| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:HIP1-ALK |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: HIP1-ALK | FusionPDB ID: 36380 | FusionGDB2.0 ID: 36380 | Hgene | Tgene | Gene symbol | HIP1 | ALK | Gene ID | 3092 | 238 |
| Gene name | huntingtin interacting protein 1 | ALK receptor tyrosine kinase | |
| Synonyms | HIP-I|ILWEQ|SHON|SHONbeta|SHONgamma | CD246|NBLST3 | |
| Cytomap | 7q11.23 | 2p23.2-p23.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | huntingtin-interacting protein 1huntingtin-interacting protein I | ALK tyrosine kinase receptorCD246 antigenanaplastic lymphoma receptor tyrosine kinasemutant anaplastic lymphoma kinase | |
| Modification date | 20200313 | 20200329 | |
| UniProtAcc | O75146 | Q96BT7 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000336926, ENST00000434438, ENST00000479835, | ENST00000431873, ENST00000498037, ENST00000389048, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 6 X 9 X 5=270 | 56 X 74 X 20=82880 |
| # samples | 7 | 57 | |
| ** MAII score | log2(7/270*10)=-1.94753258010586 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(57/82880*10)=-7.18391827352181 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: HIP1 [Title/Abstract] AND ALK [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | HIP1(75183402)-ALK(29446405), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | HIP1 | GO:0006915 | apoptotic process | 11788820 |
| Hgene | HIP1 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process | 11788820 |
| Hgene | HIP1 | GO:0042981 | regulation of apoptotic process | 11788820 |
| Hgene | HIP1 | GO:0048268 | clathrin coat assembly | 11577110|11889126 |
| Hgene | HIP1 | GO:0050821 | protein stabilization | 14732715 |
| Hgene | HIP1 | GO:0097190 | apoptotic signaling pathway | 11007801 |
| Tgene | ALK | GO:0016310 | phosphorylation | 9174053 |
| Tgene | ALK | GO:0046777 | protein autophosphorylation | 9174053 |
 Fusion gene breakpoints across HIP1 (5'-gene) Fusion gene breakpoints across HIP1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 Fusion gene breakpoints across ALK (3'-gene) Fusion gene breakpoints across ALK (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerKB3 | . | . | HIP1 | chr7 | 75168642 | - | ALK | chr2 | 29446394 | - |
| ChimerKB3 | . | . | HIP1 | chr7 | 75172169 | - | ALK | chr2 | 29446394 | - |
| ChimerKB3 | . | . | HIP1 | chr7 | 75183411 | - | ALK | chr2 | 29446394 | - |
| ChiTaRS5.0 | N/A | KF360988 | HIP1 | chr7 | 75183402 | - | ALK | chr2 | 29446405 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000336926 | HIP1 | chr7 | 75168642 | - | ENST00000389048 | ALK | chr2 | 29446394 | - | 5229 | 3088 | 27 | 4778 | 1583 |
| ENST00000434438 | HIP1 | chr7 | 75168642 | - | ENST00000389048 | ALK | chr2 | 29446394 | - | 5070 | 2929 | 21 | 4619 | 1532 |
| ENST00000336926 | HIP1 | chr7 | 75172169 | - | ENST00000389048 | ALK | chr2 | 29446394 | - | 5058 | 2917 | 27 | 4607 | 1526 |
| ENST00000434438 | HIP1 | chr7 | 75172169 | - | ENST00000389048 | ALK | chr2 | 29446394 | - | 4899 | 2758 | 21 | 4448 | 1475 |
| ENST00000336926 | HIP1 | chr7 | 75183411 | - | ENST00000389048 | ALK | chr2 | 29446394 | - | 4326 | 2185 | 27 | 3875 | 1282 |
