| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:NPM1-ALK |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: NPM1-ALK | FusionPDB ID: 59963 | FusionGDB2.0 ID: 59963 | Hgene | Tgene | Gene symbol | NPM1 | ALK | Gene ID | 4869 | 238 |
| Gene name | nucleophosmin 1 | ALK receptor tyrosine kinase | |
| Synonyms | B23|NPM | CD246|NBLST3 | |
| Cytomap | 5q35.1 | 2p23.2-p23.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | nucleophosminnucleolar protein NO38nucleophosmin (nucleolar phosphoprotein B23, numatrin)nucleophosmin/nucleoplasmin family, member 1testicular tissue protein Li 128 | ALK tyrosine kinase receptorCD246 antigenanaplastic lymphoma receptor tyrosine kinasemutant anaplastic lymphoma kinase | |
| Modification date | 20200329 | 20200329 | |
| UniProtAcc | P06748 | Q96BT7 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000296930, ENST00000351986, ENST00000393820, ENST00000517671, | ENST00000389048, ENST00000431873, ENST00000498037, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 21 X 25 X 6=3150 | 56 X 74 X 20=82880 |
| # samples | 32 | 57 | |
| ** MAII score | log2(32/3150*10)=-3.29920801838728 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(57/82880*10)=-7.18391827352181 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: NPM1 [Title/Abstract] AND ALK [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | NPM1(170818803)-ALK(29446396), # samples:4 | ||
| Anticipated loss of major functional domain due to fusion event. | NPM1-ALK seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. NPM1-ALK seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. NPM1-ALK seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. NPM1-ALK seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. NPM1-ALK seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. NPM1-ALK seems lost the major protein functional domain in Hgene partner, which is a epigenetic factor due to the frame-shifted ORF. NPM1-ALK seems lost the major protein functional domain in Hgene partner, which is a tumor suppressor due to the frame-shifted ORF. NPM1-ALK seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. NPM1-ALK seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. NPM1-ALK seems lost the major protein functional domain in Tgene partner, which is a kinase due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | NPM1 | GO:0006281 | DNA repair | 19188445 |
| Hgene | NPM1 | GO:0006334 | nucleosome assembly | 11602260 |
| Hgene | NPM1 | GO:0006913 | nucleocytoplasmic transport | 16041368 |
| Hgene | NPM1 | GO:0008104 | protein localization | 18420587 |
| Hgene | NPM1 | GO:0008284 | positive regulation of cell proliferation | 22528486 |
| Hgene | NPM1 | GO:0032071 | regulation of endodeoxyribonuclease activity | 19188445 |
| Hgene | NPM1 | GO:0034644 | cellular response to UV | 19160485 |
| Hgene | NPM1 | GO:0043066 | negative regulation of apoptotic process | 12882984 |
| Hgene | NPM1 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation | 12882984 |
| Hgene | NPM1 | GO:0045727 | positive regulation of translation | 12882984 |
| Hgene | NPM1 | GO:0045893 | positive regulation of transcription, DNA-templated | 22528486 |
| Hgene | NPM1 | GO:0045944 | positive regulation of transcription by RNA polymerase II | 19160485 |
| Hgene | NPM1 | GO:0060699 | regulation of endoribonuclease activity | 19188445 |
| Hgene | NPM1 | GO:0060735 | regulation of eIF2 alpha phosphorylation by dsRNA | 12882984 |
| Hgene | NPM1 | GO:1902751 | positive regulation of cell cycle G2/M phase transition | 22528486 |
| Tgene | ALK | GO:0016310 | phosphorylation | 9174053 |
