| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:ARID1A-MAST2 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: ARID1A-MAST2 | FusionPDB ID: 6380 | FusionGDB2.0 ID: 6380 | Hgene | Tgene | Gene symbol | ARID1A | MAST2 | Gene ID | 8289 | 23139 |
| Gene name | AT-rich interaction domain 1A | microtubule associated serine/threonine kinase 2 | |
| Synonyms | B120|BAF250|BAF250a|BM029|C1orf4|CSS2|ELD|MRD14|OSA1|P270|SMARCF1|hELD|hOSA1 | MAST205|MTSSK | |
| Cytomap | 1p36.11 | 1p34.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | AT-rich interactive domain-containing protein 1AARID domain-containing protein 1AAT rich interactive domain 1A (SWI-like)BRG1-associated factor 250aOSA1 nuclear proteinSWI-like proteinSWI/SNF complex protein p270SWI/SNF-related, matrix-associated, | microtubule-associated serine/threonine-protein kinase 2microtubule associated testis specific serine/threonine protein kinase | |
| Modification date | 20200329 | 20200313 | |
| UniProtAcc | O14497 | Q6P0Q8 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000324856, ENST00000457599, ENST00000374152, ENST00000540690, | ENST00000477968, ENST00000361297, ENST00000372009, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 29 X 19 X 15=8265 | 14 X 16 X 10=2240 |
| # samples | 45 | 17 | |
| ** MAII score | log2(45/8265*10)=-4.19901791296264 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(17/2240*10)=-3.71989208080727 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: ARID1A [Title/Abstract] AND MAST2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | |||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ARID1A | GO:0006337 | nucleosome disassembly | 8895581 |
| Hgene | ARID1A | GO:0006338 | chromatin remodeling | 11726552 |
| Hgene | ARID1A | GO:0030520 | intracellular estrogen receptor signaling pathway | 12200431 |
| Hgene | ARID1A | GO:0030521 | androgen receptor signaling pathway | 12200431 |
| Hgene | ARID1A | GO:0042921 | glucocorticoid receptor signaling pathway | 12200431 |
| Hgene | ARID1A | GO:0045893 | positive regulation of transcription, DNA-templated | 12200431 |
 Fusion gene breakpoints across ARID1A (5'-gene) Fusion gene breakpoints across ARID1A (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 Fusion gene breakpoints across MAST2 (3'-gene) Fusion gene breakpoints across MAST2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerKB3 | . | . | ARID1A | chr1 | 27024031 | + | MAST2 | chr1 | 46290104 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000324856 | ARID1A | chr1 | 27024031 | + | ENST00000372009 | MAST2 | chr1 | 46290104 | + | 6158 | 1508 | 371 | 6157 | 1929 |
| ENST00000324856 | ARID1A | chr1 | 27024031 | + | ENST00000361297 | MAST2 | chr1 | 46290104 | + | 6786 | 1508 | 371 | 6727 | 2118 |
| ENST00000457599 | ARID1A | chr1 | 27024031 | + | ENST00000372009 | MAST2 | chr1 | 46290104 | + | 5787 | 1137 | 0 | 5786 | 1928 |
