| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:ATF7IP-PDGFRB |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: ATF7IP-PDGFRB | FusionPDB ID: 7486 | FusionGDB2.0 ID: 7486 | Hgene | Tgene | Gene symbol | ATF7IP | PDGFRB | Gene ID | 55729 | 5159 |
| Gene name | activating transcription factor 7 interacting protein | platelet derived growth factor receptor beta | |
| Synonyms | AM|ATF-IP|MCAF|MCAF1|p621 | CD140B|IBGC4|IMF1|JTK12|KOGS|PDGFR|PDGFR-1|PDGFR1|PENTT | |
| Cytomap | 12p13.1 | 5q32 | |
| Type of gene | protein-coding | protein-coding | |
| Description | activating transcription factor 7-interacting protein 1ATF-interacting proteinATF7-interacting proteinATFa-associated modulatorMBD1-containing chromatin-associated factor 1 | platelet-derived growth factor receptor betaActivated tyrosine kinase PDGFRBCD140 antigen-like family member BNDEL1-PDGFRBPDGF-R-betaPDGFR-betabeta-type platelet-derived growth factor receptorplatelet-derived growth factor receptor 1platelet-deriv | |
| Modification date | 20200313 | 20200329 | |
| UniProtAcc | Q5U623 | P09619 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000261168, ENST00000536444, ENST00000540793, ENST00000543189, ENST00000544627, ENST00000541654, | ENST00000523456, ENST00000261799, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 16 X 15 X 8=1920 | 28 X 26 X 6=4368 |
| # samples | 20 | 15 | |
| ** MAII score | log2(20/1920*10)=-3.26303440583379 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(15/4368*10)=-4.86393845042397 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: ATF7IP [Title/Abstract] AND PDGFRB [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | ATF7IP(14634119)-PDGFRB(149506177), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | ATF7IP-PDGFRB seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ATF7IP-PDGFRB seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ATF7IP-PDGFRB seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ATF7IP-PDGFRB seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ATF7IP-PDGFRB seems lost the major protein functional domain in Hgene partner, which is a epigenetic factor due to the frame-shifted ORF. ATF7IP-PDGFRB seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. ATF7IP-PDGFRB seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. ATF7IP-PDGFRB seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. ATF7IP-PDGFRB seems lost the major protein functional domain in Tgene partner, which is a kinase due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ATF7IP | GO:0006306 | DNA methylation | 12665582 |
| Hgene | ATF7IP | GO:0045892 | negative regulation of transcription, DNA-templated | 12665582 |
| Hgene | ATF7IP | GO:0045893 | positive regulation of transcription, DNA-templated | 12665582 |
| Hgene | ATF7IP | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly | 12665582 |