| ENST00000434438 | HIP1 | chr7 | 75183411 | - | ENST00000389048 | ALK | chr2 | 29446394 | - | 4320 | 2179 | 21 | 3869 | 1282 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >36380_36380_1_HIP1-ALK_HIP1_chr7_75168642_ENST00000336926_ALK_chr2_29446394_ENST00000389048_length(amino acids)=1583AA_BP=1020 MDRMASSMKQVPNPLPKVLSRRGVGAGLEAAERESFERTQTVSINKAINTQEVAVKEKHARTCILGTHHEKGAQTFWSVVNRLPLSSNAV LCWKFCHVFHKLLRDGHPNVLKDSLRYRNELSDMSRMWGHLSEGYGQLCSIYLKLLRTKMEYHTKNPRFPGNLQMSDRQLDEAGESDVNN FFQLTVEMFDYLECELNLFQTVFNSLDMSRSVSVTAAGQCRLAPLIQVILDCSHLYDYTVKLLFKLHSCLPADTLQGHRDRFMEQFTKLK DLFYRSSNLQYFKRLIQIPQLPENPPNFLRASALSEHISPVVVIPAEASSPDSEPVLEKDDLMDMDASQQNLFDNKFDDIFGSSFSSDPF NFNSQNGVNKDEKDHLIERLYREISGLKAQLENMKTESQRVVLQLKGHVSELEADLAEQQHLRQQAADDCEFLRAELDELRRQREDTEKA QRSLSEIERKAQANEQRYSKLKEKYSELVQNHADLLRKNAEVTKQVSMARQAQVDLEREKKELEDSLERISDQGQRKTQEQLEVLESLKQ ELATSQRELQVLQGSLETSAQSEANWAAEFAELEKERDSLVSGAAHREEELSALRKELQDTQLKLASTEESMCQLAKDQRKMLLVGSRKA AEQVIQDALNQLEEPPLISCAGSADHLLSTVTSISSCIEQLEKSWSQYLACPEDISGLLHSITLLAHLTSDAIAHGATTCLRAPPEPADS LTEACKQYGRETLAYLASLEEEGSLENADSTAMRNCLSKIKAIGEELLPRGLDIKQEELGDLVDKEMAATSAAIETATARIEEMLSKSRA GDTGVKLEVNERILGCCTSLMQAIQVLIVASKDLQREIVESGRGTASPKEFYAKNSRWTEGLISASKAVGWGATVMVDAADLVVQGRGKF EELMVCSHEIAASTAQLVAASKVKADKDSPNLAQLQQASRGVNQATAGVVASTISGKSQIEETDNMDFSSMTLTQIKRQEMDSQVRVLEL ENELQKERQKLGELRKKHYELAGVAEGWEEVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNIT LIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSF LRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMP PEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCT QDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVN MAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMK -------------------------------------------------------------- >36380_36380_2_HIP1-ALK_HIP1_chr7_75168642_ENST00000434438_ALK_chr2_29446394_ENST00000389048_length(amino acids)=1532AA_BP=969 MDRMASSMKQVPNPLPKVLSRRGVGAGLEAAERESFERTQTVSINKAINTQEVAVKEKHARTCILGTHHEKGAQTFWSVVNRLPLSSNAV LCWKFCHVFHKLLRDGHPNVLKDSLRYRNELSDMSRMWGHLSEGYGQLCSIYLKLLRTKMEYHTKNPRFPGNLQMSDRQLDEAGESDVNN FFQLTVEMFDYLECELNLFQTVFNSLDMSRSVSVTAAGQCRLAPLIQVILDCSHLYDYTVKLLFKLHSCLPADTLQGHRDRFMEQFTKLK DLFYRSSNLQYFKRLIQIPQLPENPPNFLRASALSEHISPVVVIPAEASSPDSEPVLEKDDLMDMDASQQNLFDNKFDDIFGSSFSSDPF NFNSQNGVNKDEKDHLIERLYREISGLKAQLENMKTESQRVVLQLKGHVSELEADLAEQQHLRQQAADDCEFLRAELDELRRQREDTEKA QRSLSEIERKAQANEQRYSKLKEKYSELVQNHADLLRKNAEVTKQVSMARQAQVDLEREKKELEDSLERISDQGQRKTQEQLEVLESLKQ ELATSQRELQVLQGSLETSAQSEANWAAEFAELEKERDSLVSGAAHREEELSALRKELQDTQLKLASTEESMCQLAKDQRKMLLVGSRKA AEQVIQDALNQLEEPPLISCAGSADHLLSTVTSISSCIEQLEKSWSQYLACPEDISGLLHSITLLAHLTSDAIAHGATTCLRAPPEPADS LTEACKQYGRETLAYLASLEEEGSLENADSTAMRNCLSKIKAIGEELLPRGLDIKQEELGDLVDKEMAATSAAIETATARIEGTASPKEF YAKNSRWTEGLISASKAVGWGATVMVDAADLVVQGRGKFEELMVCSHEIAASTAQLVAASKVKADKDSPNLAQLQQASRGVNQATAGVVA STISGKSQIEETDNMDFSSMTLTQIKRQEMDSQVRVLELENELQKERQKLGELRKKHYELAGVAEGWEEVYRRKHQELQAMQMELQSPEY KLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEA LIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLT CPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRM DPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSP AAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPH DRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYEDTILKSKNSMNQP -------------------------------------------------------------- >36380_36380_3_HIP1-ALK_HIP1_chr7_75172169_ENST00000336926_ALK_chr2_29446394_ENST00000389048_length(amino acids)=1526AA_BP=963 MDRMASSMKQVPNPLPKVLSRRGVGAGLEAAERESFERTQTVSINKAINTQEVAVKEKHARTCILGTHHEKGAQTFWSVVNRLPLSSNAV LCWKFCHVFHKLLRDGHPNVLKDSLRYRNELSDMSRMWGHLSEGYGQLCSIYLKLLRTKMEYHTKNPRFPGNLQMSDRQLDEAGESDVNN FFQLTVEMFDYLECELNLFQTVFNSLDMSRSVSVTAAGQCRLAPLIQVILDCSHLYDYTVKLLFKLHSCLPADTLQGHRDRFMEQFTKLK DLFYRSSNLQYFKRLIQIPQLPENPPNFLRASALSEHISPVVVIPAEASSPDSEPVLEKDDLMDMDASQQNLFDNKFDDIFGSSFSSDPF NFNSQNGVNKDEKDHLIERLYREISGLKAQLENMKTESQRVVLQLKGHVSELEADLAEQQHLRQQAADDCEFLRAELDELRRQREDTEKA QRSLSEIERKAQANEQRYSKLKEKYSELVQNHADLLRKNAEVTKQVSMARQAQVDLEREKKELEDSLERISDQGQRKTQEQLEVLESLKQ ELATSQRELQVLQGSLETSAQSEANWAAEFAELEKERDSLVSGAAHREEELSALRKELQDTQLKLASTEESMCQLAKDQRKMLLVGSRKA AEQVIQDALNQLEEPPLISCAGSADHLLSTVTSISSCIEQLEKSWSQYLACPEDISGLLHSITLLAHLTSDAIAHGATTCLRAPPEPADS LTEACKQYGRETLAYLASLEEEGSLENADSTAMRNCLSKIKAIGEELLPRGLDIKQEELGDLVDKEMAATSAAIETATARIEEMLSKSRA