCKVYSDAQPHIQWLKHVEVNGSKVGPDGTPYVTVLKTAGANTTDKELEVLSLHNVTFEDAGEYTCLAGNSIGFSH HSAWLVVLPAEEELVEADEAGSVYAGILSYGVGFFLFILVVAAVTLCRLRSPPKKGLGSPTVHKISRFPLKRQVSLESNASMSSNTPLVR IARLSSGEGPTLANVSELELPADPKWELSRARLTLGKPLGEGCFGQVVMAEAIGIDKDRAAKPVTVAVKMLKDDATDKDLSDLVSEMEMM KMIGKHKNIINLLGACTQGGPLYVLVEYAAKGNLREFLRARRPPGLDYSFDTCKPPEEQLTFKDLVSCAYQVARGMEYLASQKCIHRDLA ARNVLVTEDNVMKIADFGLARDVHNLDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLLWEIFTLGGSPYPGIPVEELFKLLKE GHRMDKPANCTHDLYMIMRECWHAAPSQRPTFKQLVEDLDRVLTVTSTDNVMEQFNPGLRNLINLGKNYEKAVNAMILAGKAYYDGVAKI GEIATGSPVSTELGHVLIEISSTHKKLNESLDENFKKFHKEIIHELEKKIELDVKYMNATLKRYQTEHKNKLESLEKSQAELKKIRRKSQ GSRNALKYEHKEIEYVETVTSRQSEIQKFIADGCKEALLEEKRRFCFLVDKHCGFANHIHYYHLQSAELLNSKLPRWQETCVDAIKVPEK IMNMIEEIKTPASTPVSGTPQASPMIERSNVVRKDYDTLSKCSPKMPPAPSGRAYTSPLIDMFNNPATAAPNSQRVNNSTGTSEDPSLQR SVSVATGLNMMKKQKVKTIFPHTAGSNKTLLSFAQGDVITLLIPEEKDGWLYGEHDVSKARGWFPSSYTKLLEENETEAVTVPTPSPTPV RSISTVNLSENSSVVIPPPDYLECLSMGAAADRRADSARTTSTFKAPASKPETAAPNDANGTAKPPFLSGENPFATVKLRPTVTNDRSAP -------------------------------------------------------------- >30301_30301_6_FGFR3-BAIAP2L1_FGFR3_chr4_1808661_ENST00000481110_BAIAP2L1_chr7_97949610_ENST00000005260_length(amino acids)=1186AA_BP=747 MPEDAAAPAPAMGAPACALALCVAVAIVAGASSESLGTEQRVVGRAAEVPGPEPGQQEQLVFGSGDAVELSCPPPGGGPMGPTVWVKDGT GLVPSERVLVGPQRLQVLNASHEDSGAYSCRQRLTQRVLCHFSVRVTDAPSSGDDEDGEDEAEDTGVDTGAPYWTRPERMDKKLLAVPAA NTVRFRCPAAGNPTPSISWLKNGREFRGEHRIGGIKLRHQQWSLVMESVVPSDRGNYTCVVENKFGSIRQTYTLDVLERSPHRPILQAGL PANQTAVLGSDVEFHCKVYSDAQPHIQWLKHVEVNGSKVGPDGTPYVTVLKTAGANTTDKELEVLSLHNVTFEDAGEYTCLAGNSIGFSH HSAWLVVLPAEEELVEADEAGSVYAGILSYGVGFFLFILVVAAVTLCRLRSPPKKGLGSPTVHKISRFPLKRQQVSLESNASMSSNTPLV RIARLSSGEGPTLANVSELELPADPKWELSRARLTLGKPLGEGCFGQVVMAEAIGIDKDRAAKPVTVAVKMLKDDATDKDLSDLVSEMEM MKMIGKHKNIINLLGACTQGGPLYVLVEYAAKGNLREFLRARRPPGLDYSFDTCKPPEEQLTFKDLVSCAYQVARGMEYLASQKCIHRDL AARNVLVTEDNVMKIADFGLARDVHNLDYYKKTTNLVLWGPALGDLHAGGLPVPRHPCGGALQAAEGGPPHGQARQLHTRPVHDHAGVLA CRALPEAHLQAAGGGPGPCPYRDVHRRHVLIEISSTHKKLNESLDENFKKFHKEIIHELEKKIELDVKYMNATLKRYQTEHKNKLESLEK SQAELKKIRRKSQGSRNALKYEHKEIEYVETVTSRQSEIQKFIADGCKEALLEEKRRFCFLVDKHCGFANHIHYYHLQSAELLNSKLPRW QETCVDAIKVPEKIMNMIEEIKTPASTPVSGTPQASPMIERSNVVRKDYDTLSKCSPKMPPAPSGRAYTSPLIDMFNNPATAAPNSQRVN NSTGTSEDPSLQRSVSVATGLNMMKKQKVKTIFPHTAGSNKTLLSFAQGDVITLLIPEEKDGWLYGEHDVSKARGWFPSSYTKLLEENET EAVTVPTPSPTPVRSISTVNLSENSSVVIPPPDYLECLSMGAAADRRADSARTTSTFKAPASKPETAAPNDANGTAKPPFLSGENPFATV -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr4:1808661/chr7:97991744) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| FGFR3 | BAIAP2L1 |