| Tgene | PDGFRB | GO:0007165 | signal transduction | 10821867 |
| Tgene | PDGFRB | GO:0010863 | positive regulation of phospholipase C activity | 1653029 |
| Tgene | PDGFRB | GO:0018108 | peptidyl-tyrosine phosphorylation | 1653029|2536956|2850496 |
| Tgene | PDGFRB | GO:0030335 | positive regulation of cell migration | 17470632 |
| Tgene | PDGFRB | GO:0032516 | positive regulation of phosphoprotein phosphatase activity | 7691811 |
| Tgene | PDGFRB | GO:0038091 | positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway | 17470632 |
| Tgene | PDGFRB | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity | 1314164 |
| Tgene | PDGFRB | GO:0046777 | protein autophosphorylation | 1314164|2536956|2850496 |
| Tgene | PDGFRB | GO:0048008 | platelet-derived growth factor receptor signaling pathway | 1314164|2536956 |
| Tgene | PDGFRB | GO:0060326 | cell chemotaxis | 2554309|17991872 |
 Fusion gene breakpoints across ATF7IP (5'-gene) Fusion gene breakpoints across ATF7IP (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
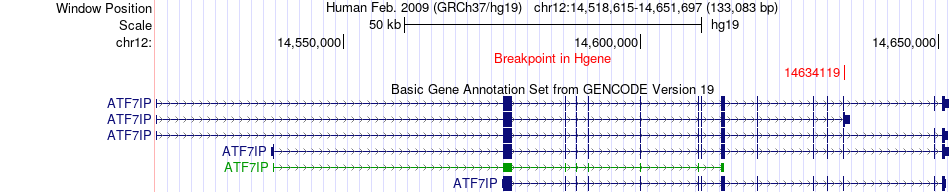 |
 Fusion gene breakpoints across PDGFRB (3'-gene) Fusion gene breakpoints across PDGFRB (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
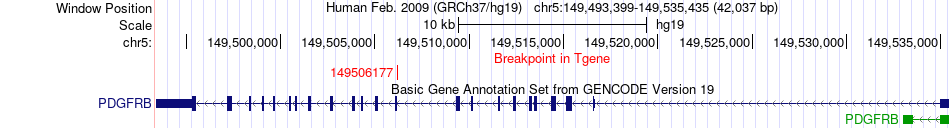 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerKB4 | . | . | ATF7IP | chr12 | 14613428 | + | PDGFRB | chr5 | 14613428 | - |
| ChimerKB4 | . | . | ATF7IP | chr12 | 14633936 | + | PDGFRB | chr5 | 14633936 | - |
| ChiTaRS5.0 | N/A | HZ039234 | ATF7IP | chr12 | 14634119 | + | PDGFRB | chr5 | 149506177 | - |
| ChiTaRS5.0 | N/A | HZ039235 | ATF7IP | chr12 | 14634119 | + | PDGFRB | chr5 | 149506177 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000261168 | ATF7IP | chr12 | 14634119 | + | ENST00000261799 | PDGFRB | chr5 | 149506177 | - | 7101 | 3433 | 153 | 5174 | 1673 |
| ENST00000536444 | ATF7IP | chr12 | 14634119 | + | ENST00000261799 | PDGFRB | chr5 | 149506177 | - | 7051 | 3383 | 106 | 5124 | 1672 |
| ENST00000544627 | ATF7IP | chr12 | 14634119 | + | ENST00000261799 | PDGFRB | chr5 | 149506177 | - | 7292 | 3624 | 320 | 5365 | 1681 |
| ENST00000540793 | ATF7IP | chr12 | 14634119 | + | ENST00000261799 | PDGFRB | chr5 | 149506177 | - | 7103 | 3435 | 155 | 5176 | 1673 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000261168 | ENST00000261799 | ATF7IP | chr12 | 14634119 | + | PDGFRB | chr5 | 149506177 | - | 0.001302262 | 0.99869776 |
| ENST00000536444 | ENST00000261799 | ATF7IP | chr12 | 14634119 | + | PDGFRB | chr5 | 149506177 | - | 0.001150028 | 0.99885 |
| ENST00000544627 | ENST00000261799 | ATF7IP | chr12 | 14634119 | + | PDGFRB | chr5 | 149506177 | - | 0.001398285 | 0.99860173 |
