| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:LMNA-RAF1 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: LMNA-RAF1 | FusionPDB ID: 45796 | FusionGDB2.0 ID: 45796 | Hgene | Tgene | Gene symbol | LMNA | RAF1 | Gene ID | 4000 | 6037 |
| Gene name | lamin A/C | ribonuclease A family member 3 | |
| Synonyms | CDCD1|CDDC|CMD1A|CMT2B1|EMD2|FPL|FPLD|FPLD2|HGPS|IDC|LDP1|LFP|LGMD1B|LMN1|LMNC|LMNL1|MADA|PRO1 | ECP|RAF1|RNS3 | |
| Cytomap | 1q22 | 14q11.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | lamin70 kDa laminepididymis secretory sperm binding proteinlamin A/C-like 1mandibuloacral dysplasia type Aprelamin-A/Crenal carcinoma antigen NY-REN-32 | eosinophil cationic proteinRNase 3cytotoxic ribonucleaseribonuclease 3ribonuclease, RNase A family, 3 | |
| Modification date | 20200329 | 20200313 | |
| UniProtAcc | P02545 | P04049 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000496738, ENST00000361308, ENST00000368297, ENST00000368301, ENST00000392353, ENST00000368299, ENST00000368300, ENST00000448611, ENST00000473598, ENST00000347559, | ENST00000251849, ENST00000442415, ENST00000534997, ENST00000542177, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 16 X 16 X 9=2304 | 19 X 16 X 7=2128 |
| # samples | 21 | 18 | |
| ** MAII score | log2(21/2304*10)=-3.45567948377619 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(18/2128*10)=-3.56342933917152 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: LMNA [Title/Abstract] AND RAF1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | LMNA(156107534)-RAF1(12641914), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | LMNA-RAF1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. LMNA-RAF1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. LMNA-RAF1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. LMNA-RAF1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. LMNA-RAF1 seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. LMNA-RAF1 seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. LMNA-RAF1 seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. LMNA-RAF1 seems lost the major protein functional domain in Tgene partner, which is a kinase due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | LMNA | GO:0090343 | positive regulation of cell aging | 20458013 |
| Tgene | RAF1 | GO:0002227 | innate immune response in mucosa | 12860195 |
| Tgene | RAF1 | GO:0019731 | antibacterial humoral response | 12860195 |
| Tgene | RAF1 | GO:0043152 | induction of bacterial agglutination | 23992292 |
| Tgene | RAF1 | GO:0045087 | innate immune response | 23992292 |
| Tgene | RAF1 | GO:0050829 | defense response to Gram-negative bacterium | 23992292 |
| Tgene | RAF1 | GO:0050830 | defense response to Gram-positive bacterium | 12860195|23992292 |
| Tgene | RAF1 | GO:0061844 | antimicrobial humoral immune response mediated by antimicrobial peptide | 12860195 |
 Fusion gene breakpoints across LMNA (5'-gene) Fusion gene breakpoints across LMNA (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
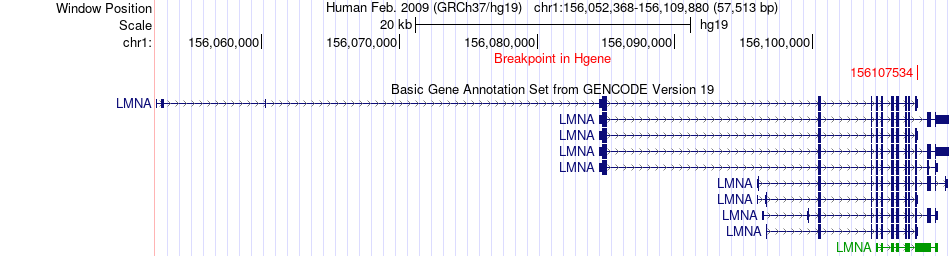 |
 Fusion gene breakpoints across RAF1 (3'-gene) Fusion gene breakpoints across RAF1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
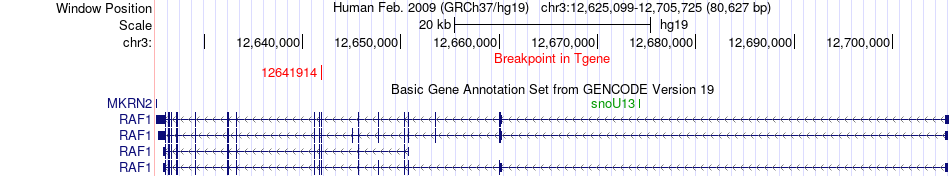 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SKCM | TCGA-EB-A5SF-01A | LMNA | chr1 | 156107534 | + | RAF1 | chr3 | 12641914 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000368300 | LMNA | chr1 | 156107534 | + | ENST00000251849 | RAF1 | chr3 | 12641914 | - | 3936 | 1910 | 83 | 3022 | 979 |
| ENST00000368300 | LMNA | chr1 | 156107534 | + | ENST00000442415 | RAF1 | chr3 | 12641914 | - | 3739 | 1910 | 83 | 3022 | 979 |
| ENST00000368300 | LMNA | chr1 | 156107534 | + | ENST00000534997 | RAF1 | chr3 | 12641914 | - | 3270 | 1910 | 83 | 3022 | 979 |
| ENST00000368300 | LMNA | chr1 | 156107534 | + | ENST00000542177 | RAF1 | chr3 | 12641914 | - | 3204 | 1910 | 83 | 3022 | 979 |
| ENST00000368299 | LMNA | chr1 | 156107534 | + | ENST00000251849 | RAF1 | chr3 | 12641914 | - | 3921 | 1895 | 68 | 3007 | 979 |
| ENST00000368299 | LMNA | chr1 | 156107534 | + | ENST00000442415 | RAF1 | chr3 | 12641914 | - | 3724 | 1895 | 68 | 3007 | 979 |
| ENST00000368299 | LMNA | chr1 | 156107534 | + | ENST00000534997 | RAF1 | chr3 | 12641914 | - | 3255 | 1895 | 68 | 3007 | 979 |
| ENST00000368299 | LMNA | chr1 | 156107534 | + | ENST00000542177 | RAF1 | chr3 | 12641914 | - | 3189 | 1895 | 68 | 3007 | 979 |
| ENST00000448611 | LMNA | chr1 | 156107534 | + | ENST00000251849 | RAF1 | chr3 | 12641914 | - | 3432 | 1406 | 8 | 2518 | 836 |
| ENST00000448611 | LMNA | chr1 | 156107534 | + | ENST00000442415 | RAF1 | chr3 | 12641914 | - | 3235 | 1406 | 8 | 2518 | 836 |
| ENST00000448611 | LMNA | chr1 | 156107534 | + | ENST00000534997 | RAF1 | chr3 | 12641914 | - | 2766 | 1406 | 8 | 2518 | 836 |
| ENST00000448611 | LMNA | chr1 | 156107534 | + | ENST00000542177 | RAF1 | chr3 | 12641914 | - | 2700 | 1406 | 8 | 2518 | 836 |
| ENST00000473598 | LMNA | chr1 | 156107534 | + | ENST00000251849 | RAF1 | chr3 | 12641914 | - | 3547 | 1521 | 84 | 2633 | 849 |
| ENST00000473598 | LMNA | chr1 | 156107534 | + | ENST00000442415 | RAF1 | chr3 | 12641914 | - | 3350 | 1521 | 84 | 2633 | 849 |