| Tgene | ALK | GO:0046777 | protein autophosphorylation | 9174053 |
 Fusion gene breakpoints across NPM1 (5'-gene) Fusion gene breakpoints across NPM1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
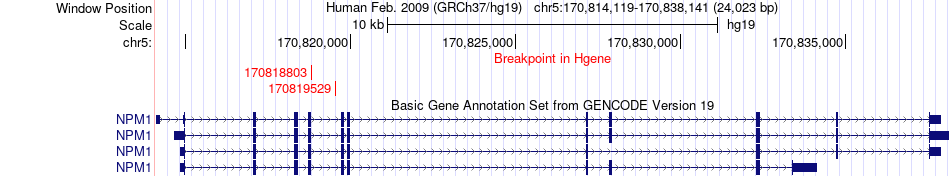 |
 Fusion gene breakpoints across ALK (3'-gene) Fusion gene breakpoints across ALK (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
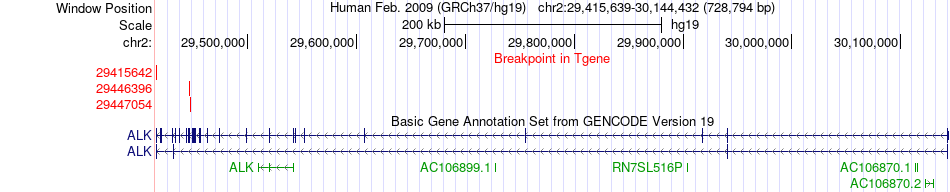 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | anaplastic large cell lymphoma;Hodgkin disease;npn Hodgkin lymphoma | D45915 | NPM1 | chr5 | 170818803 | ALK | chr2 | 29415642 | ||
| ChimerKB3 | . | . | NPM1 | chr5 | 170818803 | + | ALK | chr2 | 29415642 | - |
| ChimerKB3 | . | . | NPM1 | chr5 | 170818803 | + | ALK | chr2 | 29416090 | - |
| ChimerKB3 | . | . | NPM1 | chr5 | 170818803 | + | ALK | chr2 | 29432744 | - |
| ChimerKB3 | . | . | NPM1 | chr5 | 170819820 | + | ALK | chr2 | 29446394 | - |
| ChimerKB4 | . | . | NPM1 | chr5 | 170818803 | + | ALK | chr2 | 29416090 | - |
| ChiTaRS5.0 | N/A | D45915 | NPM1 | chr5 | 170818803 | + | ALK | chr2 | 29446396 | - |
| ChiTaRS5.0 | N/A | DI348757 | NPM1 | chr5 | 170818803 | + | ALK | chr2 | 29446396 | - |
| ChiTaRS5.0 | N/A | HW349670 | NPM1 | chr5 | 170818803 | + | ALK | chr2 | 29446396 | - |
| ChiTaRS5.0 | N/A | S82740 | NPM1 | chr5 | 170819529 | + | ALK | chr2 | 29447054 | - |
| ChiTaRS5.0 | N/A | U04946 | NPM1 | chr5 | 170818803 | + | ALK | chr2 | 29446396 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000517671 | NPM1 | chr5 | 170818803 | + | ENST00000389048 | ALK | chr2 | 29446396 | - | 2628 | 487 | 42 | 2177 | 711 |
| ENST00000296930 | NPM1 | chr5 | 170818803 | + | ENST00000389048 | ALK | chr2 | 29446396 | - | 2794 | 653 | 70 | 2343 | 757 |
| ENST00000351986 | NPM1 | chr5 | 170818803 | + | ENST00000389048 | ALK | chr2 | 29446396 | - | 2613 | 472 | 42 | 2162 | 706 |
| ENST00000393820 | NPM1 | chr5 | 170818803 | + | ENST00000389048 | ALK | chr2 | 29446396 | - | 2591 | 450 | 20 | 2140 | 706 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000517671 | ENST00000389048 | NPM1 | chr5 | 170818803 | + | ALK | chr2 | 29446396 | - | 0.002362132 | 0.99763787 |
| ENST00000296930 | ENST00000389048 | NPM1 | chr5 | 170818803 | + | ALK | chr2 | 29446396 | - | 0.001957753 | 0.9980422 |
| ENST00000351986 | ENST00000389048 | NPM1 | chr5 | 170818803 | + | ALK | chr2 | 29446396 | - | 0.002376777 | 0.99762326 |
| ENST00000393820 | ENST00000389048 | NPM1 | chr5 | 170818803 | + | ALK | chr2 | 29446396 | - | 0.002370775 | 0.9976292 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >59963_59963_1_NPM1-ALK_NPM1_chr5_170818803_ENST00000296930_ALK_chr2_29446396_ENST00000389048_length(amino acids)=757AA_BP=194 MKSACAGRLRYGGGRASEHARGGGTWEALARSSGSIYKRGEPASFPWCDSVLRGCSLEQRSFISVRLLSYLSACRHPMEDSMDMDMSPLR PQNYLFGCELKADKDYHFKVDNDENEHQLSLRTVSLGAGAKDELHIVEAEAMNYEGSPIKVTLATLKMSVQPTVSLGGFEITPPVVLRLK CGSGPVHISGQHLVVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEG QVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAM LDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTW SFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGP LVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVH GSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVN -------------------------------------------------------------- >59963_59963_2_NPM1-ALK_NPM1_chr5_170818803_ENST00000351986_ALK_chr2_29446396_ENST00000389048_length(amino acids)=706AA_BP=143 MRGCSLEQRSFISVRLLSYLSACRHPMEDSMDMDMSPLRPQNYLFGCELKADKDYHFKVDNDENEHQLSLRTVSLGAGAKDELHIVEAEA MNYEGSPIKVTLATLKMSVQPTVSLGGFEITPPVVLRLKCGSGPVHISGQHLVVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNP NYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIG VSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMA RDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQ CWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAK KPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPN -------------------------------------------------------------- >59963_59963_3_NPM1-ALK_NPM1_chr5_170818803_ENST00000393820_ALK_chr2_29446396_ENST00000389048_length(amino acids)=706AA_BP=143 MRGCSLEQRSFISVRLLSYLSACRHPMEDSMDMDMSPLRPQNYLFGCELKADKDYHFKVDNDENEHQLSLRTVSLGAGAKDELHIVEAEA MNYEGSPIKVTLATLKMSVQPTVSLGGFEITPPVVLRLKCGSGPVHISGQHLVVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNP NYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIG VSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMA RDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQ CWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAK KPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPN -------------------------------------------------------------- >59963_59963_4_NPM1-ALK_NPM1_chr5_170818803_ENST00000517671_ALK_chr2_29446396_ENST00000389048_length(amino acids)=711AA_BP=148 MAPKLRKDKDFGDVFSGRTELKNKVPACRHPMEDSMDMDMSPLRPQNYLFGCELKADKDYHFKVDNDENEHQLSLRTVSLGAGAKDELHI VEAEAMNYEGSPIKVTLATLKMSVQPTVSLGGFEITPPVVLRLKCGSGPVHISGQHLVVYRRKHQELQAMQMELQSPEYKLSKLRTSTIM TDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNI VRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIG DFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVY RIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSS GKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSC -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr5:170818803/chr2:29446396) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| NPM1 | ALK |
| FUNCTION: Involved in diverse cellular processes such as ribosome biogenesis, centrosome duplication, protein chaperoning, histone assembly, cell proliferation, and regulation of tumor suppressors p53/TP53 and ARF. Binds ribosome presumably to drive ribosome nuclear export. Associated with nucleolar ribonucleoprotein structures and bind single-stranded nucleic acids. Acts as a chaperonin for the core histones H3, H2B and H4. Stimulates APEX1 endonuclease activity on apurinic/apyrimidinic (AP) double-stranded DNA but inhibits APEX1 endonuclease activity on AP single-stranded RNA. May exert a control of APEX1 endonuclease activity within nucleoli devoted to repair AP on rDNA and the removal of oxidized rRNA molecules. In concert with BRCA2, regulates centrosome duplication. Regulates centriole duplication: phosphorylation by PLK2 is able to trigger centriole replication. Negatively regulates the activation of EIF2AK2/PKR and suppresses apoptosis through inhibition of EIF2AK2/PKR autophosphorylation. Antagonizes the inhibitory effect of ATF5 on cell proliferation and relieves ATF5-induced G2/M blockade (PubMed:22528486). In complex with MYC enhances the transcription of MYC target genes (PubMed:25956029). {ECO:0000269|PubMed:12882984, ECO:0000269|PubMed:16107701, ECO:0000269|PubMed:17015463, ECO:0000269|PubMed:18809582, ECO:0000269|PubMed:19188445, ECO:0000269|PubMed:20352051, ECO:0000269|PubMed:21084279, ECO:0000269|PubMed:22002061, ECO:0000269|PubMed:22528486, ECO:0000269|PubMed:25956029}. | FUNCTION: Catalyzes the methylation of 5-carboxymethyl uridine to 5-methylcarboxymethyl uridine at the wobble position of the anticodon loop in tRNA via its methyltransferase domain (PubMed:20123966, PubMed:20308323, PubMed:31079898). Catalyzes the last step in the formation of 5-methylcarboxymethyl uridine at the