| ENST00000540793 | ENST00000261799 | ATF7IP | chr12 | 14634119 | + | PDGFRB | chr5 | 149506177 | - | 0.001135758 | 0.9988643 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >7486_7486_1_ATF7IP-PDGFRB_ATF7IP_chr12_14634119_ENST00000261168_PDGFRB_chr5_149506177_ENST00000261799_length(amino acids)=1673AA_BP=1094 MDSLEEPQKKVFKARKTMRVSDRQQLEAVYKVKEELLKTDVKLLNGNHENGDLDPTSPLENMDYIKDKEEVNGIEEICFDPEGSKAEWKE TPCILSVNVKNKQDDDLNCEPLSPHNITPEPVSKLPAEPVSGDPAPGDLDAGDPASGVLASGDSTSGDPTSSEPSSSDAASGDATSGDAP SGDVSPGDATSGDATADDLSSGDPTSSDPIPGEPVPVEPISGDCAADDIASSEITSVDLASGAPASTDPASDDLASGDLSSSELASDDLA TGELASDELTSESTFDRTFEPKSVPVCEPVPEIDNIEPSSNKDDDFLEKNGADEKLEQIQSKDSLDEKNKADNNIDANEETLETDDTTIC SDRPPENEKKVEEDIITELALGEDAISSSMEIDQGEKNEDETSADLVETINENVIEDNKSENILENTDSMETDEIIPILEKLAPSEDELT CFSKTSLLPIDETNPDLEEKMESSFGSPSKQESSESLPKEAFLVLSDEEDISGEKDESEVISQNETCSPAEVESNEKDNKPEEEEQVIHE DDERPSEKNEFSRRKRSKSEDMDNVQSKRRRYMEEEYEAEFQVKITAKGDINQKLQKVIQWLLEEKLCALQCAVFDKTLAELKTRVEKIE CNKRHKTVLTELQAKIARLTKRFEAAKEDLKKRHEHPPNPPVSPGKTVNDVNSNNNMSYRNAGTVRQMLESKRNVSESAPPSFQTPVNTV SSTNLVTPPAVVSSQPKLQTPVTSGSLTATSVLPAPNTATVVATTQVPSGNPQPTISLQPLPVILHVPVAVSSQPQLLQSHPGTLVTNQP SGNVEFISVQSPPTVSGLTKNPVSLPSLPNPTKPNNVPSVPSPSIQRNPTASAAPLGTTLAVQAVPTAHSIVQATRTSLPTVGPSGLYSP STNRGPIQMKIPISAFSTSSAAEQNSNTTPRIENQTNKTIDASVSKKAADSTSQCGKATGSDSSGVIDLTMDDEESGASQDPKKLNHTPV STMSSSQPVSRPLQPIQPAPPLQPSGVPTSGPSQTTIHLLPTAPTTVNVTHRPVTQVTTRLPVPRAPANHQVVYTTLPAPPAQAPLRGTV MQAPAVRQVNPQNTLPFKVVVISAILALVVLTIISLIILIMLWQKKPRYEIRWKVIESVSSDGHEYIYVDPMQLPYDSTWELPRDQLVLG RTLGSGAFGQVVEATAHGLSHSQATMKVAVKMLKSTARSSEKQALMSELKIMSHLGPHLNVVNLLGACTKGGPIYIITEYCRYGDLVDYL HRNKHTFLQHHSDKRRPPSAELYSNALPVGLPLPSHVSLTGESDGGYMDMSKDESVDYVPMLDMKGDVKYADIESSNYMAPYDNYVPSAP ERTCRATLINESPVLSYMDLVGFSYQVANGMEFLASKNCVHRDLAARNVLICEGKLVKICDFGLARDIMRDSNYISKGSTFLPLKWMAPE SIFNSLYTTLSDVWSFGILLWEIFTLGGTPYPELPMNEQFYNAIKRGYRMAQPAHASDEIYEIMQKCWEEKFEIRPPFSQLVLLLERLLG EGYKKKYQQVDEEFLRSDHPAILRSQARLPGFHGLRSPLDTSSVLYTAVQPNEGDNDYIIPLPDPKPEVADEGPLEGSPSLASSTLNEVN -------------------------------------------------------------- >7486_7486_2_ATF7IP-PDGFRB_ATF7IP_chr12_14634119_ENST00000536444_PDGFRB_chr5_149506177_ENST00000261799_length(amino acids)=1672AA_BP=1093 MDSLEEPQKKVFKARKTMRVSDRQQLEAVYKVKEELLKTDVKLLNGNHENGDLDPTSPLENMDYIKDKEEVNGIEEICFDPEGSKAEWKE TPCILSVNVKNKQDDDLNCEPLSPHNITPEPVSKLPAEPVSGDPAPGDLDAGDPASGVLASGDSTSGDPTSSEPSSSDAASGDATSGDAP SGDVSPGDATSGDATADDLSSGDPTSSDPIPGEPVPVEPISGDCAADDIASSEITSVDLASGAPASTDPASDDLASGDLSSSELASDDLA TGELASDELTSESTFDRTFEPKSVPVCEPVPEIDNIEPSSNKDDDFLEKNGADEKLEQIQSKDSLDEKNKADNNIDANEETLETDDTTIC SDRPPENEKKVEEDIITELALGEDAISSSMEIDQGEKNEDETSADLVETINENVIEDNKSENILENTDSMETDEIIPILEKLAPSEDELT CFSKTSLLPIDETNPDLEEKMESSFGSPSKQESSESLPKEAFLVLSDEEDISGEKDESEVISQNETCSPEVESNEKDNKPEEEEQVIHED DERPSEKNEFSRRKRSKSEDMDNVQSKRRRYMEEEYEAEFQVKITAKGDINQKLQKVIQWLLEEKLCALQCAVFDKTLAELKTRVEKIEC NKRHKTVLTELQAKIARLTKRFEAAKEDLKKRHEHPPNPPVSPGKTVNDVNSNNNMSYRNAGTVRQMLESKRNVSESAPPSFQTPVNTVS STNLVTPPAVVSSQPKLQTPVTSGSLTATSVLPAPNTATVVATTQVPSGNPQPTISLQPLPVILHVPVAVSSQPQLLQSHPGTLVTNQPS GNVEFISVQSPPTVSGLTKNPVSLPSLPNPTKPNNVPSVPSPSIQRNPTASAAPLGTTLAVQAVPTAHSIVQATRTSLPTVGPSGLYSPS