| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:ITK-SYK |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: ITK-SYK | FusionPDB ID: 40601 | FusionGDB2.0 ID: 40601 | Hgene | Tgene | Gene symbol | ITK | SYK | Gene ID | 3702 | 6850 |
| Gene name | IL2 inducible T cell kinase | spleen associated tyrosine kinase | |
| Synonyms | EMT|LPFS1|LYK|PSCTK2 | p72-Syk | |
| Cytomap | 5q33.3 | 9q22.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | tyrosine-protein kinase ITK/TSKIL-2-inducible T-cell kinaseT-cell-specific kinasehomolog of mouse T-cell itk/tskinterleukin-2-inducible T-cell kinasekinase EMTtyrosine-protein kinase LYK | tyrosine-protein kinase SYKspleen tyrosine kinase | |
| Modification date | 20200327 | 20200329 | |
| UniProtAcc | Q08881 | P43405 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000422843, ENST00000519749, | ENST00000375746, ENST00000476708, ENST00000375747, ENST00000375751, ENST00000375754, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 3 X 3 X 2=18 | 9 X 9 X 6=486 |
| # samples | 2 | 9 | |
| ** MAII score | log2(2/18*10)=0.15200309344505 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(9/486*10)=-2.43295940727611 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: ITK [Title/Abstract] AND SYK [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | |||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | SYK | GO:0006468 | protein phosphorylation | 17681949 |
| Tgene | SYK | GO:0007159 | leukocyte cell-cell adhesion | 12885943 |
| Tgene | SYK | GO:0030593 | neutrophil chemotaxis | 12885943 |
 Fusion gene breakpoints across ITK (5'-gene) Fusion gene breakpoints across ITK (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 Fusion gene breakpoints across SYK (3'-gene) Fusion gene breakpoints across SYK (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerKB3 | . | . | ITK | chr5 | 156644917 | + | SYK | chr9 | 93636485 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000422843 | ITK | chr5 | 156644917 | + | ENST00000375751 | SYK | chr9 | 93636485 | + | 4589 | 647 | 29 | 1639 | 536 |
