| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:B4GALT5-ERBB4 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: B4GALT5-ERBB4 | FusionPDB ID: 8762 | FusionGDB2.0 ID: 8762 | Hgene | Tgene | Gene symbol | B4GALT5 | ERBB4 | Gene ID | 9334 | 2066 |
| Gene name | beta-1,4-galactosyltransferase 5 | erb-b2 receptor tyrosine kinase 4 | |
| Synonyms | B4Gal-T5|BETA4-GALT-IV|beta4Gal-T5|beta4GalT-V|gt-V | ALS19|HER4|p180erbB4 | |
| Cytomap | 20q13.13 | 2q34 | |
| Type of gene | protein-coding | protein-coding | |
| Description | beta-1,4-galactosyltransferase 5UDP-Gal:beta-GlcNAc beta-1,4-galactosyltransferase 5UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 5UDP-galactose:beta-N-acetylglucosamine beta-1,4-galactosyltransferase 5beta-1,4-GalT IIbeta-1,4-GalT I | receptor tyrosine-protein kinase erbB-4avian erythroblastic leukemia viral (v-erb-b2) oncogene homolog 4human epidermal growth factor receptor 4proto-oncogene-like protein c-ErbB-4tyrosine kinase-type cell surface receptor HER4v-erb-a erythroblastic | |
| Modification date | 20200313 | 20200327 | |
| UniProtAcc | O43286 | Q15303 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000371711, | ENST00000484474, ENST00000342788, ENST00000402597, ENST00000436443, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 21 X 14 X 12=3528 | 18 X 16 X 8=2304 |
| # samples | 25 | 17 | |
| ** MAII score | log2(25/3528*10)=-3.81885056089543 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(17/2304*10)=-3.76053406530461 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: B4GALT5 [Title/Abstract] AND ERBB4 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | |||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | ERBB4 | GO:0007165 | signal transduction | 10572067 |
| Tgene | ERBB4 | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | 10353604|18334220 |
| Tgene | ERBB4 | GO:0016477 | cell migration | 9135143 |
| Tgene | ERBB4 | GO:0018108 | peptidyl-tyrosine phosphorylation | 18334220 |
| Tgene | ERBB4 | GO:0046777 | protein autophosphorylation | 18334220 |
 Fusion gene breakpoints across B4GALT5 (5'-gene) Fusion gene breakpoints across B4GALT5 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 Fusion gene breakpoints across ERBB4 (3'-gene) Fusion gene breakpoints across ERBB4 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerKB3 | . | . | B4GALT5 | chr20 | 48330113 | - | ERBB4 | chr2 | 212812341 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000371711 | B4GALT5 | chr20 | 48330113 | - | ENST00000436443 | ERBB4 | chr2 | 212812341 | - | 11842 | 303 | 357 | 3947 | 1196 |
