| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:HNF1B-NOTCH1 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: HNF1B-NOTCH1 | FusionPDB ID: 37008 | FusionGDB2.0 ID: 37008 | Hgene | Tgene | Gene symbol | HNF1B | NOTCH1 | Gene ID | 6928 | 4851 |
| Gene name | HNF1 homeobox B | notch receptor 1 | |
| Synonyms | FJHN|HNF-1-beta|HNF-1B|HNF1beta|HNF2|HPC11|LF-B3|LFB3|MODY5|TCF-2|TCF2|VHNF1 | AOS5|AOVD1|TAN1|hN1 | |
| Cytomap | 17q12 | 9q34.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | hepatocyte nuclear factor 1-betaHNF1 beta Ahomeoprotein LFB3transcription factor 2, hepatic | neurogenic locus notch homolog protein 1Notch homolog 1, translocation-associatednotch 1translocation-associated notch protein TAN-1 | |
| Modification date | 20200313 | 20200329 | |
| UniProtAcc | P35680 | P46531 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000225893, ENST00000427275, ENST00000560016, ENST00000561193, | ENST00000491649, ENST00000277541, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 10 X 10 X 8=800 | 9 X 11 X 9=891 |
| # samples | 10 | 11 | |
| ** MAII score | log2(10/800*10)=-3 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(11/891*10)=-3.01792190799726 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: HNF1B [Title/Abstract] AND NOTCH1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | HNF1B(36099431)-NOTCH1(139396940), # samples:2 NOTCH1(139397634)-HNF1B(36093736), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | HNF1B-NOTCH1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. HNF1B-NOTCH1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. HNF1B-NOTCH1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. HNF1B-NOTCH1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. NOTCH1-HNF1B seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. NOTCH1-HNF1B seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | HNF1B | GO:0001822 | kidney development | 21281489 |
| Hgene | HNF1B | GO:0045893 | positive regulation of transcription, DNA-templated | 16297991|21281489 |
| Hgene | HNF1B | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter | 15355349 |
| Tgene | NOTCH1 | GO:0007050 | cell cycle arrest | 11306509 |
| Tgene | NOTCH1 | GO:0007219 | Notch signaling pathway | 11306509 |
| Tgene | NOTCH1 | GO:0008284 | positive regulation of cell proliferation | 17849174 |
| Tgene | NOTCH1 | GO:0008285 | negative regulation of cell proliferation | 11306509|20616313 |
| Tgene | NOTCH1 | GO:0010629 | negative regulation of gene expression | 11306509 |
| Tgene | NOTCH1 | GO:0010812 | negative regulation of cell-substrate adhesion | 16501043 |
| Tgene | NOTCH1 | GO:0035924 | cellular response to vascular endothelial growth factor stimulus | 20616313 |
| Tgene | NOTCH1 | GO:0045944 | positive regulation of transcription by RNA polymerase II | 20616313 |
| Tgene | NOTCH1 | GO:0045967 | negative regulation of growth rate | 11306509 |
| Tgene | NOTCH1 | GO:0046579 | positive regulation of Ras protein signal transduction | 11306509 |
| Tgene | NOTCH1 | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 11306509 |
| Tgene | NOTCH1 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus | 20613903 |
| Tgene | NOTCH1 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis | 20616313 |
| Tgene | NOTCH1 | GO:2001027 | negative regulation of endothelial cell chemotaxis | 20616313 |
 Fusion gene breakpoints across HNF1B (5'-gene) Fusion gene breakpoints across HNF1B (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
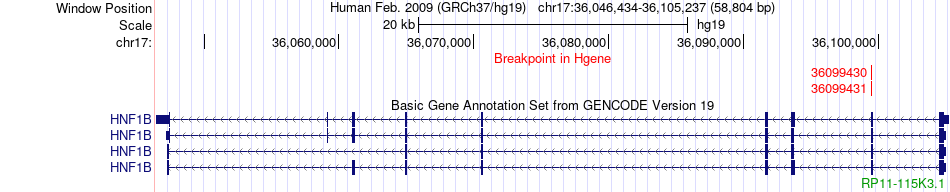 |
 Fusion gene breakpoints across NOTCH1 (3'-gene) Fusion gene breakpoints across NOTCH1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
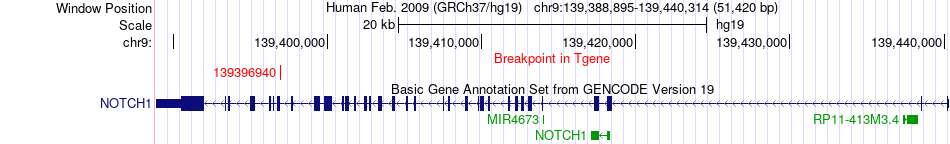 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | KIRP | TCGA-BQ-7053-01A | HNF1B | chr17 | 36099431 | - | NOTCH1 | chr9 | 139396940 | - |
| ChimerDB4 | KIRP | TCGA-BQ-7053 | HNF1B | chr17 | 36099430 | - | NOTCH1 | chr9 | 139396940 | - |
| ChimerDB4 | KIRP | TCGA-BQ-7053 | HNF1B | chr17 | 36099431 | - | NOTCH1 | chr9 | 139396940 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000225893 | HNF1B | chr17 | 36099430 | - | ENST00000277541 | NOTCH1 | chr9 | 139396940 | - | 5034 | 906 | 362 | 3406 | 1014 |
| ENST00000561193 | HNF1B | chr17 | 36099430 | - | ENST00000277541 | NOTCH1 | chr9 | 139396940 | - | 4847 | 719 | 175 | 3219 | 1014 |
| ENST00000560016 | HNF1B | chr17 | 36099430 | - | ENST00000277541 | NOTCH1 | chr9 | 139396940 | - | 4806 | 678 | 134 | 3178 | 1014 |
| ENST00000427275 | HNF1B | chr17 | 36099430 | - | ENST00000277541 | NOTCH1 | chr9 | 139396940 | - | 4806 | 678 | 134 | 3178 | 1014 |