| ENST00000457599 | ARID1A | chr1 | 27024031 | + | ENST00000361297 | MAST2 | chr1 | 46290104 | + | 6415 | 1137 | 0 | 6356 | 2118 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >6380_6380_1_ARID1A-MAST2_ARID1A_chr1_27024031_ENST00000324856_MAST2_chr1_46290104_ENST00000361297_length(amino acids)=2118AA_BP=367 MAAQVAPAAASSLGNPPPPPPSELKKAEQQQREEAGGEAAAAAAAERGEMKAAAGQESEGPAVGPPQPLGKELQDGAESNGGGGGGGAGS GGGPGAEPDLKNSNGNAGPRPALNNNLTEPPGGGGGGSSDGVGAPPHSAAAALPPPAYGFGQPYGRSPSAVAAAAAAVFHQQHGGQQSPG LAALQSGGGGGLEPYAGPQQNSHDHGFPNHQYNSYYPNRSAYPPPAPAYALSSPRGGTPGSGAAAAAGSKPPPSSSASASSSSSSFAQQR FGAMGGGGPSAAGGGTPQPTATPTLNQLLTSPSSARGYQGYPGGDYSGGPQDGGAGKGPADMASQCWGAAAAAAAAAAASGGAQQRSHHA PMSPGSSGGGGQPLARTPQDVVTGVSPLLFRKLSNPDIFSSTGKVKLQRQLSQDDCKLWRGNLASSLSGKQLLPLSSSVHSSVGQVTWQS SGEASNLVRMRNQSLGQSAPSLTAGLKELSLPRRGSFCRTSNRKSLIVTSSTSPTLPRPHSPLHGHTGNSPLDSPRNFSPNAPAHFSFVP ARRTDGRRWSLASLPSSGYGTNTPSSTVSSSCSSQEKLHQLPFQPTADELHFLTKHFSTESVPDEEGRQSPAMRPRSRSLSPGRSPVSFD SEIIMMNHVYKERFPKATAQMEERLAEFISSNTPDSVLPLADGALSFIHHQVIEMARDCLDKSRSGLITSQYFYELQDNLEKLLQDAHER SESSEVAFVMQLVKKLMIIIARPARLLECLEFDPEEFYHLLEAAEGHAKEGQGIKCDIPRYIVSQLGLTRDPLEEMAQLSSCDSPDTPET DDSIEGHGASLPSKKTPSEEDFETIKLISNGAYGAVFLVRHKSTRQRFAMKKINKQNLILRNQIQQAFVERDILTFAENPFVVSMFCSFD TKRHLCMVMEYVEGGDCATLLKNIGALPVDMVRLYFAETVLALEYLHNYGIVHRDLKPDNLLITSMGHIKLTDFGLSKIGLMSLTTNLYE GHIEKDAREFLDKQVCGTPEYIAPEVILRQGYGKPVDWWAMGIILYEFLVGCVPFFGDTPEELFGQVISDEIVWPEGDEALPPDAQDLTS KLLHQNPLERLGTGSAYEVKQHPFFTGLDWTGLLRQKAEFIPQLESEDDTSYFDTRSERYHHMDSEDEEEVSEDGCLEIRQFSSCSPRFN KVYSSMERLSLLEERRTPPPTKRSLSEEKEDHSDGLAGLKGRDRSWVIGSPEILRKRLSVSESSHTESDSSPPMTVRRRCSGLLDAPRFP EGPEEASSTLRRQPQEGIWVLTPPSGEGVSGPVTEHSGEQRPKLDEEAVGRSSGSSPAMETRGRGTSQLAEGATAKAISDLAVRRARHRL LSGDSTEKRTARPVNKVIKSASATALSLLIPSEHHTCSPLASPMSPHSQSSNPSSRDSSPSRDFLPALGSMRPPIIIHRAGKKYGFTLRA IRVYMGDSDVYTVHHMVWHVEDGGPASEAGLRQGDLITHVNGEPVHGLVHTEVVELILKSGNKVAISTTPLENTSIKVGPARKGSYKAKM ARRSKRSRGKDGQESRKRSSLFRKITKQASLLHTSRSLSSLNRSLSSGESGPGSPTHSHSLSPRSPTQGYRVTPDAVHSVGGNSSQSSSP SSSVPSSPAGSGHTRPSSLHGLAPKLQRQYRSPRRKSAGSIPLSPLAHTPSPPPPTASPQRSPSPLSGHVAQAFPTKLHLSPPLGRQLSR PKSAEPPRSPLLKRVQSAEKLAAALAASEKKLATSRKHSLDLPHSELKKELPPREVSPLEVVGARSVLSGKGALPGKGVLQPAPSRALGT LRQDRAERRESLQKQEAIREVDSSEDDTEEGPENSQGAQELSLAPHPEVSQSVAPKGAGESGEEDPFPSRDPRSLGPMVPSLLTGITLGP PRMESPSGPHRRLGSPQAIEEAASSSSAGPNLGQSGATDPIPPEGCWKAQHLHTQALTALSPSTSGLTPTSSCSPPSSTSGKLSMWSWKS LIEGPDRASPSRKATMAGGLANLQDLENTTPAQPKNLSPREQGKTQPPSAPRLAHPSYEDPSQGWLWESECAQAVKEDPALSITQVPDAS -------------------------------------------------------------- >6380_6380_2_ARID1A-MAST2_ARID1A_chr1_27024031_ENST00000324856_MAST2_chr1_46290104_ENST00000372009_length(amino acids)=1929AA_BP=367 MAAQVAPAAASSLGNPPPPPPSELKKAEQQQREEAGGEAAAAAAAERGEMKAAAGQESEGPAVGPPQPLGKELQDGAESNGGGGGGGAGS GGGPGAEPDLKNSNGNAGPRPALNNNLTEPPGGGGGGSSDGVGAPPHSAAAALPPPAYGFGQPYGRSPSAVAAAAAAVFHQQHGGQQSPG LAALQSGGGGGLEPYAGPQQNSHDHGFPNHQYNSYYPNRSAYPPPAPAYALSSPRGGTPGSGAAAAAGSKPPPSSSASASSSSSSFAQQR FGAMGGGGPSAAGGGTPQPTATPTLNQLLTSPSSARGYQGYPGGDYSGGPQDGGAGKGPADMASQCWGAAAAAAAAAAASGGAQQRSHHA PMSPGSSGGGGQPLARTPQDVVTGVSPLLFRKLSNPDIFSSTGKVKLQRQLSQDDCKLWRGNLASSLSGKQLLPLSSSVHSSVGQVTWQS SGEASNLVRMRNQSLGQSAPSLTAGLKELSLPRRGSFCRTSNRKSLIVTSSTSPTLPRPHSPLHGHTGNSPLDSPRNFSPNAPAHFSFVP ARRTDGRRWSLASLPSSGYGTNTPSSTVSSSCSSQEKLHQLPFQPTADELHFLTKHFSTESVPDEEGRQSPAMRPRSRSLSPGRSPVSFD SEIIMMNHVYKERFPKAHERSESSEVAFVMQLVKKLMIIIARPARLLECLEFDPEEFYHLLEAAEGHAKEGQGIKCDIPRYIVSQLGLTR DPLEEMAQLSSCDSPDTPETDDSIEGHGASLPSKKTPSEEDFETIKLISNGAYGAVFLVRHKSTRQRFAMKKINKQNLILRNQIQQAFVE RDILTFAENPFVVSMFCSFDTKRHLCMVMEYVEGGDCATLLKNIGALPVDMVRLYFAETVLALEYLHNYGIVHRDLKPDNLLITSMGHIK LTDFGLSKIGLMSLTTNLYEGHIEKDAREFLDKQVCGTPEYIAPEVILRQGYGKPVDWWAMGIILYEFLVGCVPFFGDTPEELFGQVISD EIVWPEGDEALPPDAQDLTSKLLHQNPLERLGTGSAYEVKQHPFFTGLDWTGLLRQKAEFIPQLESEDDTSYFDTRSERYHHMDSEDEEE