wobble position of the anticodon loop in target tRNA (PubMed:20123966, PubMed:20308323). Has a preference for tRNA(Arg) and tRNA(Glu), and does not bind tRNA(Lys)(PubMed:20308323). Binds tRNA and catalyzes the iron and alpha-ketoglutarate dependent hydroxylation of 5-methylcarboxymethyl uridine at the wobble position of the anticodon loop in tRNA via its dioxygenase domain, giving rise to 5-(S)-methoxycarbonylhydroxymethyluridine; has a preference for tRNA(Gly) (PubMed:21285950). Required for normal survival after DNA damage (PubMed:20308323). May inhibit apoptosis and promote cell survival and angiogenesis (PubMed:19293182). {ECO:0000269|PubMed:19293182, ECO:0000269|PubMed:20123966, ECO:0000269|PubMed:20308323, ECO:0000269|PubMed:21285950, ECO:0000269|PubMed:31079898}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | NPM1 | chr5:170818803 | chr2:29446396 | ENST00000296930 | + | 4 | 11 | 1_9 | 117.33333333333333 | 295.0 | Compositional bias | Note=Met-rich |
| Hgene | NPM1 | chr5:170818803 | chr2:29446396 | ENST00000351986 | + | 4 | 10 | 1_9 | 117.33333333333333 | 266.0 | Compositional bias | Note=Met-rich |
| Hgene | NPM1 | chr5:170818803 | chr2:29446396 | ENST00000393820 | + | 4 | 10 | 1_9 | 117.33333333333333 | 260.0 | Compositional bias | Note=Met-rich |
| Hgene | NPM1 | chr5:170818803 | chr2:29446396 | ENST00000517671 | + | 5 | 12 | 1_9 | 117.33333333333333 | 295.0 | Compositional bias | Note=Met-rich |
| Tgene | ALK | chr5:170818803 | chr2:29446396 | ENST00000389048 | 18 | 29 | 1116_1392 | 1057.3333333333333 | 1621.0 | Domain | Protein kinase | |
| Tgene | ALK | chr5:170818803 | chr2:29446396 | ENST00000389048 | 18 | 29 | 1197_1199 | 1057.3333333333333 | 1621.0 | Region | Note=Inhibitor binding | |
| Tgene | ALK | chr5:170818803 | chr2:29446396 | ENST00000389048 | 18 | 29 | 1060_1620 | 1057.3333333333333 | 1621.0 | Topological domain | Cytoplasmic |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | NPM1 | chr5:170818803 | chr2:29446396 | ENST00000296930 | + | 4 | 11 | 120_132 | 117.33333333333333 | 295.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | NPM1 | chr5:170818803 | chr2:29446396 | ENST00000296930 | + | 4 | 11 | 161_188 | 117.33333333333333 | 295.0 | Compositional bias | Note=Asp/Glu-rich (highly acidic) |
| Hgene | NPM1 | chr5:170818803 | chr2:29446396 | ENST00000351986 | + | 4 | 10 | 120_132 | 117.33333333333333 | 266.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | NPM1 | chr5:170818803 | chr2:29446396 | ENST00000351986 | + | 4 | 10 | 161_188 | 117.33333333333333 | 266.0 | Compositional bias | Note=Asp/Glu-rich (highly acidic) |
| Hgene | NPM1 | chr5:170818803 | chr2:29446396 | ENST00000393820 | + | 4 | 10 | 120_132 | 117.33333333333333 | 260.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | NPM1 | chr5:170818803 | chr2:29446396 | ENST00000393820 | + | 4 | 10 | 161_188 | 117.33333333333333 | 260.0 | Compositional bias | Note=Asp/Glu-rich (highly acidic) |
| Hgene | NPM1 | chr5:170818803 | chr2:29446396 | ENST00000517671 | + | 5 | 12 | 120_132 | 117.33333333333333 | 295.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | NPM1 | chr5:170818803 | chr2:29446396 | ENST00000517671 | + | 5 | 12 | 161_188 | 117.33333333333333 | 295.0 | Compositional bias | Note=Asp/Glu-rich (highly acidic) |
| Hgene | NPM1 | chr5:170818803 | chr2:29446396 | ENST00000296930 | + | 4 | 11 | 152_157 | 117.33333333333333 | 295.0 | Motif | Nuclear localization signal |
| Hgene | NPM1 | chr5:170818803 | chr2:29446396 | ENST00000296930 | + | 4 | 11 | 191_197 | 117.33333333333333 | 295.0 | Motif | Nuclear localization signal |