TNRGPIQMKIPISAFSTSSAAEQNSNTTPRIENQTNKTIDASVSKKAADSTSQCGKATGSDSSGVIDLTMDDEESGASQDPKKLNHTPVS TMSSSQPVSRPLQPIQPAPPLQPSGVPTSGPSQTTIHLLPTAPTTVNVTHRPVTQVTTRLPVPRAPANHQVVYTTLPAPPAQAPLRGTVM QAPAVRQVNPQNTLPFKVVVISAILALVVLTIISLIILIMLWQKKPRYEIRWKVIESVSSDGHEYIYVDPMQLPYDSTWELPRDQLVLGR TLGSGAFGQVVEATAHGLSHSQATMKVAVKMLKSTARSSEKQALMSELKIMSHLGPHLNVVNLLGACTKGGPIYIITEYCRYGDLVDYLH RNKHTFLQHHSDKRRPPSAELYSNALPVGLPLPSHVSLTGESDGGYMDMSKDESVDYVPMLDMKGDVKYADIESSNYMAPYDNYVPSAPE RTCRATLINESPVLSYMDLVGFSYQVANGMEFLASKNCVHRDLAARNVLICEGKLVKICDFGLARDIMRDSNYISKGSTFLPLKWMAPES IFNSLYTTLSDVWSFGILLWEIFTLGGTPYPELPMNEQFYNAIKRGYRMAQPAHASDEIYEIMQKCWEEKFEIRPPFSQLVLLLERLLGE GYKKKYQQVDEEFLRSDHPAILRSQARLPGFHGLRSPLDTSSVLYTAVQPNEGDNDYIIPLPDPKPEVADEGPLEGSPSLASSTLNEVNT -------------------------------------------------------------- >7486_7486_3_ATF7IP-PDGFRB_ATF7IP_chr12_14634119_ENST00000540793_PDGFRB_chr5_149506177_ENST00000261799_length(amino acids)=1673AA_BP=1094 MDSLEEPQKKVFKARKTMRVSDRQQLEAVYKVKEELLKTDVKLLNGNHENGDLDPTSPLENMDYIKDKEEVNGIEEICFDPEGSKAEWKE TPCILSVNVKNKQDDDLNCEPLSPHNITPEPVSKLPAEPVSGDPAPGDLDAGDPASGVLASGDSTSGDPTSSEPSSSDAASGDATSGDAP SGDVSPGDATSGDATADDLSSGDPTSSDPIPGEPVPVEPISGDCAADDIASSEITSVDLASGAPASTDPASDDLASGDLSSSELASDDLA TGELASDELTSESTFDRTFEPKSVPVCEPVPEIDNIEPSSNKDDDFLEKNGADEKLEQIQSKDSLDEKNKADNNIDANEETLETDDTTIC SDRPPENEKKVEEDIITELALGEDAISSSMEIDQGEKNEDETSADLVETINENVIEDNKSENILENTDSMETDEIIPILEKLAPSEDELT CFSKTSLLPIDETNPDLEEKMESSFGSPSKQESSESLPKEAFLVLSDEEDISGEKDESEVISQNETCSPAEVESNEKDNKPEEEEQVIHE DDERPSEKNEFSRRKRSKSEDMDNVQSKRRRYMEEEYEAEFQVKITAKGDINQKLQKVIQWLLEEKLCALQCAVFDKTLAELKTRVEKIE CNKRHKTVLTELQAKIARLTKRFEAAKEDLKKRHEHPPNPPVSPGKTVNDVNSNNNMSYRNAGTVRQMLESKRNVSESAPPSFQTPVNTV SSTNLVTPPAVVSSQPKLQTPVTSGSLTATSVLPAPNTATVVATTQVPSGNPQPTISLQPLPVILHVPVAVSSQPQLLQSHPGTLVTNQP SGNVEFISVQSPPTVSGLTKNPVSLPSLPNPTKPNNVPSVPSPSIQRNPTASAAPLGTTLAVQAVPTAHSIVQATRTSLPTVGPSGLYSP STNRGPIQMKIPISAFSTSSAAEQNSNTTPRIENQTNKTIDASVSKKAADSTSQCGKATGSDSSGVIDLTMDDEESGASQDPKKLNHTPV STMSSSQPVSRPLQPIQPAPPLQPSGVPTSGPSQTTIHLLPTAPTTVNVTHRPVTQVTTRLPVPRAPANHQVVYTTLPAPPAQAPLRGTV MQAPAVRQVNPQNTLPFKVVVISAILALVVLTIISLIILIMLWQKKPRYEIRWKVIESVSSDGHEYIYVDPMQLPYDSTWELPRDQLVLG RTLGSGAFGQVVEATAHGLSHSQATMKVAVKMLKSTARSSEKQALMSELKIMSHLGPHLNVVNLLGACTKGGPIYIITEYCRYGDLVDYL HRNKHTFLQHHSDKRRPPSAELYSNALPVGLPLPSHVSLTGESDGGYMDMSKDESVDYVPMLDMKGDVKYADIESSNYMAPYDNYVPSAP ERTCRATLINESPVLSYMDLVGFSYQVANGMEFLASKNCVHRDLAARNVLICEGKLVKICDFGLARDIMRDSNYISKGSTFLPLKWMAPE SIFNSLYTTLSDVWSFGILLWEIFTLGGTPYPELPMNEQFYNAIKRGYRMAQPAHASDEIYEIMQKCWEEKFEIRPPFSQLVLLLERLLG EGYKKKYQQVDEEFLRSDHPAILRSQARLPGFHGLRSPLDTSSVLYTAVQPNEGDNDYIIPLPDPKPEVADEGPLEGSPSLASSTLNEVN -------------------------------------------------------------- >7486_7486_4_ATF7IP-PDGFRB_ATF7IP_chr12_14634119_ENST00000544627_PDGFRB_chr5_149506177_ENST00000261799_length(amino acids)=1681AA_BP=1102 MHQDQRFRMDSLEEPQKKVFKARKTMRVSDRQQLEAVYKVKEELLKTDVKLLNGNHENGDLDPTSPLENMDYIKDKEEVNGIEEICFDPE