VSEDGCLEIRQFSSCSPRFNKVYSSMERLSLLEERRTPPPTKRSLSEEKEDHSDGLAGLKGRDRSWVIGSPEILRKRLSVSESSHTESDS SPPMTVRRRCSGLLDAPRFPEGPEEASSTLRRQPQEGIWVLTPPSGEGVSGPVTEHSGEQRPKLDEEAVGRSSGSSPAMETRGRGTSQLA EGATAKAISDLAVRRARHRLLSGDSTEKRTARPVNKVIKSASATALSLLIPSEHHTCSPLASPMSPHSQSSNPSSRDSSPYGFTLRAIRV YMGDSDVYTVHHMVWHVEDGGPASEAGLRQGDLITHVNGEPVHGLVHTEVVELILKSGNKVAISTTPLENTSIKVGPARKGSYKAKMARR SKRSRGKDGQESRKRSSLFRKITKQASLLHTSRSLSSLNRSLSSGESGPGSPTHSHSLSPRSPTQGYRVTPDAVHSELSRPKSAEPPRSP LLKRVQSAEKLAAALAASEKKLATSRKHSLDLPHSELKKELPPREVSPLEVVGARSVLSGKGALPGKGVLQPAPSRALGTLRQDRAERRE SLQKQEAIREVDSSEDDTEEGPENSQGAQELSLAPHPEVSQSVAPKGAGESGEEDPFPSRDPRSLGPMVPSLLTGITLGPPRMESPSGPH RRLGSPQAIEEAASSSSAGPNLGQSGATDPIPPEGCWKAQHLHTQALTALSPSTSGLTPTSSCSPPSSTSGKLSMWSWKSLIEGPDRASP SRKATMAGGLANLQDLENTTPAQPKNLSPREQGKTQPPSAPRLAHPSYEDPSQGWLWESECAQAVKEDPALSITQVPDASGDRRQDVPCR -------------------------------------------------------------- >6380_6380_3_ARID1A-MAST2_ARID1A_chr1_27024031_ENST00000457599_MAST2_chr1_46290104_ENST00000361297_length(amino acids)=2118AA_BP=367 MAAQVAPAAASSLGNPPPPPPSELKKAEQQQREEAGGEAAAAAAAERGEMKAAAGQESEGPAVGPPQPLGKELQDGAESNGGGGGGGAGS GGGPGAEPDLKNSNGNAGPRPALNNNLTEPPGGGGGGSSDGVGAPPHSAAAALPPPAYGFGQPYGRSPSAVAAAAAAVFHQQHGGQQSPG LAALQSGGGGGLEPYAGPQQNSHDHGFPNHQYNSYYPNRSAYPPPAPAYALSSPRGGTPGSGAAAAAGSKPPPSSSASASSSSSSFAQQR FGAMGGGGPSAAGGGTPQPTATPTLNQLLTSPSSARGYQGYPGGDYSGGPQDGGAGKGPADMASQCWGAAAAAAAAAAASGGAQQRSHHA PMSPGSSGGGGQPLARTPQDVVTGVSPLLFRKLSNPDIFSSTGKVKLQRQLSQDDCKLWRGNLASSLSGKQLLPLSSSVHSSVGQVTWQS SGEASNLVRMRNQSLGQSAPSLTAGLKELSLPRRGSFCRTSNRKSLIVTSSTSPTLPRPHSPLHGHTGNSPLDSPRNFSPNAPAHFSFVP ARRTDGRRWSLASLPSSGYGTNTPSSTVSSSCSSQEKLHQLPFQPTADELHFLTKHFSTESVPDEEGRQSPAMRPRSRSLSPGRSPVSFD SEIIMMNHVYKERFPKATAQMEERLAEFISSNTPDSVLPLADGALSFIHHQVIEMARDCLDKSRSGLITSQYFYELQDNLEKLLQDAHER SESSEVAFVMQLVKKLMIIIARPARLLECLEFDPEEFYHLLEAAEGHAKEGQGIKCDIPRYIVSQLGLTRDPLEEMAQLSSCDSPDTPET DDSIEGHGASLPSKKTPSEEDFETIKLISNGAYGAVFLVRHKSTRQRFAMKKINKQNLILRNQIQQAFVERDILTFAENPFVVSMFCSFD TKRHLCMVMEYVEGGDCATLLKNIGALPVDMVRLYFAETVLALEYLHNYGIVHRDLKPDNLLITSMGHIKLTDFGLSKIGLMSLTTNLYE GHIEKDAREFLDKQVCGTPEYIAPEVILRQGYGKPVDWWAMGIILYEFLVGCVPFFGDTPEELFGQVISDEIVWPEGDEALPPDAQDLTS KLLHQNPLERLGTGSAYEVKQHPFFTGLDWTGLLRQKAEFIPQLESEDDTSYFDTRSERYHHMDSEDEEEVSEDGCLEIRQFSSCSPRFN KVYSSMERLSLLEERRTPPPTKRSLSEEKEDHSDGLAGLKGRDRSWVIGSPEILRKRLSVSESSHTESDSSPPMTVRRRCSGLLDAPRFP EGPEEASSTLRRQPQEGIWVLTPPSGEGVSGPVTEHSGEQRPKLDEEAVGRSSGSSPAMETRGRGTSQLAEGATAKAISDLAVRRARHRL LSGDSTEKRTARPVNKVIKSASATALSLLIPSEHHTCSPLASPMSPHSQSSNPSSRDSSPSRDFLPALGSMRPPIIIHRAGKKYGFTLRA IRVYMGDSDVYTVHHMVWHVEDGGPASEAGLRQGDLITHVNGEPVHGLVHTEVVELILKSGNKVAISTTPLENTSIKVGPARKGSYKAKM ARRSKRSRGKDGQESRKRSSLFRKITKQASLLHTSRSLSSLNRSLSSGESGPGSPTHSHSLSPRSPTQGYRVTPDAVHSVGGNSSQSSSP SSSVPSSPAGSGHTRPSSLHGLAPKLQRQYRSPRRKSAGSIPLSPLAHTPSPPPPTASPQRSPSPLSGHVAQAFPTKLHLSPPLGRQLSR PKSAEPPRSPLLKRVQSAEKLAAALAASEKKLATSRKHSLDLPHSELKKELPPREVSPLEVVGARSVLSGKGALPGKGVLQPAPSRALGT LRQDRAERRESLQKQEAIREVDSSEDDTEEGPENSQGAQELSLAPHPEVSQSVAPKGAGESGEEDPFPSRDPRSLGPMVPSLLTGITLGP PRMESPSGPHRRLGSPQAIEEAASSSSAGPNLGQSGATDPIPPEGCWKAQHLHTQALTALSPSTSGLTPTSSCSPPSSTSGKLSMWSWKS LIEGPDRASPSRKATMAGGLANLQDLENTTPAQPKNLSPREQGKTQPPSAPRLAHPSYEDPSQGWLWESECAQAVKEDPALSITQVPDAS -------------------------------------------------------------- >6380_6380_4_ARID1A-MAST2_ARID1A_chr1_27024031_ENST00000457599_MAST2_chr1_46290104_ENST00000372009_length(amino acids)=1928AA_BP=367 