| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:HHAT-KIF5B |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: HHAT-KIF5B | FusionPDB ID: 36266 | FusionGDB2.0 ID: 36266 | Hgene | Tgene | Gene symbol | HHAT | KIF5B | Gene ID | 55733 | 3799 |
| Gene name | hedgehog acyltransferase | kinesin family member 5B | |
| Synonyms | MART2|SKI1|Skn | HEL-S-61|KINH|KNS|KNS1|UKHC | |
| Cytomap | 1q32.2 | 10p11.22 | |
| Type of gene | protein-coding | protein-coding | |
| Description | protein-cysteine N-palmitoyltransferase HHATmelanoma antigen recognized by T-cells 2skinny hedgehog protein 1 | kinesin-1 heavy chainconventional kinesin heavy chainepididymis secretory protein Li 61kinesin 1 (110-120kD)kinesin heavy chainubiquitous kinesin heavy chain | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q9HCP6 | P33176 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000261458, ENST00000308852, ENST00000367010, ENST00000391905, ENST00000413764, ENST00000537898, ENST00000541565, ENST00000545154, ENST00000545781, ENST00000367009, | ENST00000493889, ENST00000302418, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 8 X 8 X 3=192 | 11 X 12 X 5=660 |
| # samples | 8 | 12 | |
| ** MAII score | log2(8/192*10)=-1.26303440583379 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(12/660*10)=-2.4594316186373 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: HHAT [Title/Abstract] AND KIF5B [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | KIF5B(32337392)-HHAT(210637849), # samples:3 HHAT(210591669)-KIF5B(32329385), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | HHAT-KIF5B seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. HHAT-KIF5B seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. HHAT-KIF5B seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. HHAT-KIF5B seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. KIF5B-HHAT seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | KIF5B | GO:0042391 | regulation of membrane potential | 19675065 |
| Tgene | KIF5B | GO:0043268 | positive regulation of potassium ion transport | 19675065 |
| Tgene | KIF5B | GO:0047496 | vesicle transport along microtubule | 28426968 |
| Tgene | KIF5B | GO:1903078 | positive regulation of protein localization to plasma membrane | 19675065 |
 Fusion gene breakpoints across HHAT (5'-gene) Fusion gene breakpoints across HHAT (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
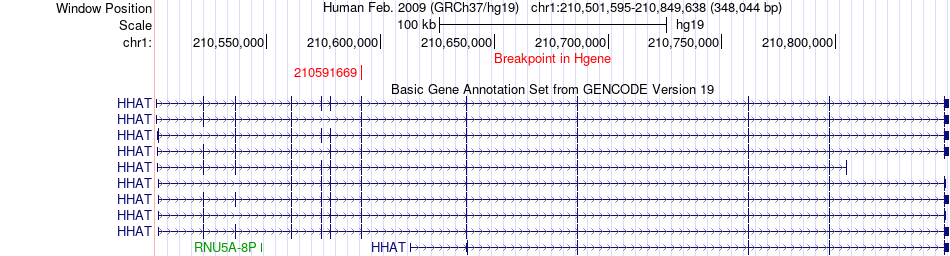 |
 Fusion gene breakpoints across KIF5B (3'-gene) Fusion gene breakpoints across KIF5B (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
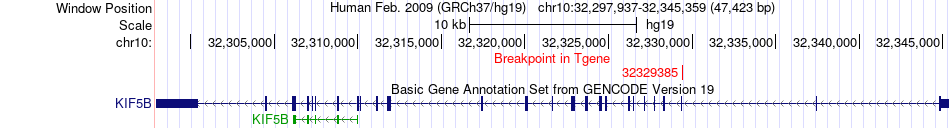 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | PRAD | TCGA-VN-A88L-01A | HHAT | chr1 | 210591669 | - | KIF5B | chr10 | 32329385 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000541565 | HHAT | chr1 | 210591669 | - | ENST00000302418 | KIF5B | chr10 | 32329385 | - | 5826 | 621 | 176 | 3298 | 1040 |
| ENST00000413764 | HHAT | chr1 | 210591669 | - | ENST00000302418 | KIF5B | chr10 | 32329385 | - | 6237 | 1032 | 176 | 3709 | 1177 |
| ENST00000391905 | HHAT | chr1 | 210591669 | - | ENST00000302418 | KIF5B | chr10 | 32329385 | - | 6303 | 1098 | 167 | 3775 | 1202 |
| ENST00000537898 | HHAT | chr1 | 210591669 | - | ENST00000302418 | KIF5B | chr10 | 32329385 | - | 6108 | 903 | 167 | 3580 | 1137 |
| ENST00000545154 | HHAT | chr1 | 210591669 | - | ENST00000302418 | KIF5B | chr10 | 32329385 | - | 6169 | 964 | 105 | 3641 | 1178 |
| ENST00000545781 | HHAT | chr1 | 210591669 | - | ENST00000302418 | KIF5B | chr10 | 32329385 | - | 6087 | 882 | 23 | 3559 | 1178 |
| ENST00000308852 | HHAT | chr1 | 210591669 | - | ENST00000302418 | KIF5B | chr10 | 32329385 | - | 6188 | 983 | 262 | 3660 | 1132 |
| ENST00000261458 | HHAT | chr1 | 210591669 | - | ENST00000302418 | KIF5B | chr10 | 32329385 | - | 6256 | 1051 | 120 | 3728 | 1202 |
| ENST00000367010 | HHAT | chr1 | 210591669 | - | ENST00000302418 | KIF5B | chr10 | 32329385 | - | 6288 | 1083 | 74 | 3760 | 1228 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000541565 | ENST00000302418 | HHAT | chr1 | 210591669 | - | KIF5B | chr10 | 32329385 | - | 0.000204426 | 0.9997956 |
| ENST00000413764 | ENST00000302418 | HHAT | chr1 | 210591669 | - | KIF5B | chr10 | 32329385 | - | 0.000162574 | 0.9998374 |
| ENST00000391905 | ENST00000302418 | HHAT | chr1 | 210591669 | - | KIF5B | chr10 | 32329385 | - | 0.000166545 | 0.99983346 |
| ENST00000537898 | ENST00000302418 | HHAT | chr1 | 210591669 | - | KIF5B | chr10 | 32329385 | - | 0.000174086 | 0.9998259 |
| ENST00000545154 | ENST00000302418 | HHAT | chr1 | 210591669 | - | KIF5B | chr10 | 32329385 | - | 0.000131846 | 0.99986815 |
| ENST00000545781 | ENST00000302418 | HHAT | chr1 | 210591669 | - | KIF5B | chr10 | 32329385 | - | 0.000147436 | 0.99985254 |
| ENST00000308852 | ENST00000302418 | HHAT | chr1 | 210591669 | - | KIF5B | chr10 | 32329385 | - | 0.000164287 | 0.9998357 |
| ENST00000261458 | ENST00000302418 | HHAT | chr1 | 210591669 | - | KIF5B | chr10 | 32329385 | - | 0.000156336 | 0.9998436 |