GDTGVKLEVNERILGCCTSLMQAIQVLIVASKDLQREIVESGRGTASPKEFYAKNSRWTEGLISASKAVGWGATVMVDAADLVVQGRGKF EELMVCSHEIAASTAQLVAASKVKADKDSPNLAQLQQASRGVNQATAGVVASTISGKSQIEETVYRRKHQELQAMQMELQSPEYKLSKLR TSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKF NHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGR VAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNC PGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPL PTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLG -------------------------------------------------------------- >36380_36380_4_HIP1-ALK_HIP1_chr7_75172169_ENST00000434438_ALK_chr2_29446394_ENST00000389048_length(amino acids)=1475AA_BP=912 MDRMASSMKQVPNPLPKVLSRRGVGAGLEAAERESFERTQTVSINKAINTQEVAVKEKHARTCILGTHHEKGAQTFWSVVNRLPLSSNAV LCWKFCHVFHKLLRDGHPNVLKDSLRYRNELSDMSRMWGHLSEGYGQLCSIYLKLLRTKMEYHTKNPRFPGNLQMSDRQLDEAGESDVNN FFQLTVEMFDYLECELNLFQTVFNSLDMSRSVSVTAAGQCRLAPLIQVILDCSHLYDYTVKLLFKLHSCLPADTLQGHRDRFMEQFTKLK DLFYRSSNLQYFKRLIQIPQLPENPPNFLRASALSEHISPVVVIPAEASSPDSEPVLEKDDLMDMDASQQNLFDNKFDDIFGSSFSSDPF NFNSQNGVNKDEKDHLIERLYREISGLKAQLENMKTESQRVVLQLKGHVSELEADLAEQQHLRQQAADDCEFLRAELDELRRQREDTEKA QRSLSEIERKAQANEQRYSKLKEKYSELVQNHADLLRKNAEVTKQVSMARQAQVDLEREKKELEDSLERISDQGQRKTQEQLEVLESLKQ ELATSQRELQVLQGSLETSAQSEANWAAEFAELEKERDSLVSGAAHREEELSALRKELQDTQLKLASTEESMCQLAKDQRKMLLVGSRKA AEQVIQDALNQLEEPPLISCAGSADHLLSTVTSISSCIEQLEKSWSQYLACPEDISGLLHSITLLAHLTSDAIAHGATTCLRAPPEPADS LTEACKQYGRETLAYLASLEEEGSLENADSTAMRNCLSKIKAIGEELLPRGLDIKQEELGDLVDKEMAATSAAIETATARIEGTASPKEF YAKNSRWTEGLISASKAVGWGATVMVDAADLVVQGRGKFEELMVCSHEIAASTAQLVAASKVKADKDSPNLAQLQQASRGVNQATAGVVA STISGKSQIEETVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQV SGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLD LLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSF GVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLV EEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGS RNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYG -------------------------------------------------------------- >36380_36380_5_HIP1-ALK_HIP1_chr7_75183411_ENST00000336926_ALK_chr2_29446394_ENST00000389048_length(amino acids)=1282AA_BP=720 MDRMASSMKQVPNPLPKVLSRRGVGAGLEAAERESFERTQTVSINKAINTQEVAVKEKHARTCILGTHHEKGAQTFWSVVNRLPLSSNAV LCWKFCHVFHKLLRDGHPNVLKDSLRYRNELSDMSRMWGHLSEGYGQLCSIYLKLLRTKMEYHTKNPRFPGNLQMSDRQLDEAGESDVNN FFQLTVEMFDYLECELNLFQTVFNSLDMSRSVSVTAAGQCRLAPLIQVILDCSHLYDYTVKLLFKLHSCLPADTLQGHRDRFMEQFTKLK DLFYRSSNLQYFKRLIQIPQLPENPPNFLRASALSEHISPVVVIPAEASSPDSEPVLEKDDLMDMDASQQNLFDNKFDDIFGSSFSSDPF NFNSQNGVNKDEKDHLIERLYREISGLKAQLENMKTESQRVVLQLKGHVSELEADLAEQQHLRQQAADDCEFLRAELDELRRQREDTEKA QRSLSEIERKAQANEQRYSKLKEKYSELVQNHADLLRKNAEVTKQVSMARQAQVDLEREKKELEDSLERISDQGQRKTQEQLEVLESLKQ ELATSQRELQVLQGSLETSAQSEANWAAEFAELEKERDSLVSGAAHREEELSALRKELQDTQLKLASTEESMCQLAKDQRKMLLVGSRKA AEQVIQDALNQLEEPPLISCAGSADHLLSTVTSISSCIEQLEKSWSQYLACPEDISGLLHSITLLAHLTSDAIAHGATTCLRAPPEPADL YRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVA VKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQ YLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYM PYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDP EGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYG SWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATA -------------------------------------------------------------- >36380_36380_6_HIP1-ALK_HIP1_chr7_75183411_ENST00000434438_ALK_chr2_29446394_ENST00000389048_length(amino acids)=1282AA_BP=720 MDRMASSMKQVPNPLPKVLSRRGVGAGLEAAERESFERTQTVSINKAINTQEVAVKEKHARTCILGTHHEKGAQTFWSVVNRLPLSSNAV LCWKFCHVFHKLLRDGHPNVLKDSLRYRNELSDMSRMWGHLSEGYGQLCSIYLKLLRTKMEYHTKNPRFPGNLQMSDRQLDEAGESDVNN FFQLTVEMFDYLECELNLFQTVFNSLDMSRSVSVTAAGQCRLAPLIQVILDCSHLYDYTVKLLFKLHSCLPADTLQGHRDRFMEQFTKLK