MAAQVAPAAASSLGNPPPPPPSELKKAEQQQREEAGGEAAAAAAAERGEMKAAAGQESEGPAVGPPQPLGKELQDGAESNGGGGGGGAGS GGGPGAEPDLKNSNGNAGPRPALNNNLTEPPGGGGGGSSDGVGAPPHSAAAALPPPAYGFGQPYGRSPSAVAAAAAAVFHQQHGGQQSPG LAALQSGGGGGLEPYAGPQQNSHDHGFPNHQYNSYYPNRSAYPPPAPAYALSSPRGGTPGSGAAAAAGSKPPPSSSASASSSSSSFAQQR FGAMGGGGPSAAGGGTPQPTATPTLNQLLTSPSSARGYQGYPGGDYSGGPQDGGAGKGPADMASQCWGAAAAAAAAAAASGGAQQRSHHA PMSPGSSGGGGQPLARTPQDVVTGVSPLLFRKLSNPDIFSSTGKVKLQRQLSQDDCKLWRGNLASSLSGKQLLPLSSSVHSSVGQVTWQS SGEASNLVRMRNQSLGQSAPSLTAGLKELSLPRRGSFCRTSNRKSLIVTSSTSPTLPRPHSPLHGHTGNSPLDSPRNFSPNAPAHFSFVP ARRTDGRRWSLASLPSSGYGTNTPSSTVSSSCSSQEKLHQLPFQPTADELHFLTKHFSTESVPDEEGRQSPAMRPRSRSLSPGRSPVSFD SEIIMMNHVYKERFPKAHERSESSEVAFVMQLVKKLMIIIARPARLLECLEFDPEEFYHLLEAAEGHAKEGQGIKCDIPRYIVSQLGLTR DPLEEMAQLSSCDSPDTPETDDSIEGHGASLPSKKTPSEEDFETIKLISNGAYGAVFLVRHKSTRQRFAMKKINKQNLILRNQIQQAFVE RDILTFAENPFVVSMFCSFDTKRHLCMVMEYVEGGDCATLLKNIGALPVDMVRLYFAETVLALEYLHNYGIVHRDLKPDNLLITSMGHIK LTDFGLSKIGLMSLTTNLYEGHIEKDAREFLDKQVCGTPEYIAPEVILRQGYGKPVDWWAMGIILYEFLVGCVPFFGDTPEELFGQVISD EIVWPEGDEALPPDAQDLTSKLLHQNPLERLGTGSAYEVKQHPFFTGLDWTGLLRQKAEFIPQLESEDDTSYFDTRSERYHHMDSEDEEE VSEDGCLEIRQFSSCSPRFNKVYSSMERLSLLEERRTPPPTKRSLSEEKEDHSDGLAGLKGRDRSWVIGSPEILRKRLSVSESSHTESDS SPPMTVRRRCSGLLDAPRFPEGPEEASSTLRRQPQEGIWVLTPPSGEGVSGPVTEHSGEQRPKLDEEAVGRSSGSSPAMETRGRGTSQLA EGATAKAISDLAVRRARHRLLSGDSTEKRTARPVNKVIKSASATALSLLIPSEHHTCSPLASPMSPHSQSSNPSSRDSSPYGFTLRAIRV YMGDSDVYTVHHMVWHVEDGGPASEAGLRQGDLITHVNGEPVHGLVHTEVVELILKSGNKVAISTTPLENTSIKVGPARKGSYKAKMARR SKRSRGKDGQESRKRSSLFRKITKQASLLHTSRSLSSLNRSLSSGESGPGSPTHSHSLSPRSPTQGYRVTPDAVHSELSRPKSAEPPRSP LLKRVQSAEKLAAALAASEKKLATSRKHSLDLPHSELKKELPPREVSPLEVVGARSVLSGKGALPGKGVLQPAPSRALGTLRQDRAERRE SLQKQEAIREVDSSEDDTEEGPENSQGAQELSLAPHPEVSQSVAPKGAGESGEEDPFPSRDPRSLGPMVPSLLTGITLGPPRMESPSGPH RRLGSPQAIEEAASSSSAGPNLGQSGATDPIPPEGCWKAQHLHTQALTALSPSTSGLTPTSSCSPPSSTSGKLSMWSWKSLIEGPDRASP SRKATMAGGLANLQDLENTTPAQPKNLSPREQGKTQPPSAPRLAHPSYEDPSQGWLWESECAQAVKEDPALSITQVPDASGDRRQDVPCR -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:/chr1:) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ARID1A | MAST2 |
| FUNCTION: Involved in transcriptional activation and repression of select genes by chromatin remodeling (alteration of DNA-nucleosome topology). Component of SWI/SNF chromatin remodeling complexes that carry out key enzymatic activities, changing chromatin structure by altering DNA-histone contacts within a nucleosome in an ATP-dependent manner. Binds DNA non-specifically. Belongs to the neural progenitors-specific chromatin remodeling complex (npBAF complex) and the neuron-specific chromatin remodeling complex (nBAF complex). During neural development a switch from a stem/progenitor to a postmitotic chromatin remodeling mechanism occurs as neurons exit the cell cycle and become committed to their adult state. The transition from proliferating neural stem/progenitor cells to postmitotic neurons requires a switch in subunit composition of the npBAF and nBAF complexes. As neural progenitors exit mitosis and differentiate into neurons, npBAF complexes which contain ACTL6A/BAF53A and PHF10/BAF45A, are exchanged for homologous alternative ACTL6B/BAF53B and DPF1/BAF45B or DPF3/BAF45C subunits in neuron-specific complexes (nBAF). The npBAF complex is essential for the self-renewal/proliferative capacity of the multipotent neural stem cells. The nBAF complex along with CREST