| ENST00000473598 | LMNA | chr1 | 156107534 | + | ENST00000534997 | RAF1 | chr3 | 12641914 | - | 2881 | 1521 | 84 | 2633 | 849 |
| ENST00000473598 | LMNA | chr1 | 156107534 | + | ENST00000542177 | RAF1 | chr3 | 12641914 | - | 2815 | 1521 | 84 | 2633 | 849 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000368300 | ENST00000251849 | LMNA | chr1 | 156107534 | + | RAF1 | chr3 | 12641914 | - | 0.00582304 | 0.994177 |
| ENST00000368300 | ENST00000442415 | LMNA | chr1 | 156107534 | + | RAF1 | chr3 | 12641914 | - | 0.008715062 | 0.9912849 |
| ENST00000368300 | ENST00000534997 | LMNA | chr1 | 156107534 | + | RAF1 | chr3 | 12641914 | - | 0.00924723 | 0.9907528 |
| ENST00000368300 | ENST00000542177 | LMNA | chr1 | 156107534 | + | RAF1 | chr3 | 12641914 | - | 0.00951526 | 0.9904847 |
| ENST00000368299 | ENST00000251849 | LMNA | chr1 | 156107534 | + | RAF1 | chr3 | 12641914 | - | 0.005980463 | 0.9940195 |
| ENST00000368299 | ENST00000442415 | LMNA | chr1 | 156107534 | + | RAF1 | chr3 | 12641914 | - | 0.008941601 | 0.9910584 |
| ENST00000368299 | ENST00000534997 | LMNA | chr1 | 156107534 | + | RAF1 | chr3 | 12641914 | - | 0.009536465 | 0.9904635 |
| ENST00000368299 | ENST00000542177 | LMNA | chr1 | 156107534 | + | RAF1 | chr3 | 12641914 | - | 0.009830482 | 0.99016947 |
| ENST00000448611 | ENST00000251849 | LMNA | chr1 | 156107534 | + | RAF1 | chr3 | 12641914 | - | 0.014983132 | 0.98501694 |
| ENST00000448611 | ENST00000442415 | LMNA | chr1 | 156107534 | + | RAF1 | chr3 | 12641914 | - | 0.019259676 | 0.9807403 |
| ENST00000448611 | ENST00000534997 | LMNA | chr1 | 156107534 | + | RAF1 | chr3 | 12641914 | - | 0.019979503 | 0.9800205 |
| ENST00000448611 | ENST00000542177 | LMNA | chr1 | 156107534 | + | RAF1 | chr3 | 12641914 | - | 0.020669507 | 0.9793305 |
| ENST00000473598 | ENST00000251849 | LMNA | chr1 | 156107534 | + | RAF1 | chr3 | 12641914 | - | 0.005441572 | 0.99455845 |
| ENST00000473598 | ENST00000442415 | LMNA | chr1 | 156107534 | + | RAF1 | chr3 | 12641914 | - | 0.008506982 | 0.99149305 |
| ENST00000473598 | ENST00000534997 | LMNA | chr1 | 156107534 | + | RAF1 | chr3 | 12641914 | - | 0.009186047 | 0.9908139 |
| ENST00000473598 | ENST00000542177 | LMNA | chr1 | 156107534 | + | RAF1 | chr3 | 12641914 | - | 0.009586508 | 0.9904135 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >45796_45796_1_LMNA-RAF1_LMNA_chr1_156107534_ENST00000368299_RAF1_chr3_12641914_ENST00000251849_length(amino acids)=979AA_BP=74 MPGASREPAGRRTPTPSSLCPSTRAPRPFRDPCPAGSAANLPAMETPSQRRATRSGAQASSTPLSPTRITRLQEKEDLQELNDRLAVYID RVRSLETENAGLRLRITESEEVVSREVSGIKAAYEAELGDARKTLDSVAKERARLQLELSKVREEFKELKARNTKKEGDLIAAQARLKDL EALLNSKEAALSTALSEKRTLEGELHDLRGQVAKLEAALGEAKKQLQDEMLRRVDAENRLQTMKEELDFQKNIYSEELRETKRRHETRLV EIDNGKQREFESRLADALQELRAQHEDQVEQYKKELEKTYSAKLDNARQSAERNSNLVGAAHEELQQSRIRIDSLSAQLSQLQKQLAAKE AKLRDLEDSLARERDTSRRLLAEKEREMAEMRARMQQQLDEYQELLDIKLALDMEIHAYRKLLEGEEERLRLSPSPTSQRSRGRASSHSS QTQGGGSVTKKRKLESTESRSSFSQHARTSGRVAVEEVDEEGKFVRLRNKSNEDQSMGNWQIKRQNGDDPLLTYRFPPKFTLKAGQVVTI WAAGAGATHSPPTDLVWKAQNTWGCGNSLRTALINSTGEEVAMRKLVRSVTVVEDDEDEDGDDLLHHHHDAIRSHSESASPSALSSSPNN LSPTGWSQPKTPVPAQRERAPVSGTQEKNKIRPRGQRDSSYYWEIEASEVMLSTRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQFQA FRNEVAVLRKTRHVNILLFMGYMTKDNLAIVTQWCEGSSLYKHLHVQETKFQMFQLIDIARQTAQGMDYLHAKNIIHRDMKSNNIFLHEG LTVKIGDFGLATVKSRWSGSQQVEQPTGSVLWMAPEVIRMQDNNPFSFQSDVYSYGIVLYELMTGELPYSHINNRDQIIFMVGRGYASPD -------------------------------------------------------------- >45796_45796_2_LMNA-RAF1_LMNA_chr1_156107534_ENST00000368299_RAF1_chr3_12641914_ENST00000442415_length(amino acids)=979AA_BP=74 MPGASREPAGRRTPTPSSLCPSTRAPRPFRDPCPAGSAANLPAMETPSQRRATRSGAQASSTPLSPTRITRLQEKEDLQELNDRLAVYID RVRSLETENAGLRLRITESEEVVSREVSGIKAAYEAELGDARKTLDSVAKERARLQLELSKVREEFKELKARNTKKEGDLIAAQARLKDL EALLNSKEAALSTALSEKRTLEGELHDLRGQVAKLEAALGEAKKQLQDEMLRRVDAENRLQTMKEELDFQKNIYSEELRETKRRHETRLV EIDNGKQREFESRLADALQELRAQHEDQVEQYKKELEKTYSAKLDNARQSAERNSNLVGAAHEELQQSRIRIDSLSAQLSQLQKQLAAKE AKLRDLEDSLARERDTSRRLLAEKEREMAEMRARMQQQLDEYQELLDIKLALDMEIHAYRKLLEGEEERLRLSPSPTSQRSRGRASSHSS QTQGGGSVTKKRKLESTESRSSFSQHARTSGRVAVEEVDEEGKFVRLRNKSNEDQSMGNWQIKRQNGDDPLLTYRFPPKFTLKAGQVVTI WAAGAGATHSPPTDLVWKAQNTWGCGNSLRTALINSTGEEVAMRKLVRSVTVVEDDEDEDGDDLLHHHHDAIRSHSESASPSALSSSPNN LSPTGWSQPKTPVPAQRERAPVSGTQEKNKIRPRGQRDSSYYWEIEASEVMLSTRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQFQA FRNEVAVLRKTRHVNILLFMGYMTKDNLAIVTQWCEGSSLYKHLHVQETKFQMFQLIDIARQTAQGMDYLHAKNIIHRDMKSNNIFLHEG LTVKIGDFGLATVKSRWSGSQQVEQPTGSVLWMAPEVIRMQDNNPFSFQSDVYSYGIVLYELMTGELPYSHINNRDQIIFMVGRGYASPD -------------------------------------------------------------- >45796_45796_3_LMNA-RAF1_LMNA_chr1_156107534_ENST00000368299_RAF1_chr3_12641914_ENST00000534997_length(amino acids)=979AA_BP=74 MPGASREPAGRRTPTPSSLCPSTRAPRPFRDPCPAGSAANLPAMETPSQRRATRSGAQASSTPLSPTRITRLQEKEDLQELNDRLAVYID RVRSLETENAGLRLRITESEEVVSREVSGIKAAYEAELGDARKTLDSVAKERARLQLELSKVREEFKELKARNTKKEGDLIAAQARLKDL EALLNSKEAALSTALSEKRTLEGELHDLRGQVAKLEAALGEAKKQLQDEMLRRVDAENRLQTMKEELDFQKNIYSEELRETKRRHETRLV EIDNGKQREFESRLADALQELRAQHEDQVEQYKKELEKTYSAKLDNARQSAERNSNLVGAAHEELQQSRIRIDSLSAQLSQLQKQLAAKE AKLRDLEDSLARERDTSRRLLAEKEREMAEMRARMQQQLDEYQELLDIKLALDMEIHAYRKLLEGEEERLRLSPSPTSQRSRGRASSHSS