| ENST00000371711 | B4GALT5 | chr20 | 48330113 | - | ENST00000342788 | ERBB4 | chr2 | 212812341 | - | 11890 | 303 | 357 | 3995 | 1212 |
| ENST00000371711 | B4GALT5 | chr20 | 48330113 | - | ENST00000402597 | ERBB4 | chr2 | 212812341 | - | 3966 | 303 | 357 | 3965 | 1202 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >8762_8762_1_B4GALT5-ERBB4_B4GALT5_chr20_48330113_ENST00000371711_ERBB4_chr2_212812341_ENST00000342788_length(amino acids)=1212AA_BP= MPLENLRIIRGTKLYEDRYALAIFLNYRKDGNFGLQELGLKNLTEILNGGVYVDQNKFLCYADTIHWQDIVRNPWPSNLTLVSTNGSSGC GRCHKSCTGRCWGPTENHCQTLTRTVCAEQCDGRCYGPYVSDCCHRECAGGCSGPKDTDCFACMNFNDSGACVTQCPQTFVYNPTTFQLE HNFNAKYTYGAFCVKKCPHNFVVDSSSCVRACPSSKMEVEENGIKMCKPCTDICPKACDGIGTGSLMSAQTVDSSNIDKFINCTKINGNL IFLVTGIHGDPYNAIEAIDPEKLNVFRTVREITGFLNIQSWPPNMTDFSVFSNLVTIGGRVLYSGLSLLILKQQGITSLQFQSLKEISAG NIYITDNSNLCYYHTINWTTLFSTINQRIVIRDNRKAENCTAEGMVCNHLCSSDGCWGPGPDQCLSCRRFSRGRICIESCNLYDGEFREF ENGSICVECDPQCEKMEDGLLTCHGPGPDNCTKCSHFKDGPNCVEKCPDGLQGANSFIFKYADPDRECHPCHPNCTQGCNGPTSHDCIYY PWTGHSTLPQHARTPLIAAGVIGGLFILVIVGLTFAVYVRRKSIKKKRALRRFLETELVEPLTPSGTAPNQAQLRILKETELKRVKVLGS GAFGTVYKGIWVPEGETVKIPVAIKILNETTGPKANVEFMDEALIMASMDHPHLVRLLGVCLSPTIQLVTQLMPHGCLLEYVHEHKDNIG SQLLLNWCVQIAKGMMYLEERRLVHRDLAARNVLVKSPNHVKITDFGLARLLEGDEKEYNADGGKMPIKWMALECIHYRKFTHQSDVWSY GVTIWELMTFGGKPYDGIPTREIPDLLEKGERLPQPPICTIDVYMVMVKCWMIDADSRPKFKELAAEFSRMARDPQRYLVIQGDDRMKLP SPNDSKFFQNLLDEEDLEDMMDAEEYLVPQAFNIPPPIYTSRARIDSNRSEIGHSPPPAYTPMSGNQFVYRDGGFAAEQGVSVPYRAPTS TIPEAPVAQGATAEIFDDSCCNGTLRKPVAPHVQEDSSTQRYSADPTVFAPERSPRGELDEEGYMTPMRDKPKQEYLNPVEENPFVSRRK NGDLQALDNPEYHNASNGPPKAEDEYVNEPLYLNTFANTLGKAEYLKNNILSMPEKAKKAFDNPDYWNHSLPPRSTLQHPDYLQEYSTKY -------------------------------------------------------------- >8762_8762_2_B4GALT5-ERBB4_B4GALT5_chr20_48330113_ENST00000371711_ERBB4_chr2_212812341_ENST00000402597_length(amino acids)=1202AA_BP= MPLENLRIIRGTKLYEDRYALAIFLNYRKDGNFGLQELGLKNLTEILNGGVYVDQNKFLCYADTIHWQDIVRNPWPSNLTLVSTNGSSGC GRCHKSCTGRCWGPTENHCQTLTRTVCAEQCDGRCYGPYVSDCCHRECAGGCSGPKDTDCFACMNFNDSGACVTQCPQTFVYNPTTFQLE HNFNAKYTYGAFCVKKCPHNFVVDSSSCVRACPSSKMEVEENGIKMCKPCTDICPKACDGIGTGSLMSAQTVDSSNIDKFINCTKINGNL IFLVTGIHGDPYNAIEAIDPEKLNVFRTVREITGFLNIQSWPPNMTDFSVFSNLVTIGGRVLYSGLSLLILKQQGITSLQFQSLKEISAG NIYITDNSNLCYYHTINWTTLFSTINQRIVIRDNRKAENCTAEGMVCNHLCSSDGCWGPGPDQCLSCRRFSRGRICIESCNLYDGEFREF ENGSICVECDPQCEKMEDGLLTCHGPGPDNCTKCSHFKDGPNCVEKCPDGLQGANSFIFKYADPDRECHPCHPNCTQGCIGSSIEDCIGL MDRTPLIAAGVIGGLFILVIVGLTFAVYVRRKSIKKKRALRRFLETELVEPLTPSGTAPNQAQLRILKETELKRVKVLGSGAFGTVYKGI WVPEGETVKIPVAIKILNETTGPKANVEFMDEALIMASMDHPHLVRLLGVCLSPTIQLVTQLMPHGCLLEYVHEHKDNIGSQLLLNWCVQ IAKGMMYLEERRLVHRDLAARNVLVKSPNHVKITDFGLARLLEGDEKEYNADGGKMPIKWMALECIHYRKFTHQSDVWSYGVTIWELMTF GGKPYDGIPTREIPDLLEKGERLPQPPICTIDVYMVMVKCWMIDADSRPKFKELAAEFSRMARDPQRYLVIQGDDRMKLPSPNDSKFFQN LLDEEDLEDMMDAEEYLVPQAFNIPPPIYTSRARIDSNRSEIGHSPPPAYTPMSGNQFVYRDGGFAAEQGVSVPYRAPTSTIPEAPVAQG ATAEIFDDSCCNGTLRKPVAPHVQEDSSTQRYSADPTVFAPERSPRGELDEEGYMTPMRDKPKQEYLNPVEENPFVSRRKNGDLQALDNP EYHNASNGPPKAEDEYVNEPLYLNTFANTLGKAEYLKNNILSMPEKAKKAFDNPDYWNHSLPPRSTLQHPDYLQEYSTKYFYKQNGRIRP -------------------------------------------------------------- >8762_8762_3_B4GALT5-ERBB4_B4GALT5_chr20_48330113_ENST00000371711_ERBB4_chr2_212812341_ENST00000436443_length(amino acids)=1196AA_BP= MPLENLRIIRGTKLYEDRYALAIFLNYRKDGNFGLQELGLKNLTEILNGGVYVDQNKFLCYADTIHWQDIVRNPWPSNLTLVSTNGSSGC GRCHKSCTGRCWGPTENHCQTLTRTVCAEQCDGRCYGPYVSDCCHRECAGGCSGPKDTDCFACMNFNDSGACVTQCPQTFVYNPTTFQLE HNFNAKYTYGAFCVKKCPHNFVVDSSSCVRACPSSKMEVEENGIKMCKPCTDICPKACDGIGTGSLMSAQTVDSSNIDKFINCTKINGNL IFLVTGIHGDPYNAIEAIDPEKLNVFRTVREITGFLNIQSWPPNMTDFSVFSNLVTIGGRVLYSGLSLLILKQQGITSLQFQSLKEISAG NIYITDNSNLCYYHTINWTTLFSTINQRIVIRDNRKAENCTAEGMVCNHLCSSDGCWGPGPDQCLSCRRFSRGRICIESCNLYDGEFREF ENGSICVECDPQCEKMEDGLLTCHGPGPDNCTKCSHFKDGPNCVEKCPDGLQGANSFIFKYADPDRECHPCHPNCTQGCNGPTSHDCIYY PWTGHSTLPQHARTPLIAAGVIGGLFILVIVGLTFAVYVRRKSIKKKRALRRFLETELVEPLTPSGTAPNQAQLRILKETELKRVKVLGS GAFGTVYKGIWVPEGETVKIPVAIKILNETTGPKANVEFMDEALIMASMDHPHLVRLLGVCLSPTIQLVTQLMPHGCLLEYVHEHKDNIG SQLLLNWCVQIAKGMMYLEERRLVHRDLAARNVLVKSPNHVKITDFGLARLLEGDEKEYNADGGKMPIKWMALECIHYRKFTHQSDVWSY GVTIWELMTFGGKPYDGIPTREIPDLLEKGERLPQPPICTIDVYMVMVKCWMIDADSRPKFKELAAEFSRMARDPQRYLVIQGDDRMKLP SPNDSKFFQNLLDEEDLEDMMDAEEYLVPQAFNIPPPIYTSRARIDSNRNQFVYRDGGFAAEQGVSVPYRAPTSTIPEAPVAQGATAEIF DDSCCNGTLRKPVAPHVQEDSSTQRYSADPTVFAPERSPRGELDEEGYMTPMRDKPKQEYLNPVEENPFVSRRKNGDLQALDNPEYHNAS NGPPKAEDEYVNEPLYLNTFANTLGKAEYLKNNILSMPEKAKKAFDNPDYWNHSLPPRSTLQHPDYLQEYSTKYFYKQNGRIRPIVAENP -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr20:/chr2:) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| B4GALT5 | ERBB4 |
| FUNCTION: Catalyzes the synthesis of lactosylceramide (LacCer) via the transfer of galactose from UDP-galactose to glucosylceramide (GlcCer) (PubMed:24498430). LacCer is the starting point in the biosynthesis of all gangliosides (membrane-bound glycosphingolipids) which play pivotal roles in the CNS including neuronal maturation and axonal and myelin formation (By similarity). Plays a role in the glycosylation of BMPR1A and regulation of its protein stability (By similarity). Essential for extraembryonic development during early