| ENST00000367010 | ENST00000302418 | HHAT | chr1 | 210591669 | - | KIF5B | chr10 | 32329385 | - | 0.000172395 | 0.9998276 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >36266_36266_1_HHAT-KIF5B_HHAT_chr1_210591669_ENST00000261458_KIF5B_chr10_32329385_ENST00000302418_length(amino acids)=1202AA_BP=310 MPSPPPGTARRNSQRRHREPSCQGAMLPRWELALYLLASLGFHFYSFYEVYKVSREHEEELDQEFELETDTLFGGLKKDATDFEWSFWME WGKQWLVWLLLGHMVVSQMATLLARKHRPWILMLYGMWACWCVLGTPGVAMVLLHTTISFCVAQFRSQLLTWLCSLLLLSTLRLQGVEEV KRRWYKTENEYYLLQFTLTVRCLYYTSFSLELCWQQLPAASTSYSFPWMLAYVFYYPVLHNGPILSFSEFIKQMQQQEHDSLKASLCVLA LGLGRLLCWWWLAELMAHLMYMHAIYSSIPLLETVSCWTLDVLEGYNGTIFAYGQTSSGKTHTMEGKLHDPEGMGIIPRIVQDIFNYIYS MDENLEFHIKVSYFEIYLDKIRDLLDVSKTNLSVHEDKNRVPYVKGCTERFVCSPDEVMDTIDEGKSNRHVAVTNMNEHSSRSHSIFLIN VKQENTQTEQKLSGKLYLVDLAGSEKVSKTGAEGAVLDEAKNINKSLSALGNVISALAEGSTYVPYRDSKMTRILQDSLGGNCRTTIVIC CSPSSYNESETKSTLLFGQRAKTIKNTVCVNVELTAEQWKKKYEKEKEKNKILRNTIQWLENELNRWRNGETVPIDEQFDKEKANLEAFT VDKDITLTNDKPATAIGVIGNFTDAERRKCEEEIAKLYKQLDDKDEEINQQSQLVEKLKTQMLDQEELLASTRRDQDNMQAELNRLQAEN DASKEEVKEVLQALEELAVNYDQKSQEVEDKTKEYELLSDELNQKSATLASIDAELQKLKEMTNHQKKRAAEMMASLLKDLAEIGIAVGN NDVKQPEGTGMIDEEFTVARLYISKMKSEVKTMVKRCKQLESTQTESNKKMEENEKELAACQLRISQHEAKIKSLTEYLQNVEQKKRQLE ESVDALSEELVQLRAQEKVHEMEKEHLNKVQTANEVKQAVEQQIQSHRETHQKQISSLRDEVEAKAKLITDLQDQNQKMMLEQERLRVEH EKLKATDQEKSRKLHELTVMQDRREQARQDLKGLEETVAKELQTLHNLRKLFVQDLATRVKKSAEIDSDDTGGSAAQKQKISFLENNLEQ LTKVHKQLVRDNADLRCELPKLEKRLRATAERVKALESALKEAKENASRDRKRYQQEVDRIKEAVRSKNMARRGHSAQIAKPIRPGQHPA -------------------------------------------------------------- >36266_36266_2_HHAT-KIF5B_HHAT_chr1_210591669_ENST00000308852_KIF5B_chr10_32329385_ENST00000302418_length(amino acids)=1132AA_BP=240 MKFTKSPEDATDFEWSFWMEWGKQWLVWLLLGHMVVSQMATLLARKHRPWILMLYGMWACWCVLGTPGVAMVLLHTTISFCVAQFRSQLL TWLCSLLLLSTLRLQGVEEVKRRWYKTENEYYLLQFTLTVRCLYYTSFSLELCWQQLPAASTSYSFPWMLAYVFYYPVLHNGPILSFSEF IKQMQQQEHDSLKASLCVLALGLGRLLCWWWLAELMAHLMYMHAIYSSIPLLETVSCWTLDVLEGYNGTIFAYGQTSSGKTHTMEGKLHD PEGMGIIPRIVQDIFNYIYSMDENLEFHIKVSYFEIYLDKIRDLLDVSKTNLSVHEDKNRVPYVKGCTERFVCSPDEVMDTIDEGKSNRH VAVTNMNEHSSRSHSIFLINVKQENTQTEQKLSGKLYLVDLAGSEKVSKTGAEGAVLDEAKNINKSLSALGNVISALAEGSTYVPYRDSK MTRILQDSLGGNCRTTIVICCSPSSYNESETKSTLLFGQRAKTIKNTVCVNVELTAEQWKKKYEKEKEKNKILRNTIQWLENELNRWRNG ETVPIDEQFDKEKANLEAFTVDKDITLTNDKPATAIGVIGNFTDAERRKCEEEIAKLYKQLDDKDEEINQQSQLVEKLKTQMLDQEELLA STRRDQDNMQAELNRLQAENDASKEEVKEVLQALEELAVNYDQKSQEVEDKTKEYELLSDELNQKSATLASIDAELQKLKEMTNHQKKRA AEMMASLLKDLAEIGIAVGNNDVKQPEGTGMIDEEFTVARLYISKMKSEVKTMVKRCKQLESTQTESNKKMEENEKELAACQLRISQHEA KIKSLTEYLQNVEQKKRQLEESVDALSEELVQLRAQEKVHEMEKEHLNKVQTANEVKQAVEQQIQSHRETHQKQISSLRDEVEAKAKLIT DLQDQNQKMMLEQERLRVEHEKLKATDQEKSRKLHELTVMQDRREQARQDLKGLEETVAKELQTLHNLRKLFVQDLATRVKKSAEIDSDD TGGSAAQKQKISFLENNLEQLTKVHKQLVRDNADLRCELPKLEKRLRATAERVKALESALKEAKENASRDRKRYQQEVDRIKEAVRSKNM -------------------------------------------------------------- >36266_36266_3_HHAT-KIF5B_HHAT_chr1_210591669_ENST00000367010_KIF5B_chr10_32329385_ENST00000302418_length(amino acids)=1228AA_BP=336 MASSPASGTAAGSCELWSSWQYRRCACLPAVTPGTLKNSQRRHREPSCQGAMLPRWELALYLLASLGFHFYSFYEVYKVSREHEEELDQE FELETDTLFGGLKKDATDFEWSFWMEWGKQWLVWLLLGHMVVSQMATLLARKHRPWILMLYGMWACWCVLGTPGVAMVLLHTTISFCVAQ FRSQLLTWLCSLLLLSTLRLQGVEEVKRRWYKTENEYYLLQFTLTVRCLYYTSFSLELCWQQLPAASTSYSFPWMLAYVFYYPVLHNGPI LSFSEFIKQMQQQEHDSLKASLCVLALGLGRLLCWWWLAELMAHLMYMHAIYSSIPLLETVSCWTLDVLEGYNGTIFAYGQTSSGKTHTM EGKLHDPEGMGIIPRIVQDIFNYIYSMDENLEFHIKVSYFEIYLDKIRDLLDVSKTNLSVHEDKNRVPYVKGCTERFVCSPDEVMDTIDE GKSNRHVAVTNMNEHSSRSHSIFLINVKQENTQTEQKLSGKLYLVDLAGSEKVSKTGAEGAVLDEAKNINKSLSALGNVISALAEGSTYV PYRDSKMTRILQDSLGGNCRTTIVICCSPSSYNESETKSTLLFGQRAKTIKNTVCVNVELTAEQWKKKYEKEKEKNKILRNTIQWLENEL NRWRNGETVPIDEQFDKEKANLEAFTVDKDITLTNDKPATAIGVIGNFTDAERRKCEEEIAKLYKQLDDKDEEINQQSQLVEKLKTQMLD QEELLASTRRDQDNMQAELNRLQAENDASKEEVKEVLQALEELAVNYDQKSQEVEDKTKEYELLSDELNQKSATLASIDAELQKLKEMTN HQKKRAAEMMASLLKDLAEIGIAVGNNDVKQPEGTGMIDEEFTVARLYISKMKSEVKTMVKRCKQLESTQTESNKKMEENEKELAACQLR ISQHEAKIKSLTEYLQNVEQKKRQLEESVDALSEELVQLRAQEKVHEMEKEHLNKVQTANEVKQAVEQQIQSHRETHQKQISSLRDEVEA KAKLITDLQDQNQKMMLEQERLRVEHEKLKATDQEKSRKLHELTVMQDRREQARQDLKGLEETVAKELQTLHNLRKLFVQDLATRVKKSA EIDSDDTGGSAAQKQKISFLENNLEQLTKVHKQLVRDNADLRCELPKLEKRLRATAERVKALESALKEAKENASRDRKRYQQEVDRIKEA -------------------------------------------------------------- >36266_36266_4_HHAT-KIF5B_HHAT_chr1_210591669_ENST00000391905_KIF5B_chr10_32329385_ENST00000302418_length(amino acids)=1202AA_BP=310 MPSPPPGTARRNSQRRHREPSCQGAMLPRWELALYLLASLGFHFYSFYEVYKVSREHEEELDQEFELETDTLFGGLKKDATDFEWSFWME WGKQWLVWLLLGHMVVSQMATLLARKHRPWILMLYGMWACWCVLGTPGVAMVLLHTTISFCVAQFRSQLLTWLCSLLLLSTLRLQGVEEV KRRWYKTENEYYLLQFTLTVRCLYYTSFSLELCWQQLPAASTSYSFPWMLAYVFYYPVLHNGPILSFSEFIKQMQQQEHDSLKASLCVLA LGLGRLLCWWWLAELMAHLMYMHAIYSSIPLLETVSCWTLDVLEGYNGTIFAYGQTSSGKTHTMEGKLHDPEGMGIIPRIVQDIFNYIYS MDENLEFHIKVSYFEIYLDKIRDLLDVSKTNLSVHEDKNRVPYVKGCTERFVCSPDEVMDTIDEGKSNRHVAVTNMNEHSSRSHSIFLIN VKQENTQTEQKLSGKLYLVDLAGSEKVSKTGAEGAVLDEAKNINKSLSALGNVISALAEGSTYVPYRDSKMTRILQDSLGGNCRTTIVIC CSPSSYNESETKSTLLFGQRAKTIKNTVCVNVELTAEQWKKKYEKEKEKNKILRNTIQWLENELNRWRNGETVPIDEQFDKEKANLEAFT VDKDITLTNDKPATAIGVIGNFTDAERRKCEEEIAKLYKQLDDKDEEINQQSQLVEKLKTQMLDQEELLASTRRDQDNMQAELNRLQAEN DASKEEVKEVLQALEELAVNYDQKSQEVEDKTKEYELLSDELNQKSATLASIDAELQKLKEMTNHQKKRAAEMMASLLKDLAEIGIAVGN NDVKQPEGTGMIDEEFTVARLYISKMKSEVKTMVKRCKQLESTQTESNKKMEENEKELAACQLRISQHEAKIKSLTEYLQNVEQKKRQLE ESVDALSEELVQLRAQEKVHEMEKEHLNKVQTANEVKQAVEQQIQSHRETHQKQISSLRDEVEAKAKLITDLQDQNQKMMLEQERLRVEH EKLKATDQEKSRKLHELTVMQDRREQARQDLKGLEETVAKELQTLHNLRKLFVQDLATRVKKSAEIDSDDTGGSAAQKQKISFLENNLEQ LTKVHKQLVRDNADLRCELPKLEKRLRATAERVKALESALKEAKENASRDRKRYQQEVDRIKEAVRSKNMARRGHSAQIAKPIRPGQHPA -------------------------------------------------------------- >36266_36266_5_HHAT-KIF5B_HHAT_chr1_210591669_ENST00000413764_KIF5B_chr10_32329385_ENST00000302418_length(amino acids)=1177AA_BP=285 MLPRWELALYLLASLGFHFYSFYEVYKVSREHEEELDQEFELETDTLFGGLKKDATDFEWSFWMEWGKQWLVWLLLGHMVVSQMATLLAR