plays a role regulating the activity of genes essential for dendrite growth (By similarity). {ECO:0000250|UniProtKB:A2BH40, ECO:0000303|PubMed:12672490, ECO:0000303|PubMed:22952240, ECO:0000303|PubMed:26601204}. | FUNCTION: Appears to link the dystrophin/utrophin network with microtubule filaments via the syntrophins. Phosphorylation of DMD or UTRN may modulate their affinities for associated proteins. Functions in a multi-protein complex in spermatid maturation. Regulates lipopolysaccharide-induced IL-12 synthesis in macrophages by forming a complex with TRAF6, resulting in the inhibition of TRAF6 NF-kappa-B activation (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
Top |
Fusion Protein Structures |
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
| PDB file (1067) | ARID1A | chr1 | 27024031 | + | MAST2 | chr1 | 46290104 | + | MAAQVAPAAASSLGNPPPPPPSELKKAEQQQREEAGGEAAAAAAAERGEM KAAAGQESEGPAVGPPQPLGKELQDGAESNGGGGGGGAGSGGGPGAEPDL KNSNGNAGPRPALNNNLTEPPGGGGGGSSDGVGAPPHSAAAALPPPAYGF GQPYGRSPSAVAAAAAAVFHQQHGGQQSPGLAALQSGGGGGLEPYAGPQQ NSHDHGFPNHQYNSYYPNRSAYPPPAPAYALSSPRGGTPGSGAAAAAGSK PPPSSSASASSSSSSFAQQRFGAMGGGGPSAAGGGTPQPTATPTLNQLLT SPSSARGYQGYPGGDYSGGPQDGGAGKGPADMASQCWGAAAAAAAAAAAS GGAQQRSHHAPMSPGSSGGGGQPLARTPQDVVTGVSPLLFRKLSNPDIFS STGKVKLQRQLSQDDCKLWRGNLASSLSGKQLLPLSSSVHSSVGQVTWQS SGEASNLVRMRNQSLGQSAPSLTAGLKELSLPRRGSFCRTSNRKSLIVTS STSPTLPRPHSPLHGHTGNSPLDSPRNFSPNAPAHFSFVPARRTDGRRWS LASLPSSGYGTNTPSSTVSSSCSSQEKLHQLPFQPTADELHFLTKHFSTE SVPDEEGRQSPAMRPRSRSLSPGRSPVSFDSEIIMMNHVYKERFPKAHER SESSEVAFVMQLVKKLMIIIARPARLLECLEFDPEEFYHLLEAAEGHAKE GQGIKCDIPRYIVSQLGLTRDPLEEMAQLSSCDSPDTPETDDSIEGHGAS LPSKKTPSEEDFETIKLISNGAYGAVFLVRHKSTRQRFAMKKINKQNLIL RNQIQQAFVERDILTFAENPFVVSMFCSFDTKRHLCMVMEYVEGGDCATL LKNIGALPVDMVRLYFAETVLALEYLHNYGIVHRDLKPDNLLITSMGHIK LTDFGLSKIGLMSLTTNLYEGHIEKDAREFLDKQVCGTPEYIAPEVILRQ GYGKPVDWWAMGIILYEFLVGCVPFFGDTPEELFGQVISDEIVWPEGDEA LPPDAQDLTSKLLHQNPLERLGTGSAYEVKQHPFFTGLDWTGLLRQKAEF IPQLESEDDTSYFDTRSERYHHMDSEDEEEVSEDGCLEIRQFSSCSPRFN KVYSSMERLSLLEERRTPPPTKRSLSEEKEDHSDGLAGLKGRDRSWVIGS PEILRKRLSVSESSHTESDSSPPMTVRRRCSGLLDAPRFPEGPEEASSTL RRQPQEGIWVLTPPSGEGVSGPVTEHSGEQRPKLDEEAVGRSSGSSPAME TRGRGTSQLAEGATAKAISDLAVRRARHRLLSGDSTEKRTARPVNKVIKS ASATALSLLIPSEHHTCSPLASPMSPHSQSSNPSSRDSSPYGFTLRAIRV YMGDSDVYTVHHMVWHVEDGGPASEAGLRQGDLITHVNGEPVHGLVHTEV VELILKSGNKVAISTTPLENTSIKVGPARKGSYKAKMARRSKRSRGKDGQ ESRKRSSLFRKITKQASLLHTSRSLSSLNRSLSSGESGPGSPTHSHSLSP RSPTQGYRVTPDAVHSELSRPKSAEPPRSPLLKRVQSAEKLAAALAASEK KLATSRKHSLDLPHSELKKELPPREVSPLEVVGARSVLSGKGALPGKGVL QPAPSRALGTLRQDRAERRESLQKQEAIREVDSSEDDTEEGPENSQGAQE LSLAPHPEVSQSVAPKGAGESGEEDPFPSRDPRSLGPMVPSLLTGITLGP PRMESPSGPHRRLGSPQAIEEAASSSSAGPNLGQSGATDPIPPEGCWKAQ HLHTQALTALSPSTSGLTPTSSCSPPSSTSGKLSMWSWKSLIEGPDRASP SRKATMAGGLANLQDLENTTPAQPKNLSPREQGKTQPPSAPRLAHPSYED PSQGWLWESECAQAVKEDPALSITQVPDASGDRRQDVPCRGCPLTQKSEP | 1928 |
| PDB file (1068) | ARID1A | chr1 | 27024031 | + | MAST2 | chr1 | 46290104 | + | MAAQVAPAAASSLGNPPPPPPSELKKAEQQQREEAGGEAAAAAAAERGEM KAAAGQESEGPAVGPPQPLGKELQDGAESNGGGGGGGAGSGGGPGAEPDL KNSNGNAGPRPALNNNLTEPPGGGGGGSSDGVGAPPHSAAAALPPPAYGF GQPYGRSPSAVAAAAAAVFHQQHGGQQSPGLAALQSGGGGGLEPYAGPQQ NSHDHGFPNHQYNSYYPNRSAYPPPAPAYALSSPRGGTPGSGAAAAAGSK PPPSSSASASSSSSSFAQQRFGAMGGGGPSAAGGGTPQPTATPTLNQLLT SPSSARGYQGYPGGDYSGGPQDGGAGKGPADMASQCWGAAAAAAAAAAAS GGAQQRSHHAPMSPGSSGGGGQPLARTPQDVVTGVSPLLFRKLSNPDIFS