| ENST00000422843 | ITK | chr5 | 156644917 | + | ENST00000375754 | SYK | chr9 | 93636485 | + | 4589 | 647 | 29 | 1639 | 536 |
| ENST00000422843 | ITK | chr5 | 156644917 | + | ENST00000375747 | SYK | chr9 | 93636485 | + | 4589 | 647 | 29 | 1639 | 536 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >40601_40601_1_ITK-SYK_ITK_chr5_156644917_ENST00000422843_SYK_chr9_93636485_ENST00000375747_length(amino acids)=536AA_BP=206 MPLSSSLKINCLHIAFFAPKLFPLVVLRGDAQGAPPFKNWIMNNFILLEEQLIKKSQQKRRTSPSNFKVRFFVLTKASLAYFEDRHGKKR TLKGSIELSRIKCVEIVKSDISIPCHYKYPFQVVHDNYLLYVFAPDRESRQRWVLALKEETRNNNSLVPKYHPNFWMDGKWRCCSQLEKL ATGCAQYDPTKNASKKPLPPTPEDNRSSPAQGNRQESTVSFNPYEPELAPWAADKGPQREALPMDTEVYESPYADPEEIRPKEVYLDRKL LTLEDKELGSGNFGTVKKGYYQMKKVVKTVAVKILKNEANDPALKDELLAEANVMQQLDNPYIVRMIGICEAESWMLVMEMAELGPLNKY LQQNRHVKDKNIIELVHQVSMGMKYLEESNFVHRDLAARNVLLVTQHYAKISDFGLSKALRADENYYKAQTHGKWPVKWYAPECINYYKF -------------------------------------------------------------- >40601_40601_2_ITK-SYK_ITK_chr5_156644917_ENST00000422843_SYK_chr9_93636485_ENST00000375751_length(amino acids)=536AA_BP=206 MPLSSSLKINCLHIAFFAPKLFPLVVLRGDAQGAPPFKNWIMNNFILLEEQLIKKSQQKRRTSPSNFKVRFFVLTKASLAYFEDRHGKKR TLKGSIELSRIKCVEIVKSDISIPCHYKYPFQVVHDNYLLYVFAPDRESRQRWVLALKEETRNNNSLVPKYHPNFWMDGKWRCCSQLEKL ATGCAQYDPTKNASKKPLPPTPEDNRSSPAQGNRQESTVSFNPYEPELAPWAADKGPQREALPMDTEVYESPYADPEEIRPKEVYLDRKL LTLEDKELGSGNFGTVKKGYYQMKKVVKTVAVKILKNEANDPALKDELLAEANVMQQLDNPYIVRMIGICEAESWMLVMEMAELGPLNKY LQQNRHVKDKNIIELVHQVSMGMKYLEESNFVHRDLAARNVLLVTQHYAKISDFGLSKALRADENYYKAQTHGKWPVKWYAPECINYYKF -------------------------------------------------------------- >40601_40601_3_ITK-SYK_ITK_chr5_156644917_ENST00000422843_SYK_chr9_93636485_ENST00000375754_length(amino acids)=536AA_BP=206 MPLSSSLKINCLHIAFFAPKLFPLVVLRGDAQGAPPFKNWIMNNFILLEEQLIKKSQQKRRTSPSNFKVRFFVLTKASLAYFEDRHGKKR