GSKAEWKETPCILSVNVKNKQDDDLNCEPLSPHNITPEPVSKLPAEPVSGDPAPGDLDAGDPASGVLASGDSTSGDPTSSEPSSSDAASG DATSGDAPSGDVSPGDATSGDATADDLSSGDPTSSDPIPGEPVPVEPISGDCAADDIASSEITSVDLASGAPASTDPASDDLASGDLSSS ELASDDLATGELASDELTSESTFDRTFEPKSVPVCEPVPEIDNIEPSSNKDDDFLEKNGADEKLEQIQSKDSLDEKNKADNNIDANEETL ETDDTTICSDRPPENEKKVEEDIITELALGEDAISSSMEIDQGEKNEDETSADLVETINENVIEDNKSENILENTDSMETDEIIPILEKL APSEDELTCFSKTSLLPIDETNPDLEEKMESSFGSPSKQESSESLPKEAFLVLSDEEDISGEKDESEVISQNETCSPAEVESNEKDNKPE EEEQVIHEDDERPSEKNEFSRRKRSKSEDMDNVQSKRRRYMEEEYEAEFQVKITAKGDINQKLQKVIQWLLEEKLCALQCAVFDKTLAEL KTRVEKIECNKRHKTVLTELQAKIARLTKRFEAAKEDLKKRHEHPPNPPVSPGKTVNDVNSNNNMSYRNAGTVRQMLESKRNVSESAPPS FQTPVNTVSSTNLVTPPAVVSSQPKLQTPVTSGSLTATSVLPAPNTATVVATTQVPSGNPQPTISLQPLPVILHVPVAVSSQPQLLQSHP GTLVTNQPSGNVEFISVQSPPTVSGLTKNPVSLPSLPNPTKPNNVPSVPSPSIQRNPTASAAPLGTTLAVQAVPTAHSIVQATRTSLPTV GPSGLYSPSTNRGPIQMKIPISAFSTSSAAEQNSNTTPRIENQTNKTIDASVSKKAADSTSQCGKATGSDSSGVIDLTMDDEESGASQDP KKLNHTPVSTMSSSQPVSRPLQPIQPAPPLQPSGVPTSGPSQTTIHLLPTAPTTVNVTHRPVTQVTTRLPVPRAPANHQVVYTTLPAPPA QAPLRGTVMQAPAVRQVNPQNTLPFKVVVISAILALVVLTIISLIILIMLWQKKPRYEIRWKVIESVSSDGHEYIYVDPMQLPYDSTWEL PRDQLVLGRTLGSGAFGQVVEATAHGLSHSQATMKVAVKMLKSTARSSEKQALMSELKIMSHLGPHLNVVNLLGACTKGGPIYIITEYCR YGDLVDYLHRNKHTFLQHHSDKRRPPSAELYSNALPVGLPLPSHVSLTGESDGGYMDMSKDESVDYVPMLDMKGDVKYADIESSNYMAPY DNYVPSAPERTCRATLINESPVLSYMDLVGFSYQVANGMEFLASKNCVHRDLAARNVLICEGKLVKICDFGLARDIMRDSNYISKGSTFL PLKWMAPESIFNSLYTTLSDVWSFGILLWEIFTLGGTPYPELPMNEQFYNAIKRGYRMAQPAHASDEIYEIMQKCWEEKFEIRPPFSQLV LLLERLLGEGYKKKYQQVDEEFLRSDHPAILRSQARLPGFHGLRSPLDTSSVLYTAVQPNEGDNDYIIPLPDPKPEVADEGPLEGSPSLA -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr12:14634119/chr5:149506177) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ATF7IP | PDGFRB |
| FUNCTION: Recruiter that couples transcriptional factors to general transcription apparatus and thereby modulates transcription regulation and chromatin formation. Can both act as an activator or a repressor depending on the context. Mediates MBD1-dependent transcriptional repression, probably by recruiting complexes containing SETDB1. The complex formed with MBD1 and SETDB1 represses transcription and probably couples DNA methylation and histone H3 'Lys-9' trimethylation (H3K9me3) activity (Probable). {ECO:0000305}. | FUNCTION: Tyrosine-protein kinase that acts as cell-surface receptor for homodimeric PDGFB and PDGFD and for heterodimers formed by PDGFA and PDGFB, and plays an essential role in the regulation of embryonic development, cell proliferation, survival, differentiation, chemotaxis and migration. Plays an essential role in blood vessel development by promoting proliferation, migration and recruitment of pericytes and smooth muscle cells to endothelial cells. Plays a role in the migration of vascular smooth muscle cells and the formation of neointima at vascular injury sites. Required for normal development of the cardiovascular system. Required for normal recruitment of