| ENST00000225893 | HNF1B | chr17 | 36099431 | - | ENST00000277541 | NOTCH1 | chr9 | 139396940 | - | 5034 | 906 | 362 | 3406 | 1014 |
| ENST00000561193 | HNF1B | chr17 | 36099431 | - | ENST00000277541 | NOTCH1 | chr9 | 139396940 | - | 4847 | 719 | 175 | 3219 | 1014 |
| ENST00000560016 | HNF1B | chr17 | 36099431 | - | ENST00000277541 | NOTCH1 | chr9 | 139396940 | - | 4806 | 678 | 134 | 3178 | 1014 |
| ENST00000427275 | HNF1B | chr17 | 36099431 | - | ENST00000277541 | NOTCH1 | chr9 | 139396940 | - | 4806 | 678 | 134 | 3178 | 1014 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000225893 | ENST00000277541 | HNF1B | chr17 | 36099430 | - | NOTCH1 | chr9 | 139396940 | - | 0.004016955 | 0.995983 |
| ENST00000561193 | ENST00000277541 | HNF1B | chr17 | 36099430 | - | NOTCH1 | chr9 | 139396940 | - | 0.003301837 | 0.99669814 |
| ENST00000560016 | ENST00000277541 | HNF1B | chr17 | 36099430 | - | NOTCH1 | chr9 | 139396940 | - | 0.003162839 | 0.99683714 |
| ENST00000427275 | ENST00000277541 | HNF1B | chr17 | 36099430 | - | NOTCH1 | chr9 | 139396940 | - | 0.003162839 | 0.99683714 |
| ENST00000225893 | ENST00000277541 | HNF1B | chr17 | 36099431 | - | NOTCH1 | chr9 | 139396940 | - | 0.004016955 | 0.995983 |
| ENST00000561193 | ENST00000277541 | HNF1B | chr17 | 36099431 | - | NOTCH1 | chr9 | 139396940 | - | 0.003301837 | 0.99669814 |
| ENST00000560016 | ENST00000277541 | HNF1B | chr17 | 36099431 | - | NOTCH1 | chr9 | 139396940 | - | 0.003162839 | 0.99683714 |
| ENST00000427275 | ENST00000277541 | HNF1B | chr17 | 36099431 | - | NOTCH1 | chr9 | 139396940 | - | 0.003162839 | 0.99683714 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >37008_37008_1_HNF1B-NOTCH1_HNF1B_chr17_36099430_ENST00000225893_NOTCH1_chr9_139396940_ENST00000277541_length(amino acids)=1014AA_BP=182 MVSKLTSLQQELLSALLSSGVTKEVLVQALEELLPSPNFGVKLETLPLSPGSGAEPDTKPVFHTLTNGHAKGRLSGDEGSEDGDDYDTPP ILKELQALNTEEAAEQRAEVDRMLSEDPWRAAKMIKGYMQQHNIPQREVVDVTGLNQSHLSQHLNKGTPMKTQKRAALYTWYVRKQREIL RRETVEPPPPAQLHFMYVAAAAFVLLFFVGCGVLLSRKRRRQHGQLWFPEGFKVSEASKKKRREPLGEDSVGLKPLKNASDGALMDDNQN EWGDEDLETKKFRFEEPVVLPDLDDQTDHRQWTQQHLDAADLRMSAMAPTPPQGEVDADCMDVNVRGPDGFTPLMIASCSGGGLETGNSE EEEDAPAVISDFIYQGASLHNQTDRTGETALHLAARYSRSDAAKRLLEASADANIQDNMGRTPLHAAVSADAQGVFQILIRNRATDLDAR MHDGTTPLILAARLAVEGMLEDLINSHADVNAVDDLGKSALHWAAAVNNVDAAVVLLKNGANKDMQNNREETPLFLAAREGSYETAKVLL DHFANRDITDHMDRLPRDIAQERMHHDIVRLLDEYNLVRSPQLHGAPLGGTPTLSPPLCSPNGYLGSLKPGVQGKKVRKPSSKGLACGSK EAKDLKARRKKSQDGKGCLLDSSGMLSPVDSLESPHGYLSDVASPPLLPSPFQQSPSVPLNHLPGMPDTHLGIGHLNVAAKPEMAALGGG GRLAFETGPPRLSHLPVASGTSTVLGSSSGGALNFTVGGSTSLNGQCEWLSRLQSGMVPNQYNPLRGSVAPGPLSTQAPSLQHGMVGPLH SSLAASALSQMMSYQGLPSTRLATQPHLVQTQQVQPQNLQMQQQNLQPANIQQQQSLQPPPPPPQPHLGVSSAASGHLGRSFLSGEPSQA DVQPLGPSSLAVHTILPQESPALPTSLPSSLVPPVTAAQFLTPPSQHSYSSPVDNTPSHQLQVPEHPFLTPSPESPDQWSSSSPHSNVSD -------------------------------------------------------------- >37008_37008_2_HNF1B-NOTCH1_HNF1B_chr17_36099430_ENST00000427275_NOTCH1_chr9_139396940_ENST00000277541_length(amino acids)=1014AA_BP=182 MVSKLTSLQQELLSALLSSGVTKEVLVQALEELLPSPNFGVKLETLPLSPGSGAEPDTKPVFHTLTNGHAKGRLSGDEGSEDGDDYDTPP ILKELQALNTEEAAEQRAEVDRMLSEDPWRAAKMIKGYMQQHNIPQREVVDVTGLNQSHLSQHLNKGTPMKTQKRAALYTWYVRKQREIL RRETVEPPPPAQLHFMYVAAAAFVLLFFVGCGVLLSRKRRRQHGQLWFPEGFKVSEASKKKRREPLGEDSVGLKPLKNASDGALMDDNQN EWGDEDLETKKFRFEEPVVLPDLDDQTDHRQWTQQHLDAADLRMSAMAPTPPQGEVDADCMDVNVRGPDGFTPLMIASCSGGGLETGNSE EEEDAPAVISDFIYQGASLHNQTDRTGETALHLAARYSRSDAAKRLLEASADANIQDNMGRTPLHAAVSADAQGVFQILIRNRATDLDAR MHDGTTPLILAARLAVEGMLEDLINSHADVNAVDDLGKSALHWAAAVNNVDAAVVLLKNGANKDMQNNREETPLFLAAREGSYETAKVLL DHFANRDITDHMDRLPRDIAQERMHHDIVRLLDEYNLVRSPQLHGAPLGGTPTLSPPLCSPNGYLGSLKPGVQGKKVRKPSSKGLACGSK EAKDLKARRKKSQDGKGCLLDSSGMLSPVDSLESPHGYLSDVASPPLLPSPFQQSPSVPLNHLPGMPDTHLGIGHLNVAAKPEMAALGGG GRLAFETGPPRLSHLPVASGTSTVLGSSSGGALNFTVGGSTSLNGQCEWLSRLQSGMVPNQYNPLRGSVAPGPLSTQAPSLQHGMVGPLH SSLAASALSQMMSYQGLPSTRLATQPHLVQTQQVQPQNLQMQQQNLQPANIQQQQSLQPPPPPPQPHLGVSSAASGHLGRSFLSGEPSQA DVQPLGPSSLAVHTILPQESPALPTSLPSSLVPPVTAAQFLTPPSQHSYSSPVDNTPSHQLQVPEHPFLTPSPESPDQWSSSSPHSNVSD -------------------------------------------------------------- >37008_37008_3_HNF1B-NOTCH1_HNF1B_chr17_36099430_ENST00000560016_NOTCH1_chr9_139396940_ENST00000277541_length(amino acids)=1014AA_BP=182 MVSKLTSLQQELLSALLSSGVTKEVLVQALEELLPSPNFGVKLETLPLSPGSGAEPDTKPVFHTLTNGHAKGRLSGDEGSEDGDDYDTPP ILKELQALNTEEAAEQRAEVDRMLSEDPWRAAKMIKGYMQQHNIPQREVVDVTGLNQSHLSQHLNKGTPMKTQKRAALYTWYVRKQREIL RRETVEPPPPAQLHFMYVAAAAFVLLFFVGCGVLLSRKRRRQHGQLWFPEGFKVSEASKKKRREPLGEDSVGLKPLKNASDGALMDDNQN EWGDEDLETKKFRFEEPVVLPDLDDQTDHRQWTQQHLDAADLRMSAMAPTPPQGEVDADCMDVNVRGPDGFTPLMIASCSGGGLETGNSE EEEDAPAVISDFIYQGASLHNQTDRTGETALHLAARYSRSDAAKRLLEASADANIQDNMGRTPLHAAVSADAQGVFQILIRNRATDLDAR MHDGTTPLILAARLAVEGMLEDLINSHADVNAVDDLGKSALHWAAAVNNVDAAVVLLKNGANKDMQNNREETPLFLAAREGSYETAKVLL