| Hgene | NPM1 | chr5:170818803 | chr2:29446396 | ENST00000351986 | + | 4 | 10 | 152_157 | 117.33333333333333 | 266.0 | Motif | Nuclear localization signal |
| Hgene | NPM1 | chr5:170818803 | chr2:29446396 | ENST00000351986 | + | 4 | 10 | 191_197 | 117.33333333333333 | 266.0 | Motif | Nuclear localization signal |
| Hgene | NPM1 | chr5:170818803 | chr2:29446396 | ENST00000393820 | + | 4 | 10 | 152_157 | 117.33333333333333 | 260.0 | Motif | Nuclear localization signal |
| Hgene | NPM1 | chr5:170818803 | chr2:29446396 | ENST00000393820 | + | 4 | 10 | 191_197 | 117.33333333333333 | 260.0 | Motif | Nuclear localization signal |
| Hgene | NPM1 | chr5:170818803 | chr2:29446396 | ENST00000517671 | + | 5 | 12 | 152_157 | 117.33333333333333 | 295.0 | Motif | Nuclear localization signal |
| Hgene | NPM1 | chr5:170818803 | chr2:29446396 | ENST00000517671 | + | 5 | 12 | 191_197 | 117.33333333333333 | 295.0 | Motif | Nuclear localization signal |
| Hgene | NPM1 | chr5:170818803 | chr2:29446396 | ENST00000296930 | + | 4 | 11 | 243_294 | 117.33333333333333 | 295.0 | Region | Note=Required for nucleolar localization |
| Hgene | NPM1 | chr5:170818803 | chr2:29446396 | ENST00000351986 | + | 4 | 10 | 243_294 | 117.33333333333333 | 266.0 | Region | Note=Required for nucleolar localization |
| Hgene | NPM1 | chr5:170818803 | chr2:29446396 | ENST00000393820 | + | 4 | 10 | 243_294 | 117.33333333333333 | 260.0 | Region | Note=Required for nucleolar localization |
| Hgene | NPM1 | chr5:170818803 | chr2:29446396 | ENST00000517671 | + | 5 | 12 | 243_294 | 117.33333333333333 | 295.0 | Region | Note=Required for nucleolar localization |
| Tgene | ALK | chr5:170818803 | chr2:29446396 | ENST00000389048 | 18 | 29 | 816_940 | 1057.3333333333333 | 1621.0 | Compositional bias | Note=Gly-rich | |
| Tgene | ALK | chr5:170818803 | chr2:29446396 | ENST00000389048 | 18 | 29 | 264_427 | 1057.3333333333333 | 1621.0 | Domain | MAM 1 | |
| Tgene | ALK | chr5:170818803 | chr2:29446396 | ENST00000389048 | 18 | 29 | 437_473 | 1057.3333333333333 | 1621.0 | Domain | Note=LDL-receptor class A | |
| Tgene | ALK | chr5:170818803 | chr2:29446396 | ENST00000389048 | 18 | 29 | 478_636 | 1057.3333333333333 | 1621.0 | Domain | MAM 2 | |
| Tgene | ALK | chr5:170818803 | chr2:29446396 | ENST00000389048 | 18 | 29 | 19_1038 | 1057.3333333333333 | 1621.0 | Topological domain | Extracellular | |
| Tgene | ALK | chr5:170818803 | chr2:29446396 | ENST00000389048 | 18 | 29 | 1039_1059 | 1057.3333333333333 | 1621.0 | Transmembrane | Helical |
Top |
Fusion Protein Structures |
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
| PDB file >>>1525_NPM1_170818803_ALK_29446396_ranked_0.pdb | NPM1 | 170818803 | 170818803 | ENST00000389048 | ALK | chr2 | 29446396 | - | MKSACAGRLRYGGGRASEHARGGGTWEALARSSGSIYKRGEPASFPWCDSVLRGCSLEQRSFISVRLLSYLSACRHPMEDSMDMDMSPLR PQNYLFGCELKADKDYHFKVDNDENEHQLSLRTVSLGAGAKDELHIVEAEAMNYEGSPIKVTLATLKMSVQPTVSLGGFEITPPVVLRLK CGSGPVHISGQHLVVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEG QVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAM LDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTW SFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGP LVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVH GSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVN | 757 |
Top |
pLDDT score distribution |
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
NPM1_pLDDT.png |
ALK_pLDDT.png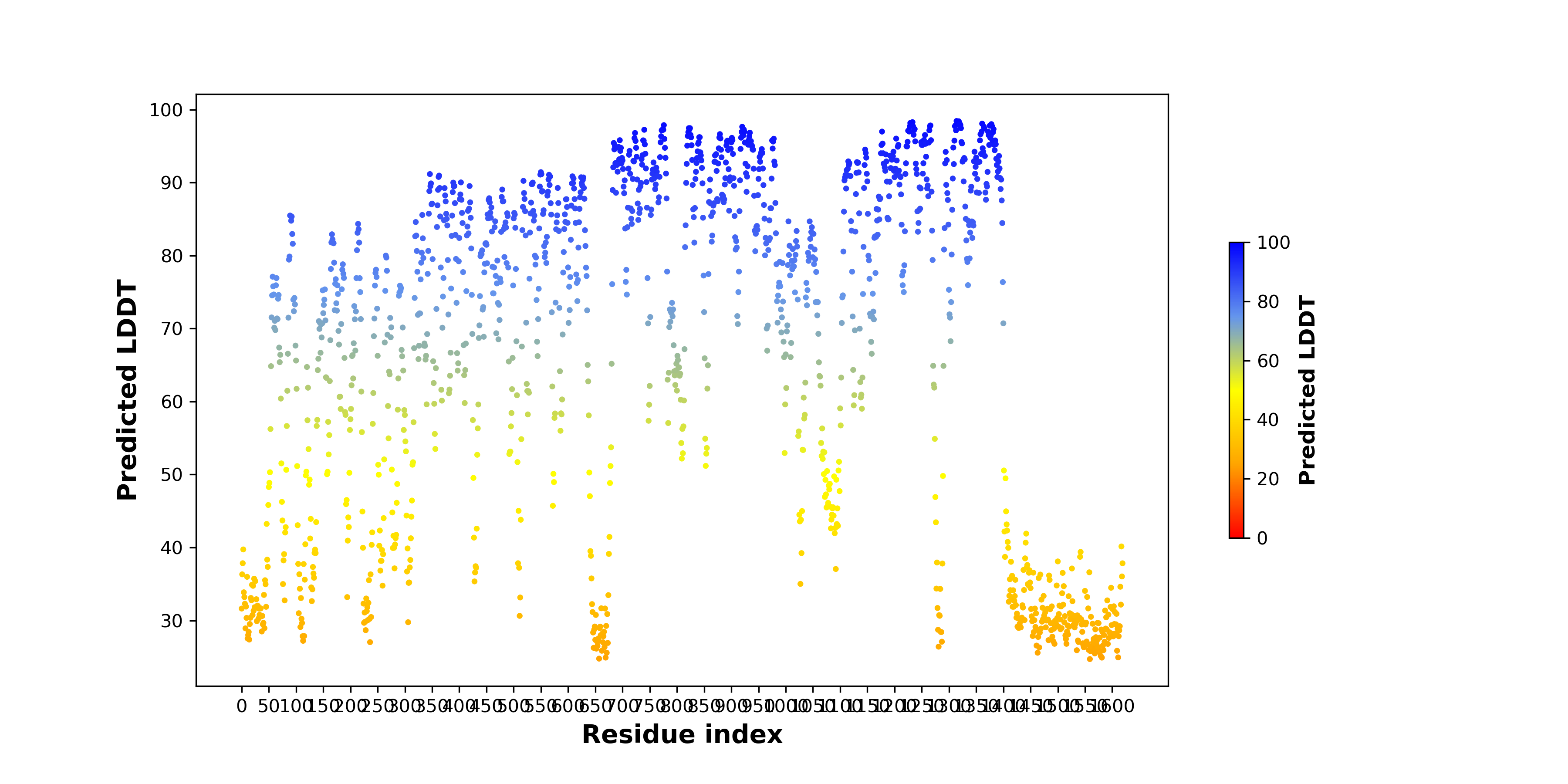 |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
 |
Top |