pericytes (mesangial cells) in the kidney glomerulus, and for normal formation of a branched network of capillaries in kidney glomeruli. Promotes rearrangement of the actin cytoskeleton and the formation of membrane ruffles. Binding of its cognate ligands - homodimeric PDGFB, heterodimers formed by PDGFA and PDGFB or homodimeric PDGFD -leads to the activation of several signaling cascades; the response depends on the nature of the bound ligand and is modulated by the formation of heterodimers between PDGFRA and PDGFRB. Phosphorylates PLCG1, PIK3R1, PTPN11, RASA1/GAP, CBL, SHC1 and NCK1. Activation of PLCG1 leads to the production of the cellular signaling molecules diacylglycerol and inositol 1,4,5-trisphosphate, mobilization of cytosolic Ca(2+) and the activation of protein kinase C. Phosphorylation of PIK3R1, the regulatory subunit of phosphatidylinositol 3-kinase, leads to the activation of the AKT1 signaling pathway. Phosphorylation of SHC1, or of the C-terminus of PTPN11, creates a binding site for GRB2, resulting in the activation of HRAS, RAF1 and down-stream MAP kinases, including MAPK1/ERK2 and/or MAPK3/ERK1. Promotes phosphorylation and activation of SRC family kinases. Promotes phosphorylation of PDCD6IP/ALIX and STAM. Receptor signaling is down-regulated by protein phosphatases that dephosphorylate the receptor and its down-stream effectors, and by rapid internalization of the activated receptor. {ECO:0000269|PubMed:11297552, ECO:0000269|PubMed:11331881, ECO:0000269|PubMed:1314164, ECO:0000269|PubMed:1396585, ECO:0000269|PubMed:1653029, ECO:0000269|PubMed:1709159, ECO:0000269|PubMed:1846866, ECO:0000269|PubMed:20494825, ECO:0000269|PubMed:20529858, ECO:0000269|PubMed:21098708, ECO:0000269|PubMed:21679854, ECO:0000269|PubMed:21733313, ECO:0000269|PubMed:2554309, ECO:0000269|PubMed:26599395, ECO:0000269|PubMed:2835772, ECO:0000269|PubMed:2850496, ECO:0000269|PubMed:7685273, ECO:0000269|PubMed:7691811, ECO:0000269|PubMed:7692233, ECO:0000269|PubMed:8195171}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ATF7IP | chr12:14634119 | chr5:149506177 | ENST00000261168 | + | 13 | 15 | 617_665 | 1093.3333333333333 | 1271.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | ATF7IP | chr12:14634119 | chr5:149506177 | ENST00000540793 | + | 12 | 14 | 617_665 | 1093.3333333333333 | 1271.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | ATF7IP | chr12:14634119 | chr5:149506177 | ENST00000261168 | + | 13 | 15 | 349_580 | 1093.3333333333333 | 1271.0 | Compositional bias | Note=Glu-rich |
| Hgene | ATF7IP | chr12:14634119 | chr5:149506177 | ENST00000540793 | + | 12 | 14 | 349_580 | 1093.3333333333333 | 1271.0 | Compositional bias | Note=Glu-rich |
| Hgene | ATF7IP | chr12:14634119 | chr5:149506177 | ENST00000261168 | + | 13 | 15 | 553_571 | 1093.3333333333333 | 1271.0 | Motif | Nuclear localization signal |