| FUNCTION: Tyrosine-protein kinase that acts as cell-surface receptor for fibroblast growth factors and plays an essential role in the regulation of cell proliferation, differentiation and apoptosis. Plays an essential role in the regulation of chondrocyte differentiation, proliferation and apoptosis, and is required for normal skeleton development. Regulates both osteogenesis and postnatal bone mineralization by osteoblasts. Promotes apoptosis in chondrocytes, but can also promote cancer cell proliferation. Required for normal development of the inner ear. Phosphorylates PLCG1, CBL and FRS2. Ligand binding leads to the activation of several signaling cascades. Activation of PLCG1 leads to the production of the cellular signaling molecules diacylglycerol and inositol 1,4,5-trisphosphate. Phosphorylation of FRS2 triggers recruitment of GRB2, GAB1, PIK3R1 and SOS1, and mediates activation of RAS, MAPK1/ERK2, MAPK3/ERK1 and the MAP kinase signaling pathway, as well as of the AKT1 signaling pathway. Plays a role in the regulation of vitamin D metabolism. Mutations that lead to constitutive kinase activation or impair normal FGFR3 maturation, internalization and degradation lead to aberrant signaling. Over-expressed or constitutively activated FGFR3 promotes activation of PTPN11/SHP2, STAT1, STAT5A and STAT5B. Secreted isoform 3 retains its capacity to bind FGF1 and FGF2 and hence may interfere with FGF signaling. {ECO:0000269|PubMed:10611230, ECO:0000269|PubMed:11294897, ECO:0000269|PubMed:11703096, ECO:0000269|PubMed:14534538, ECO:0000269|PubMed:16410555, ECO:0000269|PubMed:16597617, ECO:0000269|PubMed:17145761, ECO:0000269|PubMed:17311277, ECO:0000269|PubMed:17509076, ECO:0000269|PubMed:17561467, ECO:0000269|PubMed:19088846, ECO:0000269|PubMed:19286672, ECO:0000269|PubMed:8663044}. | FUNCTION: May function as adapter protein. Involved in the formation of clusters of actin bundles. Plays a role in the reorganization of the actin cytoskeleton in response to bacterial infection. {ECO:0000269|PubMed:17430976, ECO:0000269|PubMed:19366662, ECO:0000269|PubMed:22921828}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000260795 | + | 16 | 17 | 151_244 | 758.0 | 807.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000260795 | + | 16 | 17 | 24_126 | 758.0 | 807.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000260795 | + | 16 | 17 | 253_355 | 758.0 | 807.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000260795 | + | 16 | 17 | 472_761 | 758.0 | 807.0 | Domain | Protein kinase |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000340107 | + | 17 | 18 | 151_244 | 760.0 | 809.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000340107 | + | 17 | 18 | 24_126 | 760.0 | 809.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000340107 | + | 17 | 18 | 253_355 | 760.0 | 809.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000340107 | + | 17 | 18 | 472_761 | 760.0 | 809.0 | Domain | Protein kinase |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000352904 | + | 14 | 15 | 151_244 | 646.0 | 695.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000352904 | + | 14 | 15 | 24_126 | 646.0 | 695.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000352904 | + | 14 | 15 | 253_355 | 646.0 | 695.