KHRPWILMLYGMWACWCVLGTPGVAMVLLHTTISFCVAQFRSQLLTWLCSLLLLSTLRLQGVEEVKRRWYKTENEYYLLQFTLTVRCLYY TSFSLELCWQQLPAASTSYSFPWMLAYVFYYPVLHNGPILSFSEFIKQMQQQEHDSLKASLCVLALGLGRLLCWWWLAELMAHLMYMHAI YSSIPLLETVSCWTLDVLEGYNGTIFAYGQTSSGKTHTMEGKLHDPEGMGIIPRIVQDIFNYIYSMDENLEFHIKVSYFEIYLDKIRDLL DVSKTNLSVHEDKNRVPYVKGCTERFVCSPDEVMDTIDEGKSNRHVAVTNMNEHSSRSHSIFLINVKQENTQTEQKLSGKLYLVDLAGSE KVSKTGAEGAVLDEAKNINKSLSALGNVISALAEGSTYVPYRDSKMTRILQDSLGGNCRTTIVICCSPSSYNESETKSTLLFGQRAKTIK NTVCVNVELTAEQWKKKYEKEKEKNKILRNTIQWLENELNRWRNGETVPIDEQFDKEKANLEAFTVDKDITLTNDKPATAIGVIGNFTDA ERRKCEEEIAKLYKQLDDKDEEINQQSQLVEKLKTQMLDQEELLASTRRDQDNMQAELNRLQAENDASKEEVKEVLQALEELAVNYDQKS QEVEDKTKEYELLSDELNQKSATLASIDAELQKLKEMTNHQKKRAAEMMASLLKDLAEIGIAVGNNDVKQPEGTGMIDEEFTVARLYISK MKSEVKTMVKRCKQLESTQTESNKKMEENEKELAACQLRISQHEAKIKSLTEYLQNVEQKKRQLEESVDALSEELVQLRAQEKVHEMEKE HLNKVQTANEVKQAVEQQIQSHRETHQKQISSLRDEVEAKAKLITDLQDQNQKMMLEQERLRVEHEKLKATDQEKSRKLHELTVMQDRRE QARQDLKGLEETVAKELQTLHNLRKLFVQDLATRVKKSAEIDSDDTGGSAAQKQKISFLENNLEQLTKVHKQLVRDNADLRCELPKLEKR LRATAERVKALESALKEAKENASRDRKRYQQEVDRIKEAVRSKNMARRGHSAQIAKPIRPGQHPAASPTHPSAIRGGGAFVQNSQPVAVR -------------------------------------------------------------- >36266_36266_6_HHAT-KIF5B_HHAT_chr1_210591669_ENST00000537898_KIF5B_chr10_32329385_ENST00000302418_length(amino acids)=1137AA_BP=245 MPSPPPGTARRNSQRRHREPSCQGAMLPRWELALYLLASLGFHFYSFYEVYKVSREHEEELDQEFELETDTLFGGLKKDATDFEWSFWME WGKQWLVWLLLGHMVVSQMATLLARKRRWYKTENEYYLLQFTLTVRCLYYTSFSLELCWQQLPAASTSYSFPWMLAYVFYYPVLHNGPIL SFSEFIKQMQQQEHDSLKASLCVLALGLGRLLCWWWLAELMAHLMYMHAIYSSIPLLETVSCWTLDVLEGYNGTIFAYGQTSSGKTHTME GKLHDPEGMGIIPRIVQDIFNYIYSMDENLEFHIKVSYFEIYLDKIRDLLDVSKTNLSVHEDKNRVPYVKGCTERFVCSPDEVMDTIDEG KSNRHVAVTNMNEHSSRSHSIFLINVKQENTQTEQKLSGKLYLVDLAGSEKVSKTGAEGAVLDEAKNINKSLSALGNVISALAEGSTYVP YRDSKMTRILQDSLGGNCRTTIVICCSPSSYNESETKSTLLFGQRAKTIKNTVCVNVELTAEQWKKKYEKEKEKNKILRNTIQWLENELN RWRNGETVPIDEQFDKEKANLEAFTVDKDITLTNDKPATAIGVIGNFTDAERRKCEEEIAKLYKQLDDKDEEINQQSQLVEKLKTQMLDQ EELLASTRRDQDNMQAELNRLQAENDASKEEVKEVLQALEELAVNYDQKSQEVEDKTKEYELLSDELNQKSATLASIDAELQKLKEMTNH QKKRAAEMMASLLKDLAEIGIAVGNNDVKQPEGTGMIDEEFTVARLYISKMKSEVKTMVKRCKQLESTQTESNKKMEENEKELAACQLRI SQHEAKIKSLTEYLQNVEQKKRQLEESVDALSEELVQLRAQEKVHEMEKEHLNKVQTANEVKQAVEQQIQSHRETHQKQISSLRDEVEAK AKLITDLQDQNQKMMLEQERLRVEHEKLKATDQEKSRKLHELTVMQDRREQARQDLKGLEETVAKELQTLHNLRKLFVQDLATRVKKSAE IDSDDTGGSAAQKQKISFLENNLEQLTKVHKQLVRDNADLRCELPKLEKRLRATAERVKALESALKEAKENASRDRKRYQQEVDRIKEAV -------------------------------------------------------------- >36266_36266_7_HHAT-KIF5B_HHAT_chr1_210591669_ENST00000541565_KIF5B_chr10_32329385_ENST00000302418_length(amino acids)=1040AA_BP=148 MLPRWELALYLLASLGFHFYSFYEVYKVSREHEEELDQEFELETDTLFGGLKKDATDFEWSFWMEWGKQWLVWLLLGHMVVSQMATLLAR KMQQQEHDSLKASLCVLALGLGRLLCWWWLAELMAHLMYMHAIYSSIPLLETVSCWTLDVLEGYNGTIFAYGQTSSGKTHTMEGKLHDPE GMGIIPRIVQDIFNYIYSMDENLEFHIKVSYFEIYLDKIRDLLDVSKTNLSVHEDKNRVPYVKGCTERFVCSPDEVMDTIDEGKSNRHVA VTNMNEHSSRSHSIFLINVKQENTQTEQKLSGKLYLVDLAGSEKVSKTGAEGAVLDEAKNINKSLSALGNVISALAEGSTYVPYRDSKMT RILQDSLGGNCRTTIVICCSPSSYNESETKSTLLFGQRAKTIKNTVCVNVELTAEQWKKKYEKEKEKNKILRNTIQWLENELNRWRNGET VPIDEQFDKEKANLEAFTVDKDITLTNDKPATAIGVIGNFTDAERRKCEEEIAKLYKQLDDKDEEINQQSQLVEKLKTQMLDQEELLAST RRDQDNMQAELNRLQAENDASKEEVKEVLQALEELAVNYDQKSQEVEDKTKEYELLSDELNQKSATLASIDAELQKLKEMTNHQKKRAAE MMASLLKDLAEIGIAVGNNDVKQPEGTGMIDEEFTVARLYISKMKSEVKTMVKRCKQLESTQTESNKKMEENEKELAACQLRISQHEAKI KSLTEYLQNVEQKKRQLEESVDALSEELVQLRAQEKVHEMEKEHLNKVQTANEVKQAVEQQIQSHRETHQKQISSLRDEVEAKAKLITDL QDQNQKMMLEQERLRVEHEKLKATDQEKSRKLHELTVMQDRREQARQDLKGLEETVAKELQTLHNLRKLFVQDLATRVKKSAEIDSDDTG GSAAQKQKISFLENNLEQLTKVHKQLVRDNADLRCELPKLEKRLRATAERVKALESALKEAKENASRDRKRYQQEVDRIKEAVRSKNMAR -------------------------------------------------------------- >36266_36266_8_HHAT-KIF5B_HHAT_chr1_210591669_ENST00000545154_KIF5B_chr10_32329385_ENST00000302418_length(amino acids)=1178AA_BP=286 MSLGLGSAERGVLGTRGARERCRRRRPGQPGEHEEELDQEFELETDTLFGGLKKDATDFEWSFWMEWGKQWLVWLLLGHMVVSQMATLLA RKHRPWILMLYGMWACWCVLGTPGVAMVLLHTTISFCVAQFRSQLLTWLCSLLLLSTLRLQGVEEVKRRWYKTENEYYLLQFTLTVRCLY YTSFSLELCWQQLPAASTSYSFPWMLAYVFYYPVLHNGPILSFSEFIKQMQQQEHDSLKASLCVLALGLGRLLCWWWLAELMAHLMYMHA IYSSIPLLETVSCWTLDVLEGYNGTIFAYGQTSSGKTHTMEGKLHDPEGMGIIPRIVQDIFNYIYSMDENLEFHIKVSYFEIYLDKIRDL LDVSKTNLSVHEDKNRVPYVKGCTERFVCSPDEVMDTIDEGKSNRHVAVTNMNEHSSRSHSIFLINVKQENTQTEQKLSGKLYLVDLAGS EKVSKTGAEGAVLDEAKNINKSLSALGNVISALAEGSTYVPYRDSKMTRILQDSLGGNCRTTIVICCSPSSYNESETKSTLLFGQRAKTI KNTVCVNVELTAEQWKKKYEKEKEKNKILRNTIQWLENELNRWRNGETVPIDEQFDKEKANLEAFTVDKDITLTNDKPATAIGVIGNFTD AERRKCEEEIAKLYKQLDDKDEEINQQSQLVEKLKTQMLDQEELLASTRRDQDNMQAELNRLQAENDASKEEVKEVLQALEELAVNYDQK SQEVEDKTKEYELLSDELNQKSATLASIDAELQKLKEMTNHQKKRAAEMMASLLKDLAEIGIAVGNNDVKQPEGTGMIDEEFTVARLYIS KMKSEVKTMVKRCKQLESTQTESNKKMEENEKELAACQLRISQHEAKIKSLTEYLQNVEQKKRQLEESVDALSEELVQLRAQEKVHEMEK EHLNKVQTANEVKQAVEQQIQSHRETHQKQISSLRDEVEAKAKLITDLQDQNQKMMLEQERLRVEHEKLKATDQEKSRKLHELTVMQDRR EQARQDLKGLEETVAKELQTLHNLRKLFVQDLATRVKKSAEIDSDDTGGSAAQKQKISFLENNLEQLTKVHKQLVRDNADLRCELPKLEK RLRATAERVKALESALKEAKENASRDRKRYQQEVDRIKEAVRSKNMARRGHSAQIAKPIRPGQHPAASPTHPSAIRGGGAFVQNSQPVAV -------------------------------------------------------------- >36266_36266_9_HHAT-KIF5B_HHAT_chr1_210591669_ENST00000545781_KIF5B_chr10_32329385_ENST00000302418_length(amino acids)=1178AA_BP=286 MLLQRAAPGERGWRPGEARSRRRCRWDSEVPKEGCWELAARVNVAVAAARDSPEDATDFEWSFWMEWGKQWLVWLLLGHMVVSQMATLLA RKHRPWILMLYGMWACWCVLGTPGVAMVLLHTTISFCVAQFRSQLLTWLCSLLLLSTLRLQGVEEVKRRWYKTENEYYLLQFTLTVRCLY YTSFSLELCWQQLPAASTSYSFPWMLAYVFYYPVLHNGPILSFSEFIKQMQQQEHDSLKASLCVLALGLGRLLCWWWLAELMAHLMYMHA