| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:IL6ST-LAMC2 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: IL6ST-LAMC2 | FusionPDB ID: 39584 | FusionGDB2.0 ID: 39584 | Hgene | Tgene | Gene symbol | IL6ST | LAMC2 | Gene ID | 3572 | 3918 |
| Gene name | interleukin 6 signal transducer | laminin subunit gamma 2 | |
| Synonyms | CD130|CDW130|GP130|HIES4|IL-6RB|sGP130 | B2T|BM600|CSF|EBR2|EBR2A|LAMB2T|LAMNB2 | |
| Cytomap | 5q11.2 | 1q25.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | interleukin-6 receptor subunit betaCD130 antigenIL-6 receptor subunit betaIL-6R subunit betagp130 of the rheumatoid arthritis antigenic peptide-bearing soluble formgp130, oncostatin M receptorinterleukin receptor beta chainmembrane glycoprotein 130 | laminin subunit gamma-2BM600-100kDaCSF 140 kDa subunitcell-scattering factor 140 kDa subunitepiligrin subunit gammakalinin subunit gammaladsin 140 kDa subunitlaminin B2t chainlaminin, gamma 2large adhesive scatter factor 140 kDa subunitnicein su | |
| Modification date | 20200327 | 20200315 | |
| UniProtAcc | P40189 | Q13753 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000381287, ENST00000522633, ENST00000536319, ENST00000336909, ENST00000381294, ENST00000381298, ENST00000502326, ENST00000381286, ENST00000381293, ENST00000396816, ENST00000577363, | ENST00000264144, ENST00000493293, ENST00000461729, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 16 X 11 X 5=880 | 6 X 5 X 4=120 |
| # samples | 19 | 6 | |
| ** MAII score | log2(19/880*10)=-2.21150410519371 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(6/120*10)=-1 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: IL6ST [Title/Abstract] AND LAMC2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | IL6ST(55247292)-LAMC2(183177016), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | IL6ST-LAMC2 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. IL6ST-LAMC2 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. IL6ST-LAMC2 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. IL6ST-LAMC2 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. IL6ST-LAMC2 seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. IL6ST-LAMC2 seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | IL6ST | GO:0008284 | positive regulation of cell proliferation | 2261637 |
| Hgene | IL6ST | GO:0019221 | cytokine-mediated signaling pathway | 2261637 |
| Hgene | IL6ST | GO:0034097 | response to cytokine | 2261637|8999038 |
| Hgene | IL6ST | GO:0048861 | leukemia inhibitory factor signaling pathway | 8999038|12643274 |
| Hgene | IL6ST | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway | 12643274 |
 Fusion gene breakpoints across IL6ST (5'-gene) Fusion gene breakpoints across IL6ST (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
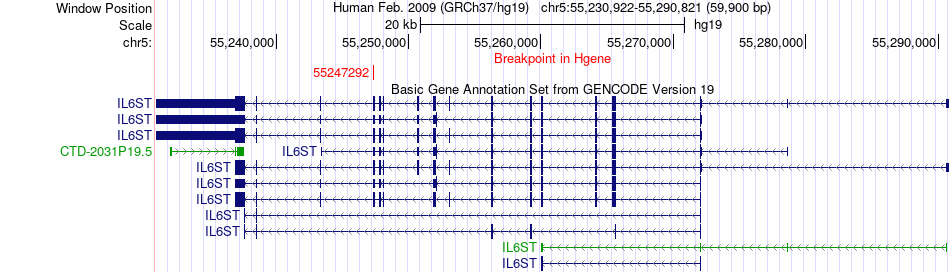 |
 Fusion gene breakpoints across LAMC2 (3'-gene) Fusion gene breakpoints across LAMC2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
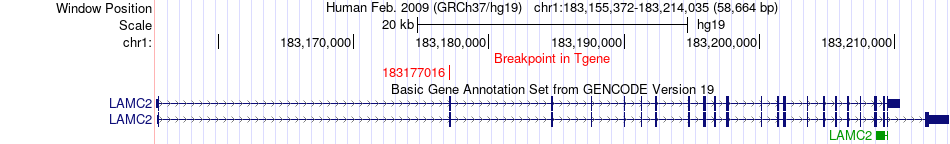 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | PRAD | TCGA-YL-A9WL-01A | IL6ST | chr5 | 55247292 | - | LAMC2 | chr1 | 183177016 | + |
| ChimerDB4 | PRAD | TCGA-YL-A9WL | IL6ST | chr5 | 55247292 | - | LAMC2 | chr1 | 183177016 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000381298 | IL6ST | chr5 | 55247292 | - | ENST00000493293 | LAMC2 | chr1 | 183177016 | + | 6272 | 2153 | 313 | 5409 | 1698 |
| ENST00000381298 | IL6ST | chr5 | 55247292 | - | ENST00000264144 | LAMC2 | chr1 | 183177016 | + | 7156 | 2153 | 313 | 5655 | 1780 |
| ENST00000336909 | IL6ST | chr5 | 55247292 | - | ENST00000493293 | LAMC2 | chr1 | 183177016 | + | 5995 | 1876 | 6 | 5132 | 1708 |
| ENST00000336909 | IL6ST | chr5 | 55247292 | - | ENST00000264144 | LAMC2 | chr1 | 183177016 | + | 6879 | 1876 | 6 | 5378 | 1790 |
| ENST00000381294 | IL6ST | chr5 | 55247292 | - | ENST00000493293 | LAMC2 | chr1 | 183177016 | + | 5776 | 1657 | 0 | 4913 | 1637 |
| ENST00000381294 | IL6ST | chr5 | 55247292 | - | ENST00000264144 | LAMC2 | chr1 | 183177016 | + | 6660 | 1657 | 0 | 5159 | 1719 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000381298 | ENST00000493293 | IL6ST | chr5 | 55247292 | - | LAMC2 | chr1 | 183177016 | + | 0.000504235 | 0.99949574 |
| ENST00000381298 | ENST00000264144 | IL6ST | chr5 | 55247292 | - | LAMC2 | chr1 | 183177016 | + | 0.000886583 | 0.9991134 |
| ENST00000336909 | ENST00000493293 | IL6ST | chr5 | 55247292 | - | LAMC2 | chr1 | 183177016 | + | 0.000374627 | 0.9996253 |
| ENST00000336909 | ENST00000264144 | IL6ST | chr5 | 55247292 | - | LAMC2 | chr1 | 183177016 | + | 0.0006308 | 0.9993692 |