DLFYRSSNLQYFKRLIQIPQLPENPPNFLRASALSEHISPVVVIPAEASSPDSEPVLEKDDLMDMDASQQNLFDNKFDDIFGSSFSSDPF NFNSQNGVNKDEKDHLIERLYREISGLKAQLENMKTESQRVVLQLKGHVSELEADLAEQQHLRQQAADDCEFLRAELDELRRQREDTEKA QRSLSEIERKAQANEQRYSKLKEKYSELVQNHADLLRKNAEVTKQVSMARQAQVDLEREKKELEDSLERISDQGQRKTQEQLEVLESLKQ ELATSQRELQVLQGSLETSAQSEANWAAEFAELEKERDSLVSGAAHREEELSALRKELQDTQLKLASTEESMCQLAKDQRKMLLVGSRKA AEQVIQDALNQLEEPPLISCAGSADHLLSTVTSISSCIEQLEKSWSQYLACPEDISGLLHSITLLAHLTSDAIAHGATTCLRAPPEPADL YRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVA VKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQ YLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYM PYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDP EGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYG SWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATA -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr7:75183402/chr2:29446405) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| HIP1 | ALK |
| FUNCTION: Component of clathrin-coated pits and vesicles, that may link the endocytic machinery to the actin cytoskeleton. Binds 3-phosphoinositides (via ENTH domain). May act through the ENTH domain to promote cell survival by stabilizing receptor tyrosine kinases following ligand-induced endocytosis. {ECO:0000269|PubMed:11889126, ECO:0000269|PubMed:14732715}. | FUNCTION: Catalyzes the methylation of 5-carboxymethyl uridine to 5-methylcarboxymethyl uridine at the wobble position of the anticodon loop in tRNA via its methyltransferase domain (PubMed:20123966, PubMed:20308323, PubMed:31079898). Catalyzes the last step in the formation of 5-methylcarboxymethyl uridine at the wobble position of the anticodon loop in target tRNA (PubMed:20123966, PubMed:20308323). Has a preference for tRNA(Arg) and tRNA(Glu), and does not bind tRNA(Lys)(PubMed:20308323). Binds tRNA and catalyzes the iron and alpha-ketoglutarate dependent hydroxylation of 5-methylcarboxymethyl uridine at the wobble position of the anticodon loop in tRNA via its dioxygenase domain, giving rise to 5-(S)-methoxycarbonylhydroxymethyluridine; has a preference for tRNA(Gly) (PubMed:21285950). Required for normal survival after DNA damage (PubMed:20308323). May inhibit apoptosis and promote cell survival and angiogenesis (PubMed:19293182). {ECO:0000269|PubMed:19293182, ECO:0000269|PubMed:20123966, ECO:0000269|PubMed:20308323, ECO:0000269|PubMed:21285950, ECO:0000269|PubMed:31079898}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
Top |
Fusion Protein Structures |
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
| PDB file (876)CIF file (876) >>>876.cif | HIP1 | chr7 | 75183411 | - | ALK | chr2 | 29446394 | - | MDRMASSMKQVPNPLPKVLSRRGVGAGLEAAERESFERTQTVSINKAINT QEVAVKEKHARTCILGTHHEKGAQTFWSVVNRLPLSSNAVLCWKFCHVFH KLLRDGHPNVLKDSLRYRNELSDMSRMWGHLSEGYGQLCSIYLKLLRTKM EYHTKNPRFPGNLQMSDRQLDEAGESDVNNFFQLTVEMFDYLECELNLFQ TVFNSLDMSRSVSVTAAGQCRLAPLIQVILDCSHLYDYTVKLLFKLHSCL PADTLQGHRDRFMEQFTKLKDLFYRSSNLQYFKRLIQIPQLPENPPNFLR ASALSEHISPVVVIPAEASSPDSEPVLEKDDLMDMDASQQNLFDNKFDDI FGSSFSSDPFNFNSQNGVNKDEKDHLIERLYREISGLKAQLENMKTESQR VVLQLKGHVSELEADLAEQQHLRQQAADDCEFLRAELDELRRQREDTEKA QRSLSEIERKAQANEQRYSKLKEKYSELVQNHADLLRKNAEVTKQVSMAR QAQVDLEREKKELEDSLERISDQGQRKTQEQLEVLESLKQELATSQRELQ VLQGSLETSAQSEANWAAEFAELEKERDSLVSGAAHREEELSALRKELQD TQLKLASTEESMCQLAKDQRKMLLVGSRKAAEQVIQDALNQLEEPPLISC AGSADHLLSTVTSISSCIEQLEKSWSQYLACPEDISGLLHSITLLAHLTS DAIAHGATTCLRAPPEPADLYRRKHQELQAMQMELQSPEYKLSKLRTSTI MTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGM PNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQ SLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQ YLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGGC AMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEV LEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQ DPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSP AAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSE LHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGS CTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQ | 1282 |