QTQGGGSVTKKRKLESTESRSSFSQHARTSGRVAVEEVDEEGKFVRLRNKSNEDQSMGNWQIKRQNGDDPLLTYRFPPKFTLKAGQVVTI WAAGAGATHSPPTDLVWKAQNTWGCGNSLRTALINSTGEEVAMRKLVRSVTVVEDDEDEDGDDLLHHHHDAIRSHSESASPSALSSSPNN LSPTGWSQPKTPVPAQRERAPVSGTQEKNKIRPRGQRDSSYYWEIEASEVMLSTRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQFQA FRNEVAVLRKTRHVNILLFMGYMTKDNLAIVTQWCEGSSLYKHLHVQETKFQMFQLIDIARQTAQGMDYLHAKNIIHRDMKSNNIFLHEG LTVKIGDFGLATVKSRWSGSQQVEQPTGSVLWMAPEVIRMQDNNPFSFQSDVYSYGIVLYELMTGELPYSHINNRDQIIFMVGRGYASPD -------------------------------------------------------------- >45796_45796_4_LMNA-RAF1_LMNA_chr1_156107534_ENST00000368299_RAF1_chr3_12641914_ENST00000542177_length(amino acids)=979AA_BP=74 MPGASREPAGRRTPTPSSLCPSTRAPRPFRDPCPAGSAANLPAMETPSQRRATRSGAQASSTPLSPTRITRLQEKEDLQELNDRLAVYID RVRSLETENAGLRLRITESEEVVSREVSGIKAAYEAELGDARKTLDSVAKERARLQLELSKVREEFKELKARNTKKEGDLIAAQARLKDL EALLNSKEAALSTALSEKRTLEGELHDLRGQVAKLEAALGEAKKQLQDEMLRRVDAENRLQTMKEELDFQKNIYSEELRETKRRHETRLV EIDNGKQREFESRLADALQELRAQHEDQVEQYKKELEKTYSAKLDNARQSAERNSNLVGAAHEELQQSRIRIDSLSAQLSQLQKQLAAKE AKLRDLEDSLARERDTSRRLLAEKEREMAEMRARMQQQLDEYQELLDIKLALDMEIHAYRKLLEGEEERLRLSPSPTSQRSRGRASSHSS QTQGGGSVTKKRKLESTESRSSFSQHARTSGRVAVEEVDEEGKFVRLRNKSNEDQSMGNWQIKRQNGDDPLLTYRFPPKFTLKAGQVVTI WAAGAGATHSPPTDLVWKAQNTWGCGNSLRTALINSTGEEVAMRKLVRSVTVVEDDEDEDGDDLLHHHHDAIRSHSESASPSALSSSPNN LSPTGWSQPKTPVPAQRERAPVSGTQEKNKIRPRGQRDSSYYWEIEASEVMLSTRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQFQA FRNEVAVLRKTRHVNILLFMGYMTKDNLAIVTQWCEGSSLYKHLHVQETKFQMFQLIDIARQTAQGMDYLHAKNIIHRDMKSNNIFLHEG LTVKIGDFGLATVKSRWSGSQQVEQPTGSVLWMAPEVIRMQDNNPFSFQSDVYSYGIVLYELMTGELPYSHINNRDQIIFMVGRGYASPD -------------------------------------------------------------- >45796_45796_5_LMNA-RAF1_LMNA_chr1_156107534_ENST00000368300_RAF1_chr3_12641914_ENST00000251849_length(amino acids)=979AA_BP=74 MPGASREPAGRRTPTPSSLCPSTRAPRPFRDPCPAGSAANLPAMETPSQRRATRSGAQASSTPLSPTRITRLQEKEDLQELNDRLAVYID RVRSLETENAGLRLRITESEEVVSREVSGIKAAYEAELGDARKTLDSVAKERARLQLELSKVREEFKELKARNTKKEGDLIAAQARLKDL EALLNSKEAALSTALSEKRTLEGELHDLRGQVAKLEAALGEAKKQLQDEMLRRVDAENRLQTMKEELDFQKNIYSEELRETKRRHETRLV EIDNGKQREFESRLADALQELRAQHEDQVEQYKKELEKTYSAKLDNARQSAERNSNLVGAAHEELQQSRIRIDSLSAQLSQLQKQLAAKE AKLRDLEDSLARERDTSRRLLAEKEREMAEMRARMQQQLDEYQELLDIKLALDMEIHAYRKLLEGEEERLRLSPSPTSQRSRGRASSHSS QTQGGGSVTKKRKLESTESRSSFSQHARTSGRVAVEEVDEEGKFVRLRNKSNEDQSMGNWQIKRQNGDDPLLTYRFPPKFTLKAGQVVTI WAAGAGATHSPPTDLVWKAQNTWGCGNSLRTALINSTGEEVAMRKLVRSVTVVEDDEDEDGDDLLHHHHDAIRSHSESASPSALSSSPNN LSPTGWSQPKTPVPAQRERAPVSGTQEKNKIRPRGQRDSSYYWEIEASEVMLSTRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQFQA FRNEVAVLRKTRHVNILLFMGYMTKDNLAIVTQWCEGSSLYKHLHVQETKFQMFQLIDIARQTAQGMDYLHAKNIIHRDMKSNNIFLHEG LTVKIGDFGLATVKSRWSGSQQVEQPTGSVLWMAPEVIRMQDNNPFSFQSDVYSYGIVLYELMTGELPYSHINNRDQIIFMVGRGYASPD -------------------------------------------------------------- >45796_45796_6_LMNA-RAF1_LMNA_chr1_156107534_ENST00000368300_RAF1_chr3_12641914_ENST00000442415_length(amino acids)=979AA_BP=74 MPGASREPAGRRTPTPSSLCPSTRAPRPFRDPCPAGSAANLPAMETPSQRRATRSGAQASSTPLSPTRITRLQEKEDLQELNDRLAVYID RVRSLETENAGLRLRITESEEVVSREVSGIKAAYEAELGDARKTLDSVAKERARLQLELSKVREEFKELKARNTKKEGDLIAAQARLKDL EALLNSKEAALSTALSEKRTLEGELHDLRGQVAKLEAALGEAKKQLQDEMLRRVDAENRLQTMKEELDFQKNIYSEELRETKRRHETRLV EIDNGKQREFESRLADALQELRAQHEDQVEQYKKELEKTYSAKLDNARQSAERNSNLVGAAHEELQQSRIRIDSLSAQLSQLQKQLAAKE AKLRDLEDSLARERDTSRRLLAEKEREMAEMRARMQQQLDEYQELLDIKLALDMEIHAYRKLLEGEEERLRLSPSPTSQRSRGRASSHSS QTQGGGSVTKKRKLESTESRSSFSQHARTSGRVAVEEVDEEGKFVRLRNKSNEDQSMGNWQIKRQNGDDPLLTYRFPPKFTLKAGQVVTI WAAGAGATHSPPTDLVWKAQNTWGCGNSLRTALINSTGEEVAMRKLVRSVTVVEDDEDEDGDDLLHHHHDAIRSHSESASPSALSSSPNN LSPTGWSQPKTPVPAQRERAPVSGTQEKNKIRPRGQRDSSYYWEIEASEVMLSTRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQFQA FRNEVAVLRKTRHVNILLFMGYMTKDNLAIVTQWCEGSSLYKHLHVQETKFQMFQLIDIARQTAQGMDYLHAKNIIHRDMKSNNIFLHEG LTVKIGDFGLATVKSRWSGSQQVEQPTGSVLWMAPEVIRMQDNNPFSFQSDVYSYGIVLYELMTGELPYSHINNRDQIIFMVGRGYASPD -------------------------------------------------------------- >45796_45796_7_LMNA-RAF1_LMNA_chr1_156107534_ENST00000368300_RAF1_chr3_12641914_ENST00000534997_length(amino acids)=979AA_BP=74 MPGASREPAGRRTPTPSSLCPSTRAPRPFRDPCPAGSAANLPAMETPSQRRATRSGAQASSTPLSPTRITRLQEKEDLQELNDRLAVYID RVRSLETENAGLRLRITESEEVVSREVSGIKAAYEAELGDARKTLDSVAKERARLQLELSKVREEFKELKARNTKKEGDLIAAQARLKDL EALLNSKEAALSTALSEKRTLEGELHDLRGQVAKLEAALGEAKKQLQDEMLRRVDAENRLQTMKEELDFQKNIYSEELRETKRRHETRLV EIDNGKQREFESRLADALQELRAQHEDQVEQYKKELEKTYSAKLDNARQSAERNSNLVGAAHEELQQSRIRIDSLSAQLSQLQKQLAAKE AKLRDLEDSLARERDTSRRLLAEKEREMAEMRARMQQQLDEYQELLDIKLALDMEIHAYRKLLEGEEERLRLSPSPTSQRSRGRASSHSS QTQGGGSVTKKRKLESTESRSSFSQHARTSGRVAVEEVDEEGKFVRLRNKSNEDQSMGNWQIKRQNGDDPLLTYRFPPKFTLKAGQVVTI WAAGAGATHSPPTDLVWKAQNTWGCGNSLRTALINSTGEEVAMRKLVRSVTVVEDDEDEDGDDLLHHHHDAIRSHSESASPSALSSSPNN LSPTGWSQPKTPVPAQRERAPVSGTQEKNKIRPRGQRDSSYYWEIEASEVMLSTRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQFQA FRNEVAVLRKTRHVNILLFMGYMTKDNLAIVTQWCEGSSLYKHLHVQETKFQMFQLIDIARQTAQGMDYLHAKNIIHRDMKSNNIFLHEG LTVKIGDFGLATVKSRWSGSQQVEQPTGSVLWMAPEVIRMQDNNPFSFQSDVYSYGIVLYELMTGELPYSHINNRDQIIFMVGRGYASPD -------------------------------------------------------------- >45796_45796_8_LMNA-RAF1_LMNA_chr1_156107534_ENST00000368300_RAF1_chr3_12641914_ENST00000542177_length(amino acids)=979AA_BP=74 MPGASREPAGRRTPTPSSLCPSTRAPRPFRDPCPAGSAANLPAMETPSQRRATRSGAQASSTPLSPTRITRLQEKEDLQELNDRLAVYID