| ENST00000381294 | ENST00000493293 | IL6ST | chr5 | 55247292 | - | LAMC2 | chr1 | 183177016 | + | 0.000435555 | 0.99956447 |
| ENST00000381294 | ENST00000264144 | IL6ST | chr5 | 55247292 | - | LAMC2 | chr1 | 183177016 | + | 0.000700934 | 0.99929905 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >39584_39584_1_IL6ST-LAMC2_IL6ST_chr5_55247292_ENST00000336909_LAMC2_chr1_183177016_ENST00000264144_length(amino acids)=1790AA_BP=623 MCLLQKYPRKMLTLQTWLVQALFIFLTTESTGELLDPCGYISPESPVVQLHSNFTAVCVLKEKCMDYFHVNANYIVWKTNHFTIPKEQYT IINRTASSVTFTDIASLNIQLTCNILTFGQLEQNVYGITIISGLPPEKPKNLSCIVNEGKKMRCEWDGGRETHLETNFTLKSEWATHKFA DCKAKRDTPTSCTVDYSTVYFVNIEVWVEAENALGKVTSDHINFDPVYKVKPNPPHNLSVINSEELSSILKLTWTNPSIKSVIILKYNIQ YRTKDASTWSQIPPEDTASTRSSFTVQDLKPFTEYVFRIRCMKEDGKGYWSDWSEEASGITYEDRPSKAPSFWYKIDPSHTQGYRTVQLV WKTLPPFEANGKILDYEVTLTRWKSHLQNYTVNATKLTVNLTNDRYLATLTVRNLVGKSDAAVLTIPACDFQATHPVMDLKAFPKDNMLW VEWTTPRESVKKYILEWCVLSDKAPCITDWQQEDGTVHRTYLRGNLAESKCYLITVTPVYADGPGSPESIKAYLKQAPPSKGPTVRTKKV GKNEAVLEWDQLPVDVQNGFIRNYTIFYRTIIGNETAVNVDSSHTEYTLSSLTSDTLYMVRMAAYTDEGGKDGPEFTFTTPKFVCDCNGK SRQCIFDRELHRQTGNGFRCLNCNDNTDGIHCEKCKNGFYRHRERDRCLPCNCNSKGSLSARCDNSGRCSCKPGVTGARCDRCLPGFHML TDAGCTQDQRLLDSKCDCDPAGIAGPCDAGRCVCKPAVTGERCDRCRSGYYNLDGGNPEGCTQCFCYGHSASCRSSAEYSVHKITSTFHQ DVDGWKAVQRNGSPAKLQWSQRHQDVFSSAQRLDPVYFVAPAKFLGNQQVSYGQSLSFDYRVDRGGRHPSAHDVILEGAGLRITAPLMPL GKTLPCGLTKTYTFRLNEHPSNNWSPQLSYFEYRRLLRNLTALRIRATYGEYSTGYIDNVTLISARPVSGAPAPWVEQCICPVGYKGQFC QDCASGYKRDSARLGPFGTCIPCNCQGGGACDPDTGDCYSGDENPDIECADCPIGFYNDPHDPRSCKPCPCHNGFSCSVMPETEEVVCNN CPPGVTGARCELCADGYFGDPFGEHGPVRPCQPCQCNNNVDPSASGNCDRLTGRCLKCIHNTAGIYCDQCKAGYFGDPLAPNPADKCRAC NCNPMGSEPVGCRSDGTCVCKPGFGGPNCEHGAFSCPACYNQVKIQMDQFMQQLQRMEALISKAQGGDGVVPDTELEGRMQQAEQALQDI LRDAQISEGASRSLGLQLAKVRSQENSYQSRLDDLKMTVERVRALGSQYQNRVRDTHRLITQMQLSLAESEASLGNTNIPASDHYVGPNG FKSLAQEATRLAESHVESASNMEQLTRETEDYSKQALSLVRKALHEGVGSGSGSPDGAVVQGLVEKLEKTKSLAQQLTREATQAEIEADR SYQHSLRLLDSVSRLQGVSDQSFQVEEAKRIKQKADSLSSLVTRHMDEFKRTQKNLGNWKEEAQQLLQNGKSGREKSDQLLSRANLAKSR AQEALSMGNATFYEVESILKNLREFDLQVDNRKAEAEEAMKRLSYISQKVSDASDKTQQAERALGSAAADAQRAKNGAGEALEISSEIEQ EIGSLNLEANVTADGALAMEKGLASLKSEMREVEGELERKELEFDTNMDAVQMVITEAQKVDTRAKNAGVTIQDTLNTLDGLLHLMDQPL -------------------------------------------------------------- >39584_39584_2_IL6ST-LAMC2_IL6ST_chr5_55247292_ENST00000336909_LAMC2_chr1_183177016_ENST00000493293_length(amino acids)=1708AA_BP=623 MCLLQKYPRKMLTLQTWLVQALFIFLTTESTGELLDPCGYISPESPVVQLHSNFTAVCVLKEKCMDYFHVNANYIVWKTNHFTIPKEQYT IINRTASSVTFTDIASLNIQLTCNILTFGQLEQNVYGITIISGLPPEKPKNLSCIVNEGKKMRCEWDGGRETHLETNFTLKSEWATHKFA DCKAKRDTPTSCTVDYSTVYFVNIEVWVEAENALGKVTSDHINFDPVYKVKPNPPHNLSVINSEELSSILKLTWTNPSIKSVIILKYNIQ YRTKDASTWSQIPPEDTASTRSSFTVQDLKPFTEYVFRIRCMKEDGKGYWSDWSEEASGITYEDRPSKAPSFWYKIDPSHTQGYRTVQLV WKTLPPFEANGKILDYEVTLTRWKSHLQNYTVNATKLTVNLTNDRYLATLTVRNLVGKSDAAVLTIPACDFQATHPVMDLKAFPKDNMLW VEWTTPRESVKKYILEWCVLSDKAPCITDWQQEDGTVHRTYLRGNLAESKCYLITVTPVYADGPGSPESIKAYLKQAPPSKGPTVRTKKV GKNEAVLEWDQLPVDVQNGFIRNYTIFYRTIIGNETAVNVDSSHTEYTLSSLTSDTLYMVRMAAYTDEGGKDGPEFTFTTPKFVCDCNGK SRQCIFDRELHRQTGNGFRCLNCNDNTDGIHCEKCKNGFYRHRERDRCLPCNCNSKGSLSARCDNSGRCSCKPGVTGARCDRCLPGFHML TDAGCTQDQRLLDSKCDCDPAGIAGPCDAGRCVCKPAVTGERCDRCRSGYYNLDGGNPEGCTQCFCYGHSASCRSSAEYSVHKITSTFHQ DVDGWKAVQRNGSPAKLQWSQRHQDVFSSAQRLDPVYFVAPAKFLGNQQVSYGQSLSFDYRVDRGGRHPSAHDVILEGAGLRITAPLMPL GKTLPCGLTKTYTFRLNEHPSNNWSPQLSYFEYRRLLRNLTALRIRATYGEYSTGYIDNVTLISARPVSGAPAPWVEQCICPVGYKGQFC QDCASGYKRDSARLGPFGTCIPCNCQGGGACDPDTGDCYSGDENPDIECADCPIGFYNDPHDPRSCKPCPCHNGFSCSVMPETEEVVCNN CPPGVTGARCELCADGYFGDPFGEHGPVRPCQPCQCNNNVDPSASGNCDRLTGRCLKCIHNTAGIYCDQCKAGYFGDPLAPNPADKCRAC NCNPMGSEPVGCRSDGTCVCKPGFGGPNCEHGAFSCPACYNQVKIQMDQFMQQLQRMEALISKAQGGDGVVPDTELEGRMQQAEQALQDI LRDAQISEGASRSLGLQLAKVRSQENSYQSRLDDLKMTVERVRALGSQYQNRVRDTHRLITQMQLSLAESEASLGNTNIPASDHYVGPNG FKSLAQEATRLAESHVESASNMEQLTRETEDYSKQALSLVRKALHEGVGSGSGSPDGAVVQGLVEKLEKTKSLAQQLTREATQAEIEADR SYQHSLRLLDSVSRLQGVSDQSFQVEEAKRIKQKADSLSSLVTRHMDEFKRTQKNLGNWKEEAQQLLQNGKSGREKSDQLLSRANLAKSR AQEALSMGNATFYEVESILKNLREFDLQVDNRKAEAEEAMKRLSYISQKVSDASDKTQQAERALGSAAADAQRAKNGAGEALEISSEIEQ -------------------------------------------------------------- >39584_39584_3_IL6ST-LAMC2_IL6ST_chr5_55247292_ENST00000381294_LAMC2_chr1_183177016_ENST00000264144_length(amino acids)=1719AA_BP=552 MLTLQTWLVQALFIFLTTESTGELLDPCGYISPESPVVQLHSNFTAVCVLKEKCMDYFHVNANYIVWKTNHFTIPKEQYTIINRTASSVT FTDIASLNIQLTCNILTFGQLEQNVYGITIISGLPPEKPKNLSCIVNEGKKMRCEWDGGRETHLETNFTLKSEWATHKFADCKAKRDTPT SCTVDYSTVYFVNIEVWVEAENALGKVTSDHINFDPVYKVKPNPPHNLSVINSEELSSILKLTWTNPSIKSVIILKYNIQYRTKDASTWS