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000412135 | + | 15 | 16 | 151_244 | 646.0 | 695.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000412135 | + | 15 | 16 | 24_126 | 646.0 | 695.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000412135 | + | 15 | 16 | 253_355 | 646.0 | 695.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000440486 | + | 17 | 18 | 151_244 | 758.0 | 807.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000440486 | + | 17 | 18 | 24_126 | 758.0 | 807.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000440486 | + | 17 | 18 | 253_355 | 758.0 | 807.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000440486 | + | 17 | 18 | 472_761 | 758.0 | 807.0 | Domain | Protein kinase |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000260795 | + | 16 | 17 | 478_486 | 758.0 | 807.0 | Nucleotide binding | ATP |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000340107 | + | 17 | 18 | 478_486 | 760.0 | 809.0 | Nucleotide binding | ATP |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000352904 | + | 14 | 15 | 478_486 | 646.0 | 695.0 | Nucleotide binding | ATP |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000412135 | + | 15 | 16 | 478_486 | 646.0 | 695.0 | Nucleotide binding | ATP |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000440486 | + | 17 | 18 | 478_486 | 758.0 | 807.0 | Nucleotide binding | ATP |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000260795 | + | 16 | 17 | 23_375 | 758.0 | 807.0 | Topological domain | Extracellular |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000340107 | + | 17 | 18 | 23_375 | 760.0 | 809.0 | Topological domain | Extracellular |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000352904 | + | 14 | 15 | 23_375 | 646.0 | 695.0 | Topological domain | Extracellular |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000412135 | + | 15 | 16 | 23_375 | 646.0 | 695.0 | Topological domain | Extracellular |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000440486 | + | 17 | 18 | 23_375 | 758.0 | 807.0 | Topological domain | Extracellular |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000260795 | + | 16 | 17 | 376_396 | 758.0 | 807.0 | Transmembrane | Helical |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000340107 | + | 17 | 18 | 376_396 | 760.0 | 809.0 | Transmembrane | Helical |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000352904 | + | 14 | 15 | 376_396 | 646.0 | 695.0 | Transmembrane | Helical |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000412135 | + | 15 | 16 | 376_396 | 646.0 | 695.0 | Transmembrane | Helical |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000440486 | + | 17 | 18 | 376_396 | 758.0 | 807.0 | Transmembrane | Helical |
| Tgene | BAIAP2L1 | chr4:1808661 | chr7:97991744 | ENST00000005260 | 0 | 14 | 115_154 | 17.0 | 512.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | BAIAP2L1 | chr4:1808661 | chr7:97991744 | ENST00000005260 | 0 | 14 | 339_402 | 17.0 | 512.0 | Domain | SH3 | |