STGKVKLQRQLSQDDCKLWRGNLASSLSGKQLLPLSSSVHSSVGQVTWQS SGEASNLVRMRNQSLGQSAPSLTAGLKELSLPRRGSFCRTSNRKSLIVTS STSPTLPRPHSPLHGHTGNSPLDSPRNFSPNAPAHFSFVPARRTDGRRWS LASLPSSGYGTNTPSSTVSSSCSSQEKLHQLPFQPTADELHFLTKHFSTE SVPDEEGRQSPAMRPRSRSLSPGRSPVSFDSEIIMMNHVYKERFPKAHER SESSEVAFVMQLVKKLMIIIARPARLLECLEFDPEEFYHLLEAAEGHAKE GQGIKCDIPRYIVSQLGLTRDPLEEMAQLSSCDSPDTPETDDSIEGHGAS LPSKKTPSEEDFETIKLISNGAYGAVFLVRHKSTRQRFAMKKINKQNLIL RNQIQQAFVERDILTFAENPFVVSMFCSFDTKRHLCMVMEYVEGGDCATL LKNIGALPVDMVRLYFAETVLALEYLHNYGIVHRDLKPDNLLITSMGHIK LTDFGLSKIGLMSLTTNLYEGHIEKDAREFLDKQVCGTPEYIAPEVILRQ GYGKPVDWWAMGIILYEFLVGCVPFFGDTPEELFGQVISDEIVWPEGDEA LPPDAQDLTSKLLHQNPLERLGTGSAYEVKQHPFFTGLDWTGLLRQKAEF IPQLESEDDTSYFDTRSERYHHMDSEDEEEVSEDGCLEIRQFSSCSPRFN KVYSSMERLSLLEERRTPPPTKRSLSEEKEDHSDGLAGLKGRDRSWVIGS PEILRKRLSVSESSHTESDSSPPMTVRRRCSGLLDAPRFPEGPEEASSTL RRQPQEGIWVLTPPSGEGVSGPVTEHSGEQRPKLDEEAVGRSSGSSPAME TRGRGTSQLAEGATAKAISDLAVRRARHRLLSGDSTEKRTARPVNKVIKS ASATALSLLIPSEHHTCSPLASPMSPHSQSSNPSSRDSSPYGFTLRAIRV YMGDSDVYTVHHMVWHVEDGGPASEAGLRQGDLITHVNGEPVHGLVHTEV VELILKSGNKVAISTTPLENTSIKVGPARKGSYKAKMARRSKRSRGKDGQ ESRKRSSLFRKITKQASLLHTSRSLSSLNRSLSSGESGPGSPTHSHSLSP RSPTQGYRVTPDAVHSELSRPKSAEPPRSPLLKRVQSAEKLAAALAASEK KLATSRKHSLDLPHSELKKELPPREVSPLEVVGARSVLSGKGALPGKGVL QPAPSRALGTLRQDRAERRESLQKQEAIREVDSSEDDTEEGPENSQGAQE LSLAPHPEVSQSVAPKGAGESGEEDPFPSRDPRSLGPMVPSLLTGITLGP PRMESPSGPHRRLGSPQAIEEAASSSSAGPNLGQSGATDPIPPEGCWKAQ HLHTQALTALSPSTSGLTPTSSCSPPSSTSGKLSMWSWKSLIEGPDRASP SRKATMAGGLANLQDLENTTPAQPKNLSPREQGKTQPPSAPRLAHPSYED PSQGWLWESECAQAVKEDPALSITQVPDASGDRRQDVPCRGCPLTQKSEP | 1929 |
| PDB file (1099) | ARID1A | chr1 | 27024031 | + | MAST2 | chr1 | 46290104 | + | MAAQVAPAAASSLGNPPPPPPSELKKAEQQQREEAGGEAAAAAAAERGEM KAAAGQESEGPAVGPPQPLGKELQDGAESNGGGGGGGAGSGGGPGAEPDL KNSNGNAGPRPALNNNLTEPPGGGGGGSSDGVGAPPHSAAAALPPPAYGF GQPYGRSPSAVAAAAAAVFHQQHGGQQSPGLAALQSGGGGGLEPYAGPQQ NSHDHGFPNHQYNSYYPNRSAYPPPAPAYALSSPRGGTPGSGAAAAAGSK PPPSSSASASSSSSSFAQQRFGAMGGGGPSAAGGGTPQPTATPTLNQLLT SPSSARGYQGYPGGDYSGGPQDGGAGKGPADMASQCWGAAAAAAAAAAAS GGAQQRSHHAPMSPGSSGGGGQPLARTPQDVVTGVSPLLFRKLSNPDIFS STGKVKLQRQLSQDDCKLWRGNLASSLSGKQLLPLSSSVHSSVGQVTWQS SGEASNLVRMRNQSLGQSAPSLTAGLKELSLPRRGSFCRTSNRKSLIVTS STSPTLPRPHSPLHGHTGNSPLDSPRNFSPNAPAHFSFVPARRTDGRRWS LASLPSSGYGTNTPSSTVSSSCSSQEKLHQLPFQPTADELHFLTKHFSTE SVPDEEGRQSPAMRPRSRSLSPGRSPVSFDSEIIMMNHVYKERFPKATAQ MEERLAEFISSNTPDSVLPLADGALSFIHHQVIEMARDCLDKSRSGLITS QYFYELQDNLEKLLQDAHERSESSEVAFVMQLVKKLMIIIARPARLLECL EFDPEEFYHLLEAAEGHAKEGQGIKCDIPRYIVSQLGLTRDPLEEMAQLS SCDSPDTPETDDSIEGHGASLPSKKTPSEEDFETIKLISNGAYGAVFLVR HKSTRQRFAMKKINKQNLILRNQIQQAFVERDILTFAENPFVVSMFCSFD TKRHLCMVMEYVEGGDCATLLKNIGALPVDMVRLYFAETVLALEYLHNYG IVHRDLKPDNLLITSMGHIKLTDFGLSKIGLMSLTTNLYEGHIEKDAREF LDKQVCGTPEYIAPEVILRQGYGKPVDWWAMGIILYEFLVGCVPFFGDTP EELFGQVISDEIVWPEGDEALPPDAQDLTSKLLHQNPLERLGTGSAYEVK QHPFFTGLDWTGLLRQKAEFIPQLESEDDTSYFDTRSERYHHMDSEDEEE VSEDGCLEIRQFSSCSPRFNKVYSSMERLSLLEERRTPPPTKRSLSEEKE DHSDGLAGLKGRDRSWVIGSPEILRKRLSVSESSHTESDSSPPMTVRRRC SGLLDAPRFPEGPEEASSTLRRQPQEGIWVLTPPSGEGVSGPVTEHSGEQ RPKLDEEAVGRSSGSSPAMETRGRGTSQLAEGATAKAISDLAVRRARHRL LSGDSTEKRTARPVNKVIKSASATALSLLIPSEHHTCSPLASPMSPHSQS SNPSSRDSSPSRDFLPALGSMRPPIIIHRAGKKYGFTLRAIRVYMGDSDV YTVHHMVWHVEDGGPASEAGLRQGDLITHVNGEPVHGLVHTEVVELILKS GNKVAISTTPLENTSIKVGPARKGSYKAKMARRSKRSRGKDGQESRKRSS LFRKITKQASLLHTSRSLSSLNRSLSSGESGPGSPTHSHSLSPRSPTQGY RVTPDAVHSVGGNSSQSSSPSSSVPSSPAGSGHTRPSSLHGLAPKLQRQY RSPRRKSAGSIPLSPLAHTPSPPPPTASPQRSPSPLSGHVAQAFPTKLHL SPPLGRQLSRPKSAEPPRSPLLKRVQSAEKLAAALAASEKKLATSRKHSL DLPHSELKKELPPREVSPLEVVGARSVLSGKGALPGKGVLQPAPSRALGT LRQDRAERRESLQKQEAIREVDSSEDDTEEGPENSQGAQELSLAPHPEVS QSVAPKGAGESGEEDPFPSRDPRSLGPMVPSLLTGITLGPPRMESPSGPH RRLGSPQAIEEAASSSSAGPNLGQSGATDPIPPEGCWKAQHLHTQALTAL SPSTSGLTPTSSCSPPSSTSGKLSMWSWKSLIEGPDRASPSRKATMAGGL ANLQDLENTTPAQPKNLSPREQGKTQPPSAPRLAHPSYEDPSQGWLWESE CAQAVKEDPALSITQVPDASGDRRQDVPCRGCPLTQKSEPSLRRGQEPGG | 2118 |