embryogenesis (By similarity). {ECO:0000250|UniProtKB:Q9JMK0, ECO:0000269|PubMed:24498430}. | FUNCTION: Tyrosine-protein kinase that plays an essential role as cell surface receptor for neuregulins and EGF family members and regulates development of the heart, the central nervous system and the mammary gland, gene transcription, cell proliferation, differentiation, migration and apoptosis. Required for normal cardiac muscle differentiation during embryonic development, and for postnatal cardiomyocyte proliferation. Required for normal development of the embryonic central nervous system, especially for normal neural crest cell migration and normal axon guidance. Required for mammary gland differentiation, induction of milk proteins and lactation. Acts as cell-surface receptor for the neuregulins NRG1, NRG2, NRG3 and NRG4 and the EGF family members BTC, EREG and HBEGF. Ligand binding triggers receptor dimerization and autophosphorylation at specific tyrosine residues that then serve as binding sites for scaffold proteins and effectors. Ligand specificity and signaling is modulated by alternative splicing, proteolytic processing, and by the formation of heterodimers with other ERBB family members, thereby creating multiple combinations of intracellular phosphotyrosines that trigger ligand- and context-specific cellular responses. Mediates phosphorylation of SHC1 and activation of the MAP kinases MAPK1/ERK2 and MAPK3/ERK1. Isoform JM-A CYT-1 and isoform JM-B CYT-1 phosphorylate PIK3R1, leading to the activation of phosphatidylinositol 3-kinase and AKT1 and protect cells against apoptosis. Isoform JM-A CYT-1 and isoform JM-B CYT-1 mediate reorganization of the actin cytoskeleton and promote cell migration in response to NRG1. Isoform JM-A CYT-2 and isoform JM-B CYT-2 lack the phosphotyrosine that mediates interaction with PIK3R1, and hence do not phosphorylate PIK3R1, do not protect cells against apoptosis, and do not promote reorganization of the actin cytoskeleton and cell migration. Proteolytic processing of isoform JM-A CYT-1 and isoform JM-A CYT-2 gives rise to the corresponding soluble intracellular domains (4ICD) that translocate to the nucleus, promote nuclear import of STAT5A, activation of STAT5A, mammary epithelium differentiation, cell proliferation and activation of gene expression. The ERBB4 soluble intracellular domains (4ICD) colocalize with STAT5A at the CSN2 promoter to regulate transcription of milk proteins during lactation. The ERBB4 soluble intracellular domains can also translocate to mitochondria