DHFANRDITDHMDRLPRDIAQERMHHDIVRLLDEYNLVRSPQLHGAPLGGTPTLSPPLCSPNGYLGSLKPGVQGKKVRKPSSKGLACGSK EAKDLKARRKKSQDGKGCLLDSSGMLSPVDSLESPHGYLSDVASPPLLPSPFQQSPSVPLNHLPGMPDTHLGIGHLNVAAKPEMAALGGG GRLAFETGPPRLSHLPVASGTSTVLGSSSGGALNFTVGGSTSLNGQCEWLSRLQSGMVPNQYNPLRGSVAPGPLSTQAPSLQHGMVGPLH SSLAASALSQMMSYQGLPSTRLATQPHLVQTQQVQPQNLQMQQQNLQPANIQQQQSLQPPPPPPQPHLGVSSAASGHLGRSFLSGEPSQA DVQPLGPSSLAVHTILPQESPALPTSLPSSLVPPVTAAQFLTPPSQHSYSSPVDNTPSHQLQVPEHPFLTPSPESPDQWSSSSPHSNVSD -------------------------------------------------------------- >37008_37008_4_HNF1B-NOTCH1_HNF1B_chr17_36099430_ENST00000561193_NOTCH1_chr9_139396940_ENST00000277541_length(amino acids)=1014AA_BP=182 MVSKLTSLQQELLSALLSSGVTKEVLVQALEELLPSPNFGVKLETLPLSPGSGAEPDTKPVFHTLTNGHAKGRLSGDEGSEDGDDYDTPP ILKELQALNTEEAAEQRAEVDRMLSEDPWRAAKMIKGYMQQHNIPQREVVDVTGLNQSHLSQHLNKGTPMKTQKRAALYTWYVRKQREIL RRETVEPPPPAQLHFMYVAAAAFVLLFFVGCGVLLSRKRRRQHGQLWFPEGFKVSEASKKKRREPLGEDSVGLKPLKNASDGALMDDNQN EWGDEDLETKKFRFEEPVVLPDLDDQTDHRQWTQQHLDAADLRMSAMAPTPPQGEVDADCMDVNVRGPDGFTPLMIASCSGGGLETGNSE EEEDAPAVISDFIYQGASLHNQTDRTGETALHLAARYSRSDAAKRLLEASADANIQDNMGRTPLHAAVSADAQGVFQILIRNRATDLDAR MHDGTTPLILAARLAVEGMLEDLINSHADVNAVDDLGKSALHWAAAVNNVDAAVVLLKNGANKDMQNNREETPLFLAAREGSYETAKVLL DHFANRDITDHMDRLPRDIAQERMHHDIVRLLDEYNLVRSPQLHGAPLGGTPTLSPPLCSPNGYLGSLKPGVQGKKVRKPSSKGLACGSK EAKDLKARRKKSQDGKGCLLDSSGMLSPVDSLESPHGYLSDVASPPLLPSPFQQSPSVPLNHLPGMPDTHLGIGHLNVAAKPEMAALGGG GRLAFETGPPRLSHLPVASGTSTVLGSSSGGALNFTVGGSTSLNGQCEWLSRLQSGMVPNQYNPLRGSVAPGPLSTQAPSLQHGMVGPLH SSLAASALSQMMSYQGLPSTRLATQPHLVQTQQVQPQNLQMQQQNLQPANIQQQQSLQPPPPPPQPHLGVSSAASGHLGRSFLSGEPSQA DVQPLGPSSLAVHTILPQESPALPTSLPSSLVPPVTAAQFLTPPSQHSYSSPVDNTPSHQLQVPEHPFLTPSPESPDQWSSSSPHSNVSD -------------------------------------------------------------- >37008_37008_5_HNF1B-NOTCH1_HNF1B_chr17_36099431_ENST00000225893_NOTCH1_chr9_139396940_ENST00000277541_length(amino acids)=1014AA_BP=182 MVSKLTSLQQELLSALLSSGVTKEVLVQALEELLPSPNFGVKLETLPLSPGSGAEPDTKPVFHTLTNGHAKGRLSGDEGSEDGDDYDTPP ILKELQALNTEEAAEQRAEVDRMLSEDPWRAAKMIKGYMQQHNIPQREVVDVTGLNQSHLSQHLNKGTPMKTQKRAALYTWYVRKQREIL RRETVEPPPPAQLHFMYVAAAAFVLLFFVGCGVLLSRKRRRQHGQLWFPEGFKVSEASKKKRREPLGEDSVGLKPLKNASDGALMDDNQN EWGDEDLETKKFRFEEPVVLPDLDDQTDHRQWTQQHLDAADLRMSAMAPTPPQGEVDADCMDVNVRGPDGFTPLMIASCSGGGLETGNSE EEEDAPAVISDFIYQGASLHNQTDRTGETALHLAARYSRSDAAKRLLEASADANIQDNMGRTPLHAAVSADAQGVFQILIRNRATDLDAR MHDGTTPLILAARLAVEGMLEDLINSHADVNAVDDLGKSALHWAAAVNNVDAAVVLLKNGANKDMQNNREETPLFLAAREGSYETAKVLL DHFANRDITDHMDRLPRDIAQERMHHDIVRLLDEYNLVRSPQLHGAPLGGTPTLSPPLCSPNGYLGSLKPGVQGKKVRKPSSKGLACGSK EAKDLKARRKKSQDGKGCLLDSSGMLSPVDSLESPHGYLSDVASPPLLPSPFQQSPSVPLNHLPGMPDTHLGIGHLNVAAKPEMAALGGG GRLAFETGPPRLSHLPVASGTSTVLGSSSGGALNFTVGGSTSLNGQCEWLSRLQSGMVPNQYNPLRGSVAPGPLSTQAPSLQHGMVGPLH SSLAASALSQMMSYQGLPSTRLATQPHLVQTQQVQPQNLQMQQQNLQPANIQQQQSLQPPPPPPQPHLGVSSAASGHLGRSFLSGEPSQA DVQPLGPSSLAVHTILPQESPALPTSLPSSLVPPVTAAQFLTPPSQHSYSSPVDNTPSHQLQVPEHPFLTPSPESPDQWSSSSPHSNVSD -------------------------------------------------------------- >37008_37008_6_HNF1B-NOTCH1_HNF1B_chr17_36099431_ENST00000427275_NOTCH1_chr9_139396940_ENST00000277541_length(amino acids)=1014AA_BP=182 MVSKLTSLQQELLSALLSSGVTKEVLVQALEELLPSPNFGVKLETLPLSPGSGAEPDTKPVFHTLTNGHAKGRLSGDEGSEDGDDYDTPP ILKELQALNTEEAAEQRAEVDRMLSEDPWRAAKMIKGYMQQHNIPQREVVDVTGLNQSHLSQHLNKGTPMKTQKRAALYTWYVRKQREIL RRETVEPPPPAQLHFMYVAAAAFVLLFFVGCGVLLSRKRRRQHGQLWFPEGFKVSEASKKKRREPLGEDSVGLKPLKNASDGALMDDNQN EWGDEDLETKKFRFEEPVVLPDLDDQTDHRQWTQQHLDAADLRMSAMAPTPPQGEVDADCMDVNVRGPDGFTPLMIASCSGGGLETGNSE EEEDAPAVISDFIYQGASLHNQTDRTGETALHLAARYSRSDAAKRLLEASADANIQDNMGRTPLHAAVSADAQGVFQILIRNRATDLDAR MHDGTTPLILAARLAVEGMLEDLINSHADVNAVDDLGKSALHWAAAVNNVDAAVVLLKNGANKDMQNNREETPLFLAAREGSYETAKVLL DHFANRDITDHMDRLPRDIAQERMHHDIVRLLDEYNLVRSPQLHGAPLGGTPTLSPPLCSPNGYLGSLKPGVQGKKVRKPSSKGLACGSK EAKDLKARRKKSQDGKGCLLDSSGMLSPVDSLESPHGYLSDVASPPLLPSPFQQSPSVPLNHLPGMPDTHLGIGHLNVAAKPEMAALGGG GRLAFETGPPRLSHLPVASGTSTVLGSSSGGALNFTVGGSTSLNGQCEWLSRLQSGMVPNQYNPLRGSVAPGPLSTQAPSLQHGMVGPLH SSLAASALSQMMSYQGLPSTRLATQPHLVQTQQVQPQNLQMQQQNLQPANIQQQQSLQPPPPPPQPHLGVSSAASGHLGRSFLSGEPSQA DVQPLGPSSLAVHTILPQESPALPTSLPSSLVPPVTAAQFLTPPSQHSYSSPVDNTPSHQLQVPEHPFLTPSPESPDQWSSSSPHSNVSD -------------------------------------------------------------- >37008_37008_7_HNF1B-NOTCH1_HNF1B_chr17_36099431_ENST00000560016_NOTCH1_chr9_139396940_ENST00000277541_length(amino acids)=1014AA_BP=182 MVSKLTSLQQELLSALLSSGVTKEVLVQALEELLPSPNFGVKLETLPLSPGSGAEPDTKPVFHTLTNGHAKGRLSGDEGSEDGDDYDTPP ILKELQALNTEEAAEQRAEVDRMLSEDPWRAAKMIKGYMQQHNIPQREVVDVTGLNQSHLSQHLNKGTPMKTQKRAALYTWYVRKQREIL RRETVEPPPPAQLHFMYVAAAAFVLLFFVGCGVLLSRKRRRQHGQLWFPEGFKVSEASKKKRREPLGEDSVGLKPLKNASDGALMDDNQN EWGDEDLETKKFRFEEPVVLPDLDDQTDHRQWTQQHLDAADLRMSAMAPTPPQGEVDADCMDVNVRGPDGFTPLMIASCSGGGLETGNSE