| Tgene | BAIAP2L1 | chr4:1808661 | chr7:97991744 | ENST00000005260 | 0 | 14 | 483_511 | 17.0 | 512.0 | Region | Note=Binds F-actin |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000352904 | + | 14 | 15 | 472_761 | 646.0 | 695.0 | Domain | Protein kinase |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000412135 | + | 15 | 16 | 472_761 | 646.0 | 695.0 | Domain | Protein kinase |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000260795 | + | 16 | 17 | 397_806 | 758.0 | 807.0 | Topological domain | Cytoplasmic |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000340107 | + | 17 | 18 | 397_806 | 760.0 | 809.0 | Topological domain | Cytoplasmic |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000352904 | + | 14 | 15 | 397_806 | 646.0 | 695.0 | Topological domain | Cytoplasmic |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000412135 | + | 15 | 16 | 397_806 | 646.0 | 695.0 | Topological domain | Cytoplasmic |
| Hgene | FGFR3 | chr4:1808661 | chr7:97991744 | ENST00000440486 | + | 17 | 18 | 397_806 | 758.0 | 807.0 | Topological domain | Cytoplasmic |
| Tgene | BAIAP2L1 | chr4:1808661 | chr7:97991744 | ENST00000005260 | 0 | 14 | 1_249 | 17.0 | 512.0 | Domain | IMD |
Top |
Fusion Protein Structures |
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
| PDB file (829) >>>829.pdbFusion protein BP residue: 657 CIF file (829) >>>829.cif | FGFR3 | chr4 | 1808661 | + | BAIAP2L1 | chr7 | 97991744 | - | MPEDAAAPAPAMGAPACALALCVAVAIVAGASSESLGTEQRVVGRAAEVP GPEPGQQEQLVFGSGDAVELSCPPPGGGPMGPTVWVKDGTGLVPSERVLV GPQRLQVLNASHEDSGAYSCRQRLTQRVLCHFSVRVTDAPSSGDDEDGED EAEDTGVDTGAPYWTRPERMDKKLLAVPAANTVRFRCPAAGNPTPSISWL KNGREFRGEHRIGGIKLRHQQWSLVMESVVPSDRGNYTCVVENKFGSIRQ TYTLDVLERSPHRPILQAGLPANQTAVLGSDVEFHCKVYSDAQPHIQWLK HVEVNGSKVGPDGTPYVTVLKVSLESNASMSSNTPLVRIARLSSGEGPTL ANVSELELPADPKWELSRARLTLGKPLGEGCFGQVVMAEAIGIDKDRAAK PVTVAVKMLKDDATDKDLSDLVSEMEMMKMIGKHKNIINLLGACTQGGPL YVLVEYAAKGNLREFLRARRPPGLDYSFDTCKPPEEQLTFKDLVSCAYQV ARGMEYLASQKCIHRDLAARNVLVTEDNVMKIADFGLARDVHNLDYYKKT TNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLLWEIFTLGGSPYPGIPVE ELFKLLKEGHRMDKPANCTHDLYMIMRECWHAAPSQRPTFKQLVEDLDRV LTVTSTDNVMEQFNPGLRNLINLGKNYEKAVNAMILAGKAYYDGVAKIGE IATGSPVSTELGHVLIEISSTHKKLNESLDENFKKFHKEIIHELEKKIEL DVKYMNATLKRYQTEHKNKLESLEKSQAELKKIRRKSQGSRNALKYEHKE IEYVETVTSRQSEIQKFIADGCKEALLEEKRRFCFLVDKHCGFANHIHYY HLQSAELLNSKLPRWQETCVDAIKVPEKIMNMIEEIKTPASTPVSGTPQA SPMIERSNVVRKDYDTLSKCSPKMPPAPSGRAYTSPLIDMFNNPATAAPN SQRVNNSTGTSEDPSLQRSVSVATGLNMMKKQKVKTIFPHTAGSNKTLLS FAQGDVITLLIPEEKDGWLYGEHDVSKARGWFPSSYTKLLEENETEAVTV PTPSPTPVRSISTVNLSENSSVVIPPPDYLECLSMGAAADRRADSARTTS TFKAPASKPETAAPNDANGTAKPPFLSGENPFATVKLRPTVTNDRSAPII | 1151 |
| 3D view using mol* of 829 (AA BP:657) | ||||||||||
 | ||||||||||