QIPPEDTASTRSSFTVQDLKPFTEYVFRIRCMKEDGKGYWSDWSEEASGITYEDRPSKAPSFWYKIDPSHTQGYRTVQLVWKTLPPFEAN GKILDYEVTLTRWKSHLQNYTVNATKLTVNLTNDRYLATLTVRNLVGKSDAAVLTIPACDFQGNLAESKCYLITVTPVYADGPGSPESIK AYLKQAPPSKGPTVRTKKVGKNEAVLEWDQLPVDVQNGFIRNYTIFYRTIIGNETAVNVDSSHTEYTLSSLTSDTLYMVRMAAYTDEGGK DGPEFTFTTPKFVCDCNGKSRQCIFDRELHRQTGNGFRCLNCNDNTDGIHCEKCKNGFYRHRERDRCLPCNCNSKGSLSARCDNSGRCSC KPGVTGARCDRCLPGFHMLTDAGCTQDQRLLDSKCDCDPAGIAGPCDAGRCVCKPAVTGERCDRCRSGYYNLDGGNPEGCTQCFCYGHSA SCRSSAEYSVHKITSTFHQDVDGWKAVQRNGSPAKLQWSQRHQDVFSSAQRLDPVYFVAPAKFLGNQQVSYGQSLSFDYRVDRGGRHPSA HDVILEGAGLRITAPLMPLGKTLPCGLTKTYTFRLNEHPSNNWSPQLSYFEYRRLLRNLTALRIRATYGEYSTGYIDNVTLISARPVSGA PAPWVEQCICPVGYKGQFCQDCASGYKRDSARLGPFGTCIPCNCQGGGACDPDTGDCYSGDENPDIECADCPIGFYNDPHDPRSCKPCPC HNGFSCSVMPETEEVVCNNCPPGVTGARCELCADGYFGDPFGEHGPVRPCQPCQCNNNVDPSASGNCDRLTGRCLKCIHNTAGIYCDQCK AGYFGDPLAPNPADKCRACNCNPMGSEPVGCRSDGTCVCKPGFGGPNCEHGAFSCPACYNQVKIQMDQFMQQLQRMEALISKAQGGDGVV PDTELEGRMQQAEQALQDILRDAQISEGASRSLGLQLAKVRSQENSYQSRLDDLKMTVERVRALGSQYQNRVRDTHRLITQMQLSLAESE ASLGNTNIPASDHYVGPNGFKSLAQEATRLAESHVESASNMEQLTRETEDYSKQALSLVRKALHEGVGSGSGSPDGAVVQGLVEKLEKTK SLAQQLTREATQAEIEADRSYQHSLRLLDSVSRLQGVSDQSFQVEEAKRIKQKADSLSSLVTRHMDEFKRTQKNLGNWKEEAQQLLQNGK SGREKSDQLLSRANLAKSRAQEALSMGNATFYEVESILKNLREFDLQVDNRKAEAEEAMKRLSYISQKVSDASDKTQQAERALGSAAADA QRAKNGAGEALEISSEIEQEIGSLNLEANVTADGALAMEKGLASLKSEMREVEGELERKELEFDTNMDAVQMVITEAQKVDTRAKNAGVT IQDTLNTLDGLLHLMDQPLSVDEEGLVLLEQKLSRAKTQINSQLRPMMSELEERARQQRGHLHLLETSIDGILADVKNLENIRDNLPPGC -------------------------------------------------------------- >39584_39584_4_IL6ST-LAMC2_IL6ST_chr5_55247292_ENST00000381294_LAMC2_chr1_183177016_ENST00000493293_length(amino acids)=1637AA_BP=552 MLTLQTWLVQALFIFLTTESTGELLDPCGYISPESPVVQLHSNFTAVCVLKEKCMDYFHVNANYIVWKTNHFTIPKEQYTIINRTASSVT FTDIASLNIQLTCNILTFGQLEQNVYGITIISGLPPEKPKNLSCIVNEGKKMRCEWDGGRETHLETNFTLKSEWATHKFADCKAKRDTPT SCTVDYSTVYFVNIEVWVEAENALGKVTSDHINFDPVYKVKPNPPHNLSVINSEELSSILKLTWTNPSIKSVIILKYNIQYRTKDASTWS QIPPEDTASTRSSFTVQDLKPFTEYVFRIRCMKEDGKGYWSDWSEEASGITYEDRPSKAPSFWYKIDPSHTQGYRTVQLVWKTLPPFEAN GKILDYEVTLTRWKSHLQNYTVNATKLTVNLTNDRYLATLTVRNLVGKSDAAVLTIPACDFQGNLAESKCYLITVTPVYADGPGSPESIK AYLKQAPPSKGPTVRTKKVGKNEAVLEWDQLPVDVQNGFIRNYTIFYRTIIGNETAVNVDSSHTEYTLSSLTSDTLYMVRMAAYTDEGGK DGPEFTFTTPKFVCDCNGKSRQCIFDRELHRQTGNGFRCLNCNDNTDGIHCEKCKNGFYRHRERDRCLPCNCNSKGSLSARCDNSGRCSC KPGVTGARCDRCLPGFHMLTDAGCTQDQRLLDSKCDCDPAGIAGPCDAGRCVCKPAVTGERCDRCRSGYYNLDGGNPEGCTQCFCYGHSA SCRSSAEYSVHKITSTFHQDVDGWKAVQRNGSPAKLQWSQRHQDVFSSAQRLDPVYFVAPAKFLGNQQVSYGQSLSFDYRVDRGGRHPSA HDVILEGAGLRITAPLMPLGKTLPCGLTKTYTFRLNEHPSNNWSPQLSYFEYRRLLRNLTALRIRATYGEYSTGYIDNVTLISARPVSGA PAPWVEQCICPVGYKGQFCQDCASGYKRDSARLGPFGTCIPCNCQGGGACDPDTGDCYSGDENPDIECADCPIGFYNDPHDPRSCKPCPC HNGFSCSVMPETEEVVCNNCPPGVTGARCELCADGYFGDPFGEHGPVRPCQPCQCNNNVDPSASGNCDRLTGRCLKCIHNTAGIYCDQCK AGYFGDPLAPNPADKCRACNCNPMGSEPVGCRSDGTCVCKPGFGGPNCEHGAFSCPACYNQVKIQMDQFMQQLQRMEALISKAQGGDGVV PDTELEGRMQQAEQALQDILRDAQISEGASRSLGLQLAKVRSQENSYQSRLDDLKMTVERVRALGSQYQNRVRDTHRLITQMQLSLAESE ASLGNTNIPASDHYVGPNGFKSLAQEATRLAESHVESASNMEQLTRETEDYSKQALSLVRKALHEGVGSGSGSPDGAVVQGLVEKLEKTK SLAQQLTREATQAEIEADRSYQHSLRLLDSVSRLQGVSDQSFQVEEAKRIKQKADSLSSLVTRHMDEFKRTQKNLGNWKEEAQQLLQNGK SGREKSDQLLSRANLAKSRAQEALSMGNATFYEVESILKNLREFDLQVDNRKAEAEEAMKRLSYISQKVSDASDKTQQAERALGSAAADA QRAKNGAGEALEISSEIEQEIGSLNLEANVTADGALAMEKGLASLKSEMREVEGELERKELEFDTNMDAVQMVITEAQKVDTRAKNAGVT -------------------------------------------------------------- >39584_39584_5_IL6ST-LAMC2_IL6ST_chr5_55247292_ENST00000381298_LAMC2_chr1_183177016_ENST00000264144_length(amino acids)=1780AA_BP=613 MLTLQTWLVQALFIFLTTESTGELLDPCGYISPESPVVQLHSNFTAVCVLKEKCMDYFHVNANYIVWKTNHFTIPKEQYTIINRTASSVT FTDIASLNIQLTCNILTFGQLEQNVYGITIISGLPPEKPKNLSCIVNEGKKMRCEWDGGRETHLETNFTLKSEWATHKFADCKAKRDTPT SCTVDYSTVYFVNIEVWVEAENALGKVTSDHINFDPVYKVKPNPPHNLSVINSEELSSILKLTWTNPSIKSVIILKYNIQYRTKDASTWS QIPPEDTASTRSSFTVQDLKPFTEYVFRIRCMKEDGKGYWSDWSEEASGITYEDRPSKAPSFWYKIDPSHTQGYRTVQLVWKTLPPFEAN GKILDYEVTLTRWKSHLQNYTVNATKLTVNLTNDRYLATLTVRNLVGKSDAAVLTIPACDFQATHPVMDLKAFPKDNMLWVEWTTPRESV KKYILEWCVLSDKAPCITDWQQEDGTVHRTYLRGNLAESKCYLITVTPVYADGPGSPESIKAYLKQAPPSKGPTVRTKKVGKNEAVLEWD QLPVDVQNGFIRNYTIFYRTIIGNETAVNVDSSHTEYTLSSLTSDTLYMVRMAAYTDEGGKDGPEFTFTTPKFVCDCNGKSRQCIFDREL HRQTGNGFRCLNCNDNTDGIHCEKCKNGFYRHRERDRCLPCNCNSKGSLSARCDNSGRCSCKPGVTGARCDRCLPGFHMLTDAGCTQDQR LLDSKCDCDPAGIAGPCDAGRCVCKPAVTGERCDRCRSGYYNLDGGNPEGCTQCFCYGHSASCRSSAEYSVHKITSTFHQDVDGWKAVQR NGSPAKLQWSQRHQDVFSSAQRLDPVYFVAPAKFLGNQQVSYGQSLSFDYRVDRGGRHPSAHDVILEGAGLRITAPLMPLGKTLPCGLTK TYTFRLNEHPSNNWSPQLSYFEYRRLLRNLTALRIRATYGEYSTGYIDNVTLISARPVSGAPAPWVEQCICPVGYKGQFCQDCASGYKRD SARLGPFGTCIPCNCQGGGACDPDTGDCYSGDENPDIECADCPIGFYNDPHDPRSCKPCPCHNGFSCSVMPETEEVVCNNCPPGVTGARC