TLKGSIELSRIKCVEIVKSDISIPCHYKYPFQVVHDNYLLYVFAPDRESRQRWVLALKEETRNNNSLVPKYHPNFWMDGKWRCCSQLEKL ATGCAQYDPTKNASKKPLPPTPEDNRSSPAQGNRQESTVSFNPYEPELAPWAADKGPQREALPMDTEVYESPYADPEEIRPKEVYLDRKL LTLEDKELGSGNFGTVKKGYYQMKKVVKTVAVKILKNEANDPALKDELLAEANVMQQLDNPYIVRMIGICEAESWMLVMEMAELGPLNKY LQQNRHVKDKNIIELVHQVSMGMKYLEESNFVHRDLAARNVLLVTQHYAKISDFGLSKALRADENYYKAQTHGKWPVKWYAPECINYYKF -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr5:/chr9:) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ITK | SYK |
| FUNCTION: Tyrosine kinase that plays an essential role in regulation of the adaptive immune response. Regulates the development, function and differentiation of conventional T-cells and nonconventional NKT-cells. When antigen presenting cells (APC) activate T-cell receptor (TCR), a series of phosphorylation lead to the recruitment of ITK to the cell membrane, in the vicinity of the stimulated TCR receptor, where it is phosphorylated by LCK. Phosphorylation leads to ITK autophosphorylation and full activation. Once activated, phosphorylates PLCG1, leading to the activation of this lipase and subsequent cleavage of its substrates. In turn, the endoplasmic reticulum releases calcium in the cytoplasm and the nuclear activator of activated T-cells (NFAT) translocates into the nucleus to perform its transcriptional duty. Phosphorylates 2 essential adapter proteins: the linker for activation of T-cells/LAT protein and LCP2. Then, a large number of signaling molecules such as VAV1 are recruited and ultimately lead to lymphokine production, T-cell proliferation and differentiation (PubMed:12186560, PubMed:12682224, PubMed:21725281). Required for TCR-mediated calcium response in gamma-delta T-cells, may also be involved in the modulation of the transcriptomic signature in the Vgamma2-positive subset of immature gamma-delta T-cells (By similarity). Phosphorylates TBX21 at 'Tyr-530' and mediates its interaction