| PDB file (843) >>>843.pdbFusion protein BP residue: 747 CIF file (843) >>>843.cif | FGFR3 | chr4 | 1808661 | + | BAIAP2L1 | chr7 | 97949610 | - | MPEDAAAPAPAMGAPACALALCVAVAIVAGASSESLGTEQRVVGRAAEVP GPEPGQQEQLVFGSGDAVELSCPPPGGGPMGPTVWVKDGTGLVPSERVLV GPQRLQVLNASHEDSGAYSCRQRLTQRVLCHFSVRVTDAPSSGDDEDGED EAEDTGVDTGAPYWTRPERMDKKLLAVPAANTVRFRCPAAGNPTPSISWL KNGREFRGEHRIGGIKLRHQQWSLVMESVVPSDRGNYTCVVENKFGSIRQ TYTLDVLERSPHRPILQAGLPANQTAVLGSDVEFHCKVYSDAQPHIQWLK HVEVNGSKVGPDGTPYVTVLKTAGANTTDKELEVLSLHNVTFEDAGEYTC LAGNSIGFSHHSAWLVVLPAEEELVEADEAGSVYAGILSYGVGFFLFILV VAAVTLCRLRSPPKKGLGSPTVHKISRFPLKRQQVSLESNASMSSNTPLV RIARLSSGEGPTLANVSELELPADPKWELSRARLTLGKPLGEGCFGQVVM AEAIGIDKDRAAKPVTVAVKMLKDDATDKDLSDLVSEMEMMKMIGKHKNI INLLGACTQGGPLYVLVEYAAKGNLREFLRARRPPGLDYSFDTCKPPEEQ LTFKDLVSCAYQVARGMEYLASQKCIHRDLAARNVLVTEDNVMKIADFGL ARDVHNLDYYKKTTNLVLWGPALGDLHAGGLPVPRHPCGGALQAAEGGPP HGQARQLHTRPVHDHAGVLACRALPEAHLQAAGGGPGPCPYRDVHRRHVL IEISSTHKKLNESLDENFKKFHKEIIHELEKKIELDVKYMNATLKRYQTE HKNKLESLEKSQAELKKIRRKSQGSRNALKYEHKEIEYVETVTSRQSEIQ KFIADGCKEALLEEKRRFCFLVDKHCGFANHIHYYHLQSAELLNSKLPRW QETCVDAIKVPEKIMNMIEEIKTPASTPVSGTPQASPMIERSNVVRKDYD TLSKCSPKMPPAPSGRAYTSPLIDMFNNPATAAPNSQRVNNSTGTSEDPS LQRSVSVATGLNMMKKQKVKTIFPHTAGSNKTLLSFAQGDVITLLIPEEK DGWLYGEHDVSKARGWFPSSYTKLLEENETEAVTVPTPSPTPVRSISTVN LSENSSVVIPPPDYLECLSMGAAADRRADSARTTSTFKAPASKPETAAPN | 1186 |
| 3D view using mol* of 843 (AA BP:747) | ||||||||||
 | ||||||||||
| PDB file (871)CIF file (871) >>>871.cif | FGFR3 | chr4 | 1808661 | + | BAIAP2L1 | chr7 | 97991744 | - | MPEDAAAPAPAMGAPACALALCVAVAIVAGASSESLGTEQRVVGRAAEVP GPEPGQQEQLVFGSGDAVELSCPPPGGGPMGPTVWVKDGTGLVPSERVLV GPQRLQVLNASHEDSGAYSCRQRLTQRVLCHFSVRVTDAPSSGDDEDGED EAEDTGVDTGAPYWTRPERMDKKLLAVPAANTVRFRCPAAGNPTPSISWL KNGREFRGEHRIGGIKLRHQQWSLVMESVVPSDRGNYTCVVENKFGSIRQ TYTLDVLERSPHRPILQAGLPANQTAVLGSDVEFHCKVYSDAQPHIQWLK HVEVNGSKVGPDGTPYVTVLKTAGANTTDKELEVLSLHNVTFEDAGEYTC LAGNSIGFSHHSAWLVVLPAEEELVEADEAGSVYAGILSYGVGFFLFILV VAAVTLCRLRSPPKKGLGSPTVHKISRFPLKRQVSLESNASMSSNTPLVR IARLSSGEGPTLANVSELELPADPKWELSRARLTLGKPLGEGCFGQVVMA EAIGIDKDRAAKPVTVAVKMLKDDATDKDLSDLVSEMEMMKMIGKHKNII NLLGACTQGGPLYVLVEYAAKGNLREFLRARRPPGLDYSFDTCKPPEEQL TFKDLVSCAYQVARGMEYLASQKCIHRDLAARNVLVTEDNVMKIADFGLA RDVHNLDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLLWEIFT LGGSPYPGIPVEELFKLLKEGHRMDKPANCTHDLYMIMRECWHAAPSQRP TFKQLVEDLDRVLTVTSTDNVMEQFNPGLRNLINLGKNYEKAVNAMILAG KAYYDGVAKIGEIATGSPVSTELGHVLIEISSTHKKLNESLDENFKKFHK EIIHELEKKIELDVKYMNATLKRYQTEHKNKLESLEKSQAELKKIRRKSQ GSRNALKYEHKEIEYVETVTSRQSEIQKFIADGCKEALLEEKRRFCFLVD KHCGFANHIHYYHLQSAELLNSKLPRWQETCVDAIKVPEKIMNMIEEIKT PASTPVSGTPQASPMIERSNVVRKDYDTLSKCSPKMPPAPSGRAYTSPLI DMFNNPATAAPNSQRVNNSTGTSEDPSLQRSVSVATGLNMMKKQKVKTIF PHTAGSNKTLLSFAQGDVITLLIPEEKDGWLYGEHDVSKARGWFPSSYTK LLEENETEAVTVPTPSPTPVRSISTVNLSENSSVVIPPPDYLECLSMGAA ADRRADSARTTSTFKAPASKPETAAPNDANGTAKPPFLSGENPFATVKLR | 1263 |
| 3D view using mol* of 871 (AA BP:) | ||||||||||
 | ||||||||||