ELCADGYFGDPFGEHGPVRPCQPCQCNNNVDPSASGNCDRLTGRCLKCIHNTAGIYCDQCKAGYFGDPLAPNPADKCRACNCNPMGSEPV GCRSDGTCVCKPGFGGPNCEHGAFSCPACYNQVKIQMDQFMQQLQRMEALISKAQGGDGVVPDTELEGRMQQAEQALQDILRDAQISEGA SRSLGLQLAKVRSQENSYQSRLDDLKMTVERVRALGSQYQNRVRDTHRLITQMQLSLAESEASLGNTNIPASDHYVGPNGFKSLAQEATR LAESHVESASNMEQLTRETEDYSKQALSLVRKALHEGVGSGSGSPDGAVVQGLVEKLEKTKSLAQQLTREATQAEIEADRSYQHSLRLLD SVSRLQGVSDQSFQVEEAKRIKQKADSLSSLVTRHMDEFKRTQKNLGNWKEEAQQLLQNGKSGREKSDQLLSRANLAKSRAQEALSMGNA TFYEVESILKNLREFDLQVDNRKAEAEEAMKRLSYISQKVSDASDKTQQAERALGSAAADAQRAKNGAGEALEISSEIEQEIGSLNLEAN VTADGALAMEKGLASLKSEMREVEGELERKELEFDTNMDAVQMVITEAQKVDTRAKNAGVTIQDTLNTLDGLLHLMDQPLSVDEEGLVLL -------------------------------------------------------------- >39584_39584_6_IL6ST-LAMC2_IL6ST_chr5_55247292_ENST00000381298_LAMC2_chr1_183177016_ENST00000493293_length(amino acids)=1698AA_BP=613 MLTLQTWLVQALFIFLTTESTGELLDPCGYISPESPVVQLHSNFTAVCVLKEKCMDYFHVNANYIVWKTNHFTIPKEQYTIINRTASSVT FTDIASLNIQLTCNILTFGQLEQNVYGITIISGLPPEKPKNLSCIVNEGKKMRCEWDGGRETHLETNFTLKSEWATHKFADCKAKRDTPT SCTVDYSTVYFVNIEVWVEAENALGKVTSDHINFDPVYKVKPNPPHNLSVINSEELSSILKLTWTNPSIKSVIILKYNIQYRTKDASTWS QIPPEDTASTRSSFTVQDLKPFTEYVFRIRCMKEDGKGYWSDWSEEASGITYEDRPSKAPSFWYKIDPSHTQGYRTVQLVWKTLPPFEAN GKILDYEVTLTRWKSHLQNYTVNATKLTVNLTNDRYLATLTVRNLVGKSDAAVLTIPACDFQATHPVMDLKAFPKDNMLWVEWTTPRESV KKYILEWCVLSDKAPCITDWQQEDGTVHRTYLRGNLAESKCYLITVTPVYADGPGSPESIKAYLKQAPPSKGPTVRTKKVGKNEAVLEWD QLPVDVQNGFIRNYTIFYRTIIGNETAVNVDSSHTEYTLSSLTSDTLYMVRMAAYTDEGGKDGPEFTFTTPKFVCDCNGKSRQCIFDREL HRQTGNGFRCLNCNDNTDGIHCEKCKNGFYRHRERDRCLPCNCNSKGSLSARCDNSGRCSCKPGVTGARCDRCLPGFHMLTDAGCTQDQR LLDSKCDCDPAGIAGPCDAGRCVCKPAVTGERCDRCRSGYYNLDGGNPEGCTQCFCYGHSASCRSSAEYSVHKITSTFHQDVDGWKAVQR NGSPAKLQWSQRHQDVFSSAQRLDPVYFVAPAKFLGNQQVSYGQSLSFDYRVDRGGRHPSAHDVILEGAGLRITAPLMPLGKTLPCGLTK TYTFRLNEHPSNNWSPQLSYFEYRRLLRNLTALRIRATYGEYSTGYIDNVTLISARPVSGAPAPWVEQCICPVGYKGQFCQDCASGYKRD SARLGPFGTCIPCNCQGGGACDPDTGDCYSGDENPDIECADCPIGFYNDPHDPRSCKPCPCHNGFSCSVMPETEEVVCNNCPPGVTGARC ELCADGYFGDPFGEHGPVRPCQPCQCNNNVDPSASGNCDRLTGRCLKCIHNTAGIYCDQCKAGYFGDPLAPNPADKCRACNCNPMGSEPV GCRSDGTCVCKPGFGGPNCEHGAFSCPACYNQVKIQMDQFMQQLQRMEALISKAQGGDGVVPDTELEGRMQQAEQALQDILRDAQISEGA SRSLGLQLAKVRSQENSYQSRLDDLKMTVERVRALGSQYQNRVRDTHRLITQMQLSLAESEASLGNTNIPASDHYVGPNGFKSLAQEATR LAESHVESASNMEQLTRETEDYSKQALSLVRKALHEGVGSGSGSPDGAVVQGLVEKLEKTKSLAQQLTREATQAEIEADRSYQHSLRLLD SVSRLQGVSDQSFQVEEAKRIKQKADSLSSLVTRHMDEFKRTQKNLGNWKEEAQQLLQNGKSGREKSDQLLSRANLAKSRAQEALSMGNA TFYEVESILKNLREFDLQVDNRKAEAEEAMKRLSYISQKVSDASDKTQQAERALGSAAADAQRAKNGAGEALEISSEIEQEIGSLNLEAN -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr5:55247292/chr1:183177016) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| IL6ST | LAMC2 |
| FUNCTION: Signal-transducing molecule (PubMed:2261637). The receptor systems for IL6, LIF, OSM, CNTF, IL11, CTF1 and BSF3 can utilize IL6ST for initiating signal transmission. Binding of IL6 to IL6R induces IL6ST homodimerization and formation of a high-affinity receptor complex, which activate the intracellular JAK-MAPK and JAK-STAT3 signaling pathways (PubMed:2261637, PubMed:19915009, PubMed:23294003). That causes phosphorylation of IL6ST tyrosine residues which in turn activates STAT3 (PubMed:19915009, PubMed:23294003, PubMed:25731159). In parallel, the IL6 signaling pathway induces the expression of two cytokine receptor signaling inhibitors, SOCS1 and SOCS3, which inhibit JAK and terminate the activity of the IL6 signaling pathway as a negative feedback loop (By similarity). Also activates the yes-associated protein 1 (YAP) and NOTCH pathways to control inflammation-induced epithelial regeneration, independently of STAT3 (By similarity). Mediates signals which regulate immune response, hematopoiesis, pain control and bone metabolism (By similarity). Has a role in embryonic development (By similarity). Essential for survival of motor and sensory neurons and for differentiation of astrocytes (By similarity). Required for expression of TRPA1 in nociceptive neurons (By similarity). Required for the maintenance of PTH1R expression in the osteoblast lineage and for the stimulation of PTH-induced osteoblast differentiation (By similarity). Required for normal trabecular bone mass and cortical bone composition (By similarity). {ECO:0000250|UniProtKB:Q00560, ECO:0000269|PubMed:19915009, ECO:0000269|PubMed:2261637, ECO:0000269|PubMed:23294003, ECO:0000269|PubMed:25731159, ECO:0000269|PubMed:28747427, ECO:0000269|PubMed:30309848}.; FUNCTION: [Isoform 2]: Binds to the soluble IL6:sIL6R complex (hyper-IL6), thereby blocking IL6 trans-signaling. Inhibits sIL6R-dependent acute phase response (PubMed:11121117, PubMed:21990364, PubMed:30279168). Also