with GATA3 (By similarity). {ECO:0000250|UniProtKB:Q03526, ECO:0000269|PubMed:12186560, ECO:0000269|PubMed:12682224, ECO:0000269|PubMed:21725281}. | FUNCTION: Non-receptor tyrosine kinase which mediates signal transduction downstream of a variety of transmembrane receptors including classical immunoreceptors like the B-cell receptor (BCR). Regulates several biological processes including innate and adaptive immunity, cell adhesion, osteoclast maturation, platelet activation and vascular development. Assembles into signaling complexes with activated receptors at the plasma membrane via interaction between its SH2 domains and the receptor tyrosine-phosphorylated ITAM domains. The association with the receptor can also be indirect and mediated by adapter proteins containing ITAM or partial hemITAM domains. The phosphorylation of the ITAM domains is generally mediated by SRC subfamily kinases upon engagement of the receptor. More rarely signal transduction via SYK could be ITAM-independent. Direct downstream effectors phosphorylated by SYK include VAV1, PLCG1, PI-3-kinase, LCP2 and BLNK. Initially identified as essential in B-cell receptor (BCR) signaling, it is necessary for the maturation of B-cells most probably at the pro-B to pre-B transition. Activated upon BCR engagement, it phosphorylates and activates BLNK an adapter linking the activated BCR to downstream signaling adapters and effectors. It also phosphorylates and activates PLCG1 and the PKC signaling pathway. It also phosphorylates BTK and regulates its activity in B-cell antigen receptor (BCR)-coupled signaling. In addition to its function downstream of BCR plays also a role in T-cell receptor signaling. Plays also a crucial role in the innate immune response to fungal, bacterial and viral pathogens. It is for instance activated by the membrane lectin CLEC7A. Upon stimulation by fungal proteins, CLEC7A together with SYK activates immune cells