RVRSLETENAGLRLRITESEEVVSREVSGIKAAYEAELGDARKTLDSVAKERARLQLELSKVREEFKELKARNTKKEGDLIAAQARLKDL EALLNSKEAALSTALSEKRTLEGELHDLRGQVAKLEAALGEAKKQLQDEMLRRVDAENRLQTMKEELDFQKNIYSEELRETKRRHETRLV EIDNGKQREFESRLADALQELRAQHEDQVEQYKKELEKTYSAKLDNARQSAERNSNLVGAAHEELQQSRIRIDSLSAQLSQLQKQLAAKE AKLRDLEDSLARERDTSRRLLAEKEREMAEMRARMQQQLDEYQELLDIKLALDMEIHAYRKLLEGEEERLRLSPSPTSQRSRGRASSHSS QTQGGGSVTKKRKLESTESRSSFSQHARTSGRVAVEEVDEEGKFVRLRNKSNEDQSMGNWQIKRQNGDDPLLTYRFPPKFTLKAGQVVTI WAAGAGATHSPPTDLVWKAQNTWGCGNSLRTALINSTGEEVAMRKLVRSVTVVEDDEDEDGDDLLHHHHDAIRSHSESASPSALSSSPNN LSPTGWSQPKTPVPAQRERAPVSGTQEKNKIRPRGQRDSSYYWEIEASEVMLSTRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQFQA FRNEVAVLRKTRHVNILLFMGYMTKDNLAIVTQWCEGSSLYKHLHVQETKFQMFQLIDIARQTAQGMDYLHAKNIIHRDMKSNNIFLHEG LTVKIGDFGLATVKSRWSGSQQVEQPTGSVLWMAPEVIRMQDNNPFSFQSDVYSYGIVLYELMTGELPYSHINNRDQIIFMVGRGYASPD -------------------------------------------------------------- >45796_45796_9_LMNA-RAF1_LMNA_chr1_156107534_ENST00000448611_RAF1_chr3_12641914_ENST00000251849_length(amino acids)=836AA_BP=466 MLFCSLPVCCLAMGNSEGCNTKKEGDLIAAQARLKDLEALLNSKEAALSTALSEKRTLEGELHDLRGQVAKLEAALGEAKKQLQDEMLRR VDAENRLQTMKEELDFQKNIYSEELRETKRRHETRLVEIDNGKQREFESRLADALQELRAQHEDQVEQYKKELEKTYSAKLDNARQSAER NSNLVGAAHEELQQSRIRIDSLSAQLSQLQKQLAAKEAKLRDLEDSLARERDTSRRLLAEKEREMAEMRARMQQQLDEYQELLDIKLALD MEIHAYRKLLEGEEERLRLSPSPTSQRSRGRASSHSSQTQGGGSVTKKRKLESTESRSSFSQHARTSGRVAVEEVDEEGKFVRLRNKSNE DQSMGNWQIKRQNGDDPLLTYRFPPKFTLKAGQVVTIWAAGAGATHSPPTDLVWKAQNTWGCGNSLRTALINSTGEEVAMRKLVRSVTVV EDDEDEDGDDLLHHHHDAIRSHSESASPSALSSSPNNLSPTGWSQPKTPVPAQRERAPVSGTQEKNKIRPRGQRDSSYYWEIEASEVMLS TRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQFQAFRNEVAVLRKTRHVNILLFMGYMTKDNLAIVTQWCEGSSLYKHLHVQETKFQM FQLIDIARQTAQGMDYLHAKNIIHRDMKSNNIFLHEGLTVKIGDFGLATVKSRWSGSQQVEQPTGSVLWMAPEVIRMQDNNPFSFQSDVY SYGIVLYELMTGELPYSHINNRDQIIFMVGRGYASPDLSKLYKNCPKAMKRLVADCVKKVKEERPLFPQILSSIELLQHSLPKINRSASE -------------------------------------------------------------- >45796_45796_10_LMNA-RAF1_LMNA_chr1_156107534_ENST00000448611_RAF1_chr3_12641914_ENST00000442415_length(amino acids)=836AA_BP=466 MLFCSLPVCCLAMGNSEGCNTKKEGDLIAAQARLKDLEALLNSKEAALSTALSEKRTLEGELHDLRGQVAKLEAALGEAKKQLQDEMLRR VDAENRLQTMKEELDFQKNIYSEELRETKRRHETRLVEIDNGKQREFESRLADALQELRAQHEDQVEQYKKELEKTYSAKLDNARQSAER NSNLVGAAHEELQQSRIRIDSLSAQLSQLQKQLAAKEAKLRDLEDSLARERDTSRRLLAEKEREMAEMRARMQQQLDEYQELLDIKLALD MEIHAYRKLLEGEEERLRLSPSPTSQRSRGRASSHSSQTQGGGSVTKKRKLESTESRSSFSQHARTSGRVAVEEVDEEGKFVRLRNKSNE DQSMGNWQIKRQNGDDPLLTYRFPPKFTLKAGQVVTIWAAGAGATHSPPTDLVWKAQNTWGCGNSLRTALINSTGEEVAMRKLVRSVTVV EDDEDEDGDDLLHHHHDAIRSHSESASPSALSSSPNNLSPTGWSQPKTPVPAQRERAPVSGTQEKNKIRPRGQRDSSYYWEIEASEVMLS TRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQFQAFRNEVAVLRKTRHVNILLFMGYMTKDNLAIVTQWCEGSSLYKHLHVQETKFQM FQLIDIARQTAQGMDYLHAKNIIHRDMKSNNIFLHEGLTVKIGDFGLATVKSRWSGSQQVEQPTGSVLWMAPEVIRMQDNNPFSFQSDVY SYGIVLYELMTGELPYSHINNRDQIIFMVGRGYASPDLSKLYKNCPKAMKRLVADCVKKVKEERPLFPQILSSIELLQHSLPKINRSASE -------------------------------------------------------------- >45796_45796_11_LMNA-RAF1_LMNA_chr1_156107534_ENST00000448611_RAF1_chr3_12641914_ENST00000534997_length(amino acids)=836AA_BP=466 MLFCSLPVCCLAMGNSEGCNTKKEGDLIAAQARLKDLEALLNSKEAALSTALSEKRTLEGELHDLRGQVAKLEAALGEAKKQLQDEMLRR VDAENRLQTMKEELDFQKNIYSEELRETKRRHETRLVEIDNGKQREFESRLADALQELRAQHEDQVEQYKKELEKTYSAKLDNARQSAER NSNLVGAAHEELQQSRIRIDSLSAQLSQLQKQLAAKEAKLRDLEDSLARERDTSRRLLAEKEREMAEMRARMQQQLDEYQELLDIKLALD MEIHAYRKLLEGEEERLRLSPSPTSQRSRGRASSHSSQTQGGGSVTKKRKLESTESRSSFSQHARTSGRVAVEEVDEEGKFVRLRNKSNE DQSMGNWQIKRQNGDDPLLTYRFPPKFTLKAGQVVTIWAAGAGATHSPPTDLVWKAQNTWGCGNSLRTALINSTGEEVAMRKLVRSVTVV EDDEDEDGDDLLHHHHDAIRSHSESASPSALSSSPNNLSPTGWSQPKTPVPAQRERAPVSGTQEKNKIRPRGQRDSSYYWEIEASEVMLS TRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQFQAFRNEVAVLRKTRHVNILLFMGYMTKDNLAIVTQWCEGSSLYKHLHVQETKFQM FQLIDIARQTAQGMDYLHAKNIIHRDMKSNNIFLHEGLTVKIGDFGLATVKSRWSGSQQVEQPTGSVLWMAPEVIRMQDNNPFSFQSDVY SYGIVLYELMTGELPYSHINNRDQIIFMVGRGYASPDLSKLYKNCPKAMKRLVADCVKKVKEERPLFPQILSSIELLQHSLPKINRSASE -------------------------------------------------------------- >45796_45796_12_LMNA-RAF1_LMNA_chr1_156107534_ENST00000448611_RAF1_chr3_12641914_ENST00000542177_length(amino acids)=836AA_BP=466 MLFCSLPVCCLAMGNSEGCNTKKEGDLIAAQARLKDLEALLNSKEAALSTALSEKRTLEGELHDLRGQVAKLEAALGEAKKQLQDEMLRR VDAENRLQTMKEELDFQKNIYSEELRETKRRHETRLVEIDNGKQREFESRLADALQELRAQHEDQVEQYKKELEKTYSAKLDNARQSAER NSNLVGAAHEELQQSRIRIDSLSAQLSQLQKQLAAKEAKLRDLEDSLARERDTSRRLLAEKEREMAEMRARMQQQLDEYQELLDIKLALD MEIHAYRKLLEGEEERLRLSPSPTSQRSRGRASSHSSQTQGGGSVTKKRKLESTESRSSFSQHARTSGRVAVEEVDEEGKFVRLRNKSNE DQSMGNWQIKRQNGDDPLLTYRFPPKFTLKAGQVVTIWAAGAGATHSPPTDLVWKAQNTWGCGNSLRTALINSTGEEVAMRKLVRSVTVV EDDEDEDGDDLLHHHHDAIRSHSESASPSALSSSPNNLSPTGWSQPKTPVPAQRERAPVSGTQEKNKIRPRGQRDSSYYWEIEASEVMLS TRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQFQAFRNEVAVLRKTRHVNILLFMGYMTKDNLAIVTQWCEGSSLYKHLHVQETKFQM FQLIDIARQTAQGMDYLHAKNIIHRDMKSNNIFLHEGLTVKIGDFGLATVKSRWSGSQQVEQPTGSVLWMAPEVIRMQDNNPFSFQSDVY SYGIVLYELMTGELPYSHINNRDQIIFMVGRGYASPDLSKLYKNCPKAMKRLVADCVKKVKEERPLFPQILSSIELLQHSLPKINRSASE -------------------------------------------------------------- >45796_45796_13_LMNA-RAF1_LMNA_chr1_156107534_ENST00000473598_RAF1_chr3_12641914_ENST00000251849_length(amino acids)=849AA_BP=479 