| 3D view using mol* of 876 (AA BP:) | ||||||||||
 | ||||||||||
| PDB file (938)CIF file (938) >>>938.cif | HIP1 | chr7 | 75172169 | - | ALK | chr2 | 29446394 | - | MDRMASSMKQVPNPLPKVLSRRGVGAGLEAAERESFERTQTVSINKAINT QEVAVKEKHARTCILGTHHEKGAQTFWSVVNRLPLSSNAVLCWKFCHVFH KLLRDGHPNVLKDSLRYRNELSDMSRMWGHLSEGYGQLCSIYLKLLRTKM EYHTKNPRFPGNLQMSDRQLDEAGESDVNNFFQLTVEMFDYLECELNLFQ TVFNSLDMSRSVSVTAAGQCRLAPLIQVILDCSHLYDYTVKLLFKLHSCL PADTLQGHRDRFMEQFTKLKDLFYRSSNLQYFKRLIQIPQLPENPPNFLR ASALSEHISPVVVIPAEASSPDSEPVLEKDDLMDMDASQQNLFDNKFDDI FGSSFSSDPFNFNSQNGVNKDEKDHLIERLYREISGLKAQLENMKTESQR VVLQLKGHVSELEADLAEQQHLRQQAADDCEFLRAELDELRRQREDTEKA QRSLSEIERKAQANEQRYSKLKEKYSELVQNHADLLRKNAEVTKQVSMAR QAQVDLEREKKELEDSLERISDQGQRKTQEQLEVLESLKQELATSQRELQ VLQGSLETSAQSEANWAAEFAELEKERDSLVSGAAHREEELSALRKELQD TQLKLASTEESMCQLAKDQRKMLLVGSRKAAEQVIQDALNQLEEPPLISC AGSADHLLSTVTSISSCIEQLEKSWSQYLACPEDISGLLHSITLLAHLTS DAIAHGATTCLRAPPEPADSLTEACKQYGRETLAYLASLEEEGSLENADS TAMRNCLSKIKAIGEELLPRGLDIKQEELGDLVDKEMAATSAAIETATAR IEGTASPKEFYAKNSRWTEGLISASKAVGWGATVMVDAADLVVQGRGKFE ELMVCSHEIAASTAQLVAASKVKADKDSPNLAQLQQASRGVNQATAGVVA STISGKSQIEETVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPN YCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPL QVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFIL LELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHF IHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKW MPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSG GRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINT ALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLP TTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGS RNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNV ATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEA | 1475 |
| 3D view using mol* of 938 (AA BP:) | ||||||||||
 | ||||||||||
| PDB file (950)CIF file (950) >>>950.cif | HIP1 | chr7 | 75172169 | - | ALK | chr2 | 29446394 | - | MDRMASSMKQVPNPLPKVLSRRGVGAGLEAAERESFERTQTVSINKAINT QEVAVKEKHARTCILGTHHEKGAQTFWSVVNRLPLSSNAVLCWKFCHVFH KLLRDGHPNVLKDSLRYRNELSDMSRMWGHLSEGYGQLCSIYLKLLRTKM EYHTKNPRFPGNLQMSDRQLDEAGESDVNNFFQLTVEMFDYLECELNLFQ TVFNSLDMSRSVSVTAAGQCRLAPLIQVILDCSHLYDYTVKLLFKLHSCL PADTLQGHRDRFMEQFTKLKDLFYRSSNLQYFKRLIQIPQLPENPPNFLR ASALSEHISPVVVIPAEASSPDSEPVLEKDDLMDMDASQQNLFDNKFDDI FGSSFSSDPFNFNSQNGVNKDEKDHLIERLYREISGLKAQLENMKTESQR VVLQLKGHVSELEADLAEQQHLRQQAADDCEFLRAELDELRRQREDTEKA QRSLSEIERKAQANEQRYSKLKEKYSELVQNHADLLRKNAEVTKQVSMAR QAQVDLEREKKELEDSLERISDQGQRKTQEQLEVLESLKQELATSQRELQ VLQGSLETSAQSEANWAAEFAELEKERDSLVSGAAHREEELSALRKELQD TQLKLASTEESMCQLAKDQRKMLLVGSRKAAEQVIQDALNQLEEPPLISC AGSADHLLSTVTSISSCIEQLEKSWSQYLACPEDISGLLHSITLLAHLTS DAIAHGATTCLRAPPEPADSLTEACKQYGRETLAYLASLEEEGSLENADS TAMRNCLSKIKAIGEELLPRGLDIKQEELGDLVDKEMAATSAAIETATAR IEEMLSKSRAGDTGVKLEVNERILGCCTSLMQAIQVLIVASKDLQREIVE SGRGTASPKEFYAKNSRWTEGLISASKAVGWGATVMVDAADLVVQGRGKF EELMVCSHEIAASTAQLVAASKVKADKDSPNLAQLQQASRGVNQATAGVV ASTISGKSQIEETVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNP NYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSP LQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFI LLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENH FIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVK WMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTS GGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVIN TALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPL PTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHG SRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPN VATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLE | 1526 |