blocks IL11 cluster signaling through IL11R (PubMed:30279168). {ECO:0000269|PubMed:11121117, ECO:0000269|PubMed:21990364, ECO:0000269|PubMed:30279168}. | FUNCTION: Binding to cells via a high affinity receptor, laminin is thought to mediate the attachment, migration and organization of cells into tissues during embryonic development by interacting with other extracellular matrix components. Ladsin exerts cell-scattering activity toward a wide variety of cells, including epithelial, endothelial, and fibroblastic cells. {ECO:0000269|PubMed:8265624}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000522633 | - | 12 | 13 | 725_755 | 585.6666666666666 | 299.3333333333333 | Compositional bias | Note=Ser-rich |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000536319 | - | 10 | 13 | 725_755 | 524.6666666666666 | 441.3333333333333 | Compositional bias | Note=Ser-rich |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000336909 | - | 12 | 15 | 125_216 | 613.3333333333334 | 919.0 | Domain | Fibronectin type-III 1 |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000336909 | - | 12 | 15 | 224_324 | 613.3333333333334 | 919.0 | Domain | Fibronectin type-III 2 |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000336909 | - | 12 | 15 | 26_120 | 613.3333333333334 | 919.0 | Domain | Note=Ig-like C2-type |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000336909 | - | 12 | 15 | 329_424 | 613.3333333333334 | 919.0 | Domain | Fibronectin type-III 3 |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000336909 | - | 12 | 15 | 426_517 | 613.3333333333334 | 919.0 | Domain | Fibronectin type-III 4 |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000336909 | - | 12 | 15 | 518_613 | 613.3333333333334 | 919.0 | Domain | Fibronectin type-III 5 |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381287 | - | 11 | 14 | 125_216 | 585.6666666666666 | 2415.0 | Domain | Fibronectin type-III 1 |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381287 | - | 11 | 14 | 224_324 | 585.6666666666666 | 2415.0 | Domain | Fibronectin type-III 2 |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381287 | - | 11 | 14 | 26_120 | 585.6666666666666 | 2415.0 | Domain | Note=Ig-like C2-type |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381287 | - | 11 | 14 | 329_424 | 585.6666666666666 | 2415.0 | Domain | Fibronectin type-III 3 |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381287 | - | 11 | 14 | 426_517 | 585.6666666666666 | 2415.0 | Domain | Fibronectin type-III 4 |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381294 | - | 11 | 14 | 125_216 | 552.3333333333334 | 858.0 | Domain | Fibronectin type-III 1 |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381294 | - | 11 | 14 | 224_324 | 552.3333333333334 | 858.0 | Domain | Fibronectin type-III 2 |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381294 | - | 11 | 14 | 26_120 | 552.3333333333334 | 858.0 | Domain | Note=Ig-like C2-type |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381294 | - | 11 | 14 | 329_424 | 552.3333333333334 | 858.0 | Domain | Fibronectin type-III 3 |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381294 | - | 11 | 14 | 426_517 | 552.3333333333334 | 858.0 | Domain | Fibronectin type-III 4 |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381298 | - | 14 | 17 | 125_216 | 613.3333333333334 | 919.0 | Domain | Fibronectin type-III 1 |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381298 | - | 14 | 17 | 224_324 | 613.3333333333334 | 919.0 | Domain | Fibronectin type-III 2 |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381298 | - | 14 | 17 | 26_120 | 613.3333333333334 | 919.0 | Domain | Note=Ig-like C2-type |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381298 | - | 14 | 17 | 329_424 | 613.3333333333334 | 919.0 | Domain | Fibronectin type-III 3 |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381298 | - | 14 | 17 | 426_517 | 613.3333333333334 | 919.0 | Domain | Fibronectin type-III 4 |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381298 | - | 14 | 17 | 518_613 | 613.3333333333334 | 919.0 | Domain | Fibronectin type-III 5 |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000502326 | - | 13 | 16 | 125_216 | 613.3333333333334 | 919.0 | Domain | Fibronectin type-III 1 |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000502326 | - | 13 | 16 | 224_324 | 613.3333333333334 | 919.0 | Domain | Fibronectin type-III 2 |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000502326 | - | 13 | 16 | 26_120 | 613.3333333333334 | 919.0 | Domain | Note=Ig-like C2-type |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000502326 | - | 13 | 16 | 329_424 | 613.3333333333334 | 919.0 | Domain | Fibronectin type-III 3 |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000502326 | - | 13 | 16 | 426_517 | 613.3333333333334 | 919.0 | Domain | Fibronectin type-III 4 |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000502326 | - | 13 | 16 | 518_613 | 613.3333333333334 | 919.0 | Domain | Fibronectin type-III 5 |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000522633 | - | 12 | 13 | 125_216 | 585.6666666666666 | 299.3333333333333 | Domain | Fibronectin type-III 1 |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000522633 | - | 12 | 13 | 224_324 | 585.6666666666666 | 299.3333333333333 | Domain | Fibronectin type-III 2 |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000522633 | - | 12 | 13 | 26_120 | 585.6666666666666 | 299.3333333333333 | Domain | Note=Ig-like C2-type |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000522633 | - | 12 | 13 | 329_424 | 585.6666666666666 | 299.3333333333333 | Domain | Fibronectin type-III 3 |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000522633 | - | 12 | 13 | 426_517 | 585.6666666666666 | 299.3333333333333 | Domain | Fibronectin type-III 4 |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000522633 | - | 12 | 13 | 518_613 | 585.6666666666666 | 299.3333333333333 | Domain | Fibronectin type-III 5 |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000536319 | - | 10 | 13 | 125_216 | 524.6666666666666 | 441.3333333333333 | Domain | Fibronectin type-III 1 |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000536319 | - | 10 | 13 | 224_324 | 524.6666666666666 | 441.3333333333333 | Domain | Fibronectin type-III 2 |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000536319 | - | 10 | 13 | 26_120 | 524.6666666666666 | 441.3333333333333 | Domain | Note=Ig-like C2-type |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000536319 | - | 10 | 13 | 329_424 | 524.6666666666666 | 441.3333333333333 | Domain | Fibronectin type-III 3 |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000536319 | - | 10 | 13 | 426_517 | 524.6666666666666 | 441.3333333333333 | Domain | Fibronectin type-III 4 |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000536319 | - | 10 | 13 | 518_613 | 524.6666666666666 | 441.3333333333333 | Domain | Fibronectin type-III 5 |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000336909 | - | 12 | 15 | 310_314 | 613.3333333333334 | 919.0 | Motif | Note=WSXWS motif |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381287 | - | 11 | 14 | 310_314 | 585.6666666666666 | 2415.0 | Motif | Note=WSXWS motif |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381294 | - | 11 | 14 | 310_314 | 552.3333333333334 | 858.0 | Motif | Note=WSXWS motif |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381298 | - | 14 | 17 | 310_314 | 613.3333333333334 | 919.0 | Motif | Note=WSXWS motif |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000502326 | - | 13 | 16 | 310_314 | 613.3333333333334 | 919.0 | Motif | Note=WSXWS motif |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000522633 | - | 12 | 13 | 310_314 | 585.6666666666666 | 299.3333333333333 | Motif | Note=WSXWS motif |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000522633 | - | 12 | 13 | 651_659 | 585.6666666666666 | 299.3333333333333 | Motif | Note=Box 1 motif |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000536319 | - | 10 | 13 | 310_314 | 524.6666666666666 | 441.3333333333333 | Motif | Note=WSXWS motif |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000536319 | - | 10 | 13 | 651_659 | 524.6666666666666 | 441.3333333333333 | Motif | Note=Box 1 motif |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000522633 | - | 12 | 13 | 23_619 | 585.6666666666666 | 299.3333333333333 | Topological domain | Extracellular |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000522633 | - | 12 | 13 | 642_918 | 585.6666666666666 | 299.3333333333333 | Topological domain | Cytoplasmic |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000536319 | - | 10 | 13 | 23_619 | 524.6666666666666 | 441.3333333333333 | Topological domain | Extracellular |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000536319 | - | 10 | 13 | 642_918 | 524.6666666666666 | 441.3333333333333 | Topological domain | Cytoplasmic |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000522633 | - | 12 | 13 | 620_641 | 585.6666666666666 | 299.3333333333333 | Transmembrane | Helical |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000536319 | - | 10 | 13 | 620_641 | 524.6666666666666 | 441.3333333333333 | Transmembrane | Helical |