EEEDAPAVISDFIYQGASLHNQTDRTGETALHLAARYSRSDAAKRLLEASADANIQDNMGRTPLHAAVSADAQGVFQILIRNRATDLDAR MHDGTTPLILAARLAVEGMLEDLINSHADVNAVDDLGKSALHWAAAVNNVDAAVVLLKNGANKDMQNNREETPLFLAAREGSYETAKVLL DHFANRDITDHMDRLPRDIAQERMHHDIVRLLDEYNLVRSPQLHGAPLGGTPTLSPPLCSPNGYLGSLKPGVQGKKVRKPSSKGLACGSK EAKDLKARRKKSQDGKGCLLDSSGMLSPVDSLESPHGYLSDVASPPLLPSPFQQSPSVPLNHLPGMPDTHLGIGHLNVAAKPEMAALGGG GRLAFETGPPRLSHLPVASGTSTVLGSSSGGALNFTVGGSTSLNGQCEWLSRLQSGMVPNQYNPLRGSVAPGPLSTQAPSLQHGMVGPLH SSLAASALSQMMSYQGLPSTRLATQPHLVQTQQVQPQNLQMQQQNLQPANIQQQQSLQPPPPPPQPHLGVSSAASGHLGRSFLSGEPSQA DVQPLGPSSLAVHTILPQESPALPTSLPSSLVPPVTAAQFLTPPSQHSYSSPVDNTPSHQLQVPEHPFLTPSPESPDQWSSSSPHSNVSD -------------------------------------------------------------- >37008_37008_8_HNF1B-NOTCH1_HNF1B_chr17_36099431_ENST00000561193_NOTCH1_chr9_139396940_ENST00000277541_length(amino acids)=1014AA_BP=182 MVSKLTSLQQELLSALLSSGVTKEVLVQALEELLPSPNFGVKLETLPLSPGSGAEPDTKPVFHTLTNGHAKGRLSGDEGSEDGDDYDTPP ILKELQALNTEEAAEQRAEVDRMLSEDPWRAAKMIKGYMQQHNIPQREVVDVTGLNQSHLSQHLNKGTPMKTQKRAALYTWYVRKQREIL RRETVEPPPPAQLHFMYVAAAAFVLLFFVGCGVLLSRKRRRQHGQLWFPEGFKVSEASKKKRREPLGEDSVGLKPLKNASDGALMDDNQN EWGDEDLETKKFRFEEPVVLPDLDDQTDHRQWTQQHLDAADLRMSAMAPTPPQGEVDADCMDVNVRGPDGFTPLMIASCSGGGLETGNSE EEEDAPAVISDFIYQGASLHNQTDRTGETALHLAARYSRSDAAKRLLEASADANIQDNMGRTPLHAAVSADAQGVFQILIRNRATDLDAR MHDGTTPLILAARLAVEGMLEDLINSHADVNAVDDLGKSALHWAAAVNNVDAAVVLLKNGANKDMQNNREETPLFLAAREGSYETAKVLL DHFANRDITDHMDRLPRDIAQERMHHDIVRLLDEYNLVRSPQLHGAPLGGTPTLSPPLCSPNGYLGSLKPGVQGKKVRKPSSKGLACGSK EAKDLKARRKKSQDGKGCLLDSSGMLSPVDSLESPHGYLSDVASPPLLPSPFQQSPSVPLNHLPGMPDTHLGIGHLNVAAKPEMAALGGG GRLAFETGPPRLSHLPVASGTSTVLGSSSGGALNFTVGGSTSLNGQCEWLSRLQSGMVPNQYNPLRGSVAPGPLSTQAPSLQHGMVGPLH SSLAASALSQMMSYQGLPSTRLATQPHLVQTQQVQPQNLQMQQQNLQPANIQQQQSLQPPPPPPQPHLGVSSAASGHLGRSFLSGEPSQA DVQPLGPSSLAVHTILPQESPALPTSLPSSLVPPVTAAQFLTPPSQHSYSSPVDNTPSHQLQVPEHPFLTPSPESPDQWSSSSPHSNVSD -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr17:36099431/chr9:139396940) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| HNF1B | NOTCH1 |
| FUNCTION: Transcription factor, probably binds to the inverted palindrome 5'-GTTAATNATTAAC-3'. | FUNCTION: Functions as a receptor for membrane-bound ligands Jagged-1 (JAG1), Jagged-2 (JAG2) and Delta-1 (DLL1) to regulate cell-fate determination. Upon ligand activation through the released notch intracellular domain (NICD) it forms a transcriptional activator complex with RBPJ/RBPSUH and activates genes of the enhancer of split locus. Affects the implementation of differentiation, proliferation and apoptotic programs. Involved in angiogenesis; negatively regulates endothelial cell proliferation and migration and angiogenic sprouting. Involved in the maturation of both CD4(+) and CD8(+) cells in the thymus. Important for follicular differentiation and possibly cell fate selection within the follicle. During cerebellar development, functions as a receptor for neuronal DNER and is involved in the differentiation of Bergmann glia. Represses neuronal and myogenic differentiation. May play an essential role in postimplantation development, probably in some aspect of cell specification and/or differentiation. May be involved in mesoderm development, somite formation and neurogenesis. May enhance HIF1A function by sequestering HIF1AN away from HIF1A. Required for the THBS4 function in regulating protective astrogenesis from the subventricular zone (SVZ) niche after injury. Involved in determination of left/right symmetry by modulating the balance between motile and immotile (sensory) cilia at the left-right organiser (LRO). {ECO:0000269|PubMed:20616313}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | HNF1B | chr17:36099430 | chr9:139396940 | ENST00000225893 | - | 2 | 9 | 1_32 | 181.33333333333334 | 558.0 | Domain | HNF-p1 |
| Hgene | HNF1B | chr17:36099430 | chr9:139396940 | ENST00000427275 | - | 2 | 7 | 1_32 | 181.33333333333334 | 458.0 | Domain | HNF-p1 |
| Hgene | HNF1B | chr17:36099430 | chr9:139396940 | ENST00000561193 | - | 2 | 9 | 1_32 | 181.33333333333334 | 532.0 | Domain | HNF-p1 |
| Hgene | HNF1B | chr17:36099431 | chr9:139396940 | ENST00000225893 | - | 2 | 9 | 1_32 | 181.33333333333334 | 558.0 | Domain | HNF-p1 |
| Hgene | HNF1B | chr17:36099431 | chr9:139396940 | ENST00000427275 | - | 2 | 7 | 1_32 | 181.33333333333334 | 458.0 | Domain | HNF-p1 |
| Hgene | HNF1B | chr17:36099431 | chr9:139396940 | ENST00000561193 | - | 2 | 9 | 1_32 | 181.33333333333334 | 532.0 | Domain | HNF-p1 |
| Hgene | HNF1B | chr17:36099430 | chr9:139396940 | ENST00000225893 | - | 2 | 9 | 1_31 | 181.33333333333334 | 558.0 | Region | Dimerization |
| Hgene | HNF1B | chr17:36099430 | chr9:139396940 | ENST00000427275 | - | 2 | 7 | 1_31 | 181.33333333333334 | 458.0 | Region | Dimerization |