IYSSIPLLETVSCWTLDVLEGYNGTIFAYGQTSSGKTHTMEGKLHDPEGMGIIPRIVQDIFNYIYSMDENLEFHIKVSYFEIYLDKIRDL LDVSKTNLSVHEDKNRVPYVKGCTERFVCSPDEVMDTIDEGKSNRHVAVTNMNEHSSRSHSIFLINVKQENTQTEQKLSGKLYLVDLAGS EKVSKTGAEGAVLDEAKNINKSLSALGNVISALAEGSTYVPYRDSKMTRILQDSLGGNCRTTIVICCSPSSYNESETKSTLLFGQRAKTI KNTVCVNVELTAEQWKKKYEKEKEKNKILRNTIQWLENELNRWRNGETVPIDEQFDKEKANLEAFTVDKDITLTNDKPATAIGVIGNFTD AERRKCEEEIAKLYKQLDDKDEEINQQSQLVEKLKTQMLDQEELLASTRRDQDNMQAELNRLQAENDASKEEVKEVLQALEELAVNYDQK SQEVEDKTKEYELLSDELNQKSATLASIDAELQKLKEMTNHQKKRAAEMMASLLKDLAEIGIAVGNNDVKQPEGTGMIDEEFTVARLYIS KMKSEVKTMVKRCKQLESTQTESNKKMEENEKELAACQLRISQHEAKIKSLTEYLQNVEQKKRQLEESVDALSEELVQLRAQEKVHEMEK EHLNKVQTANEVKQAVEQQIQSHRETHQKQISSLRDEVEAKAKLITDLQDQNQKMMLEQERLRVEHEKLKATDQEKSRKLHELTVMQDRR EQARQDLKGLEETVAKELQTLHNLRKLFVQDLATRVKKSAEIDSDDTGGSAAQKQKISFLENNLEQLTKVHKQLVRDNADLRCELPKLEK RLRATAERVKALESALKEAKENASRDRKRYQQEVDRIKEAVRSKNMARRGHSAQIAKPIRPGQHPAASPTHPSAIRGGGAFVQNSQPVAV -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:32337392/chr10:210637849) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| HHAT | KIF5B |
| FUNCTION: Negatively regulates N-terminal palmitoylation of SHH by HHAT/SKN. {ECO:0000250}. | FUNCTION: Microtubule-dependent motor required for normal distribution of mitochondria and lysosomes. Can induce formation of neurite-like membrane protrusions in non-neuronal cells in a ZFYVE27-dependent manner (By similarity). Regulates centrosome and nuclear positioning during mitotic entry. During the G2 phase of the cell cycle in a BICD2-dependent manner, antagonizes dynein function and drives the separation of nuclei and centrosomes (PubMed:20386726). Required for anterograde axonal transportation of MAPK8IP3/JIP3 which is essential for MAPK8IP3/JIP3 function in axon elongation (By similarity). Through binding with PLEKHM2 and ARL8B, directs lysosome movement toward microtubule plus ends (Probable). Involved in NK cell-mediated cytotoxicity. Drives the polarization of cytolytic granules and microtubule-organizing centers (MTOCs) toward the immune synapse between effector NK lymphocytes and target cells (PubMed:24088571). {ECO:0000250|UniProtKB:Q2PQA9, ECO:0000250|UniProtKB:Q61768, ECO:0000269|PubMed:20386726, ECO:0000269|PubMed:24088571, ECO:0000305|PubMed:22172677, ECO:0000305|PubMed:24088571}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000261458 | - | 7 | 12 | 203_217 | 285.3333333333333 | 494.0 | Intramembrane | Ontology_term=ECO:0000255 |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000261458 | - | 7 | 12 | 95_119 | 285.3333333333333 | 494.0 | Intramembrane | Ontology_term=ECO:0000255 |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000367010 | - | 7 | 12 | 203_217 | 285.3333333333333 | 494.0 | Intramembrane | Ontology_term=ECO:0000255 |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000367010 | - | 7 | 12 | 95_119 | 285.3333333333333 | 494.0 | Intramembrane | Ontology_term=ECO:0000255 |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000391905 | - | 7 | 12 | 203_217 | 285.3333333333333 | 519.0 | Intramembrane | Ontology_term=ECO:0000255 |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000391905 | - | 7 | 12 | 95_119 | 285.3333333333333 | 519.0 | Intramembrane | Ontology_term=ECO:0000255 |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000413764 | - | 7 | 12 | 203_217 | 285.3333333333333 | 494.0 | Intramembrane | Ontology_term=ECO:0000255 |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000413764 | - | 7 | 12 | 95_119 | 285.3333333333333 | 494.0 | Intramembrane | Ontology_term=ECO:0000255 |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000537898 | - | 6 | 11 | 203_217 | 220.33333333333334 | 429.0 | Intramembrane | Ontology_term=ECO:0000255 |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000537898 | - | 6 | 11 | 95_119 | 220.33333333333334 | 429.0 | Intramembrane | Ontology_term=ECO:0000255 |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000541565 | - | 5 | 10 | 95_119 | 148.33333333333334 | 357.0 | Intramembrane | Ontology_term=ECO:0000255 |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000545154 | - | 6 | 11 | 203_217 | 286.3333333333333 | 495.0 | Intramembrane | Ontology_term=ECO:0000255 |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000545154 | - | 6 | 11 | 95_119 | 286.3333333333333 | 495.0 | Intramembrane | Ontology_term=ECO:0000255 |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000261458 | - | 7 | 12 | 120_131 | 285.3333333333333 | 494.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000261458 | - | 7 | 12 | 149_162 | 285.3333333333333 | 494.0 | Topological domain | Lumenal |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000261458 | - | 7 | 12 | 184_202 | 285.3333333333333 | 494.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000261458 | - | 7 | 12 | 1_5 | 285.3333333333333 | 494.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000261458 | - | 7 | 12 | 218_243 | 285.3333333333333 | 494.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000261458 | - | 7 | 12 | 23_67 | 285.3333333333333 | 494.0 | Topological domain | Lumenal |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000261458 | - | 7 | 12 | 272_281 | 285.3333333333333 | 494.0 | Topological domain | Lumenal |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000261458 | - | 7 | 12 | 85_94 | 285.3333333333333 | 494.