| PDB file (872)CIF file (872) >>>872.cif | FGFR3 | chr4 | 1808661 | + | BAIAP2L1 | chr7 | 97991744 | - | MPEDAAAPAPAMGAPACALALCVAVAIVAGASSESLGTEQRVVGRAAEVP GPEPGQQEQLVFGSGDAVELSCPPPGGGPMGPTVWVKDGTGLVPSERVLV GPQRLQVLNASHEDSGAYSCRQRLTQRVLCHFSVRVTDAPSSGDDEDGED EAEDTGVDTGAPYWTRPERMDKKLLAVPAANTVRFRCPAAGNPTPSISWL KNGREFRGEHRIGGIKLRHQQWSLVMESVVPSDRGNYTCVVENKFGSIRQ TYTLDVLERSPHRPILQAGLPANQTAVLGSDVEFHCKVYSDAQPHIQWLK HVEVNGSKVGPDGTPYVTVLKSWISESVEADVRLRLANVSERDGGEYLCR ATNFIGVAEKAFWLSVHGPRAAEEELVEADEAGSVYAGILSYGVGFFLFI LVVAAVTLCRLRSPPKKGLGSPTVHKISRFPLKRQVSLESNASMSSNTPL VRIARLSSGEGPTLANVSELELPADPKWELSRARLTLGKPLGEGCFGQVV MAEAIGIDKDRAAKPVTVAVKMLKDDATDKDLSDLVSEMEMMKMIGKHKN IINLLGACTQGGPLYVLVEYAAKGNLREFLRARRPPGLDYSFDTCKPPEE QLTFKDLVSCAYQVARGMEYLASQKCIHRDLAARNVLVTEDNVMKIADFG LARDVHNLDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLLWEI FTLGGSPYPGIPVEELFKLLKEGHRMDKPANCTHDLYMIMRECWHAAPSQ RPTFKQLVEDLDRVLTVTSTDNVMEQFNPGLRNLINLGKNYEKAVNAMIL AGKAYYDGVAKIGEIATGSPVSTELGHVLIEISSTHKKLNESLDENFKKF HKEIIHELEKKIELDVKYMNATLKRYQTEHKNKLESLEKSQAELKKIRRK SQGSRNALKYEHKEIEYVETVTSRQSEIQKFIADGCKEALLEEKRRFCFL VDKHCGFANHIHYYHLQSAELLNSKLPRWQETCVDAIKVPEKIMNMIEEI KTPASTPVSGTPQASPMIERSNVVRKDYDTLSKCSPKMPPAPSGRAYTSP LIDMFNNPATAAPNSQRVNNSTGTSEDPSLQRSVSVATGLNMMKKQKVKT IFPHTAGSNKTLLSFAQGDVITLLIPEEKDGWLYGEHDVSKARGWFPSSY TKLLEENETEAVTVPTPSPTPVRSISTVNLSENSSVVIPPPDYLECLSMG AAADRRADSARTTSTFKAPASKPETAAPNDANGTAKPPFLSGENPFATVK | 1265 |
| 3D view using mol* of 872 (AA BP:) | ||||||||||
 | ||||||||||
Top |
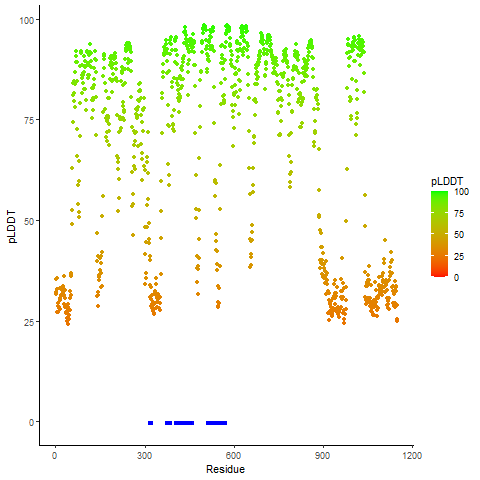
pLDDT score distribution |
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
FGFR3_pLDDT.png |
BAIAP2L1_pLDDT.png |
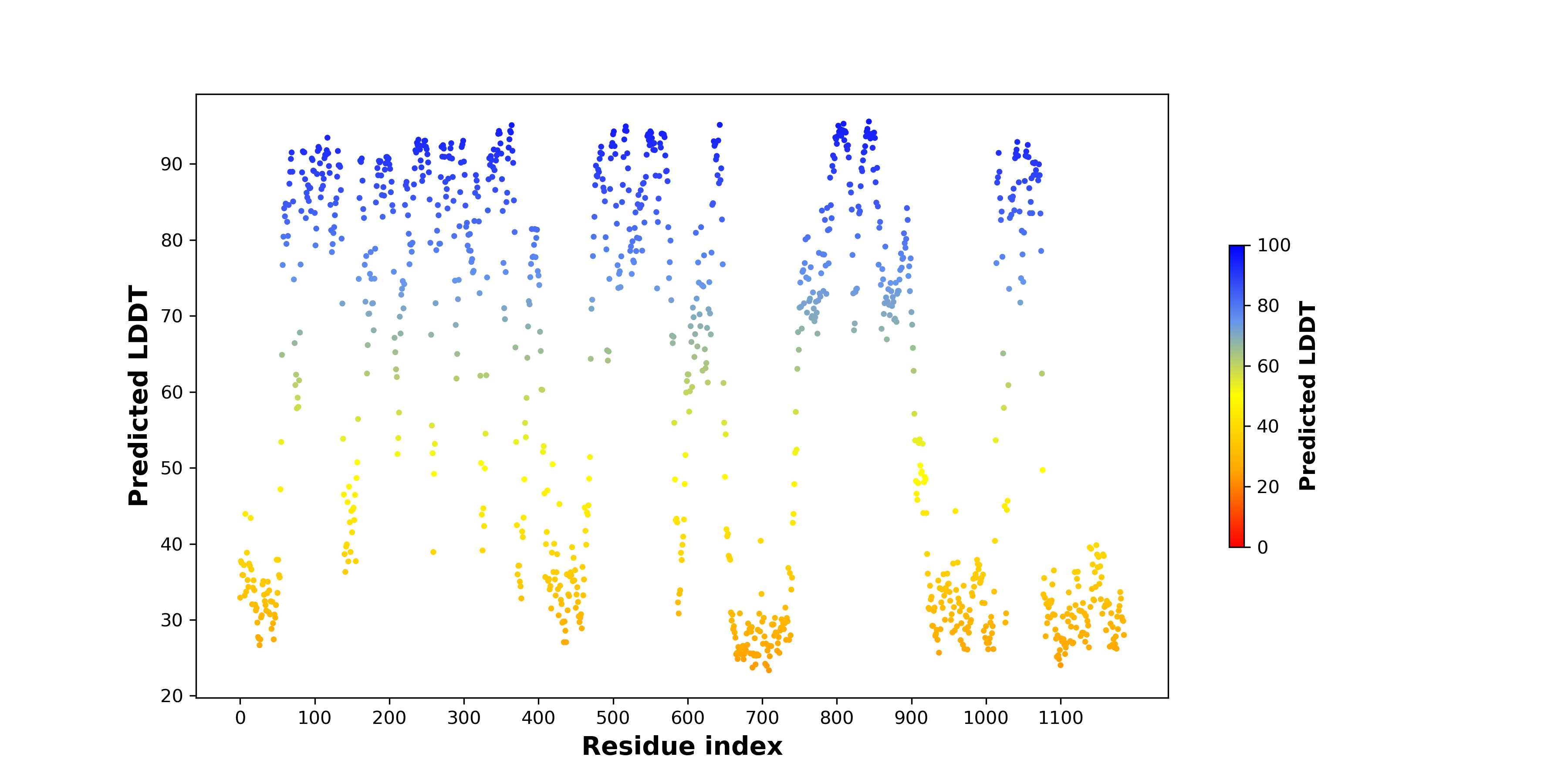
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
Top |
Ramachandran Plot of Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| Fusion AA seq ID in FusionPDB and their Ramachandran plots |
| FGFR3_BAIAP2L1_829.png |
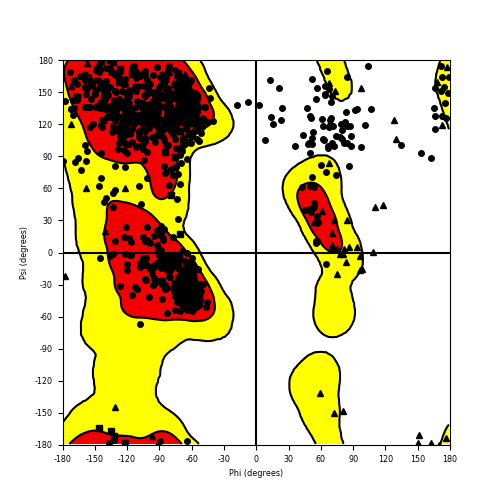 |
| FGFR3_BAIAP2L1_843.png |
 |
Top |
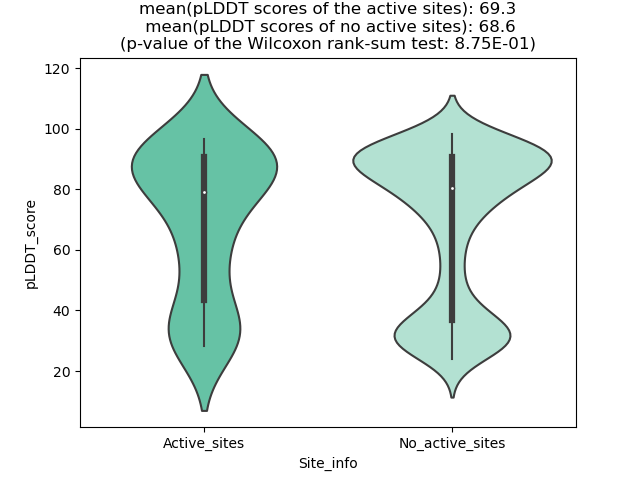
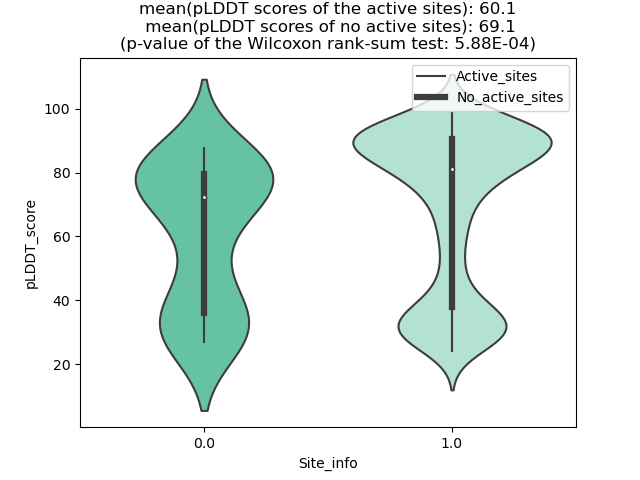
Potential Active Site Information |
 The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. |
| Fusion AA seq ID in FusionPDB | Site score | Size | D score | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
| 829 | 1.0228 | 389 | 1.039 | 1003.275 | 0.482 | 0.7322 | 0.9769 | 1.0116 | 1.044 | 0.969 | 0.8296 | Chain A: 319,320,321,322,323,375,377,378,380,381,3 82,383,384,385,405,407,408,409,416,417,420,421,424 ,428,438,454,455,456,457,460,461,515,520,521,523,5 33,534,535,536,537,539,540,543,544,545,546,547,548 ,549,550,551,552,553,554,555,556,560,564,565,566,5 68,569,570 |
| 843 | 1.0671 | 588 | 1.1334 | 1852.886 | 0.5661 | 0.6976 | 0.8934 | 1.3366 | 0.6904 | 1.9359 | 0.9596 | Chain A: 180,182,214,227,228,575,576,578,579,580,5 82,583,584,589,590,597,599,600,601,602,603,604,605 ,606,607,608,609,610,611,612,613,614,617,621,630,6 31,632,635,641,643,712,713,714,715,716,717,718,719 ,720,721,722,724,725,726,727,728,729,730,731,742,7 46,747,749,750,753,754,757,760,761,764,765,767,768 ,770,771,878,881,882,885,886,887,889,890,893,894,8 97,898,900,901,902,903,904,905,906,907,908,1176,11 77,1178,1179,1180,1181 |