| 3D view using mol* of 950 (AA BP:) | ||||||||||
 | ||||||||||
| PDB file (953)CIF file (953) >>>953.cif | HIP1 | chr7 | 75168642 | - | ALK | chr2 | 29446394 | - | MDRMASSMKQVPNPLPKVLSRRGVGAGLEAAERESFERTQTVSINKAINT QEVAVKEKHARTCILGTHHEKGAQTFWSVVNRLPLSSNAVLCWKFCHVFH KLLRDGHPNVLKDSLRYRNELSDMSRMWGHLSEGYGQLCSIYLKLLRTKM EYHTKNPRFPGNLQMSDRQLDEAGESDVNNFFQLTVEMFDYLECELNLFQ TVFNSLDMSRSVSVTAAGQCRLAPLIQVILDCSHLYDYTVKLLFKLHSCL PADTLQGHRDRFMEQFTKLKDLFYRSSNLQYFKRLIQIPQLPENPPNFLR ASALSEHISPVVVIPAEASSPDSEPVLEKDDLMDMDASQQNLFDNKFDDI FGSSFSSDPFNFNSQNGVNKDEKDHLIERLYREISGLKAQLENMKTESQR VVLQLKGHVSELEADLAEQQHLRQQAADDCEFLRAELDELRRQREDTEKA QRSLSEIERKAQANEQRYSKLKEKYSELVQNHADLLRKNAEVTKQVSMAR QAQVDLEREKKELEDSLERISDQGQRKTQEQLEVLESLKQELATSQRELQ VLQGSLETSAQSEANWAAEFAELEKERDSLVSGAAHREEELSALRKELQD TQLKLASTEESMCQLAKDQRKMLLVGSRKAAEQVIQDALNQLEEPPLISC AGSADHLLSTVTSISSCIEQLEKSWSQYLACPEDISGLLHSITLLAHLTS DAIAHGATTCLRAPPEPADSLTEACKQYGRETLAYLASLEEEGSLENADS TAMRNCLSKIKAIGEELLPRGLDIKQEELGDLVDKEMAATSAAIETATAR IEGTASPKEFYAKNSRWTEGLISASKAVGWGATVMVDAADLVVQGRGKFE ELMVCSHEIAASTAQLVAASKVKADKDSPNLAQLQQASRGVNQATAGVVA STISGKSQIEETDNMDFSSMTLTQIKRQEMDSQVRVLELENELQKERQKL GELRKKHYELAGVAEGWEEVYRRKHQELQAMQMELQSPEYKLSKLRTSTI MTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGM PNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQ SLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQ YLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGGC AMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEV LEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQ DPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSP AAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSE LHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGS CTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQ | 1532 |
| 3D view using mol* of 953 (AA BP:) | ||||||||||
 | ||||||||||
| PDB file (965)CIF file (965) >>>965.cif | HIP1 | chr7 | 75168642 | - | ALK | chr2 | 29446394 | - | MDRMASSMKQVPNPLPKVLSRRGVGAGLEAAERESFERTQTVSINKAINT QEVAVKEKHARTCILGTHHEKGAQTFWSVVNRLPLSSNAVLCWKFCHVFH KLLRDGHPNVLKDSLRYRNELSDMSRMWGHLSEGYGQLCSIYLKLLRTKM EYHTKNPRFPGNLQMSDRQLDEAGESDVNNFFQLTVEMFDYLECELNLFQ TVFNSLDMSRSVSVTAAGQCRLAPLIQVILDCSHLYDYTVKLLFKLHSCL PADTLQGHRDRFMEQFTKLKDLFYRSSNLQYFKRLIQIPQLPENPPNFLR ASALSEHISPVVVIPAEASSPDSEPVLEKDDLMDMDASQQNLFDNKFDDI FGSSFSSDPFNFNSQNGVNKDEKDHLIERLYREISGLKAQLENMKTESQR VVLQLKGHVSELEADLAEQQHLRQQAADDCEFLRAELDELRRQREDTEKA QRSLSEIERKAQANEQRYSKLKEKYSELVQNHADLLRKNAEVTKQVSMAR QAQVDLEREKKELEDSLERISDQGQRKTQEQLEVLESLKQELATSQRELQ VLQGSLETSAQSEANWAAEFAELEKERDSLVSGAAHREEELSALRKELQD TQLKLASTEESMCQLAKDQRKMLLVGSRKAAEQVIQDALNQLEEPPLISC AGSADHLLSTVTSISSCIEQLEKSWSQYLACPEDISGLLHSITLLAHLTS DAIAHGATTCLRAPPEPADSLTEACKQYGRETLAYLASLEEEGSLENADS TAMRNCLSKIKAIGEELLPRGLDIKQEELGDLVDKEMAATSAAIETATAR IEEMLSKSRAGDTGVKLEVNERILGCCTSLMQAIQVLIVASKDLQREIVE SGRGTASPKEFYAKNSRWTEGLISASKAVGWGATVMVDAADLVVQGRGKF EELMVCSHEIAASTAQLVAASKVKADKDSPNLAQLQQASRGVNQATAGVV ASTISGKSQIEETDNMDFSSMTLTQIKRQEMDSQVRVLELENELQKERQK LGELRKKHYELAGVAEGWEEVYRRKHQELQAMQMELQSPEYKLSKLRTST IMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSG MPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSL QSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGC QYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGG CAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQE VLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCT QDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERS PAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPS ELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEG SCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQ | 1583 |