| Hgene | HNF1B | chr17:36099430 | chr9:139396940 | ENST00000561193 | - | 2 | 9 | 1_31 | 181.33333333333334 | 532.0 | Region | Dimerization |
| Hgene | HNF1B | chr17:36099431 | chr9:139396940 | ENST00000225893 | - | 2 | 9 | 1_31 | 181.33333333333334 | 558.0 | Region | Dimerization |
| Hgene | HNF1B | chr17:36099431 | chr9:139396940 | ENST00000427275 | - | 2 | 7 | 1_31 | 181.33333333333334 | 458.0 | Region | Dimerization |
| Hgene | HNF1B | chr17:36099431 | chr9:139396940 | ENST00000561193 | - | 2 | 9 | 1_31 | 181.33333333333334 | 532.0 | Region | Dimerization |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1728_1731 | 1722.3333333333333 | 2556.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1740_1743 | 1722.3333333333333 | 2556.0 | Compositional bias | Note=Poly-Ala | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1901_1904 | 1722.3333333333333 | 2556.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 2259_2262 | 1722.3333333333333 | 2556.0 | Compositional bias | Note=Poly-Gly | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 2403_2406 | 1722.3333333333333 | 2556.0 | Compositional bias | Note=Poly-Gln | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 2410_2417 | 1722.3333333333333 | 2556.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 2521_2524 | 1722.3333333333333 | 2556.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1728_1731 | 1722.3333333333333 | 2556.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1740_1743 | 1722.3333333333333 | 2556.0 | Compositional bias | Note=Poly-Ala | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1901_1904 | 1722.3333333333333 | 2556.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 2259_2262 | 1722.3333333333333 | 2556.0 | Compositional bias | Note=Poly-Gly | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 2403_2406 | 1722.3333333333333 | 2556.0 | Compositional bias | Note=Poly-Gln | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 2410_2417 | 1722.3333333333333 | 2556.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 2521_2524 | 1722.3333333333333 | 2556.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1947_1955 | 1722.3333333333333 | 2556.0 | Region | HIF1AN-binding | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 2014_2022 | 1722.3333333333333 | 2556.0 | Region | HIF1AN-binding | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1947_1955 | 1722.3333333333333 | 2556.0 | Region | HIF1AN-binding | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 2014_2022 | 1722.3333333333333 | 2556.0 | Region | HIF1AN-binding | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1927_1956 | 1722.3333333333333 | 2556.0 | Repeat | ANK 1 | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1960_1990 | 1722.3333333333333 | 2556.0 | Repeat | ANK 2 | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1994_2023 | 1722.3333333333333 | 2556.0 | Repeat | ANK 3 | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 2027_2056 | 1722.3333333333333 | 2556.0 | Repeat | ANK 4 | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 2060_2089 | 1722.3333333333333 | 2556.0 | Repeat | ANK 5 | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 2095_2122 | 1722.3333333333333 | 2556.0 | Repeat | Note=ANK 6 | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1927_1956 | 1722.3333333333333 | 2556.0 | Repeat | ANK 1 | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1960_1990 | 1722.3333333333333 | 2556.0 | Repeat | ANK 2 | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1994_2023 | 1722.3333333333333 | 2556.0 | Repeat | ANK 3 | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 2027_2056 | 1722.3333333333333 | 2556.0 | Repeat | ANK 4 | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 2060_2089 | 1722.3333333333333 | 2556.0 | Repeat | ANK 5 | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 2095_2122 | 1722.3333333333333 | 2556.0 | Repeat | Note=ANK 6 | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1757_2555 | 1722.3333333333333 | 2556.0 | Topological domain | Cytoplasmic | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1757_2555 | 1722.3333333333333 | 2556.0 | Topological domain | Cytoplasmic | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1736_1756 | 1722.3333333333333 | 2556.0 | Transmembrane | Helical | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1736_1756 | 1722.3333333333333 | 2556.0 | Transmembrane | Helical |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | HNF1B | chr17:36099430 | chr9:139396940 | ENST00000225893 | - | 2 | 9 | 231_311 | 181.33333333333334 | 558.0 | DNA binding | Homeobox%3B HNF1-type |