Top |
pLDDT score distribution |
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
ARID1A_pLDDT.png |
MAST2_pLDDT.png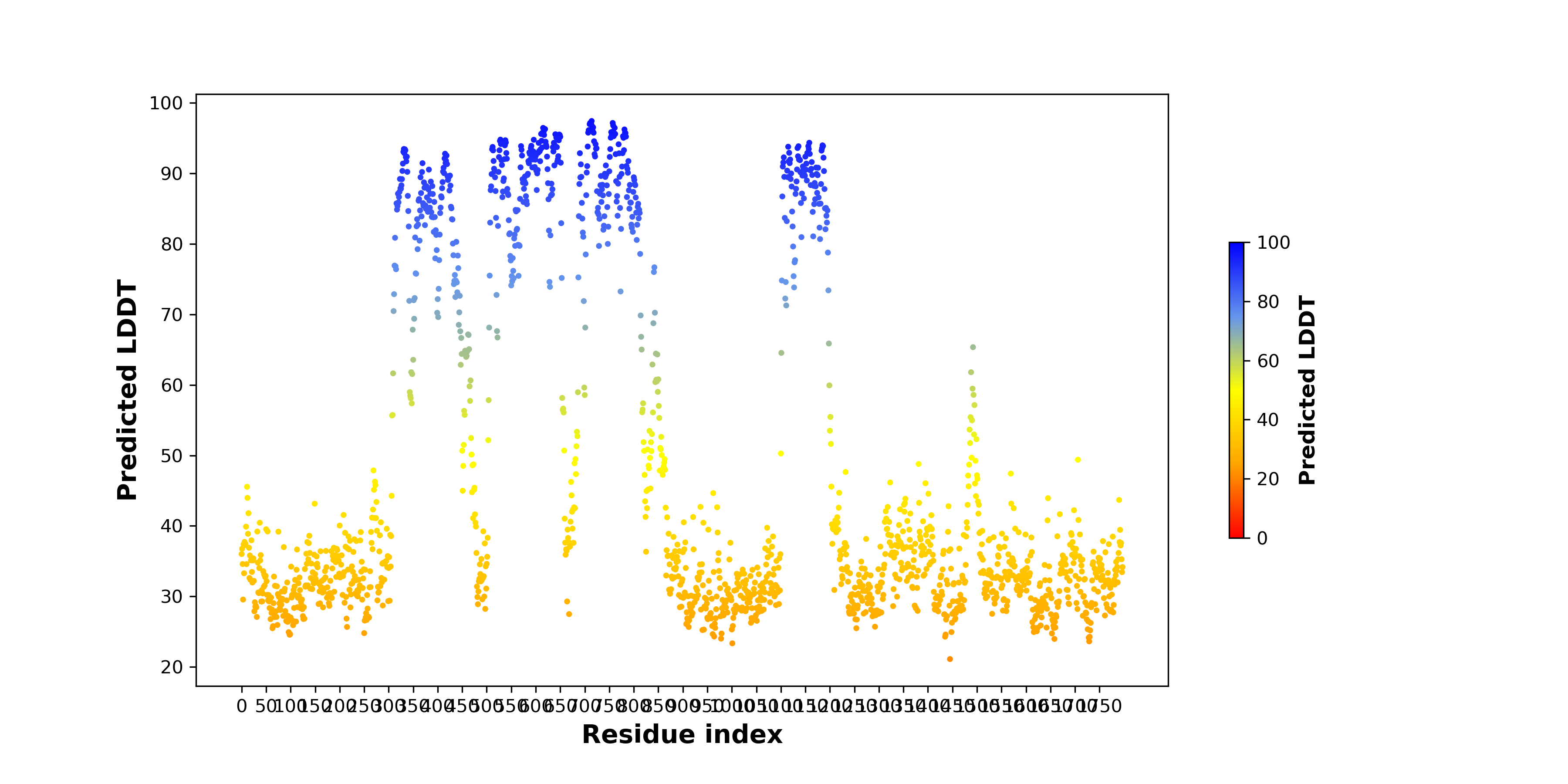 |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
ARID1A_MAST2_1099_violinplot.png (AA BP:) |
Top |
Ramachandran Plot of Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| Fusion AA seq ID in FusionPDB and their Ramachandran plots |
Top |
Potential Active Site Information |
 The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. |
| Fusion AA seq ID in FusionPDB | Site score | Size | D score | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
Top |
Potentially Interacting Small Molecules through Virtual Screening |
 The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. |
| Fusion AA seq ID in FusionPDB | ZINC ID | DrugBank ID | Drug name | Docking score | Glide gscore |
Top |
 Drug information from DrugBank of the top 20 interacting small molecules. Drug information from DrugBank of the top 20 interacting small molecules. |
| ZINC ID | DrugBank ID | Drug name | Drug type | SMILES | Drug group |
Top |
Biochemical Features of Small Molecules |
 ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) |
| ZINC ID | mol_MW | dipole | SASA | FOSA | FISA | PISA | WPSA | volume | donorHB | accptHB | IP | Human Oral Absorption | Percent Human Oral Absorption | Rule Of Five | Rule Of Three |
Top |
Drug Toxicity Information |
 Toxicity information of individual drugs using eToxPred Toxicity information of individual drugs using eToxPred |
| ZINC ID | Smile | Surface Accessibility | Toxicity |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| ARID1A | KLF1, GATA1, SMARCB1, SMARCA4, SMARCA2, ING1, SMARCE1, PBRM1, SMARCC2, SMARCC1, PGR, BCL7C, HIC1, CUL2, tat, ARID1B, SMAD2, SMAD3, CARM1, ADNP, RARA, Bcl7b, SOX2, CHD7, SMARCD1, TP53, ELAVL1, SIRT7, TFAP4, CUL3, SMARCD2, ACTL6A, SMARCD3, PHF10, ECT2, YAP1, ITCH, NEDD4, SHMT2, MOV10, NXF1, EED, DPF3, DPF2, SS18, UNK, BCL7A, BCL7B, ESR1, NCOA1, RARB, RARG, EWSR1, Bcl7c, Smarcc1, SNW1, CDC5L, CUL1, SKP1, NFATC1, NFATC2, TOP1, CHEK1, VHL, EYA2, NR3C1, AR, ALDH2, TRIM25, HNRNPL, PDCD6, RMND5A, EZH2, CHD3, CHD4, RUNX1, TNRC6A, RTF1, RBBP7, RUVBL1, MYC, FUS, HIST1H4A, HIST1H2AB, TAF15, DDIT3, FLI1, TP53BP1, BRCA1, MDC1, KIAA1429, ACTC1, ZBTB7A, H2AFX, H2AFY, CTCF, BTRC, WWP2, PLEKHA4, nsp10, ALG13, SMG7, SUPT5H, PRC1, C18orf8, SS18L1, BRD7, DPF1, ARID2, BRD9, GLTSCR1, STK11IP, ASF1A, NUP50, TERF2IP, ATG7, TEX13B, SS18L2, SIRT6, T, ELF5, ERG, ETS1, ETV4, FEV, FOS, FOXI1, GATA2, GATA3, GCM1, HNF1B, HNF4A, IRF1, IRF4, KLF3, KLF4, KLF5, LHX1, LHX2, LHX3, LHX4, MYOD1, NFIA, NFIB, NFIC, NFIX, PAX6, PAX7, PAX8, PAX9, SOX10, SOX15, SOX17, SOX9, SP7, TBR1, TEAD1, TLX1, TLX2, TLX3, BRD3, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| ARID1A |  |