and promote apoptosis. {ECO:0000269|PubMed:10348342, ECO:0000269|PubMed:10353604, ECO:0000269|PubMed:10358079, ECO:0000269|PubMed:10722704, ECO:0000269|PubMed:10867024, ECO:0000269|PubMed:11178955, ECO:0000269|PubMed:11390655, ECO:0000269|PubMed:12807903, ECO:0000269|PubMed:15534001, ECO:0000269|PubMed:15746097, ECO:0000269|PubMed:16251361, ECO:0000269|PubMed:16778220, ECO:0000269|PubMed:16837552, ECO:0000269|PubMed:17486069, ECO:0000269|PubMed:17638867, ECO:0000269|PubMed:19098003, ECO:0000269|PubMed:20858735, ECO:0000269|PubMed:8383326, ECO:0000269|PubMed:8617750, ECO:0000269|PubMed:9135143, ECO:0000269|PubMed:9168115, ECO:0000269|PubMed:9334263}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
Top |
Fusion Protein Structures |
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
| PDB file (847) >>>847.pdbFusion protein BP residue: CIF file (847) >>>847.cif | B4GALT5 | chr20 | 48330113 | - | ERBB4 | chr2 | 212812341 | - | MPLENLRIIRGTKLYEDRYALAIFLNYRKDGNFGLQELGLKNLTEILNGG VYVDQNKFLCYADTIHWQDIVRNPWPSNLTLVSTNGSSGCGRCHKSCTGR CWGPTENHCQTLTRTVCAEQCDGRCYGPYVSDCCHRECAGGCSGPKDTDC FACMNFNDSGACVTQCPQTFVYNPTTFQLEHNFNAKYTYGAFCVKKCPHN FVVDSSSCVRACPSSKMEVEENGIKMCKPCTDICPKACDGIGTGSLMSAQ TVDSSNIDKFINCTKINGNLIFLVTGIHGDPYNAIEAIDPEKLNVFRTVR EITGFLNIQSWPPNMTDFSVFSNLVTIGGRVLYSGLSLLILKQQGITSLQ FQSLKEISAGNIYITDNSNLCYYHTINWTTLFSTINQRIVIRDNRKAENC TAEGMVCNHLCSSDGCWGPGPDQCLSCRRFSRGRICIESCNLYDGEFREF ENGSICVECDPQCEKMEDGLLTCHGPGPDNCTKCSHFKDGPNCVEKCPDG LQGANSFIFKYADPDRECHPCHPNCTQGCNGPTSHDCIYYPWTGHSTLPQ HARTPLIAAGVIGGLFILVIVGLTFAVYVRRKSIKKKRALRRFLETELVE PLTPSGTAPNQAQLRILKETELKRVKVLGSGAFGTVYKGIWVPEGETVKI PVAIKILNETTGPKANVEFMDEALIMASMDHPHLVRLLGVCLSPTIQLVT QLMPHGCLLEYVHEHKDNIGSQLLLNWCVQIAKGMMYLEERRLVHRDLAA RNVLVKSPNHVKITDFGLARLLEGDEKEYNADGGKMPIKWMALECIHYRK FTHQSDVWSYGVTIWELMTFGGKPYDGIPTREIPDLLEKGERLPQPPICT IDVYMVMVKCWMIDADSRPKFKELAAEFSRMARDPQRYLVIQGDDRMKLP SPNDSKFFQNLLDEEDLEDMMDAEEYLVPQAFNIPPPIYTSRARIDSNRN QFVYRDGGFAAEQGVSVPYRAPTSTIPEAPVAQGATAEIFDDSCCNGTLR KPVAPHVQEDSSTQRYSADPTVFAPERSPRGELDEEGYMTPMRDKPKQEY LNPVEENPFVSRRKNGDLQALDNPEYHNASNGPPKAEDEYVNEPLYLNTF ANTLGKAEYLKNNILSMPEKAKKAFDNPDYWNHSLPPRSTLQHPDYLQEY | 1196 |
| 3D view using mol* of 847 (AA BP:) | ||||||||||
 | ||||||||||