MGHIWRLKAGAQMDLEAWDPHLEPDAEAMVDGNTKKEGDLIAAQARLKDLEALLNSKEAALSTALSEKRTLEGELHDLRGQVAKLEAALG EAKKQLQDEMLRRVDAENRLQTMKEELDFQKNIYSEELRETKRRHETRLVEIDNGKQREFESRLADALQELRAQHEDQVEQYKKELEKTY SAKLDNARQSAERNSNLVGAAHEELQQSRIRIDSLSAQLSQLQKQLAAKEAKLRDLEDSLARERDTSRRLLAEKEREMAEMRARMQQQLD EYQELLDIKLALDMEIHAYRKLLEGEEERLRLSPSPTSQRSRGRASSHSSQTQGGGSVTKKRKLESTESRSSFSQHARTSGRVAVEEVDE EGKFVRLRNKSNEDQSMGNWQIKRQNGDDPLLTYRFPPKFTLKAGQVVTIWAAGAGATHSPPTDLVWKAQNTWGCGNSLRTALINSTGEE VAMRKLVRSVTVVEDDEDEDGDDLLHHHHDAIRSHSESASPSALSSSPNNLSPTGWSQPKTPVPAQRERAPVSGTQEKNKIRPRGQRDSS YYWEIEASEVMLSTRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQFQAFRNEVAVLRKTRHVNILLFMGYMTKDNLAIVTQWCEGSSL YKHLHVQETKFQMFQLIDIARQTAQGMDYLHAKNIIHRDMKSNNIFLHEGLTVKIGDFGLATVKSRWSGSQQVEQPTGSVLWMAPEVIRM QDNNPFSFQSDVYSYGIVLYELMTGELPYSHINNRDQIIFMVGRGYASPDLSKLYKNCPKAMKRLVADCVKKVKEERPLFPQILSSIELL -------------------------------------------------------------- >45796_45796_14_LMNA-RAF1_LMNA_chr1_156107534_ENST00000473598_RAF1_chr3_12641914_ENST00000442415_length(amino acids)=849AA_BP=479 MGHIWRLKAGAQMDLEAWDPHLEPDAEAMVDGNTKKEGDLIAAQARLKDLEALLNSKEAALSTALSEKRTLEGELHDLRGQVAKLEAALG EAKKQLQDEMLRRVDAENRLQTMKEELDFQKNIYSEELRETKRRHETRLVEIDNGKQREFESRLADALQELRAQHEDQVEQYKKELEKTY SAKLDNARQSAERNSNLVGAAHEELQQSRIRIDSLSAQLSQLQKQLAAKEAKLRDLEDSLARERDTSRRLLAEKEREMAEMRARMQQQLD EYQELLDIKLALDMEIHAYRKLLEGEEERLRLSPSPTSQRSRGRASSHSSQTQGGGSVTKKRKLESTESRSSFSQHARTSGRVAVEEVDE EGKFVRLRNKSNEDQSMGNWQIKRQNGDDPLLTYRFPPKFTLKAGQVVTIWAAGAGATHSPPTDLVWKAQNTWGCGNSLRTALINSTGEE VAMRKLVRSVTVVEDDEDEDGDDLLHHHHDAIRSHSESASPSALSSSPNNLSPTGWSQPKTPVPAQRERAPVSGTQEKNKIRPRGQRDSS YYWEIEASEVMLSTRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQFQAFRNEVAVLRKTRHVNILLFMGYMTKDNLAIVTQWCEGSSL YKHLHVQETKFQMFQLIDIARQTAQGMDYLHAKNIIHRDMKSNNIFLHEGLTVKIGDFGLATVKSRWSGSQQVEQPTGSVLWMAPEVIRM QDNNPFSFQSDVYSYGIVLYELMTGELPYSHINNRDQIIFMVGRGYASPDLSKLYKNCPKAMKRLVADCVKKVKEERPLFPQILSSIELL -------------------------------------------------------------- >45796_45796_15_LMNA-RAF1_LMNA_chr1_156107534_ENST00000473598_RAF1_chr3_12641914_ENST00000534997_length(amino acids)=849AA_BP=479 MGHIWRLKAGAQMDLEAWDPHLEPDAEAMVDGNTKKEGDLIAAQARLKDLEALLNSKEAALSTALSEKRTLEGELHDLRGQVAKLEAALG EAKKQLQDEMLRRVDAENRLQTMKEELDFQKNIYSEELRETKRRHETRLVEIDNGKQREFESRLADALQELRAQHEDQVEQYKKELEKTY SAKLDNARQSAERNSNLVGAAHEELQQSRIRIDSLSAQLSQLQKQLAAKEAKLRDLEDSLARERDTSRRLLAEKEREMAEMRARMQQQLD EYQELLDIKLALDMEIHAYRKLLEGEEERLRLSPSPTSQRSRGRASSHSSQTQGGGSVTKKRKLESTESRSSFSQHARTSGRVAVEEVDE EGKFVRLRNKSNEDQSMGNWQIKRQNGDDPLLTYRFPPKFTLKAGQVVTIWAAGAGATHSPPTDLVWKAQNTWGCGNSLRTALINSTGEE VAMRKLVRSVTVVEDDEDEDGDDLLHHHHDAIRSHSESASPSALSSSPNNLSPTGWSQPKTPVPAQRERAPVSGTQEKNKIRPRGQRDSS YYWEIEASEVMLSTRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQFQAFRNEVAVLRKTRHVNILLFMGYMTKDNLAIVTQWCEGSSL YKHLHVQETKFQMFQLIDIARQTAQGMDYLHAKNIIHRDMKSNNIFLHEGLTVKIGDFGLATVKSRWSGSQQVEQPTGSVLWMAPEVIRM QDNNPFSFQSDVYSYGIVLYELMTGELPYSHINNRDQIIFMVGRGYASPDLSKLYKNCPKAMKRLVADCVKKVKEERPLFPQILSSIELL -------------------------------------------------------------- >45796_45796_16_LMNA-RAF1_LMNA_chr1_156107534_ENST00000473598_RAF1_chr3_12641914_ENST00000542177_length(amino acids)=849AA_BP=479 MGHIWRLKAGAQMDLEAWDPHLEPDAEAMVDGNTKKEGDLIAAQARLKDLEALLNSKEAALSTALSEKRTLEGELHDLRGQVAKLEAALG EAKKQLQDEMLRRVDAENRLQTMKEELDFQKNIYSEELRETKRRHETRLVEIDNGKQREFESRLADALQELRAQHEDQVEQYKKELEKTY SAKLDNARQSAERNSNLVGAAHEELQQSRIRIDSLSAQLSQLQKQLAAKEAKLRDLEDSLARERDTSRRLLAEKEREMAEMRARMQQQLD EYQELLDIKLALDMEIHAYRKLLEGEEERLRLSPSPTSQRSRGRASSHSSQTQGGGSVTKKRKLESTESRSSFSQHARTSGRVAVEEVDE EGKFVRLRNKSNEDQSMGNWQIKRQNGDDPLLTYRFPPKFTLKAGQVVTIWAAGAGATHSPPTDLVWKAQNTWGCGNSLRTALINSTGEE VAMRKLVRSVTVVEDDEDEDGDDLLHHHHDAIRSHSESASPSALSSSPNNLSPTGWSQPKTPVPAQRERAPVSGTQEKNKIRPRGQRDSS YYWEIEASEVMLSTRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQFQAFRNEVAVLRKTRHVNILLFMGYMTKDNLAIVTQWCEGSSL YKHLHVQETKFQMFQLIDIARQTAQGMDYLHAKNIIHRDMKSNNIFLHEGLTVKIGDFGLATVKSRWSGSQQVEQPTGSVLWMAPEVIRM QDNNPFSFQSDVYSYGIVLYELMTGELPYSHINNRDQIIFMVGRGYASPDLSKLYKNCPKAMKRLVADCVKKVKEERPLFPQILSSIELL -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:156107534/chr3:12641914) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| LMNA | RAF1 |
| FUNCTION: Lamins are components of the nuclear lamina, a fibrous layer on the nucleoplasmic side of the inner nuclear membrane, which is thought to provide a framework for the nuclear envelope and may also interact with chromatin. Lamin A and C are present in equal amounts in the lamina of mammals. Recruited by DNA repair proteins XRCC4 and IFFO1 to the DNA double-strand breaks (DSBs) to prevent chromosome translocation by immobilizing broken DNA ends (PubMed:31548606). Plays an important role in nuclear assembly, chromatin organization, nuclear membrane and telomere dynamics. Required for normal development of peripheral nervous system and skeletal muscle and for muscle satellite cell proliferation (PubMed:10080180, PubMed:22431096, PubMed:10814726, PubMed:11799477, PubMed:18551513). Required for osteoblastogenesis and bone formation (PubMed:12075506, PubMed:15317753, PubMed:18611980). Also prevents fat infiltration of muscle and bone marrow, helping to maintain the volume and strength of skeletal muscle and bone (PubMed:10587585). Required for cardiac homeostasis (PubMed:10580070, PubMed:12927431, PubMed:18611980, PubMed:23666920). {ECO:0000269|PubMed:10080180, ECO:0000269|PubMed:10580070, ECO:0000269|PubMed:10587585, ECO:0000269|PubMed:10814726, ECO:0000269|PubMed:11799477, ECO:0000269|PubMed:12075506, ECO:0000269|PubMed:12927431, ECO:0000269|PubMed:15317753, ECO:0000269|PubMed:18551513, ECO:0000269|PubMed:18611980, ECO:0000269|PubMed:22431096, ECO:0000269|PubMed:23666920, ECO:0000269|PubMed:31548606}.; FUNCTION: Prelamin-A/C can accelerate smooth muscle cell senescence. It acts to disrupt mitosis and induce DNA damage in vascular smooth muscle cells (VSMCs), leading to mitotic failure, genomic instability, and premature senescence. | FUNCTION: Serine/threonine-protein kinase that acts as a regulatory link between the membrane-associated Ras GTPases and the MAPK/ERK cascade, and this critical regulatory link functions as a switch determining cell fate decisions including proliferation, differentiation, apoptosis, survival and oncogenic transformation. RAF1 activation initiates a mitogen-activated protein kinase (MAPK) cascade that comprises a sequential phosphorylation of the dual-specific MAPK kinases (MAP2K1/MEK1 and MAP2K2/MEK2) and the extracellular signal-regulated kinases (MAPK3/ERK1 and MAPK1/ERK2). The phosphorylated form of RAF1 (on residues Ser-338 and Ser-339, by PAK1) phosphorylates BAD/Bcl2-antagonist of cell death at 'Ser-75'. Phosphorylates adenylyl cyclases: ADCY2, ADCY5 and ADCY6, resulting in their activation. Phosphorylates PPP1R12A resulting in inhibition of the phosphatase activity. Phosphorylates TNNT2/cardiac muscle troponin T. Can promote NF-kB activation and inhibit signal transducers involved in motility (ROCK2), apoptosis (MAP3K5/ASK1 and STK3/MST2), proliferation and angiogenesis (RB1). Can protect cells from apoptosis also by translocating to the mitochondria where it binds BCL2 and displaces BAD/Bcl2-antagonist of cell death. Regulates Rho signaling and migration, and is required for normal wound healing. Plays a role in the oncogenic transformation of epithelial cells via repression of the TJ protein, occludin (OCLN) by inducing the up-regulation of a transcriptional repressor SNAI2/SLUG, which induces down-regulation of OCLN. Restricts caspase activation in response to selected stimuli, notably Fas stimulation, pathogen-mediated macrophage apoptosis, and erythroid differentiation. {ECO:0000269|PubMed:11427728, ECO:0000269|PubMed:11719507, ECO:0000269|PubMed:15385642, ECO:0000269|PubMed:15618521, ECO:0000269|PubMed:15849194, ECO:0000269|PubMed:16892053, ECO:0000269|PubMed:16924233, ECO:0000269|PubMed:9360956}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000368300 | + | 10 | 12 | 31_387 | 566.0 | 665.0 | Domain | IF rod |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000368300 | + | 10 | 12 | 428_545 | 566.0 | 665.0 | Domain | LTD |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000448611 | + | 10 | 13 | 31_387 | 454.0 | 575.0 | Domain | IF rod |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000368300 | + | 10 | 12 | 417_422 | 566.0 | 665.0 | Motif | Nuclear localization signal |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000448611 | + | 10 | 13 | 417_422 | 454.0 | 575.0 | Motif | Nuclear localization signal |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000368300 | + | 10 | 12 | 1_33 | 566.0 | 665.0 | Region | Note=Head |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000368300 | + | 10 | 12 | 219_242 | 566.0 | 665.0 | Region | Note=Linker 2 |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000368300 | + | 10 | 12 | 243_383 | 566.0 | 665.0 | Region | Note=Coil 2 |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000368300 | + | 10 | 12 | 34_70 | 566.0 | 665.0 | Region | Note=Coil 1A |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000368300 | + | 10 | 12 | 71_80 | 566.0 | 665.0 | Region | Note=Linker 1 |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000368300 | + | 10 | 12 | 81_218 | 566.0 | 665.0 | Region | Note=Coil 1B |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000448611 | + | 10 | 13 | 1_33 | 454.0 | 575.0 | Region | Note=Head |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000448611 | + | 10 | 13 | 219_242 | 454.0 | 575.0 | Region | Note=Linker 2 |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000448611 | + | 10 | 13 | 243_383 | 454.0 | 575.0 | Region | Note=Coil 2 |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000448611 | + | 10 | 13 | 34_70 | 454.0 | 575.0 | Region | Note=Coil 1A |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000448611 | + | 10 | 13 | 71_80 | 454.0 | 575.0 | Region | Note=Linker 1 |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000448611 | + | 10 | 13 | 81_218 | 454.0 | 575.0 | Region | Note=Coil 1B |
| Tgene | RAF1 | chr1:156107534 | chr3:12641914 | ENST00000251849 | 6 | 17 | 349_609 | 278.0 | 649.0 | Domain | Protein kinase | |
| Tgene | RAF1 | chr1:156107534 | chr3:12641914 | ENST00000442415 | 7 | 18 | 349_609 | 298.0 | 669.0 | Domain | Protein kinase | |
| Tgene | RAF1 | chr1:156107534 | chr3:12641914 | ENST00000251849 | 6 | 17 | 355_363 | 278.0 | 649.0 | Nucleotide binding | ATP | |