| MAST2 | 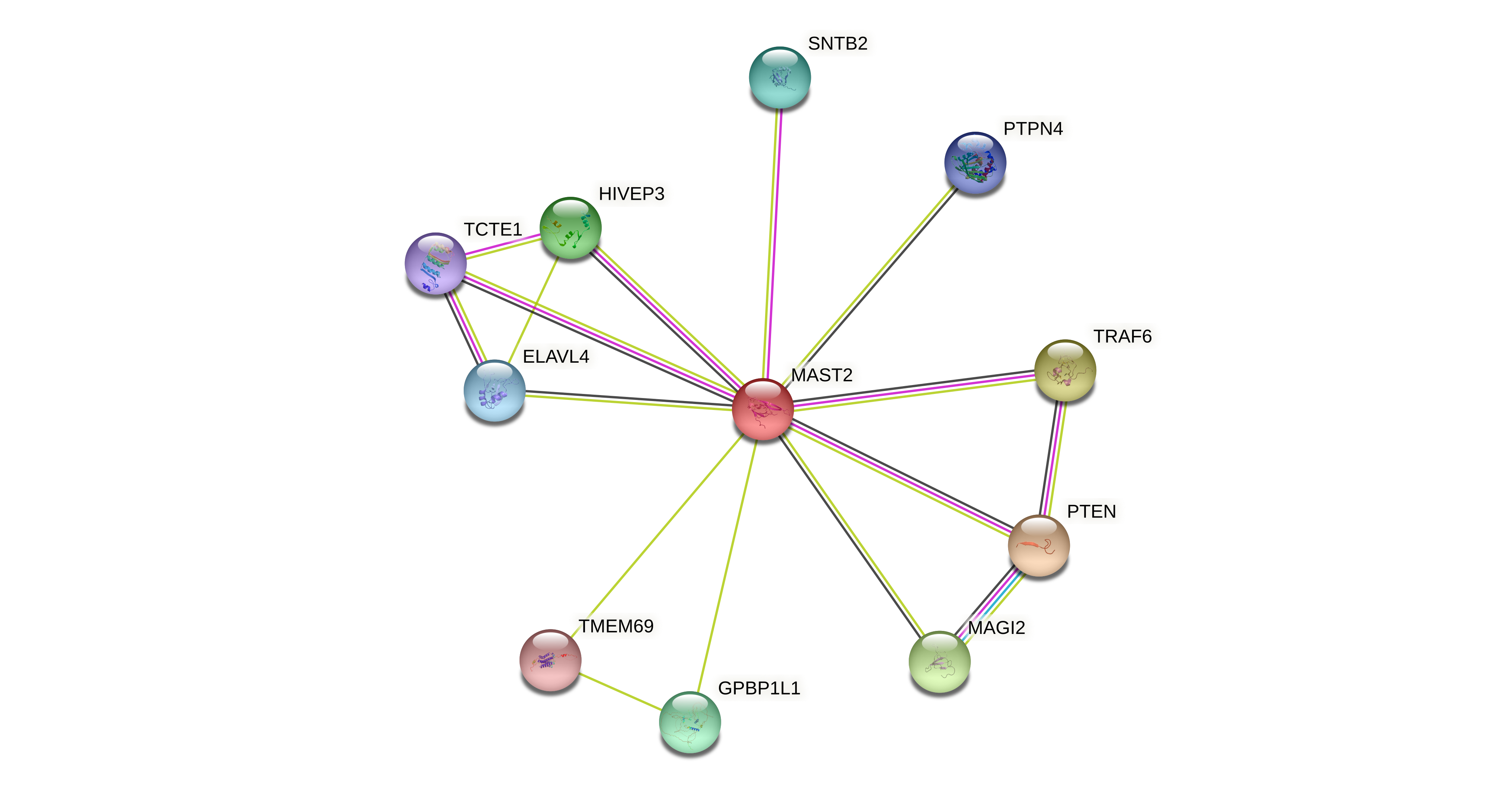 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to ARID1A-MAST2 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to ARID1A-MAST2 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
| ARID1A | MAST2 | Endometrial Endometrioid Adenocarcinoma | MyCancerGenome | |
| ARID1A | MAST2 | Lung Adenocarcinoma | MyCancerGenome | |
| ARID1A | MAST2 | Bladder Urothelial Carcinoma | MyCancerGenome | |
| ARID1A | MAST2 | Breast Invasive Ductal Carcinoma | MyCancerGenome |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | ARID1A | C0024623 | Malignant neoplasm of stomach | 3 | CTD_human |
| Hgene | ARID1A | C0038356 | Stomach Neoplasms | 3 | CTD_human |
| Hgene | ARID1A | C1708349 | Hereditary Diffuse Gastric Cancer | 3 | CTD_human |
| Hgene | ARID1A | C2239176 | Liver carcinoma | 3 | CTD_human |
| Hgene | ARID1A | C0033578 | Prostatic Neoplasms | 2 | CTD_human |
| Hgene | ARID1A | C0376358 | Malignant neoplasm of prostate | 2 | CTD_human |
| Hgene | ARID1A | C0001418 | Adenocarcinoma | 1 | CTD_human |
| Hgene | ARID1A | C0005684 | Malignant neoplasm of urinary bladder | 1 | CTD_human |
| Hgene | ARID1A | C0005695 | Bladder Neoplasm | 1 | CTD_human |
| Hgene | ARID1A | C0006413 | Burkitt Lymphoma | 1 | CTD_human |
| Hgene | ARID1A | C0007138 | Carcinoma, Transitional Cell | 1 | CTD_human |
| Hgene | ARID1A | C0009402 | Colorectal Carcinoma | 1 | CTD_human |
| Hgene | ARID1A | C0009404 | Colorectal Neoplasms | 1 | CTD_human |
| Hgene | ARID1A | C0010606 | Adenoid Cystic Carcinoma | 1 | CTD_human |
| Hgene | ARID1A | C0014170 | Endometrial Neoplasms | 1 | CTD_human |
| Hgene | ARID1A | C0027708 | Nephroblastoma | 1 | CTD_human |
| Hgene | ARID1A | C0027819 | Neuroblastoma | 1 | CTD_human |
| Hgene | ARID1A | C0036920 | Sezary Syndrome | 1 | CTD_human |
| Hgene | ARID1A | C0079772 | T-Cell Lymphoma | 1 | CTD_human |
| Hgene | ARID1A | C0079773 | Lymphoma, T-Cell, Cutaneous | 1 | CTD_human |
| Hgene | ARID1A | C0205641 | Adenocarcinoma, Basal Cell | 1 | CTD_human |
| Hgene | ARID1A | C0205642 | Adenocarcinoma, Oxyphilic | 1 | CTD_human |
| Hgene | ARID1A | C0205643 | Carcinoma, Cribriform | 1 | CTD_human |
| Hgene | ARID1A | C0205644 | Carcinoma, Granular Cell | 1 | CTD_human |
| Hgene | ARID1A | C0205645 | Adenocarcinoma, Tubular | 1 | CTD_human |
| Hgene | ARID1A | C0206656 | Embryonal Rhabdomyosarcoma | 1 | CTD_human |
| Hgene | ARID1A | C0206698 | Cholangiocarcinoma | 1 | CTD_human |
| Hgene | ARID1A | C0265338 | Coffin-Siris syndrome | 1 | CTD_human;GENOMICS_ENGLAND |
| Hgene | ARID1A | C0279628 | Adenocarcinoma Of Esophagus | 1 | CTD_human |
| Hgene | ARID1A | C0343640 | African Burkitt's lymphoma | 1 | CTD_human |
| Hgene | ARID1A | C0345905 | Intrahepatic Cholangiocarcinoma | 1 | CTD_human |
| Hgene | ARID1A | C0376407 | Granulomatous Slack Skin | 1 | CTD_human |
| Hgene | ARID1A | C0476089 | Endometrial Carcinoma | 1 | CTD_human |
| Hgene | ARID1A | C0920269 | Microsatellite Instability | 1 | CTD_human |
| Hgene | ARID1A | C1721098 | Replication Error Phenotype | 1 | CTD_human |
| Hgene | ARID1A | C2930471 | Bilateral Wilms Tumor | 1 | CTD_human |
| Hgene | ARID1A | C2931822 | Nasopharyngeal carcinoma | 1 | CTD_human |
| Hgene | ARID1A | C3805278 | Extrahepatic Cholangiocarcinoma | 1 | CTD_human |
| Hgene | ARID1A | C4721444 | Burkitt Leukemia | 1 | CTD_human |