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000367010 | - | 7 | 12 | 120_131 | 285.3333333333333 | 494.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000367010 | - | 7 | 12 | 149_162 | 285.3333333333333 | 494.0 | Topological domain | Lumenal |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000367010 | - | 7 | 12 | 184_202 | 285.3333333333333 | 494.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000367010 | - | 7 | 12 | 1_5 | 285.3333333333333 | 494.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000367010 | - | 7 | 12 | 218_243 | 285.3333333333333 | 494.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000367010 | - | 7 | 12 | 23_67 | 285.3333333333333 | 494.0 | Topological domain | Lumenal |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000367010 | - | 7 | 12 | 272_281 | 285.3333333333333 | 494.0 | Topological domain | Lumenal |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000367010 | - | 7 | 12 | 85_94 | 285.3333333333333 | 494.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000391905 | - | 7 | 12 | 120_131 | 285.3333333333333 | 519.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000391905 | - | 7 | 12 | 149_162 | 285.3333333333333 | 519.0 | Topological domain | Lumenal |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000391905 | - | 7 | 12 | 184_202 | 285.3333333333333 | 519.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000391905 | - | 7 | 12 | 1_5 | 285.3333333333333 | 519.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000391905 | - | 7 | 12 | 218_243 | 285.3333333333333 | 519.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000391905 | - | 7 | 12 | 23_67 | 285.3333333333333 | 519.0 | Topological domain | Lumenal |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000391905 | - | 7 | 12 | 272_281 | 285.3333333333333 | 519.0 | Topological domain | Lumenal |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000391905 | - | 7 | 12 | 85_94 | 285.3333333333333 | 519.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000413764 | - | 7 | 12 | 120_131 | 285.3333333333333 | 494.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000413764 | - | 7 | 12 | 149_162 | 285.3333333333333 | 494.0 | Topological domain | Lumenal |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000413764 | - | 7 | 12 | 184_202 | 285.3333333333333 | 494.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000413764 | - | 7 | 12 | 1_5 | 285.3333333333333 | 494.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000413764 | - | 7 | 12 | 218_243 | 285.3333333333333 | 494.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000413764 | - | 7 | 12 | 23_67 | 285.3333333333333 | 494.0 | Topological domain | Lumenal |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000413764 | - | 7 | 12 | 272_281 | 285.3333333333333 | 494.0 | Topological domain | Lumenal |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000413764 | - | 7 | 12 | 85_94 | 285.3333333333333 | 494.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000537898 | - | 6 | 11 | 120_131 | 220.33333333333334 | 429.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000537898 | - | 6 | 11 | 149_162 | 220.33333333333334 | 429.0 | Topological domain | Lumenal |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000537898 | - | 6 | 11 | 184_202 | 220.33333333333334 | 429.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000537898 | - | 6 | 11 | 1_5 | 220.33333333333334 | 429.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000537898 | - | 6 | 11 | 23_67 | 220.33333333333334 | 429.0 | Topological domain | Lumenal |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000537898 | - | 6 | 11 | 85_94 | 220.33333333333334 | 429.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000541565 | - | 5 | 10 | 120_131 | 148.33333333333334 | 357.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000541565 | - | 5 | 10 | 1_5 | 148.33333333333334 | 357.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000541565 | - | 5 | 10 | 23_67 | 148.33333333333334 | 357.0 | Topological domain | Lumenal |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000541565 | - | 5 | 10 | 85_94 | 148.33333333333334 | 357.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000545154 | - | 6 | 11 | 120_131 | 286.3333333333333 | 495.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000545154 | - | 6 | 11 | 149_162 | 286.3333333333333 | 495.0 | Topological domain | Lumenal |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000545154 | - | 6 | 11 | 184_202 | 286.3333333333333 | 495.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000545154 | - | 6 | 11 | 1_5 | 286.3333333333333 | 495.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000545154 | - | 6 | 11 | 218_243 | 286.3333333333333 | 495.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000545154 | - | 6 | 11 | 23_67 | 286.3333333333333 | 495.0 | Topological domain | Lumenal |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000545154 | - | 6 | 11 | 272_281 | 286.3333333333333 | 495.0 | Topological domain | Lumenal |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000545154 | - | 6 | 11 | 85_94 | 286.3333333333333 | 495.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000261458 | - | 7 | 12 | 132_148 | 285.3333333333333 | 494.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000261458 | - | 7 | 12 | 163_183 | 285.3333333333333 | 494.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000261458 | - | 7 | 12 | 244_271 | 285.3333333333333 | 494.