| Tgene | RAF1 | chr1:156107534 | chr3:12641914 | ENST00000442415 | 7 | 18 | 355_363 | 298.0 | 669.0 | Nucleotide binding | ATP |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000347559 | + | 1 | 11 | 31_387 | 0 | 635.0 | Domain | IF rod |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000347559 | + | 1 | 11 | 428_545 | 0 | 635.0 | Domain | LTD |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000361308 | + | 1 | 10 | 31_387 | 0 | 573.0 | Domain | IF rod |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000361308 | + | 1 | 10 | 428_545 | 0 | 573.0 | Domain | LTD |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000368301 | + | 1 | 13 | 31_387 | 0 | 573.0 | Domain | IF rod |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000368301 | + | 1 | 13 | 428_545 | 0 | 573.0 | Domain | LTD |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000448611 | + | 10 | 13 | 428_545 | 454.0 | 575.0 | Domain | LTD |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000347559 | + | 1 | 11 | 417_422 | 0 | 635.0 | Motif | Nuclear localization signal |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000361308 | + | 1 | 10 | 417_422 | 0 | 573.0 | Motif | Nuclear localization signal |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000368301 | + | 1 | 13 | 417_422 | 0 | 573.0 | Motif | Nuclear localization signal |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000347559 | + | 1 | 11 | 1_33 | 0 | 635.0 | Region | Note=Head |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000347559 | + | 1 | 11 | 219_242 | 0 | 635.0 | Region | Note=Linker 2 |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000347559 | + | 1 | 11 | 243_383 | 0 | 635.0 | Region | Note=Coil 2 |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000347559 | + | 1 | 11 | 34_70 | 0 | 635.0 | Region | Note=Coil 1A |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000347559 | + | 1 | 11 | 384_664 | 0 | 635.0 | Region | Note=Tail |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000347559 | + | 1 | 11 | 71_80 | 0 | 635.0 | Region | Note=Linker 1 |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000347559 | + | 1 | 11 | 81_218 | 0 | 635.0 | Region | Note=Coil 1B |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000361308 | + | 1 | 10 | 1_33 | 0 | 573.0 | Region | Note=Head |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000361308 | + | 1 | 10 | 219_242 | 0 | 573.0 | Region | Note=Linker 2 |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000361308 | + | 1 | 10 | 243_383 | 0 | 573.0 | Region | Note=Coil 2 |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000361308 | + | 1 | 10 | 34_70 | 0 | 573.0 | Region | Note=Coil 1A |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000361308 | + | 1 | 10 | 384_664 | 0 | 573.0 | Region | Note=Tail |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000361308 | + | 1 | 10 | 71_80 | 0 | 573.0 | Region | Note=Linker 1 |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000361308 | + | 1 | 10 | 81_218 | 0 | 573.0 | Region | Note=Coil 1B |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000368300 | + | 10 | 12 | 384_664 | 566.0 | 665.0 | Region | Note=Tail |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000368301 | + | 1 | 13 | 1_33 | 0 | 573.0 | Region | Note=Head |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000368301 | + | 1 | 13 | 219_242 | 0 | 573.0 | Region | Note=Linker 2 |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000368301 | + | 1 | 13 | 243_383 | 0 | 573.0 | Region | Note=Coil 2 |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000368301 | + | 1 | 13 | 34_70 | 0 | 573.0 | Region | Note=Coil 1A |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000368301 | + | 1 | 13 | 384_664 | 0 | 573.0 | Region | Note=Tail |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000368301 | + | 1 | 13 | 71_80 | 0 | 573.0 | Region | Note=Linker 1 |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000368301 | + | 1 | 13 | 81_218 | 0 | 573.0 | Region | Note=Coil 1B |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000448611 | + | 10 | 13 | 384_664 | 454.0 | 575.0 | Region | Note=Tail |
| Tgene | RAF1 | chr1:156107534 | chr3:12641914 | ENST00000251849 | 6 | 17 | 56_131 | 278.0 | 649.0 | Domain | RBD | |
| Tgene | RAF1 | chr1:156107534 | chr3:12641914 | ENST00000442415 | 7 | 18 | 56_131 | 298.0 | 669.0 | Domain | RBD | |
| Tgene | RAF1 | chr1:156107534 | chr3:12641914 | ENST00000251849 | 6 | 17 | 138_184 | 278.0 | 649.0 | Zinc finger | Phorbol-ester/DAG-type | |