| 871 | 1.071 | 311 | 1.118 | 728.875 | 0.425 | 0.737 | 0.971 | 1.104 | 0.809 | 1.364 | 0.839 | Chain A: 770,771,774,775,776,777,778,779,782,859,8 63,866,867,870,871,874,930,933,934,936,937,938,940 ,941,944,945,947,948,1243,1244,1245,1247,1248,1249 ,1250,1251,1261,1262,1263 |
| 872 | 1.091 | 226 | 1.14 | 604.023 | 0.504 | 0.761 | 0.951 | 1.263 | 0.787 | 1.606 | 1.181 | Chain A: 773,776,777,778,779,780,781,784,858,861,8 62,865,868,869,872,873,876,880,932,935,936,938,939 ,940,942,943,946,947,949,950,1244,1245,1246,1247,1 248,1249,1250,1251,1264,1265 |
Top |
Potentially Interacting Small Molecules through Virtual Screening |
 The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. |
| Fusion AA seq ID in FusionPDB | ZINC ID | DrugBank ID | Drug name | Docking score | Glide gscore |
Top |
 Drug information from DrugBank of the top 20 interacting small molecules. Drug information from DrugBank of the top 20 interacting small molecules. |
| ZINC ID | DrugBank ID | Drug name | Drug type | SMILES | Drug group |
Top |
Biochemical Features of Small Molecules |
 ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) |
| ZINC ID | mol_MW | dipole | SASA | FOSA | FISA | PISA | WPSA | volume | donorHB | accptHB | IP | Human Oral Absorption | Percent Human Oral Absorption | Rule Of Five | Rule Of Three |
Top |
Drug Toxicity Information |
 Toxicity information of individual drugs using eToxPred Toxicity information of individual drugs using eToxPred |
| ZINC ID | Smile | Surface Accessibility | Toxicity |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| FGFR3 | C6orf47, SMG7, BORA, CCDC17, HBZ, HNRNPL, NDUFS6, RNF130, ATF3, ARAP1, CHGB, CTSK, RADIL, GTF3C1, KRT8, POLA2, KIAA1377, RPL8, SLC25A6, Sh2b1, SH2B1, FGF9, FGF1, GRB2, FGF8, HSP90AA1, HSP90AB1, HSPA1B, HSPA8, CDC37, HSPA4, SOCS1, SOCS3, FGFR3, SNX11, TMEM231, ILKAP, SRPK2, PVRL1, BRD4, VHL, FNTA, CDK6, FGFR2, TMEM30B, ARRDC3, SDC2, EPHA4, KAL1, RPS6KA3, fgf3, KLB, CSPG4, HEXIM1, LARP7, MAP3K4, CDC25A, DCXR, ERBB2, BRD2, OXT, PRKAR1A, POMC, CUL4B, ATG5, MKRN2, ABCA2, CTLA4, POTEI, TNFRSF10A, MPDZ, SDC4, CLEC4A, CRLF2, FBXO2, ALG6, TM2D3, KLK1, PI15, FSTL4, PRG2, TMEM87A, PDGFRA, LY86, MFAP4, FHL3, DNASE1L1, CELA2B, FBXO6, SIRPD, CBLN4, KLK3, EDN3, PRTN3, ELSPBP1, LYPD4, GGH, GPIHBP1, IL5RA, DNAJB9, TCTN1, UQCC1, LIPG, FCGR3B, SDF2L1, EPDR1, ADAMTS12, IGLL5, CFC1, C1QB, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| FGFR3 | 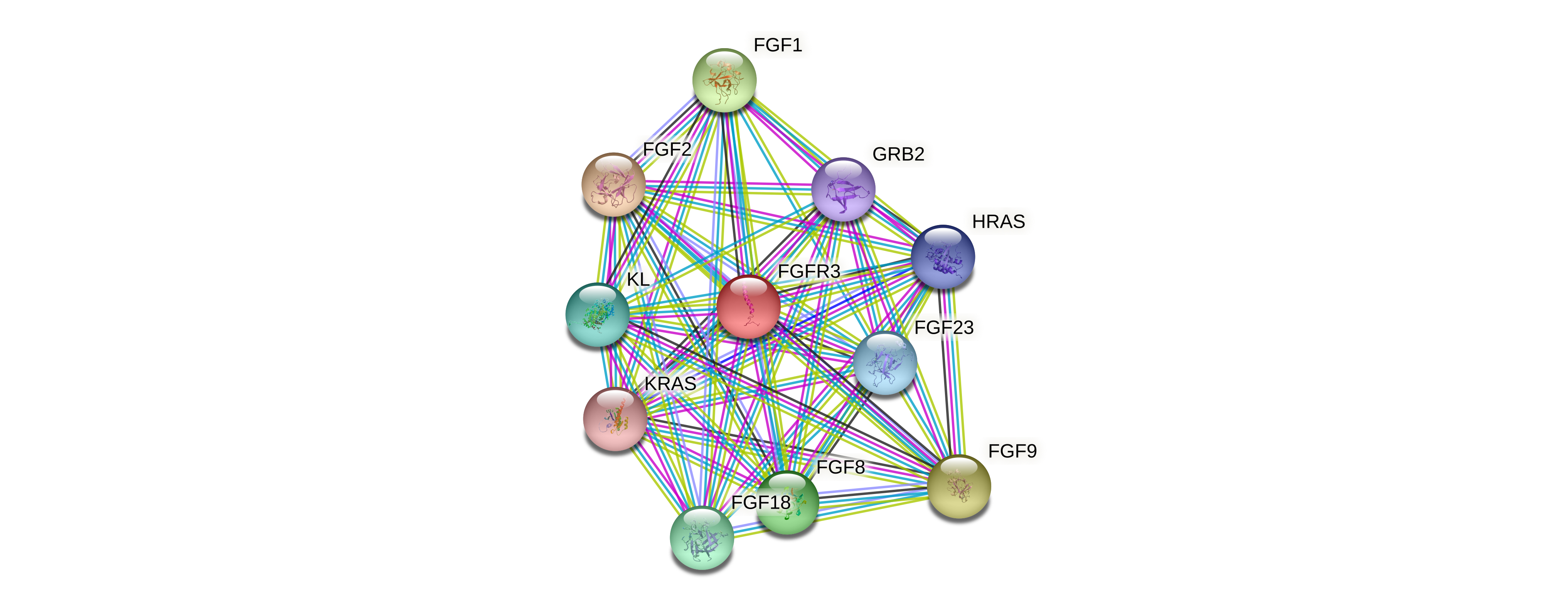 |
| BAIAP2L1 |  |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to FGFR3-BAIAP2L1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to FGFR3-BAIAP2L1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
| FGFR3 | BAIAP2L1 | Adamantinomatous Craniopharyngioma | MyCancerGenome | |
| FGFR3 | BAIAP2L1 | Breast Invasive Ductal Carcinoma | MyCancerGenome | |