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000261458 | - | 7 | 12 | 68_84 | 285.3333333333333 | 494.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000261458 | - | 7 | 12 | 6_22 | 285.3333333333333 | 494.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000367010 | - | 7 | 12 | 132_148 | 285.3333333333333 | 494.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000367010 | - | 7 | 12 | 163_183 | 285.3333333333333 | 494.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000367010 | - | 7 | 12 | 244_271 | 285.3333333333333 | 494.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000367010 | - | 7 | 12 | 68_84 | 285.3333333333333 | 494.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000367010 | - | 7 | 12 | 6_22 | 285.3333333333333 | 494.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000391905 | - | 7 | 12 | 132_148 | 285.3333333333333 | 519.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000391905 | - | 7 | 12 | 163_183 | 285.3333333333333 | 519.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000391905 | - | 7 | 12 | 244_271 | 285.3333333333333 | 519.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000391905 | - | 7 | 12 | 68_84 | 285.3333333333333 | 519.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000391905 | - | 7 | 12 | 6_22 | 285.3333333333333 | 519.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000413764 | - | 7 | 12 | 132_148 | 285.3333333333333 | 494.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000413764 | - | 7 | 12 | 163_183 | 285.3333333333333 | 494.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000413764 | - | 7 | 12 | 244_271 | 285.3333333333333 | 494.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000413764 | - | 7 | 12 | 68_84 | 285.3333333333333 | 494.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000413764 | - | 7 | 12 | 6_22 | 285.3333333333333 | 494.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000537898 | - | 6 | 11 | 132_148 | 220.33333333333334 | 429.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000537898 | - | 6 | 11 | 163_183 | 220.33333333333334 | 429.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000537898 | - | 6 | 11 | 68_84 | 220.33333333333334 | 429.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000537898 | - | 6 | 11 | 6_22 | 220.33333333333334 | 429.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000541565 | - | 5 | 10 | 132_148 | 148.33333333333334 | 357.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000541565 | - | 5 | 10 | 68_84 | 148.33333333333334 | 357.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000541565 | - | 5 | 10 | 6_22 | 148.33333333333334 | 357.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000545154 | - | 6 | 11 | 132_148 | 286.3333333333333 | 495.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000545154 | - | 6 | 11 | 163_183 | 286.3333333333333 | 495.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000545154 | - | 6 | 11 | 244_271 | 286.3333333333333 | 495.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000545154 | - | 6 | 11 | 68_84 | 286.3333333333333 | 495.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000545154 | - | 6 | 11 | 6_22 | 286.3333333333333 | 495.0 | Transmembrane | Helical |
| Tgene | KIF5B | chr1:210591669 | chr10:32329385 | ENST00000302418 | 1 | 26 | 329_914 | 71.33333333333333 | 1626.0 | Coiled coil | . | |
| Tgene | KIF5B | chr1:210591669 | chr10:32329385 | ENST00000302418 | 1 | 26 | 85_92 | 71.33333333333333 | 1626.0 | Nucleotide binding | Note=ATP | |
| Tgene | KIF5B | chr1:210591669 | chr10:32329385 | ENST00000302418 | 1 | 26 | 915_963 | 71.33333333333333 | 1626.0 | Region | Note=Globular |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000541565 | - | 5 | 10 | 203_217 | 148.33333333333334 | 357.0 | Intramembrane | Ontology_term=ECO:0000255 |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000261458 | - | 7 | 12 | 448_455 | 285.3333333333333 | 494.0 | Region | GTP-binding |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000367010 | - | 7 | 12 | 448_455 | 285.3333333333333 | 494.0 | Region | GTP-binding |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000391905 | - | 7 | 12 | 448_455 | 285.3333333333333 | 519.0 | Region | GTP-binding |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000413764 | - | 7 | 12 | 448_455 | 285.3333333333333 | 494.0 | Region | GTP-binding |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000537898 | - | 6 | 11 | 448_455 | 220.33333333333334 | 429.0 | Region | GTP-binding |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000541565 | - | 5 | 10 | 448_455 | 148.33333333333334 | 357.0 | Region | GTP-binding |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000545154 | - | 6 | 11 | 448_455 | 286.3333333333333 | 495.0 | Region | GTP-binding |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000261458 | - | 7 | 12 | 311_363 | 285.3333333333333 | 494.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000261458 | - | 7 | 12 | 381_383 | 285.3333333333333 | 494.0 | Topological domain | Lumenal |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000261458 | - | 7 | 12 | 400_427 | 285.3333333333333 | 494.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000261458 | - | 7 | 12 | 449_462 | 285.3333333333333 | 494.0 | Topological domain | Lumenal |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000261458 | - | 7 | 12 | 482_493 | 285.3333333333333 | 494.