Ramachandran Plot of Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| Fusion AA seq ID in FusionPDB and their Ramachandran plots |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| NPM1 | TCERG1, LYAR, USF2, HIST3H3, SWAP70, HAND2, NCL, OTUB1, AKT1, CDT1, RRP1B, Rrp1b, GRB2, CENPF, CASKIN1, CDC14A, PIK3R1, NUP98, PPID, GRB7, NCAPG, NUMA1, NSUN2, PADI4, TP53, BARD1, BRCA1, H2AFX, CDKN2A, HMGA2, HMGA1, HCVgp1, YY1, HIST1H3A, HIST2H2AC, HIST2H2BE, HIST1H4A, EP300, tat, PML, RARA, CTNNBL1, FANCA, FANCC, TFAP2A, HDAC1, HDAC2, SMARCA4, DOT1L, ACACA, HNRNPM, HNRNPU, DDX21, HIST1H1C, SIRT1, YBX1, PC, YBX3, H1FX, HIST1H1A, ESR1, Trp53, Cdkn2a, CDK5RAP3, ZNF668, UBC, RYK, SMN1, CD4, YWHAQ, USP36, CTCF, HDAC5, CDKN1A, NOP56, CENPA, AFF1, DYRK2, TOP1, CDK2, SREK1, SENP3, RB1, PARP1, HJURP, ARRB1, ARRB2, SIRT7, NPM3, NPM1, FBXO25, IRF1, CUL3, CUL4A, CUL5, CUL2, CUL1, COPS5, COPS6, DCUN1D1, CAND1, MDM2, PA2G4, PPP2R1A, MID1, GZF1, APEX1, COPS2, PLCG1, FBXO6, GAPDH, SIAH1, PTBP1, SRSF1, APP, RPL14, RPS6, RPL24, RPL6, NAP1L1, HNRNPR, RPS4X, PRPF6, LAMP2, SURF4, RPL10L, NAP1L4, ZYX, NUP62, NUP50, RPL5, YWHAE, FMNL1, CPSF6, HNRNPH1, PAPOLG, JUNB, MC4R, HIVEP2, CPSF1, CPSF2, CPSF4, CPSF3, FN1, VCAM1, CSNK2A1, NOS2, UBL4A, ITGA4, CBX2, CBX4, CBX8, PHB, EIF2AK2, XPO1, GZMM, PAN2, CD81, IGSF8, ICAM1, PRKCZ, KIF11, SQSTM1, FLNA, LLGL1, HSP90AA1, TRIM21, TUBA1C, HSP90AB1, PARD6B, CDC37, TUBB3, MRPL46, KCTD2, CLTC, MRPL50, VCP, KCTD5, MRPL12, HSPA5, PTRF, MRPL52, EEF1A1, KCTD17, TAB1, BAG2, ANXA2, EIF5A, PPIL4, MRPL38, STK38, FLNB, LGALS1, C1QBP, COPB1, MRPL37, MRPL49, KEAP1, PRDX4, WDR26, PSMC4, SDPR, SLC25A3, RPS12, EEF1B2, NIPSNAP1, EEF1G, MRPL39, DDX3X, RNH1, OSBPL1A, EIF3H, MRPL54, ENO1, MYCBP, MRPL18, MAP1B, PSMD1, EEF1D, DNAJC13, HSPB1, CFL1, FLNC, PPP1CC, RPLP1, CEP152, SNRPD1, ATP5A1, ARHGAP5, PRKAR1A, MRPL53, GID8, YWHAZ, CCT2, EIF3E, LAMB2, DICER1, CHCHD4, MRPL19, SNRPD2, GRWD1, TPR, PSMD4, PSMC5, PPP2CA, UNC5C, SLC25A5, PTPN14, RPS5, TTC27, BOLA2, EIF4A1, TUFM, S100A10, IMPG1, NUDT21, UTRN, MRPL44, EIF3M, MRPL43, PKM, IMPDH2, TRRAP, CHCHD6, RANBP6, EIF3I, TAB2, MRPL24, DZIP1, IPO5, AGR2, MRPS30, ICT1, PRDX6, TCP1, RCOR1, GRN, TXN, RPLP0, PCBP1, MRPL28, MRPL10, TAF3, EPAS1, CHCHD3, TXNDC12, ELMO2, SRRT, MRPL11, EIF2S1, SNRPF, KTN1, FGD6, UQCRH, RBBP4, DNAJA1, CTTN, USP15, NUDCD1, RPS28, JAK1, PROS1, PYGB, KDR, EEF2, FASN, AKAP11, CSNK2B, DNAJA2, EIF3G, DHX15, MRPS26, HNRNPA0, DOCK4, PRKCI, VASP, COPE, SAMM50, TEX15, PSME3, PPM1G, COPG1, CALU, S100A11, HSPA4, PSMD12, POLR2E, TRIM28, SLAIN2, RPL10, GOLGA2, GOLGA3, PRKCA, IPO9, PARK7, SPIN1, PPA1, BANP, DSTN, GNAI2, HOXA7, ABCC1, RELA, ACY1, TARDBP, PARK2, KPNA1, ARMCX3, LIMCH1, OSBP, TRMT61A, rev, RPA1, RPA2, RPA3, ERG, LGR4, STAU1, ELF4, AURKA, HUWE1, FUS, COX8A, NPM2, MOV10, NXF1, PHF6, CUL7, OBSL1, CCDC8, SIRT6, EBNA1BP2, NOL12, RPL10A, POP4, ZNF22, NSA2, TAF1D, NIFK, RPL26L1, NIP7, RPL4, RPF1, CCDC137, KNOP1, RBM28, RPL7A, POP1, DDX24, FTSJ3, RRS1, RPL3, ZFP62, ZNF512, DDX56, DDX27, GLYR1, MAK16, CEBPZ, RSL1D1, HP1BP3, REXO4, DDX31, MYBBP1A, NOP16, C3orf17, RRP8, GNL2, PAPD5, RSL24D1, SURF6, GTPBP4, KIAA0020, NOP2, NVL, GLTSCR2, URB1, RPL23A, GPATCH4, TEX10, RPS8, NOC3L, BRIX1, DDX54, PWP1, RBM34, NMNAT1, CENPC, SENP5, RPL36AL, PAK1IP1, ZNF800, RPL7L1, RPL37A, RPF2, PELP1, NOC2L, RPP40, RBMX2, CENPV, PPAN-P2RY11, SDAD1, RPP38, RPP25, SPTY2D1, RPP25L, POP7, RPS6KB2, UTP15, NTRK1, EWSR1, CLK1, HIST1H3E, BYSL, CHD1, DDX1, DECR1, EIF1AX, EIF4G2, FBL, FAU, HIST1H1D, HIST1H1B, RPSA, MKI67, MPG, EXOSC10, RPL7, RPL8, RPL9, RPL11, RPL13, RPL15, RPL17, RPL18, RPL18A, RPL19, RPL21, RPL22, RPL27, RPL27A, RPL29, RPL32, RPL37, RPL38, RPS2, RPS3, RPS3A, RPS9, RPS10, RPS11, RPS13, RPS14, RPS15, RPS16, RPS17, RPS18, RPS19, RPS20, RPS21, RPS23, RPS24, RPS25, RPS