inducing the production of ROS. Also activates the inflammasome and NF-kappa-B-mediated transcription of chemokines and cytokines in presence of pathogens. Regulates neutrophil degranulation and phagocytosis through activation of the MAPK signaling cascade (By similarity). Required for the stimulation of neutrophil phagocytosis by IL15 (PubMed:15123770). Also mediates the activation of dendritic cells by cell necrosis stimuli. Also involved in mast cells activation. Involved in interleukin-3/IL3-mediated signaling pathway in basophils (By similarity). Also functions downstream of receptors mediating cell adhesion. Relays for instance, integrin-mediated neutrophils and macrophages activation and P-selectin receptor/SELPG-mediated recruitment of leukocytes to inflammatory loci. Plays also a role in non-immune processes. It is for instance involved in vascular development where it may regulate blood and lymphatic vascular separation. It is also required for osteoclast development and function. Functions in the activation of platelets by collagen, mediating PLCG2 phosphorylation and activation. May be coupled to the collagen receptor by the ITAM domain-containing FCER1G. Also activated by the membrane lectin CLEC1B that is required for activation of platelets by PDPN/podoplanin. Involved in platelet adhesion being activated by ITGB3 engaged by fibrinogen. Together with CEACAM20, enhances production of the cytokine CXCL8/IL-8 via the NFKB pathway and may thus have a role in the intestinal immune response (By similarity). {ECO:0000250|UniProtKB:P48025, ECO:0000269|PubMed:12387735, ECO:0000269|PubMed:12456653, ECO:0000269|PubMed:15123770, ECO:0000269|PubMed:15388330, ECO:0000269|PubMed:19909739, ECO:0000269|PubMed:8657103, ECO:0000269|PubMed:9535867}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
Top |
Fusion Protein Structures |
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