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000367010 | - | 7 | 12 | 311_363 | 285.3333333333333 | 494.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000367010 | - | 7 | 12 | 381_383 | 285.3333333333333 | 494.0 | Topological domain | Lumenal |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000367010 | - | 7 | 12 | 400_427 | 285.3333333333333 | 494.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000367010 | - | 7 | 12 | 449_462 | 285.3333333333333 | 494.0 | Topological domain | Lumenal |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000367010 | - | 7 | 12 | 482_493 | 285.3333333333333 | 494.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000391905 | - | 7 | 12 | 311_363 | 285.3333333333333 | 519.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000391905 | - | 7 | 12 | 381_383 | 285.3333333333333 | 519.0 | Topological domain | Lumenal |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000391905 | - | 7 | 12 | 400_427 | 285.3333333333333 | 519.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000391905 | - | 7 | 12 | 449_462 | 285.3333333333333 | 519.0 | Topological domain | Lumenal |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000391905 | - | 7 | 12 | 482_493 | 285.3333333333333 | 519.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000413764 | - | 7 | 12 | 311_363 | 285.3333333333333 | 494.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000413764 | - | 7 | 12 | 381_383 | 285.3333333333333 | 494.0 | Topological domain | Lumenal |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000413764 | - | 7 | 12 | 400_427 | 285.3333333333333 | 494.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000413764 | - | 7 | 12 | 449_462 | 285.3333333333333 | 494.0 | Topological domain | Lumenal |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000413764 | - | 7 | 12 | 482_493 | 285.3333333333333 | 494.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000537898 | - | 6 | 11 | 218_243 | 220.33333333333334 | 429.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000537898 | - | 6 | 11 | 272_281 | 220.33333333333334 | 429.0 | Topological domain | Lumenal |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000537898 | - | 6 | 11 | 311_363 | 220.33333333333334 | 429.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000537898 | - | 6 | 11 | 381_383 | 220.33333333333334 | 429.0 | Topological domain | Lumenal |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000537898 | - | 6 | 11 | 400_427 | 220.33333333333334 | 429.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000537898 | - | 6 | 11 | 449_462 | 220.33333333333334 | 429.0 | Topological domain | Lumenal |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000537898 | - | 6 | 11 | 482_493 | 220.33333333333334 | 429.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000541565 | - | 5 | 10 | 149_162 | 148.33333333333334 | 357.0 | Topological domain | Lumenal |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000541565 | - | 5 | 10 | 184_202 | 148.33333333333334 | 357.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000541565 | - | 5 | 10 | 218_243 | 148.33333333333334 | 357.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000541565 | - | 5 | 10 | 272_281 | 148.33333333333334 | 357.0 | Topological domain | Lumenal |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000541565 | - | 5 | 10 | 311_363 | 148.33333333333334 | 357.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000541565 | - | 5 | 10 | 381_383 | 148.33333333333334 | 357.0 | Topological domain | Lumenal |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000541565 | - | 5 | 10 | 400_427 | 148.33333333333334 | 357.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000541565 | - | 5 | 10 | 449_462 | 148.33333333333334 | 357.0 | Topological domain | Lumenal |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000541565 | - | 5 | 10 | 482_493 | 148.33333333333334 | 357.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000545154 | - | 6 | 11 | 311_363 | 286.3333333333333 | 495.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000545154 | - | 6 | 11 | 381_383 | 286.3333333333333 | 495.0 | Topological domain | Lumenal |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000545154 | - | 6 | 11 | 400_427 | 286.3333333333333 | 495.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000545154 | - | 6 | 11 | 449_462 | 286.3333333333333 | 495.0 | Topological domain | Lumenal |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000545154 | - | 6 | 11 | 482_493 | 286.3333333333333 | 495.0 | Topological domain | Cytoplasmic |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000261458 | - | 7 | 12 | 282_310 | 285.3333333333333 | 494.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000261458 | - | 7 | 12 | 364_380 | 285.3333333333333 | 494.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000261458 | - | 7 | 12 | 384_399 | 285.3333333333333 | 494.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000261458 | - | 7 | 12 | 428_448 | 285.3333333333333 | 494.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000261458 | - | 7 | 12 | 463_481 | 285.3333333333333 | 494.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000367010 | - | 7 | 12 | 282_310 | 285.3333333333333 | 494.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000367010 | - | 7 | 12 | 364_380 | 285.3333333333333 | 494.