| Tgene | LAMC2 | chr5:55247292 | chr1:183177016 | ENST00000264144 | 0 | 23 | 1117_1193 | 26.333333333333332 | 1194.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | LAMC2 | chr5:55247292 | chr1:183177016 | ENST00000264144 | 0 | 23 | 611_718 | 26.333333333333332 | 1194.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | LAMC2 | chr5:55247292 | chr1:183177016 | ENST00000264144 | 0 | 23 | 811_1076 | 26.333333333333332 | 1194.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | LAMC2 | chr5:55247292 | chr1:183177016 | ENST00000493293 | 0 | 22 | 1117_1193 | 26.333333333333332 | 1112.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | LAMC2 | chr5:55247292 | chr1:183177016 | ENST00000493293 | 0 | 22 | 611_718 | 26.333333333333332 | 1112.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | LAMC2 | chr5:55247292 | chr1:183177016 | ENST00000493293 | 0 | 22 | 811_1076 | 26.333333333333332 | 1112.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | LAMC2 | chr5:55247292 | chr1:183177016 | ENST00000264144 | 0 | 23 | 139_186 | 26.333333333333332 | 1194.0 | Domain | Laminin EGF-like 3 | |
| Tgene | LAMC2 | chr5:55247292 | chr1:183177016 | ENST00000264144 | 0 | 23 | 187_196 | 26.333333333333332 | 1194.0 | Domain | Laminin EGF-like 4%3B first part | |
| Tgene | LAMC2 | chr5:55247292 | chr1:183177016 | ENST00000264144 | 0 | 23 | 213_381 | 26.333333333333332 | 1194.0 | Domain | Laminin IV type A | |
| Tgene | LAMC2 | chr5:55247292 | chr1:183177016 | ENST00000264144 | 0 | 23 | 28_83 | 26.333333333333332 | 1194.0 | Domain | Laminin EGF-like 1 | |
| Tgene | LAMC2 | chr5:55247292 | chr1:183177016 | ENST00000264144 | 0 | 23 | 382_415 | 26.333333333333332 | 1194.0 | Domain | Laminin EGF-like 4%3B second part | |
| Tgene | LAMC2 | chr5:55247292 | chr1:183177016 | ENST00000264144 | 0 | 23 | 416_461 | 26.333333333333332 | 1194.0 | Domain | Laminin EGF-like 5 | |
| Tgene | LAMC2 | chr5:55247292 | chr1:183177016 | ENST00000264144 | 0 | 23 | 462_516 | 26.333333333333332 | 1194.0 | Domain | Laminin EGF-like 6 | |
| Tgene | LAMC2 | chr5:55247292 | chr1:183177016 | ENST00000264144 | 0 | 23 | 517_572 | 26.333333333333332 | 1194.0 | Domain | Laminin EGF-like 7 | |
| Tgene | LAMC2 | chr5:55247292 | chr1:183177016 | ENST00000264144 | 0 | 23 | 573_602 | 26.333333333333332 | 1194.0 | Domain | Laminin EGF-like 8%3B truncated | |
| Tgene | LAMC2 | chr5:55247292 | chr1:183177016 | ENST00000264144 | 0 | 23 | 84_130 | 26.333333333333332 | 1194.0 | Domain | Laminin EGF-like 2 | |
| Tgene | LAMC2 | chr5:55247292 | chr1:183177016 | ENST00000493293 | 0 | 22 | 139_186 | 26.333333333333332 | 1112.0 | Domain | Laminin EGF-like 3 | |
| Tgene | LAMC2 | chr5:55247292 | chr1:183177016 | ENST00000493293 | 0 | 22 | 187_196 | 26.333333333333332 | 1112.0 | Domain | Laminin EGF-like 4%3B first part | |
| Tgene | LAMC2 | chr5:55247292 | chr1:183177016 | ENST00000493293 | 0 | 22 | 213_381 | 26.333333333333332 | 1112.0 | Domain | Laminin IV type A | |
| Tgene | LAMC2 | chr5:55247292 | chr1:183177016 | ENST00000493293 | 0 | 22 | 28_83 | 26.333333333333332 | 1112.0 | Domain | Laminin EGF-like 1 | |
| Tgene | LAMC2 | chr5:55247292 | chr1:183177016 | ENST00000493293 | 0 | 22 | 382_415 | 26.333333333333332 | 1112.0 | Domain | Laminin EGF-like 4%3B second part | |
| Tgene | LAMC2 | chr5:55247292 | chr1:183177016 | ENST00000493293 | 0 | 22 | 416_461 | 26.333333333333332 | 1112.0 | Domain | Laminin EGF-like 5 | |
| Tgene | LAMC2 | chr5:55247292 | chr1:183177016 | ENST00000493293 | 0 | 22 | 462_516 | 26.333333333333332 | 1112.0 | Domain | Laminin EGF-like 6 | |
| Tgene | LAMC2 | chr5:55247292 | chr1:183177016 | ENST00000493293 | 0 | 22 | 517_572 | 26.333333333333332 | 1112.0 | Domain | Laminin EGF-like 7 | |
| Tgene | LAMC2 | chr5:55247292 | chr1:183177016 | ENST00000493293 | 0 | 22 | 573_602 | 26.333333333333332 | 1112.0 | Domain | Laminin EGF-like 8%3B truncated | |
| Tgene | LAMC2 | chr5:55247292 | chr1:183177016 | ENST00000493293 | 0 | 22 | 84_130 | 26.333333333333332 | 1112.0 | Domain | Laminin EGF-like 2 | |
| Tgene | LAMC2 | chr5:55247292 | chr1:183177016 | ENST00000264144 | 0 | 23 | 603_1193 | 26.333333333333332 | 1194.0 | Region | Note=Domain II and I | |