| PDB file (367) >>>367.pdbFusion protein BP residue: 206 CIF file (367) >>>367.cif | ITK | chr5 | 156644917 | + | SYK | chr9 | 93636485 | + | MPLSSSLKINCLHIAFFAPKLFPLVVLRGDAQGAPPFKNWIMNNFILLEE QLIKKSQQKRRTSPSNFKVRFFVLTKASLAYFEDRHGKKRTLKGSIELSR IKCVEIVKSDISIPCHYKYPFQVVHDNYLLYVFAPDRESRQRWVLALKEE TRNNNSLVPKYHPNFWMDGKWRCCSQLEKLATGCAQYDPTKNASKKPLPP TPEDNRSSPAQGNRQESTVSFNPYEPELAPWAADKGPQREALPMDTEVYE SPYADPEEIRPKEVYLDRKLLTLEDKELGSGNFGTVKKGYYQMKKVVKTV AVKILKNEANDPALKDELLAEANVMQQLDNPYIVRMIGICEAESWMLVME MAELGPLNKYLQQNRHVKDKNIIELVHQVSMGMKYLEESNFVHRDLAARN VLLVTQHYAKISDFGLSKALRADENYYKAQTHGKWPVKWYAPECINYYKF SSKSDVWSFGVLMWEAFSYGQKPYRGMKGSEVTAMLEKGERMGCPAGCPR | 536 |
| 3D view using mol* of 367 (AA BP:206) | ||||||||||
 | ||||||||||
Top |
pLDDT score distribution |
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
ITK_pLDDT.png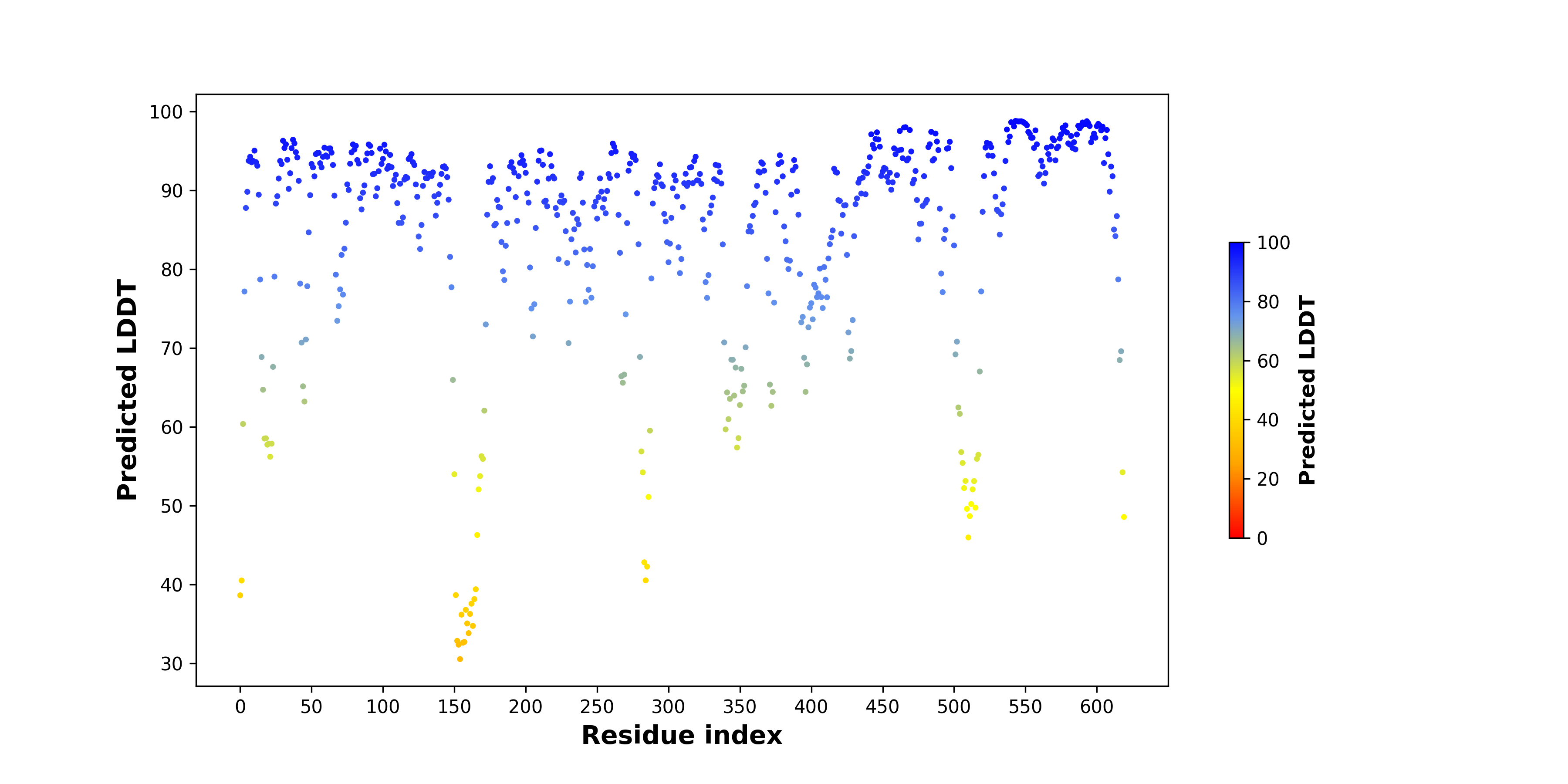 |
SYK_pLDDT.png |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
ITK_SYK_367_pLDDT.png (AA BP:206)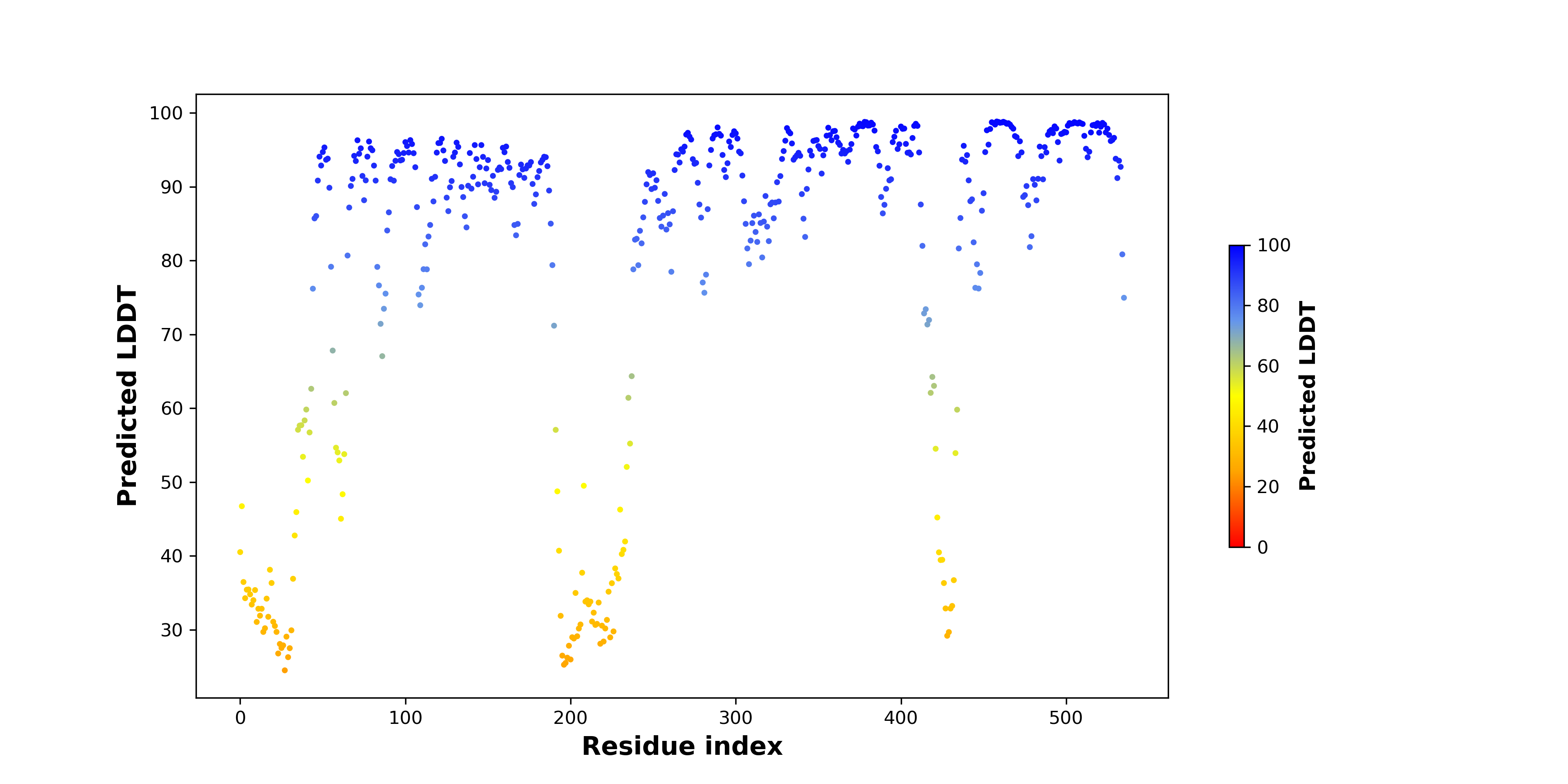 |
ITK_SYK_367_pLDDT_and_active_sites.png (AA BP:206) |
ITK_SYK_367_violinplot.png (AA BP:206) |
Top |
Ramachandran Plot of Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| Fusion AA seq ID in FusionPDB and their Ramachandran plots |
| ITK_SYK_367.png |
 |
Top |
Potential Active Site Information |
 The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. |
| Fusion AA seq ID in FusionPDB | Site score | Size | D score | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
| 367 | 0.9373 | 143 | 1.0619 | 94.325 | 0.7302 | 0.4194 | 0.3878 | 0.3613 | 0.3979 | 0.9081 | 0.8171 | Chain A: 253,258,259,261,262,263,265,266,267,270,2 91,292,293,294 |
Top |
Potentially Interacting Small Molecules through Virtual Screening |
 The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. |
| Fusion AA seq ID in FusionPDB | ZINC ID | DrugBank ID | Drug name | Docking score | Glide gscore |
Top |