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000367010 | - | 7 | 12 | 384_399 | 285.3333333333333 | 494.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000367010 | - | 7 | 12 | 428_448 | 285.3333333333333 | 494.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000367010 | - | 7 | 12 | 463_481 | 285.3333333333333 | 494.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000391905 | - | 7 | 12 | 282_310 | 285.3333333333333 | 519.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000391905 | - | 7 | 12 | 364_380 | 285.3333333333333 | 519.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000391905 | - | 7 | 12 | 384_399 | 285.3333333333333 | 519.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000391905 | - | 7 | 12 | 428_448 | 285.3333333333333 | 519.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000391905 | - | 7 | 12 | 463_481 | 285.3333333333333 | 519.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000413764 | - | 7 | 12 | 282_310 | 285.3333333333333 | 494.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000413764 | - | 7 | 12 | 364_380 | 285.3333333333333 | 494.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000413764 | - | 7 | 12 | 384_399 | 285.3333333333333 | 494.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000413764 | - | 7 | 12 | 428_448 | 285.3333333333333 | 494.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000413764 | - | 7 | 12 | 463_481 | 285.3333333333333 | 494.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000537898 | - | 6 | 11 | 244_271 | 220.33333333333334 | 429.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000537898 | - | 6 | 11 | 282_310 | 220.33333333333334 | 429.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000537898 | - | 6 | 11 | 364_380 | 220.33333333333334 | 429.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000537898 | - | 6 | 11 | 384_399 | 220.33333333333334 | 429.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000537898 | - | 6 | 11 | 428_448 | 220.33333333333334 | 429.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000537898 | - | 6 | 11 | 463_481 | 220.33333333333334 | 429.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000541565 | - | 5 | 10 | 163_183 | 148.33333333333334 | 357.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000541565 | - | 5 | 10 | 244_271 | 148.33333333333334 | 357.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000541565 | - | 5 | 10 | 282_310 | 148.33333333333334 | 357.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000541565 | - | 5 | 10 | 364_380 | 148.33333333333334 | 357.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000541565 | - | 5 | 10 | 384_399 | 148.33333333333334 | 357.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000541565 | - | 5 | 10 | 428_448 | 148.33333333333334 | 357.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000541565 | - | 5 | 10 | 463_481 | 148.33333333333334 | 357.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000545154 | - | 6 | 11 | 282_310 | 286.3333333333333 | 495.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000545154 | - | 6 | 11 | 364_380 | 286.3333333333333 | 495.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000545154 | - | 6 | 11 | 384_399 | 286.3333333333333 | 495.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000545154 | - | 6 | 11 | 428_448 | 286.3333333333333 | 495.0 | Transmembrane | Helical |
| Hgene | HHAT | chr1:210591669 | chr10:32329385 | ENST00000545154 | - | 6 | 11 | 463_481 | 286.3333333333333 | 495.0 | Transmembrane | Helical |
| Tgene | KIF5B | chr1:210591669 | chr10:32329385 | ENST00000302418 | 1 | 26 | 8_325 | 71.33333333333333 | 1626.0 | Domain | Kinesin motor |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| KIF5B | YWHAG, YWHAB, YWHAQ, YWHAZ, VDAC1, NME2, SFN, DTNB, SNAP23, SNAP25, KLC1, KLC2, KIF5C, YWHAH, USPL1, MARK2, MEPCE, FYCO1, RAPGEF2, APC, SPG20, CDC5L, GSK3B, NDRG1, SIRT7, RANBP2, KIAA0368, KLC4, ZBTB8B, KDM1A, MCCC1, EXOC7, DHX9, TWF1, SRP68, RNF168, YWHAE, CFTR, ACACA, CAST, COPE, COPS5, AKAP2, DIAPH1, ELP2, EPB41L1, NUP50, RNF20, PSMA2, PSMA4, PSMA5, PSMB3, PSMB4, PSMB5, PSMB6, PSMB7, PSME1, PTPN23, RABGAP1, SMEK2, TRIM25, STAU1, AURKB, HUWE1, PRKAR2B, VPS52, TAX1BP1, ZBTB8A, CUL7, EZH2, ABCE1, TRAK2, CCDC101, SYNE4, DTNBP1, NUP43, KPTN, AZIN1, TRIP6, CHRNA9, RPS6KB2, KIF1A, LSM2, NTRK1, MED4, L1TD1, KLC3, NHS, POU1F1, TUBA1A, OGT, SQSTM1, XPR1, NCOR1, TRAK1, KIDINS220, SLC38A10, Klc2, Klc4, Klc3, FOXQ1, Ostm1, CDH1, MYCL, COG6, EXOC1, MAP7D2, HOMER1, DLST, SOD1, BRCA1, ZNF598, EGLN3, CTNNB1, HSPA8, HDAC4, EFTUD2, ESR2, HEXIM1, PIK3R1, KDM6B, MYC, CDK9, KIAA1429, Bach1, GBF1, RFFL, HOOK1, HOOK3, BMH1, BMH2, BIRC3, LMBR1L, TRIM28, MEOX2, POU6F2, FHL2, CCDC136, LBX1, CDR2, BAG4, HSF2BP, POLR2G, GCC1, PRKAR1B, PLEKHA4, PPIB, MYO5A, KIF14, IFI16, PYHIN1, SUMO2, Rnf183, BRD4, Apc2, ORF6, SCGN, TP53, WDR5, NAA40, TPM3, SYCE1, NPEPPS, BRK1, PRPS2, HVCN1, C15orf59, HSPA2, NUP62, S100A2, CLSTN1, EP300, FGD5, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| HHAT | |
| KIF5B | 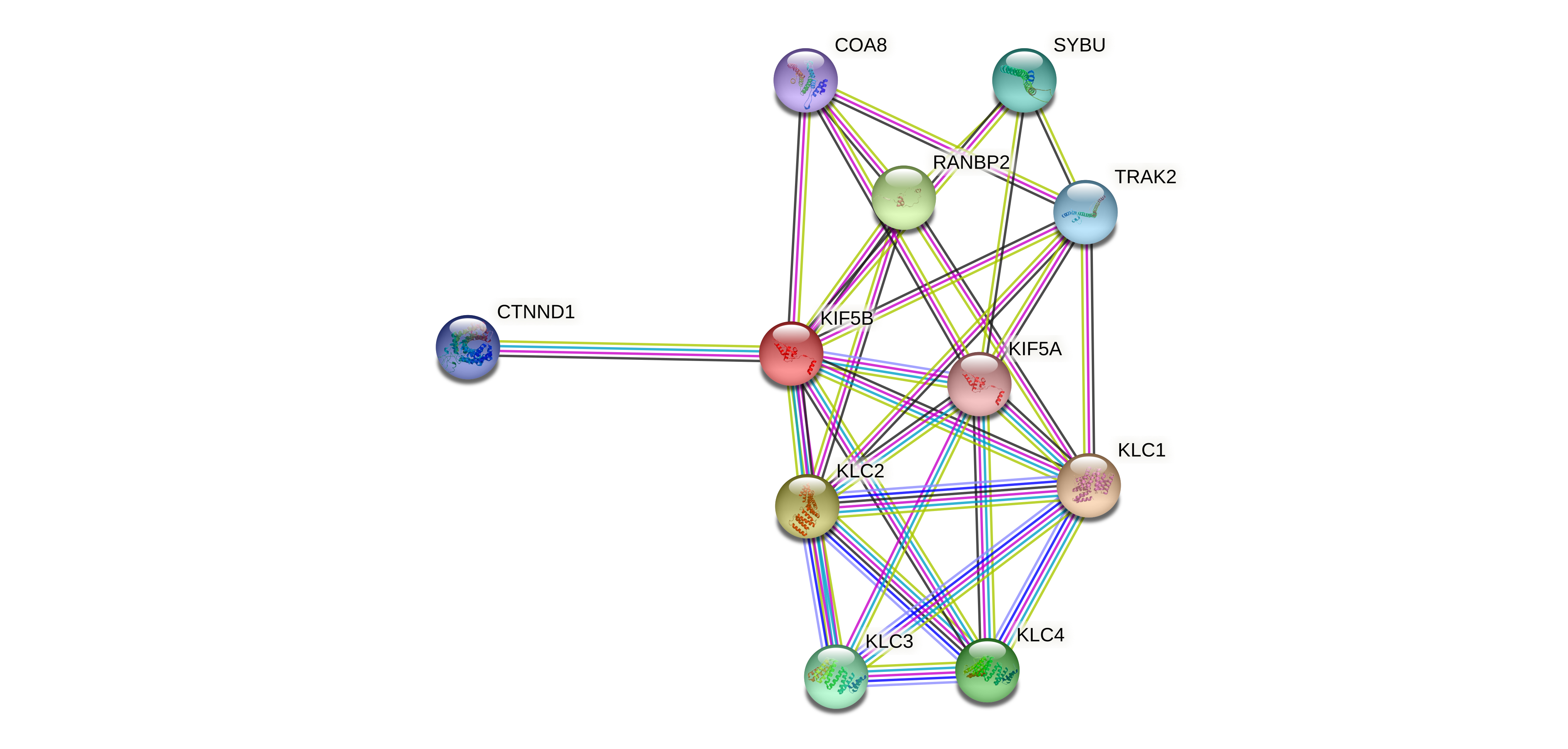 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to HHAT-KIF5B |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to HHAT-KIF5B |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Tgene | KIF5B | C0007131 | Non-Small Cell Lung Carcinoma | 1 | CTD_human |
| Tgene | KIF5B | C0011849 | Diabetes Mellitus | 1 | CTD_human |