| Hgene | HNF1B | chr17:36099430 | chr9:139396940 | ENST00000427275 | - | 2 | 7 | 231_311 | 181.33333333333334 | 458.0 | DNA binding | Homeobox%3B HNF1-type |
| Hgene | HNF1B | chr17:36099430 | chr9:139396940 | ENST00000561193 | - | 2 | 9 | 231_311 | 181.33333333333334 | 532.0 | DNA binding | Homeobox%3B HNF1-type |
| Hgene | HNF1B | chr17:36099431 | chr9:139396940 | ENST00000225893 | - | 2 | 9 | 231_311 | 181.33333333333334 | 558.0 | DNA binding | Homeobox%3B HNF1-type |
| Hgene | HNF1B | chr17:36099431 | chr9:139396940 | ENST00000427275 | - | 2 | 7 | 231_311 | 181.33333333333334 | 458.0 | DNA binding | Homeobox%3B HNF1-type |
| Hgene | HNF1B | chr17:36099431 | chr9:139396940 | ENST00000561193 | - | 2 | 9 | 231_311 | 181.33333333333334 | 532.0 | DNA binding | Homeobox%3B HNF1-type |
| Hgene | HNF1B | chr17:36099430 | chr9:139396940 | ENST00000225893 | - | 2 | 9 | 93_188 | 181.33333333333334 | 558.0 | Domain | POU-specific atypical |
| Hgene | HNF1B | chr17:36099430 | chr9:139396940 | ENST00000427275 | - | 2 | 7 | 93_188 | 181.33333333333334 | 458.0 | Domain | POU-specific atypical |
| Hgene | HNF1B | chr17:36099430 | chr9:139396940 | ENST00000561193 | - | 2 | 9 | 93_188 | 181.33333333333334 | 532.0 | Domain | POU-specific atypical |
| Hgene | HNF1B | chr17:36099431 | chr9:139396940 | ENST00000225893 | - | 2 | 9 | 93_188 | 181.33333333333334 | 558.0 | Domain | POU-specific atypical |
| Hgene | HNF1B | chr17:36099431 | chr9:139396940 | ENST00000427275 | - | 2 | 7 | 93_188 | 181.33333333333334 | 458.0 | Domain | POU-specific atypical |
| Hgene | HNF1B | chr17:36099431 | chr9:139396940 | ENST00000561193 | - | 2 | 9 | 93_188 | 181.33333333333334 | 532.0 | Domain | POU-specific atypical |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1575_1578 | 1722.3333333333333 | 2556.0 | Compositional bias | Note=Poly-Val | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1661_1664 | 1722.3333333333333 | 2556.0 | Compositional bias | Note=Poly-Arg | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1575_1578 | 1722.3333333333333 | 2556.0 | Compositional bias | Note=Poly-Val | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1661_1664 | 1722.3333333333333 | 2556.0 | Compositional bias | Note=Poly-Arg | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1021_1057 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 27 | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 102_139 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 3 | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1059_1095 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 28 | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1097_1143 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 29 | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1145_1181 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 30 | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1183_1219 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 31%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1221_1265 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 32%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1267_1305 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 33 | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1307_1346 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 34 | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1348_1384 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 35 | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1387_1426 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 36 | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 140_176 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 4 | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 178_216 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 5%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 20_58 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 1 | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 218_255 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 6 | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 257_293 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 7%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 295_333 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 8%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 335_371 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 9%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 372_410 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 10 | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 412_450 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 11%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 452_488 