27, RPS29, SKIV2L, SRP72, XPC, IFRD2, HIST1H2BC, SMARCA5, CGGBP1, EIF3A, EIF3C, PABPC4, USP10, TTC37, JADE3, ABCF2, G3BP1, GNB2L1, EMG1, IGF2BP3, PSIP1, SUPT16H, RPL35, PDCD11, LARP4B, RRP12, LARP1, RPL13A, RPL36, MTHFD1L, AHCTF1, FAM98A, SERBP1, GNL3, SND1, NOB1, RPS27L, DDX47, EIF3L, RTCB, TRMT112, DHX29, MTPAP, NAT10, TSR1, NKRF, MEPCE, BCCIP, PNO1, MRPS22, WDR18, SCAF1, NOL6, DDX50, CCDC86, MUS81, ZNF622, DHX57, H2AFV, LARP4, WBSCR22, Eif3a, Eif3e, Ktn1, Rpl35, Srp72, Rrbp1, GAN, HEMGN, TMUB1, CRY1, DNAJB4, MCM2, GLI1, Mdm2, SP1, U2AF2, RC3H1, EIF2S2, EGFR, WWP2, CD24, ZNF746, DDX51, MAGED2, HNRNPA2B1, HNRNPD, HNRNPA1, RPL23, RPS7, HIST1H2BB, HIST1H2AA, CBX3, CYLD, INO80B, LMNA, YAP1, MTF1, BRD1, HDAC6, FBXW7, CENPW, API5, CTNNB1, MAP2K3, BMP4, CCNT1, DIMT1, MATN2, TRIP4, CUL4B, UBE2M, PRPF8, EFTUD2, AAR2, PIH1D1, CHD3, RNF4, CHD4, LARP7, RNF31, TNF, FAM188B, SPDL1, HEXIM1, SNAI1, RECQL4, GPC1, REST, ZFP36L2, MYC, CDK9, Prkab1, NEK2, METTL3, METTL14, KIAA1429, RC3H2, PSMA3, ACTC1, ESR2, FAF1, RBX1, BRCA2, DISC1, NR2C2, UBQLN2, MTDH, GADD45A, AGRN, ATXN3, VRK1, VRK3, DYRK1A, SNRNP70, ITFG1, GHET1, ARAF, HMGB1, BIRC3, NFX1, BRD7, N, SOX2, PPIA, CACYBP, RNR1, CMTR1, ARIH2, PLEKHA4, RAD18, PINK1, WHSC1, FANCD2, SAMD12, LINC01554, ZC3H18, CAMK2A, FYN, PTPN12, RPS6KA3, STX7, IL7R, GPC3, RP1, KMT2A, SUZ12, BSX, LEMD1, ZCCHC7, SYNE2, ZBTB9, R3HDM4, ZNF496, PIKFYVE, COL8A1, PMS2, TRDN, PLCZ1, ASPM, TTN, SETD2, BDP1, C4orf47, CFAP54, HIST2H2BC, AIM2, SYT7, LTN1, EPRS, FST, LMOD1, MCAM, RPL34, MYT1, DYNC1I2, KIF22, ZNF181, ATAT1, TRMT10B, RBM44, ANKS3, FILIP1, FAM9A, SUPT20H, THOC2, CENPI, BICD1, CCL13, JPH2, NDST4, SARDH, SLC27A6, LECT2, HMGN4, OPA1, ITGB1, HIVEP1, PNRC1, PDE4DIP, CAPRIN2, C1orf110, MICAL1, STAB2, CEP63, NGRN, CAMSAP3, ARHGEF9, SNIP1, ORF14, ILF3, LRRC31, DUX4, CIT, ANLN, AURKB, CHMP4B, CHMP4C, ECT2, KIF14, KIF20A, KIF23, PRC1, BRD3, SP110, TRIM24, ZMYND8, LRRC59, NMRAL1, SUMO2, Rnf183, BRD4, NUPR1, RBM45, CIC, Apc2, RBM39, FBP1, ASXL1, vpr, RIN3, DNAJC19, DNAJC25, DNAJC2, OGT, DDRGK1, UFL1, CD3EAP, DDX23, HIST1H2BG, ACTG1, ALKBH4, DNAJC10, GLUD2, HIST1H2AE, HIST1H3F, HIST1H4J, HLCS, HSPA1B, MALT1, PEG10, PLD3, UBB, AIP, BCORL1, BID, CSK, LCK, PSMG1, RAB21, RBM11, VDAC1, RPL31, ZNF330, TRIM37, FZR1, WDR5, PAGE4, NUDCD2, NAA40, BGLT3, CCDC140, RPL23AP32, MTG2, NGDN, UTP18, PRPF4B, WDR74, WDR36, DKC1, DDX52, EPB41L5, ZBTB11, H2AFY, CIRH1A, SRSF5, DUSP11, DDX10, TSPYL1, IMP3, RBM19, NOM1, ZC3HAV1, RPP14, UTP14A, WDR12, DNTTIP2, MPHOSPH10, RRP1, WDR3, BUD13, RPLP2, C14orf169, ZNF771, AATF, C1orf35, OASL, NLE1, BMS1, BOP1, STAU2, TAF1C, WDR55, ZNF638, RFC1, FYTTD1, WDR43, C7orf50, SRPK2, LUC7L, FAM111A, PBRM1, NOL10, TAF1A, TBL3, ZNF770, BAZ1B, ESF1, TTF1, LAS1L, RPP30, ZNF16, RRP15, ABT1, SPRTN, ULK1, TRAF6, BTF3, SLFN11, NLRP7, RCHY1, DIDO1, CCNF, FAM129A, ATR, ATM, PSMD9, ZEB1, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| NPM1 |  |
| ALK |  |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to NPM1-ALK |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to NPM1-ALK |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | NPM1 | C0026998 | Acute Myeloid Leukemia, M1 | 6 | CTD_human;ORPHANET |
| Hgene | NPM1 | C1879321 | Acute Myeloid Leukemia (AML-M2) | 6 | CTD_human;ORPHANET |
| Hgene | NPM1 | C0023467 | Leukemia, Myelocytic, Acute | 5 | CGI;CTD_human |
| Hgene | NPM1 | C0023487 | Acute Promyelocytic Leukemia | 2 | CGI;CTD_human;ORPHANET |
| Hgene | NPM1 | C0024623 | Malignant neoplasm of stomach | 1 | CTD_human |
| Hgene | NPM1 | C0038356 | Stomach Neoplasms | 1 | CTD_human |
| Hgene | NPM1 | C0152013 | Adenocarcinoma of lung (disorder) | 1 | CTD_human |