| Tgene | RAF1 | chr1:156107534 | chr3:12641914 | ENST00000442415 | 7 | 18 | 138_184 | 298.0 | 669.0 | Zinc finger | Phorbol-ester/DAG-type |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| RAF1 | YWHAQ, YWHAG, YWHAZ, Hras, SFN, HRAS, VDAC1, PEBP4, CNKSR1, RASD2, CNKSR2, ERAS, SPRY4, SPRY2, YWHAB, OIP5, PHKA2, CCT3, MYC, AR, MRAS, KRAS, LRPAP1, RRAS2, SHOC2, TSC22D3, STK26, MAPK1, MAPK3, PRKCE, BCL2, BCL2L1, PEBP1, MAP2K1, GRB10, NRAS, MAPK8IP3, AKT1, HSP90AA1, GNG4, GNB2, VAV1, RHEB, RAP1A, SRC, FYN, RB1, RBL2, PRKG1, JAK2, YWHAH, CFLAR, CDC25A, BRAF, MAP3K1, PDGFRB, MAP3K5, MAPK7, PHB, BAG1, PAK1, PRKCZ, RRAS, PRPF6, UBE2D2, YWHAE, NR3C1, LCK, LTK, JAK1, XIAP, BIRC2, BIRC3, STUB1, Arrb2, EEF1A1, HSP82, HSPA8, HSPA5, CALM1, SGK1, NEDD4, SCNN1B, NEDD4L, STK3, BRAP, WDR83, KSR1, LATS1, MAP2K2, Rfxank, RFXANK, PPP1CC, VCP, HSPD1, BAD, BCL6, BIRC7, PIN1, MMS19, PPP2R1A, PPP2R2B, PPP2CA, HSPA4, RAF1, PPP2CB, MAPK8, RBBP8, YY1, PKM, CDK20, HIPK4, TBXA2R, SH3KBP1, ILK, GPRASP2, CDC37, IRS4, FAM96B, HUWE1, Prkab1, RANBP9, TMEM70, PACSIN3, NPLOC4, PRKAR1A, DCAF8, PURA, FYCO1, MYBPC1, COPS7A, MOV10, NXF1, EGFR, CALU, CCT8, DNAJA1, EMD, TIMM50, LOX, ARRB2, AGTR1, CASZ1, MRC2, PDLIM2, HERC2, XPO1, ACOX1, PCDH7, MAD2L1, SYNPO, TMEM63B, EEF1E1, COPS5, SS18L2, Smn1, Cep152, Aldh4a1, Ksr1, NUP133, RPGRIP1L, NCAPG2, NUP107, MSH2, LAS1L, AMER1, NUP160, EIF3D, SPATA7, NPHP4, DYNLL2, DYNLL1, IQCB1, RMDN3, TRIM25, Mapkbp1, EGLN3, RIPK4, MAPK6, CSNK1A1, HERC1, LZTR1, QKI, COPS3, TRIM28, MAEA, NANOG, ITCH, IRF7, PSMB9, MAP2K5, CDK4, CDK6, CDKN2B, FGFR4, MAP2K3, NF2, PDGFRA, TEAD2, RASSF1, ABCB5, BECN1, CBLC, GLIS2, GRM1, HGF, MAP2K6, ARNT, CCND2, CD44, CDKN2A, CDKN2C, EPHA2, ERBB2, KEAP1, LATS2, MET, NF1, TERT, DUSP2, MBP, PAK2, PLEKHA4, CRBN, ARAF, SLC25A3, TUBB4B, TUBB, HSPA6, HSP90AB3P, HSP90AB1, HSPA1A, HSPB1, BAG2, CCT7, DSG1, CCT2, CCT4, CCT6A, TCP1, AKAP8L, ATAD3A, ABCD3, SLC25A22, TMEM33, TUBA4A, HPX, MGST1, TUBB4A, TMEM161A, HSP90AB2P, CCAR2, SLC25A10, FANCD2, TUBB6, HSD17B12, SLC25A11, SLC25A1, ARL1, DNAJA2, DPM1, CCT5, AIFM1, SQSTM1, TUBG2, TUBG1, STK39, GCN1L1, ABCF2, POLD1, NUBP2, TM9SF3, NCLN, SEC61A2, SEC61A1, ALDH1A3, TUBB3, POLR2B, IARS2, TMED1, CIT, AURKB, KIF20A, BRD1, TRIM66, HAX1, CUL4A, ACTR1A, AKAP1, FLOT1, LAMTOR1, LMAN1, RAB2A, RAB4A, STX7, NPAS1, OPALIN, METTL21B, SSSCA1, FAM174A, CRKL, DDB1, DICER1, LDHB, PIK3R1, NONO, DVL2, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| LMNA | |
| RAF1 |  |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000347559 | + | 1 | 11 | 1_130 | 0 | 635.0 | MLIP |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000361308 | + | 1 | 10 | 1_130 | 0 | 573.0 | MLIP |
| Hgene | LMNA | chr1:156107534 | chr3:12641914 | ENST00000368301 | + | 1 | 13 | 1_130 | 0 | 573.0 | MLIP |
Top |
Related Drugs to LMNA-RAF1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to LMNA-RAF1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Tgene | RAF1 | C0028326 | Noonan Syndrome | 10 | CLINGEN;CTD_human;GENOMICS_ENGLAND;ORPHANET |
| Tgene | RAF1 | C0175704 | LEOPARD Syndrome | 7 | CLINGEN;CTD_human;GENOMICS_ENGLAND |
| Tgene | RAF1 | C1969057 | Noonan Syndrome 5 | 4 | CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Tgene | RAF1 | C1969056 | LEOPARD SYNDROME 2 | 3 | CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Tgene | RAF1 | C0007194 | Hypertrophic Cardiomyopathy | 2 | CTD_human |
| Tgene | RAF1 | C0041409 | Turner Syndrome, Male | 2 | CTD_human |
| Tgene | RAF1 | C1519086 | Pilomyxoid astrocytoma | 2 | ORPHANET |
| Tgene | RAF1 | C1527404 | Female Pseudo-Turner Syndrome | 2 | CTD_human |
| Tgene | RAF1 | C4551472 | Hypertrophic obstructive cardiomyopathy | 2 | CTD_human |
| Tgene | RAF1 | C4551602 | Noonan Syndrome 1 | 2 | CTD_human |
| Tgene | RAF1 | C0006142 | Malignant neoplasm of breast | 1 | CTD_human |
| Tgene | RAF1 | C0007131 | Non-Small Cell Lung Carcinoma | 1 | CTD_human |
| Tgene | RAF1 | C0007193 | Cardiomyopathy, Dilated | 1 | CTD_human |
| Tgene | RAF1 | C0017638 | Glioma | 1 | CTD_human |
| Tgene | RAF1 | C0020429 | Hyperalgesia | 1 | CTD_human |
| Tgene | RAF1 | C0022665 | Kidney Neoplasm | 1 | CTD_human |
| Tgene | RAF1 | C0023903 | Liver neoplasms | 1 | CTD_human |
| Tgene | RAF1 | C0024121 | Lung Neoplasms | 1 | CTD_human |
| Tgene | RAF1 | C0242379 | Malignant neoplasm of lung | 1 | CTD_human |
| Tgene | RAF1 | C0259783 | mixed gliomas | 1 | CTD_human |
| Tgene | RAF1 | C0340427 | Familial dilated cardiomyopathy | 1 | ORPHANET |
| Tgene | RAF1 | C0345904 | Malignant neoplasm of liver | 1 | CTD_human |
| Tgene | RAF1 | C0345967 | Malignant mesothelioma | 1 | CTD_human |
| Tgene | RAF1 | C0458247 | Allodynia | 1 | CTD_human |
| Tgene | RAF1 | C0555198 | Malignant Glioma | 1 | CTD_human |
| Tgene | RAF1 | C0587248 | Costello syndrome (disorder) | 1 | CLINGEN |
| Tgene | RAF1 | C0678222 | Breast Carcinoma | 1 | CTD_human |
| Tgene | RAF1 | C0740457 | Malignant neoplasm of kidney | 1 | CTD_human |
| Tgene | RAF1 | C0751211 | Hyperalgesia, Primary | 1 | CTD_human |
| Tgene | RAF1 | C0751212 | Hyperalgesia, Secondary | 1 | CTD_human |
| Tgene | RAF1 | C0751213 | Tactile Allodynia | 1 | CTD_human |
| Tgene | RAF1 | C0751214 | Hyperalgesia, Thermal | 1 | CTD_human |
| Tgene | RAF1 | C1257931 | Mammary Neoplasms, Human | 1 | CTD_human |
| Tgene | RAF1 | C1275081 | Cardio-facio-cutaneous syndrome | 1 | CLINGEN |
| Tgene | RAF1 | C1449563 | Cardiomyopathy, Familial Idiopathic | 1 | CTD_human |
| Tgene | RAF1 | C1458155 | Mammary Neoplasms | 1 | CTD_human |
| Tgene | RAF1 | C2936719 | Mechanical Allodynia | 1 | CTD_human |
| Tgene | RAF1 | C4014656 | CARDIOMYOPATHY, DILATED, 1NN | 1 | CTD_human;UNIPROT |
| Tgene | RAF1 | C4551484 | Leopard Syndrome 1 | 1 | CTD_human;GENOMICS_ENGLAND |
| Tgene | RAF1 | C4704874 | Mammary Carcinoma, Human | 1 | CTD_human |
| Tgene | RAF1 | C4721610 | Carcinoma, Ovarian Epithelial | 1 | CTD_human |