| FGFR3 | BAIAP2L1 | Gastric Adenocarcinoma | MyCancerGenome | |
| FGFR3 | BAIAP2L1 | Glioblastoma | MyCancerGenome | |
| FGFR3 | BAIAP2L1 | Uterine Corpus Leiomyosarcoma | MyCancerGenome |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | FGFR3 | C0001080 | Achondroplasia | 13 | CLINGEN;CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Hgene | FGFR3 | C0410529 | Hypochondroplasia (disorder) | 10 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Hgene | FGFR3 | C1864436 | Muenke Syndrome | 9 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Hgene | FGFR3 | C1868678 | THANATOPHORIC DYSPLASIA, TYPE I (disorder) | 9 | CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Hgene | FGFR3 | C2677099 | CROUZON SYNDROME WITH ACANTHOSIS NIGRICANS (disorder) | 7 | CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Hgene | FGFR3 | C0005684 | Malignant neoplasm of urinary bladder | 6 | CGI;CTD_human;UNIPROT |
| Hgene | FGFR3 | C0005695 | Bladder Neoplasm | 4 | CGI;CTD_human |
| Hgene | FGFR3 | C1300257 | Thanatophoric dysplasia, type 2 | 4 | CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Hgene | FGFR3 | C1864852 | CATSHL syndrome | 4 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Hgene | FGFR3 | C2674173 | Achondroplasia, Severe, With Developmental Delay And Acanthosis Nigricans | 3 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Hgene | FGFR3 | C0265269 | Lacrimoauriculodentodigital syndrome | 2 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Hgene | FGFR3 | C0007138 | Carcinoma, Transitional Cell | 1 | CTD_human |
| Hgene | FGFR3 | C0008924 | Cleft upper lip | 1 | CTD_human |
| Hgene | FGFR3 | C0008925 | Cleft Palate | 1 | CTD_human |
| Hgene | FGFR3 | C0014544 | Epilepsy | 1 | GENOMICS_ENGLAND |
| Hgene | FGFR3 | C0014547 | Epilepsies, Partial | 1 | GENOMICS_ENGLAND |
| Hgene | FGFR3 | C0022603 | Seborrheic keratosis | 1 | UNIPROT |
| Hgene | FGFR3 | C0026764 | Multiple Myeloma | 1 | CGI;CTD_human |
| Hgene | FGFR3 | C0036631 | Seminoma | 1 | CTD_human |
| Hgene | FGFR3 | C0039743 | Thanatophoric Dysplasia | 1 | CTD_human |
| Hgene | FGFR3 | C0152423 | Congenital small ears | 1 | GENOMICS_ENGLAND |
| Hgene | FGFR3 | C0206726 | gliosarcoma | 1 | ORPHANET |
| Hgene | FGFR3 | C0221356 | Brachycephaly | 1 | ORPHANET |
| Hgene | FGFR3 | C0265529 | Plagiocephaly | 1 | ORPHANET |
| Hgene | FGFR3 | C0334082 | NEVUS, EPIDERMAL (disorder) | 1 | CTD_human;UNIPROT |
| Hgene | FGFR3 | C0334588 | Giant Cell Glioblastoma | 1 | ORPHANET |
| Hgene | FGFR3 | C0406803 | Syringocystadenoma Papilliferum | 1 | GENOMICS_ENGLAND |
| Hgene | FGFR3 | C1336708 | Testicular Germ Cell Tumor | 1 | CTD_human;UNIPROT |
| Hgene | FGFR3 | C1837218 | Cleft palate, isolated | 1 | CTD_human |
| Hgene | FGFR3 | C4048328 | cervical cancer | 1 | CTD_human;UNIPROT |
| Tgene | BAIAP2L1 | C0003873 | Rheumatoid Arthritis | 1 | CTD_human |