| Tgene | LAMC2 | chr5:55247292 | chr1:183177016 | ENST00000493293 | 0 | 22 | 603_1193 | 26.333333333333332 | 1112.0 | Region | Note=Domain II and I |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000336909 | - | 12 | 15 | 725_755 | 613.3333333333334 | 919.0 | Compositional bias | Note=Ser-rich |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381287 | - | 11 | 14 | 725_755 | 585.6666666666666 | 2415.0 | Compositional bias | Note=Ser-rich |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381294 | - | 11 | 14 | 725_755 | 552.3333333333334 | 858.0 | Compositional bias | Note=Ser-rich |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381298 | - | 14 | 17 | 725_755 | 613.3333333333334 | 919.0 | Compositional bias | Note=Ser-rich |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000502326 | - | 13 | 16 | 725_755 | 613.3333333333334 | 919.0 | Compositional bias | Note=Ser-rich |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381287 | - | 11 | 14 | 518_613 | 585.6666666666666 | 2415.0 | Domain | Fibronectin type-III 5 |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381294 | - | 11 | 14 | 518_613 | 552.3333333333334 | 858.0 | Domain | Fibronectin type-III 5 |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000336909 | - | 12 | 15 | 651_659 | 613.3333333333334 | 919.0 | Motif | Note=Box 1 motif |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381287 | - | 11 | 14 | 651_659 | 585.6666666666666 | 2415.0 | Motif | Note=Box 1 motif |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381294 | - | 11 | 14 | 651_659 | 552.3333333333334 | 858.0 | Motif | Note=Box 1 motif |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381298 | - | 14 | 17 | 651_659 | 613.3333333333334 | 919.0 | Motif | Note=Box 1 motif |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000502326 | - | 13 | 16 | 651_659 | 613.3333333333334 | 919.0 | Motif | Note=Box 1 motif |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000336909 | - | 12 | 15 | 23_619 | 613.3333333333334 | 919.0 | Topological domain | Extracellular |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000336909 | - | 12 | 15 | 642_918 | 613.3333333333334 | 919.0 | Topological domain | Cytoplasmic |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381287 | - | 11 | 14 | 23_619 | 585.6666666666666 | 2415.0 | Topological domain | Extracellular |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381287 | - | 11 | 14 | 642_918 | 585.6666666666666 | 2415.0 | Topological domain | Cytoplasmic |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381294 | - | 11 | 14 | 23_619 | 552.3333333333334 | 858.0 | Topological domain | Extracellular |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381294 | - | 11 | 14 | 642_918 | 552.3333333333334 | 858.0 | Topological domain | Cytoplasmic |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381298 | - | 14 | 17 | 23_619 | 613.3333333333334 | 919.0 | Topological domain | Extracellular |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381298 | - | 14 | 17 | 642_918 | 613.3333333333334 | 919.0 | Topological domain | Cytoplasmic |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000502326 | - | 13 | 16 | 23_619 | 613.3333333333334 | 919.0 | Topological domain | Extracellular |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000502326 | - | 13 | 16 | 642_918 | 613.3333333333334 | 919.0 | Topological domain | Cytoplasmic |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000336909 | - | 12 | 15 | 620_641 | 613.3333333333334 | 919.0 | Transmembrane | Helical |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381287 | - | 11 | 14 | 620_641 | 585.6666666666666 | 2415.0 | Transmembrane | Helical |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381294 | - | 11 | 14 | 620_641 | 552.3333333333334 | 858.0 | Transmembrane | Helical |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000381298 | - | 14 | 17 | 620_641 | 613.3333333333334 | 919.0 | Transmembrane | Helical |
| Hgene | IL6ST | chr5:55247292 | chr1:183177016 | ENST00000502326 | - | 13 | 16 | 620_641 | 613.3333333333334 | 919.0 | Transmembrane | Helical |
Top |
Fusion Protein Structures |
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
Top |
pLDDT score distribution |
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
IL6ST_pLDDT.png |
LAMC2_pLDDT.png |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
Top |
Ramachandran Plot of Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| Fusion AA seq ID in FusionPDB and their Ramachandran plots |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| IL6ST | |
| LAMC2 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to IL6ST-LAMC2 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to IL6ST-LAMC2 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