| Hgene | ATF7IP | chr12:14634119 | chr5:149506177 | ENST00000540793 | + | 12 | 14 | 553_571 | 1093.3333333333333 | 1271.0 | Motif | Nuclear localization signal |
| Tgene | PDGFRB | chr12:14634119 | chr5:149506177 | ENST00000261799 | 9 | 23 | 600_962 | 526.3333333333334 | 1107.0 | Domain | Protein kinase | |
| Tgene | PDGFRB | chr12:14634119 | chr5:149506177 | ENST00000261799 | 9 | 23 | 606_614 | 526.3333333333334 | 1107.0 | Nucleotide binding | ATP | |
| Tgene | PDGFRB | chr12:14634119 | chr5:149506177 | ENST00000261799 | 9 | 23 | 554_1106 | 526.3333333333334 | 1107.0 | Topological domain | Cytoplasmic | |
| Tgene | PDGFRB | chr12:14634119 | chr5:149506177 | ENST00000261799 | 9 | 23 | 533_553 | 526.3333333333334 | 1107.0 | Transmembrane | Helical |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ATF7IP | chr12:14634119 | chr5:149506177 | ENST00000543189 | + | 1 | 13 | 617_665 | 0 | 1106.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | ATF7IP | chr12:14634119 | chr5:149506177 | ENST00000543189 | + | 1 | 13 | 349_580 | 0 | 1106.0 | Compositional bias | Note=Glu-rich |
| Hgene | ATF7IP | chr12:14634119 | chr5:149506177 | ENST00000261168 | + | 13 | 15 | 1160_1270 | 1093.3333333333333 | 1271.0 | Domain | Fibronectin type-III |
| Hgene | ATF7IP | chr12:14634119 | chr5:149506177 | ENST00000540793 | + | 12 | 14 | 1160_1270 | 1093.3333333333333 | 1271.0 | Domain | Fibronectin type-III |
| Hgene | ATF7IP | chr12:14634119 | chr5:149506177 | ENST00000543189 | + | 1 | 13 | 1160_1270 | 0 | 1106.0 | Domain | Fibronectin type-III |
| Hgene | ATF7IP | chr12:14634119 | chr5:149506177 | ENST00000543189 | + | 1 | 13 | 553_571 | 0 | 1106.0 | Motif | Nuclear localization signal |
| Tgene | PDGFRB | chr12:14634119 | chr5:149506177 | ENST00000261799 | 9 | 23 | 129_210 | 526.3333333333334 | 1107.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | PDGFRB | chr12:14634119 | chr5:149506177 | ENST00000261799 | 9 | 23 | 214_309 | 526.3333333333334 | 1107.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | PDGFRB | chr12:14634119 | chr5:149506177 | ENST00000261799 | 9 | 23 | 331_403 | 526.3333333333334 | 1107.0 | Domain | Note=Ig-like C2-type 4 | |
| Tgene | PDGFRB | chr12:14634119 | chr5:149506177 | ENST00000261799 | 9 | 23 | 33_120 | 526.3333333333334 | 1107.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | PDGFRB | chr12:14634119 | chr5:149506177 | ENST00000261799 | 9 | 23 | 416_524 | 526.3333333333334 | 1107.0 | Domain | Note=Ig-like C2-type 5 | |
| Tgene | PDGFRB | chr12:14634119 | chr5:149506177 | ENST00000261799 | 9 | 23 | 33_532 | 526.3333333333334 | 1107.0 | Topological domain | Extracellular |
Top |
Fusion Protein Structures |
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
Top |
pLDDT score distribution |
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
ATF7IP_pLDDT.png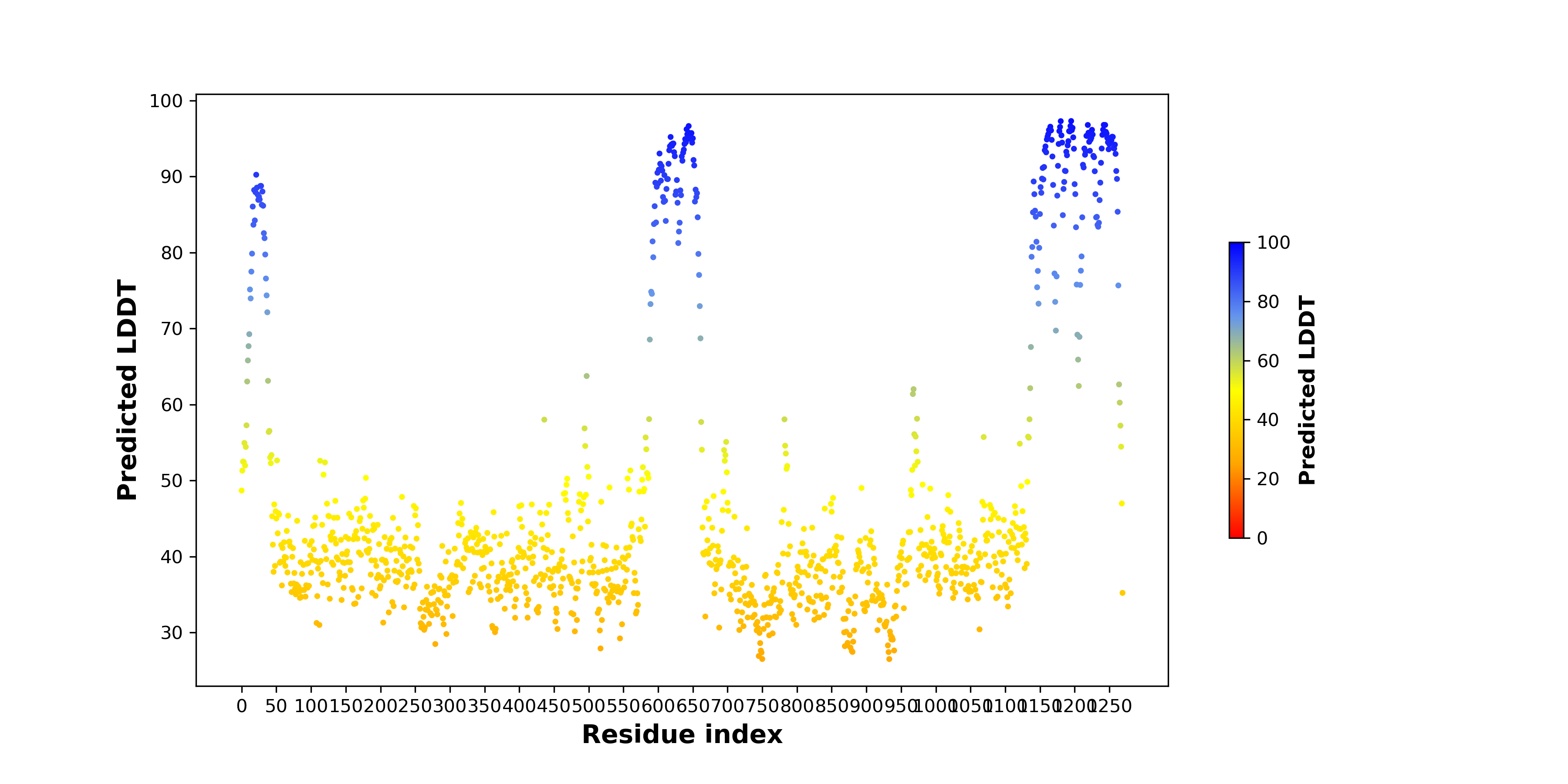 |