| 3D view using mol* of 965 (AA BP:) | ||||||||||
 | ||||||||||
Top |
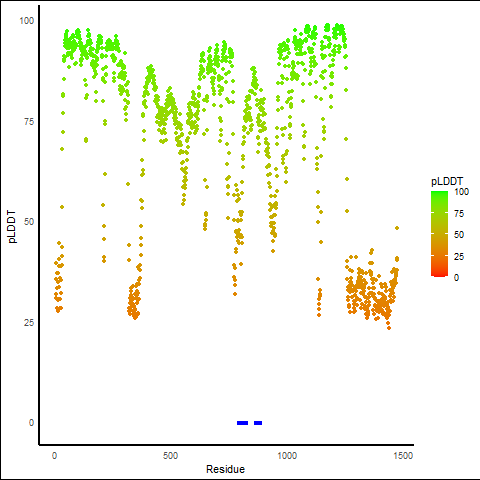
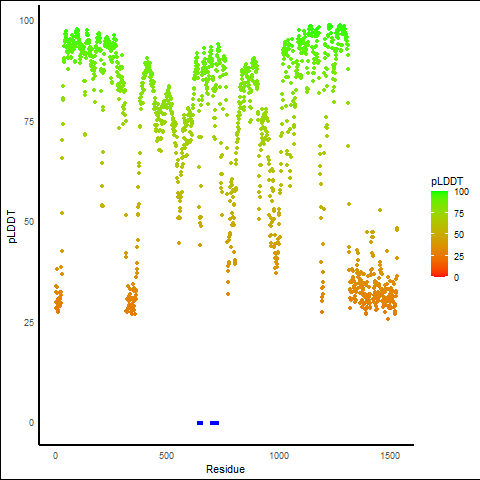
pLDDT score distribution |
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
HIP1_pLDDT.png |
ALK_pLDDT.png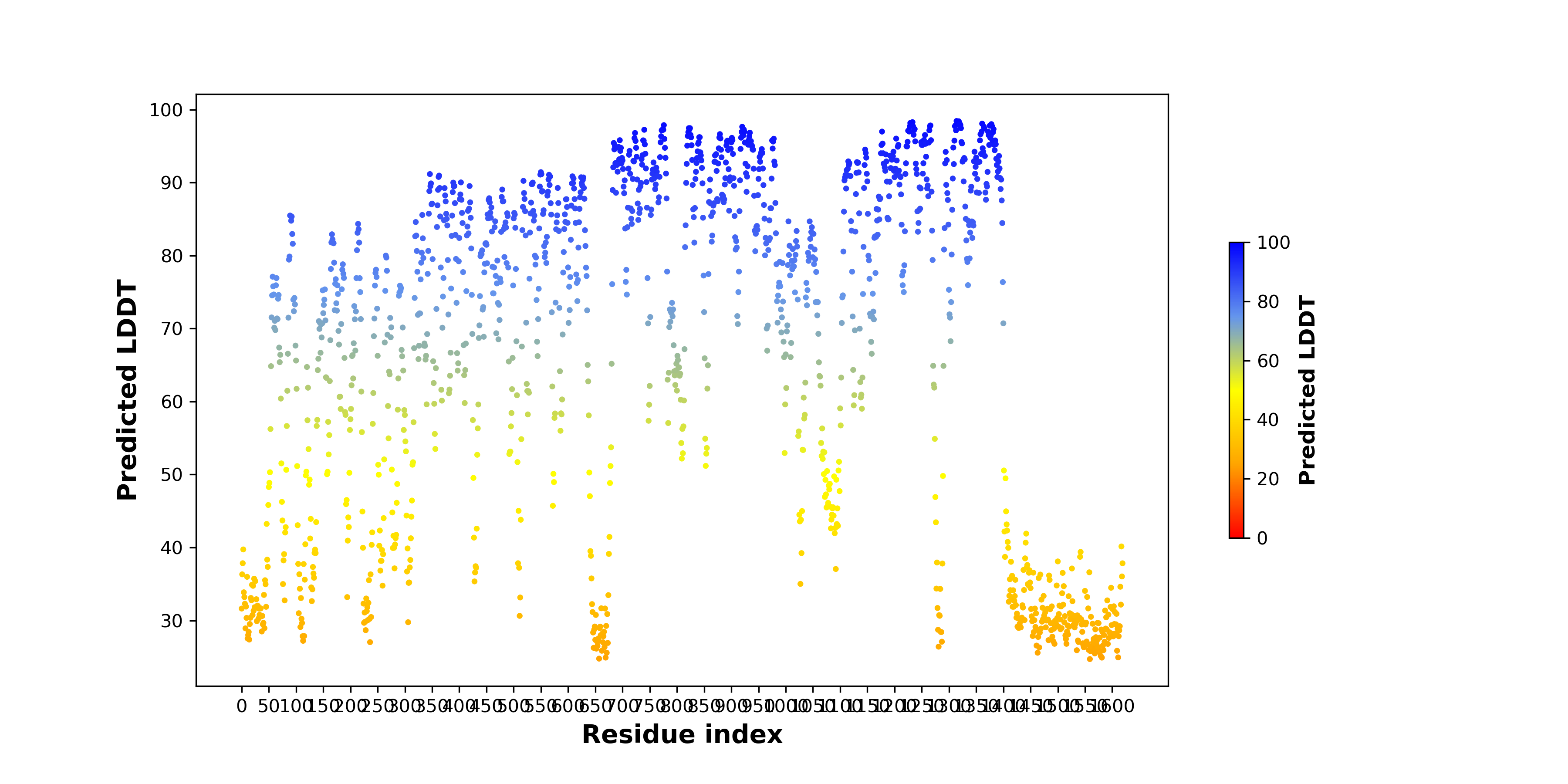 |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
Top |
Ramachandran Plot of Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| Fusion AA seq ID in FusionPDB and their Ramachandran plots |
Top |
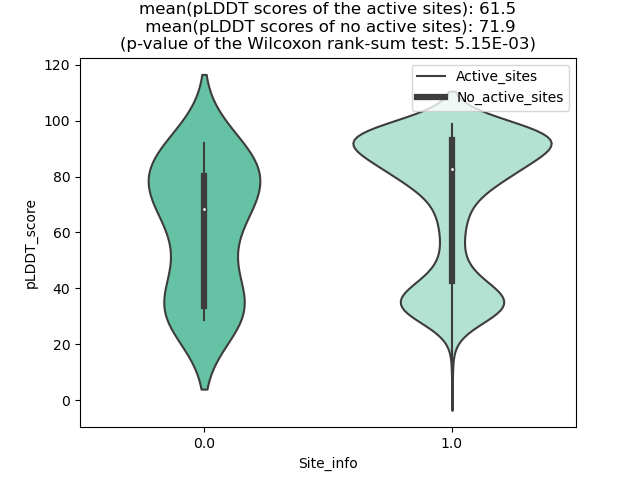
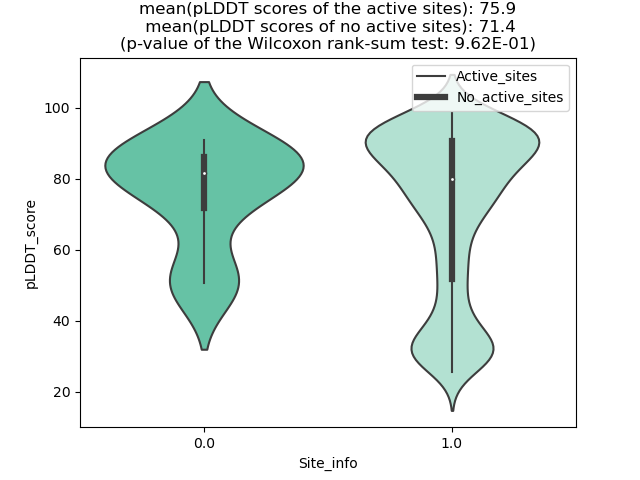
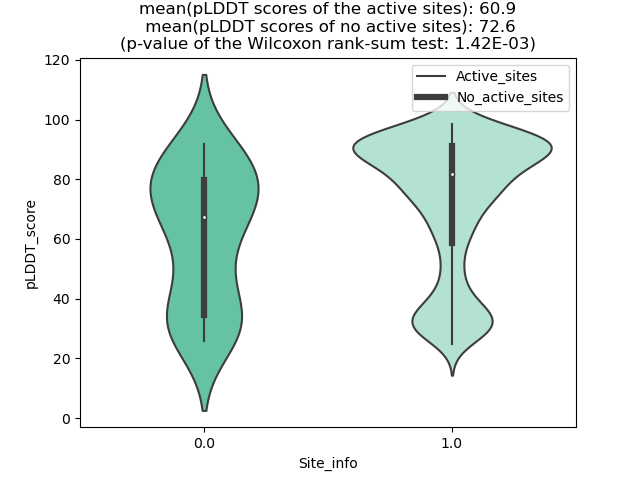
Potential Active Site Information |
 The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. |
| Fusion AA seq ID in FusionPDB | Site score | Size | D score | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
| 876 | 1.034 | 115 | 1.096 | 244.216 | 0.586 | 0.662 | 0.835 | 1.092 | 0.739 | 1.477 | 0.713 | Chain A: 910,935,939,944,945,946,947,948,949,950,9 51,952,953,955,958,962,963,964,965,966,967,968,100 1 |
| 938 | 1.14 | 135 | 1.212 | 238.042 | 0.384 | 0.782 | 1.04 | 2.252 | 0.605 | 3.722 | 0.546 | Chain A: 790,793,794,797,798,800,801,810,811,817,8 18,821,822,866,870,873,874,875,878,880,881,884 |
| 950 | 1.027 | 284 | 1.088 | 921.641 | 0.652 | 0.653 | 0.848 | 0.878 | 0.746 | 1.176 | 1.117 | Chain A: 629,630,631,632,633,634,636,637,638,641,6 42,643,645,646,647,648,649,650,651,652,656,693,696 ,700,701,703,704,705,707,708,710,711,712,722,726,7 33,878,881,882,885,888,889,892,897,898,899,901,902 ,905,906,909,910,912,913,1473,1476,1478,1479,1480, 1481,1483,1484,1485,1486 |
| 953 | 1.05 | 89 | 1.13 | 232.554 | 0.534 | 0.683 | 0.941 | 1.627 | 0.512 | 3.177 | 4.825 | Chain A: 638,641,642,643,645,646,648,650,700,703,7 04,705,707,708,710,711,722,726 |
| 965 | 1.017 | 164 | 1.047 | 389.991 | 0.509 | 0.7 | 0.94 | 0.902 | 0.962 | 0.938 | 0.681 | Chain A: 1122,1211,1236,1239,1240,1242,1243,1245,1 246,1247,1248,1249,1250,1251,1252,1254,1255,1256,1 259,1263,1264,1265,1266,1267,1268,1269,1302 |
Top |
Potentially Interacting Small Molecules through Virtual Screening |
 The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. |
| Fusion AA seq ID in FusionPDB | ZINC ID | DrugBank ID | Drug name | Docking score | Glide gscore |
Top |
 Drug information from DrugBank of the top 20 interacting small molecules. Drug information from DrugBank of the top 20 interacting small molecules. |
| ZINC ID | DrugBank ID | Drug name | Drug type | SMILES | Drug group |
Top |
Biochemical Features of Small Molecules |
 ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) |
| ZINC ID | mol_MW | dipole | SASA | FOSA | FISA | PISA | WPSA | volume | donorHB | accptHB | IP | Human Oral Absorption | Percent Human Oral Absorption | Rule Of Five | Rule Of Three |
Top |
Drug Toxicity Information |
 Toxicity information of individual drugs using eToxPred Toxicity information of individual drugs using eToxPred |
| ZINC ID | Smile | Surface Accessibility | Toxicity |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| HIP1 | CFH, PFDN1, RPS10, EIF6, HTT, HIP1R, CLTC, IFT57, CASP8, MAPK6, ITSN1, BFAR, FBXO25, HIP1, REL, DAZAP2, MAP3K1, CCDC67, NTRK1, TMEM17, GAK, MYH9, GTSE1, Calml3, SMURF1, EGFR, CPNE2, TRIM25, CLTB, Hip1r, Ap2b1, ACTA1, PDGFRB, CASP1, AP2A1, DVL2, ESR2, APAF1, CASP9, KIAA1429, MAB21L2, FAM168A, LNX1, SYT17, EHHADH, RAD23B, GLS2, FAM49B, TCEA2, CFAP53, TCEANC, RPRM, LITAF, PLEKHA4, PLEC, ESR1, SMAP2, CIT, UBASH3A, BCAR1, Rnf183, NUPR1, AP2B1, B3GAT1, CLTA, DIRAS3, GJD3, RHOB, STX6, STX7, DNAJC6, KXD1, PLCD3, ZFP1, C10orf88, UBC, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| HIP1 |  |