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 12%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 490_526 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 13%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 528_564 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 14%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 566_601 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 15%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 59_99 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 2 | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 603_639 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 16%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 641_676 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 17%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 678_714 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 18%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 716_751 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 19%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 753_789 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 20 | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 791_827 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 21%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 829_867 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 22 | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 869_905 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 23%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 907_943 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 24 | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 945_981 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 25%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 983_1019 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 26 | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1021_1057 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 27 | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 102_139 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 3 | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1059_1095 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 28 | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1097_1143 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 29 | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1145_1181 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 30 | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1183_1219 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 31%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1221_1265 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 32%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1267_1305 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 33 | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1307_1346 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 34 | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1348_1384 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 35 | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1387_1426 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 36 | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 140_176 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 4 | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 178_216 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 5%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 20_58 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 1 | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 218_255 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 6 | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 257_293 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 7%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 295_333 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 8%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 335_371 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 9%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 372_410 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 10 | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 412_450 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 11%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 452_488 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 12%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 490_526 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 13%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 528_564 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 14%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 566_601 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 15%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 59_99 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 2 | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 603_639 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 16%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 641_676 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 17%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 678_714 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 18%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 716_751 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 19%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 753_789 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 20 | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 791_827 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 21%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 829_867 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 22 | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 869_905 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 23%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 907_943 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 24 | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 945_981 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 25%3B calcium-binding | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 983_1019 | 1722.3333333333333 | 2556.0 | Domain | EGF-like 26 | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1449_1489 | 1722.3333333333333 | 2556.0 | Repeat | LNR 1 | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1490_1531 | 1722.3333333333333 | 2556.0 | Repeat | LNR 2 | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1532_1571 | 1722.3333333333333 | 2556.0 | Repeat | LNR 3 | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1449_1489 | 1722.3333333333333 | 2556.0 | Repeat | LNR 1 | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1490_1531 | 1722.3333333333333 | 2556.0 | Repeat | LNR 2 | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 1532_1571 | 1722.3333333333333 | 2556.0 | Repeat | LNR 3 | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 19_1735 | 1722.3333333333333 | 2556.0 | Topological domain | Extracellular | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 19_1735 | 1722.3333333333333 | 2556.0 | Topological domain | Extracellular |
Top |
Fusion Protein Structures |
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
| PDB file >>>1795_HNF1B_36099431_NOTCH1_139396940_ranked_0.pdb | HNF1B | 36099431 | 36099431 | ENST00000277541 | NOTCH1 | chr9 | 139396940 | - | MVSKLTSLQQELLSALLSSGVTKEVLVQALEELLPSPNFGVKLETLPLSPGSGAEPDTKPVFHTLTNGHAKGRLSGDEGSEDGDDYDTPP ILKELQALNTEEAAEQRAEVDRMLSEDPWRAAKMIKGYMQQHNIPQREVVDVTGLNQSHLSQHLNKGTPMKTQKRAALYTWYVRKQREIL RRETVEPPPPAQLHFMYVAAAAFVLLFFVGCGVLLSRKRRRQHGQLWFPEGFKVSEASKKKRREPLGEDSVGLKPLKNASDGALMDDNQN EWGDEDLETKKFRFEEPVVLPDLDDQTDHRQWTQQHLDAADLRMSAMAPTPPQGEVDADCMDVNVRGPDGFTPLMIASCSGGGLETGNSE EEEDAPAVISDFIYQGASLHNQTDRTGETALHLAARYSRSDAAKRLLEASADANIQDNMGRTPLHAAVSADAQGVFQILIRNRATDLDAR MHDGTTPLILAARLAVEGMLEDLINSHADVNAVDDLGKSALHWAAAVNNVDAAVVLLKNGANKDMQNNREETPLFLAAREGSYETAKVLL DHFANRDITDHMDRLPRDIAQERMHHDIVRLLDEYNLVRSPQLHGAPLGGTPTLSPPLCSPNGYLGSLKPGVQGKKVRKPSSKGLACGSK EAKDLKARRKKSQDGKGCLLDSSGMLSPVDSLESPHGYLSDVASPPLLPSPFQQSPSVPLNHLPGMPDTHLGIGHLNVAAKPEMAALGGG GRLAFETGPPRLSHLPVASGTSTVLGSSSGGALNFTVGGSTSLNGQCEWLSRLQSGMVPNQYNPLRGSVAPGPLSTQAPSLQHGMVGPLH SSLAASALSQMMSYQGLPSTRLATQPHLVQTQQVQPQNLQMQQQNLQPANIQQQQSLQPPPPPPQPHLGVSSAASGHLGRSFLSGEPSQA DVQPLGPSSLAVHTILPQESPALPTSLPSSLVPPVTAAQFLTPPSQHSYSSPVDNTPSHQLQVPEHPFLTPSPESPDQWSSSSPHSNVSD | 1014 |
Top |
pLDDT score distribution |
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
HNF1B_pLDDT.png |
NOTCH1_pLDDT.png |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
 |
Top |
Ramachandran Plot of Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| Fusion AA seq ID in FusionPDB and their Ramachandran plots |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| HNF1B | |
| NOTCH1 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 420_421 | 1722.3333333333333 | 2556.0 | DLL4 | |
| Tgene | NOTCH1 | chr17:36099430 | chr9:139396940 | ENST00000277541 | 26 | 34 | 448_452 | 1722.3333333333333 | 2556.0 | DLL4 | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 420_421 | 1722.3333333333333 | 2556.0 | DLL4 | |
| Tgene | NOTCH1 | chr17:36099431 | chr9:139396940 | ENST00000277541 | 26 | 34 | 448_452 | 1722.3333333333333 | 2556.0 | DLL4 |
Top |
Related Drugs to HNF1B-NOTCH1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to HNF1B-NOTCH1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