 Drug information from DrugBank of the top 20 interacting small molecules. Drug information from DrugBank of the top 20 interacting small molecules. |
| ZINC ID | DrugBank ID | Drug name | Drug type | SMILES | Drug group |
Top |
Biochemical Features of Small Molecules |
 ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) |
| ZINC ID | mol_MW | dipole | SASA | FOSA | FISA | PISA | WPSA | volume | donorHB | accptHB | IP | Human Oral Absorption | Percent Human Oral Absorption | Rule Of Five | Rule Of Three |
Top |
Drug Toxicity Information |
 Toxicity information of individual drugs using eToxPred Toxicity information of individual drugs using eToxPred |
| ZINC ID | Smile | Surface Accessibility | Toxicity |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| ITK | LCP2, KPNA2, SOCS1, PPIA, WAS, LAT, PLCG1, CD2, FYN, SRC, GRB2, KHDRBS1, ITK, HNRNPK, CBL, IKBKG, FASLG, IBTK, FGFR1, DOK1, HSP90AA1, ERBB2, ERBB3, Mbp, SPRR2A, FUS, EGFR, NSUN6, SMURF1, HSP90AB3P, HSP90AA5P, DYNC1I2, HSP90AB1, GAPDHS, CDC37, PIK3R1, TCEB2, AHNAK, AURKB, PRC1, GAB1, MET, ERBB4, C2orf44, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| ITK | 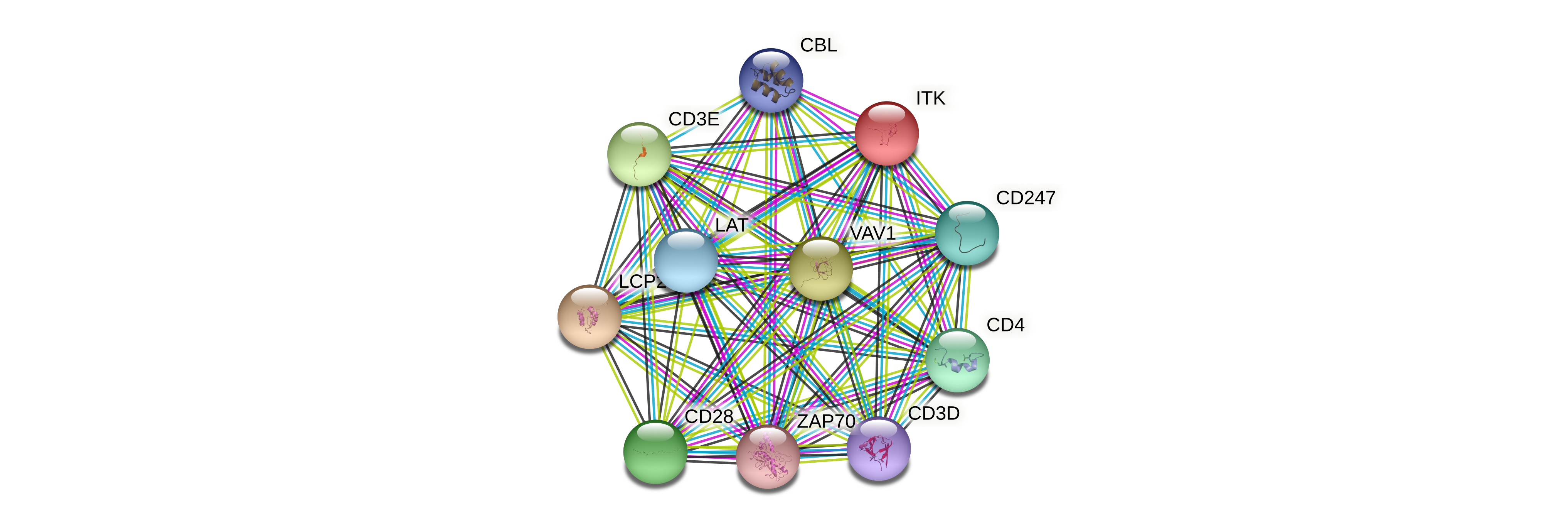 |
| SYK | 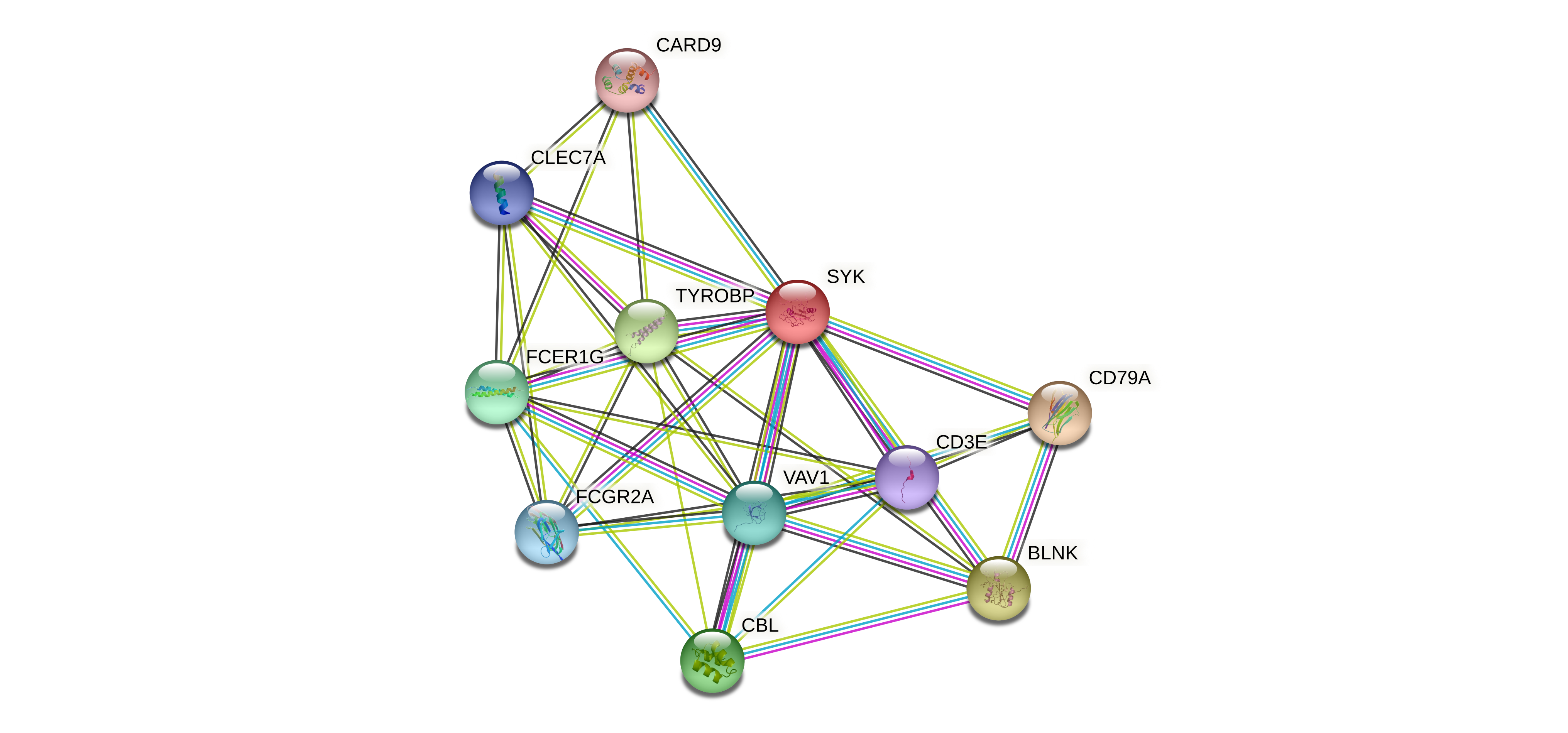 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to ITK-SYK |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
| ITK | SYK | Tofacitinib | PubMed | 31545408 |
Top |
Related Diseases to ITK-SYK |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
| ITK | SYK | Non‑Hodgkin Lymphomas | PubMed | 31545408 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | ITK | C3552634 | LYMPHOPROLIFERATIVE SYNDROME 1 | 7 | CLINGEN;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Hgene | ITK | C0494261 | Combined immunodeficiency | 1 | GENOMICS_ENGLAND |
| Hgene | ITK | C4283841 | ITK Deficiency | 1 | GENOMICS_ENGLAND |
| Tgene | SYK | C0025202 | melanoma | 1 | CTD_human |
| Tgene | SYK | C0025500 | Mesothelioma | 1 | CTD_human |