| ALK |  |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to HIP1-ALK |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
| HIP1 | ALK | Resistance To Crizotinib | PubMed | 33280926 |
Top |
Related Diseases to HIP1-ALK |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
| HIP1 | ALK | Lung Adenocarcinoma | PubMed | 33280926 |
| HIP1 | ALK | Lung Adenocarcinoma | MyCancerGenome | |
| HIP1 | ALK | Acute Myeloid Leukemia | MyCancerGenome | |
| HIP1 | ALK | Cancer Of Unknown Primary | MyCancerGenome | |
| HIP1 | ALK | Diffuse Large B-Cell Lymphoma | MyCancerGenome | |
| HIP1 | ALK | Not Otherwise Specified | MyCancerGenome | |
| HIP1 | ALK | Glioblastoma | MyCancerGenome |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | HIP1 | C0023893 | Liver Cirrhosis, Experimental | 1 | CTD_human |
| Tgene | ALK | C0007131 | Non-Small Cell Lung Carcinoma | 28 | CGI;CTD_human |
| Tgene | ALK | C0027819 | Neuroblastoma | 13 | CGI;CTD_human;ORPHANET |
| Tgene | ALK | C0152013 | Adenocarcinoma of lung (disorder) | 8 | CGI;CTD_human |
| Tgene | ALK | C2751681 | NEUROBLASTOMA, SUSCEPTIBILITY TO, 3 | 8 | CLINGEN;UNIPROT |
| Tgene | ALK | C0206180 | Ki-1+ Anaplastic Large Cell Lymphoma | 6 | CGI;CTD_human |
| Tgene | ALK | C0334121 | Inflammatory Myofibroblastic Tumor | 4 | CGI;CTD_human;ORPHANET |
| Tgene | ALK | C0018199 | Granuloma, Plasma Cell | 3 | CTD_human |
| Tgene | ALK | C0007621 | Neoplastic Cell Transformation | 2 | CTD_human |
| Tgene | ALK | C0027627 | Neoplasm Metastasis | 2 | CTD_human |
| Tgene | ALK | C0238463 | Papillary thyroid carcinoma | 2 | ORPHANET |
| Tgene | ALK | C0001973 | Alcoholic Intoxication, Chronic | 1 | PSYGENET |
| Tgene | ALK | C0006118 | Brain Neoplasms | 1 | CGI;CTD_human |
| Tgene | ALK | C0006142 | Malignant neoplasm of breast | 1 | CTD_human |
| Tgene | ALK | C0007134 | Renal Cell Carcinoma | 1 | CTD_human |
| Tgene | ALK | C0011570 | Mental Depression | 1 | PSYGENET |
| Tgene | ALK | C0011581 | Depressive disorder | 1 | PSYGENET |
| Tgene | ALK | C0027643 | Neoplasm Recurrence, Local | 1 | CTD_human |
| Tgene | ALK | C0036341 | Schizophrenia | 1 | PSYGENET |
| Tgene | ALK | C0079744 | Diffuse Large B-Cell Lymphoma | 1 | CTD_human |
| Tgene | ALK | C0085269 | Plasma Cell Granuloma, Pulmonary | 1 | CTD_human |
| Tgene | ALK | C0153633 | Malignant neoplasm of brain | 1 | CGI;CTD_human |
| Tgene | ALK | C0278601 | Inflammatory Breast Carcinoma | 1 | CTD_human |
| Tgene | ALK | C0279702 | Conventional (Clear Cell) Renal Cell Carcinoma | 1 | CTD_human |
| Tgene | ALK | C0496899 | Benign neoplasm of brain, unspecified | 1 | CTD_human |
| Tgene | ALK | C0678222 | Breast Carcinoma | 1 | CTD_human |
| Tgene | ALK | C0750974 | Brain Tumor, Primary | 1 | CTD_human |
| Tgene | ALK | C0750977 | Recurrent Brain Neoplasm | 1 | CTD_human |
| Tgene | ALK | C0750979 | Primary malignant neoplasm of brain | 1 | CTD_human |
| Tgene | ALK | C1257931 | Mammary Neoplasms, Human | 1 | CTD_human |
| Tgene | ALK | C1266042 | Chromophobe Renal Cell Carcinoma | 1 | CTD_human |
| Tgene | ALK | C1266043 | Sarcomatoid Renal Cell Carcinoma | 1 | CTD_human |
| Tgene | ALK | C1266044 | Collecting Duct Carcinoma of the Kidney | 1 | CTD_human |
| Tgene | ALK | C1306837 | Papillary Renal Cell Carcinoma | 1 | CTD_human |
| Tgene | ALK | C1332079 | Anaplastic Large Cell Lymphoma, ALK-Positive | 1 | ORPHANET |
| Tgene | ALK | C1458155 | Mammary Neoplasms | 1 | CTD_human |
| Tgene | ALK | C1527390 | Neoplasms, Intracranial | 1 | CTD_human |
| Tgene | ALK | C2931189 | Neural crest tumor | 1 | ORPHANET |
| Tgene | ALK | C3899155 | hereditary neuroblastoma | 1 | GENOMICS_ENGLAND |
| Tgene | ALK | C4704874 | Mammary Carcinoma, Human | 1 | CTD_human |