| Hgene | NPM1 | C0206182 | Lymphomatoid Papulosis | 1 | ORPHANET |
| Hgene | NPM1 | C0265965 | Dyskeratosis Congenita | 1 | CTD_human;GENOMICS_ENGLAND |
| Hgene | NPM1 | C1148551 | X-Linked Dyskeratosis Congenita | 1 | CTD_human |
| Hgene | NPM1 | C1301362 | Primary Cutaneous Anaplastic Large Cell Lymphoma | 1 | ORPHANET |
| Hgene | NPM1 | C1708349 | Hereditary Diffuse Gastric Cancer | 1 | CTD_human |
| Hgene | NPM1 | C2930974 | Acute erythroleukemia | 1 | CTD_human |
| Hgene | NPM1 | C2930975 | Acute erythroleukemia - M6a subtype | 1 | CTD_human |
| Hgene | NPM1 | C2930976 | Acute myeloid leukemia FAB-M6 | 1 | CTD_human |
| Hgene | NPM1 | C2930977 | Acute erythroleukemia - M6b subtype | 1 | CTD_human |
| Tgene | ALK | C0007131 | Non-Small Cell Lung Carcinoma | 28 | CGI;CTD_human |
| Tgene | ALK | C0027819 | Neuroblastoma | 13 | CGI;CTD_human;ORPHANET |
| Tgene | ALK | C0152013 | Adenocarcinoma of lung (disorder) | 8 | CGI;CTD_human |
| Tgene | ALK | C2751681 | NEUROBLASTOMA, SUSCEPTIBILITY TO, 3 | 8 | CLINGEN;UNIPROT |
| Tgene | ALK | C0206180 | Ki-1+ Anaplastic Large Cell Lymphoma | 6 | CGI;CTD_human |
| Tgene | ALK | C0334121 | Inflammatory Myofibroblastic Tumor | 4 | CGI;CTD_human;ORPHANET |
| Tgene | ALK | C0018199 | Granuloma, Plasma Cell | 3 | CTD_human |
| Tgene | ALK | C0007621 | Neoplastic Cell Transformation | 2 | CTD_human |
| Tgene | ALK | C0027627 | Neoplasm Metastasis | 2 | CTD_human |
| Tgene | ALK | C0238463 | Papillary thyroid carcinoma | 2 | ORPHANET |
| Tgene | ALK | C0001973 | Alcoholic Intoxication, Chronic | 1 | PSYGENET |
| Tgene | ALK | C0006118 | Brain Neoplasms | 1 | CGI;CTD_human |
| Tgene | ALK | C0006142 | Malignant neoplasm of breast | 1 | CTD_human |
| Tgene | ALK | C0007134 | Renal Cell Carcinoma | 1 | CTD_human |
| Tgene | ALK | C0011570 | Mental Depression | 1 | PSYGENET |
| Tgene | ALK | C0011581 | Depressive disorder | 1 | PSYGENET |
| Tgene | ALK | C0027643 | Neoplasm Recurrence, Local | 1 | CTD_human |
| Tgene | ALK | C0036341 | Schizophrenia | 1 | PSYGENET |
| Tgene | ALK | C0079744 | Diffuse Large B-Cell Lymphoma | 1 | CTD_human |
| Tgene | ALK | C0085269 | Plasma Cell Granuloma, Pulmonary | 1 | CTD_human |
| Tgene | ALK | C0153633 | Malignant neoplasm of brain | 1 | CGI;CTD_human |
| Tgene | ALK | C0278601 | Inflammatory Breast Carcinoma | 1 | CTD_human |
| Tgene | ALK | C0279702 | Conventional (Clear Cell) Renal Cell Carcinoma | 1 | CTD_human |
| Tgene | ALK | C0496899 | Benign neoplasm of brain, unspecified | 1 | CTD_human |
| Tgene | ALK | C0678222 | Breast Carcinoma | 1 | CTD_human |
| Tgene | ALK | C0750974 | Brain Tumor, Primary | 1 | CTD_human |
| Tgene | ALK | C0750977 | Recurrent Brain Neoplasm | 1 | CTD_human |
| Tgene | ALK | C0750979 | Primary malignant neoplasm of brain | 1 | CTD_human |
| Tgene | ALK | C1257931 | Mammary Neoplasms, Human | 1 | CTD_human |
| Tgene | ALK | C1266042 | Chromophobe Renal Cell Carcinoma | 1 | CTD_human |
| Tgene | ALK | C1266043 | Sarcomatoid Renal Cell Carcinoma | 1 | CTD_human |
| Tgene | ALK | C1266044 | Collecting Duct Carcinoma of the Kidney | 1 | CTD_human |
| Tgene | ALK | C1306837 | Papillary Renal Cell Carcinoma | 1 | CTD_human |
| Tgene | ALK | C1332079 | Anaplastic Large Cell Lymphoma, ALK-Positive | 1 | ORPHANET |
| Tgene | ALK | C1458155 | Mammary Neoplasms | 1 | CTD_human |
| Tgene | ALK | C1527390 | Neoplasms, Intracranial | 1 | CTD_human |
| Tgene | ALK | C2931189 | Neural crest tumor | 1 | ORPHANET |
| Tgene | ALK | C3899155 | hereditary neuroblastoma | 1 | GENOMICS_ENGLAND |
| Tgene | ALK | C4704874 | Mammary Carcinoma, Human | 1 | CTD_human |