PDGFRB_pLDDT.png |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
Top |
Ramachandran Plot of Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| Fusion AA seq ID in FusionPDB and their Ramachandran plots |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| ATF7IP | |
| PDGFRB |  |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | ATF7IP | chr12:14634119 | chr5:149506177 | ENST00000261168 | + | 13 | 15 | 1154_1270 | 1093.3333333333333 | 1271.0 | MBD1 |
| Hgene | ATF7IP | chr12:14634119 | chr5:149506177 | ENST00000540793 | + | 12 | 14 | 1154_1270 | 1093.3333333333333 | 1271.0 | MBD1 |
| Hgene | ATF7IP | chr12:14634119 | chr5:149506177 | ENST00000543189 | + | 1 | 13 | 1154_1270 | 0 | 1106.0 | MBD1 |
| Hgene | ATF7IP | chr12:14634119 | chr5:149506177 | ENST00000543189 | + | 1 | 13 | 562_817 | 0 | 1106.0 | SETDB1 |
| Hgene | ATF7IP | chr12:14634119 | chr5:149506177 | ENST00000543189 | + | 1 | 13 | 965_975 | 0 | 1106.0 | SUMO |
Top |
Related Drugs to ATF7IP-PDGFRB |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to ATF7IP-PDGFRB |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Tgene | PDGFRB | C3554321 | BASAL GANGLIA CALCIFICATION, IDIOPATHIC, 4 | 6 | CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Tgene | PDGFRB | C0393590 | Fahr's syndrome (disorder) | 3 | GENOMICS_ENGLAND;ORPHANET |
| Tgene | PDGFRB | C4225270 | Kosaki overgrowth syndrome | 3 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Tgene | PDGFRB | C4551572 | MYOFIBROMATOSIS, INFANTILE, 1 | 3 | GENOMICS_ENGLAND;UNIPROT |
| Tgene | PDGFRB | C0013421 | Dystonia | 2 | GENOMICS_ENGLAND |
| Tgene | PDGFRB | C0023480 | Leukemia, Myelomonocytic, Chronic | 2 | ORPHANET |
| Tgene | PDGFRB | C0023893 | Liver Cirrhosis, Experimental | 2 | CTD_human |
| Tgene | PDGFRB | C0036341 | Schizophrenia | 2 | PSYGENET |
| Tgene | PDGFRB | C0432284 | Infantile myofibromatosis | 2 | CTD_human;GENOMICS_ENGLAND;ORPHANET |
| Tgene | PDGFRB | C0004782 | Basal Ganglia Diseases | 1 | CTD_human |
| Tgene | PDGFRB | C0006663 | Calcinosis | 1 | CTD_human |
| Tgene | PDGFRB | C0015371 | Extrapyramidal Disorders | 1 | CTD_human |
| Tgene | PDGFRB | C0036337 | Schizoaffective Disorder | 1 | PSYGENET |
| Tgene | PDGFRB | C0206648 | Myofibromatosis | 1 | GENOMICS_ENGLAND |
| Tgene | PDGFRB | C0263628 | Tumoral calcinosis | 1 | CTD_human |
| Tgene | PDGFRB | C0521174 | Microcalcification | 1 | CTD_human |
| Tgene | PDGFRB | C0750951 | Lenticulostriate Disorders | 1 | CTD_human |
| Tgene | PDGFRB | C1333046 | Myeloproliferative Neoplasm, Unclassifiable | 1 | ORPHANET |
| Tgene | PDGFRB | C1866182 | Penttinen-Aula syndrome | 1 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Tgene | PDGFRB | C3472621 | Myeloid neoplasm with beta-type platelet-derived growth factor receptor gene rearrangement | 1 | ORPHANET |
| Tgene | PDGFRB | C3714756 | Intellectual Disability | 1 | GENOMICS_ENGLAND |