| PDB file (851) >>>851.pdbFusion protein BP residue: CIF file (851) >>>851.cif | B4GALT5 | chr20 | 48330113 | - | ERBB4 | chr2 | 212812341 | - | MPLENLRIIRGTKLYEDRYALAIFLNYRKDGNFGLQELGLKNLTEILNGG VYVDQNKFLCYADTIHWQDIVRNPWPSNLTLVSTNGSSGCGRCHKSCTGR CWGPTENHCQTLTRTVCAEQCDGRCYGPYVSDCCHRECAGGCSGPKDTDC FACMNFNDSGACVTQCPQTFVYNPTTFQLEHNFNAKYTYGAFCVKKCPHN FVVDSSSCVRACPSSKMEVEENGIKMCKPCTDICPKACDGIGTGSLMSAQ TVDSSNIDKFINCTKINGNLIFLVTGIHGDPYNAIEAIDPEKLNVFRTVR EITGFLNIQSWPPNMTDFSVFSNLVTIGGRVLYSGLSLLILKQQGITSLQ FQSLKEISAGNIYITDNSNLCYYHTINWTTLFSTINQRIVIRDNRKAENC TAEGMVCNHLCSSDGCWGPGPDQCLSCRRFSRGRICIESCNLYDGEFREF ENGSICVECDPQCEKMEDGLLTCHGPGPDNCTKCSHFKDGPNCVEKCPDG LQGANSFIFKYADPDRECHPCHPNCTQGCIGSSIEDCIGLMDRTPLIAAG VIGGLFILVIVGLTFAVYVRRKSIKKKRALRRFLETELVEPLTPSGTAPN QAQLRILKETELKRVKVLGSGAFGTVYKGIWVPEGETVKIPVAIKILNET TGPKANVEFMDEALIMASMDHPHLVRLLGVCLSPTIQLVTQLMPHGCLLE YVHEHKDNIGSQLLLNWCVQIAKGMMYLEERRLVHRDLAARNVLVKSPNH VKITDFGLARLLEGDEKEYNADGGKMPIKWMALECIHYRKFTHQSDVWSY GVTIWELMTFGGKPYDGIPTREIPDLLEKGERLPQPPICTIDVYMVMVKC WMIDADSRPKFKELAAEFSRMARDPQRYLVIQGDDRMKLPSPNDSKFFQN LLDEEDLEDMMDAEEYLVPQAFNIPPPIYTSRARIDSNRSEIGHSPPPAY TPMSGNQFVYRDGGFAAEQGVSVPYRAPTSTIPEAPVAQGATAEIFDDSC CNGTLRKPVAPHVQEDSSTQRYSADPTVFAPERSPRGELDEEGYMTPMRD KPKQEYLNPVEENPFVSRRKNGDLQALDNPEYHNASNGPPKAEDEYVNEP LYLNTFANTLGKAEYLKNNILSMPEKAKKAFDNPDYWNHSLPPRSTLQHP DYLQEYSTKYFYKQNGRIRPIVAENPEYLSEFSLKPGTVLPPPPYRHRNT | 1202 |
| 3D view using mol* of 851 (AA BP:) | ||||||||||
 | ||||||||||
| PDB file (857) >>>857.pdbFusion protein BP residue: CIF file (857) >>>857.cif | B4GALT5 | chr20 | 48330113 | - | ERBB4 | chr2 | 212812341 | - | MPLENLRIIRGTKLYEDRYALAIFLNYRKDGNFGLQELGLKNLTEILNGG VYVDQNKFLCYADTIHWQDIVRNPWPSNLTLVSTNGSSGCGRCHKSCTGR CWGPTENHCQTLTRTVCAEQCDGRCYGPYVSDCCHRECAGGCSGPKDTDC FACMNFNDSGACVTQCPQTFVYNPTTFQLEHNFNAKYTYGAFCVKKCPHN FVVDSSSCVRACPSSKMEVEENGIKMCKPCTDICPKACDGIGTGSLMSAQ TVDSSNIDKFINCTKINGNLIFLVTGIHGDPYNAIEAIDPEKLNVFRTVR EITGFLNIQSWPPNMTDFSVFSNLVTIGGRVLYSGLSLLILKQQGITSLQ FQSLKEISAGNIYITDNSNLCYYHTINWTTLFSTINQRIVIRDNRKAENC TAEGMVCNHLCSSDGCWGPGPDQCLSCRRFSRGRICIESCNLYDGEFREF ENGSICVECDPQCEKMEDGLLTCHGPGPDNCTKCSHFKDGPNCVEKCPDG LQGANSFIFKYADPDRECHPCHPNCTQGCNGPTSHDCIYYPWTGHSTLPQ HARTPLIAAGVIGGLFILVIVGLTFAVYVRRKSIKKKRALRRFLETELVE PLTPSGTAPNQAQLRILKETELKRVKVLGSGAFGTVYKGIWVPEGETVKI PVAIKILNETTGPKANVEFMDEALIMASMDHPHLVRLLGVCLSPTIQLVT QLMPHGCLLEYVHEHKDNIGSQLLLNWCVQIAKGMMYLEERRLVHRDLAA RNVLVKSPNHVKITDFGLARLLEGDEKEYNADGGKMPIKWMALECIHYRK FTHQSDVWSYGVTIWELMTFGGKPYDGIPTREIPDLLEKGERLPQPPICT IDVYMVMVKCWMIDADSRPKFKELAAEFSRMARDPQRYLVIQGDDRMKLP SPNDSKFFQNLLDEEDLEDMMDAEEYLVPQAFNIPPPIYTSRARIDSNRS EIGHSPPPAYTPMSGNQFVYRDGGFAAEQGVSVPYRAPTSTIPEAPVAQG ATAEIFDDSCCNGTLRKPVAPHVQEDSSTQRYSADPTVFAPERSPRGELD EEGYMTPMRDKPKQEYLNPVEENPFVSRRKNGDLQALDNPEYHNASNGPP KAEDEYVNEPLYLNTFANTLGKAEYLKNNILSMPEKAKKAFDNPDYWNHS LPPRSTLQHPDYLQEYSTKYFYKQNGRIRPIVAENPEYLSEFSLKPGTVL | 1212 |
| 3D view using mol* of 857 (AA BP:) | ||||||||||
 | ||||||||||
Top |
pLDDT score distribution |
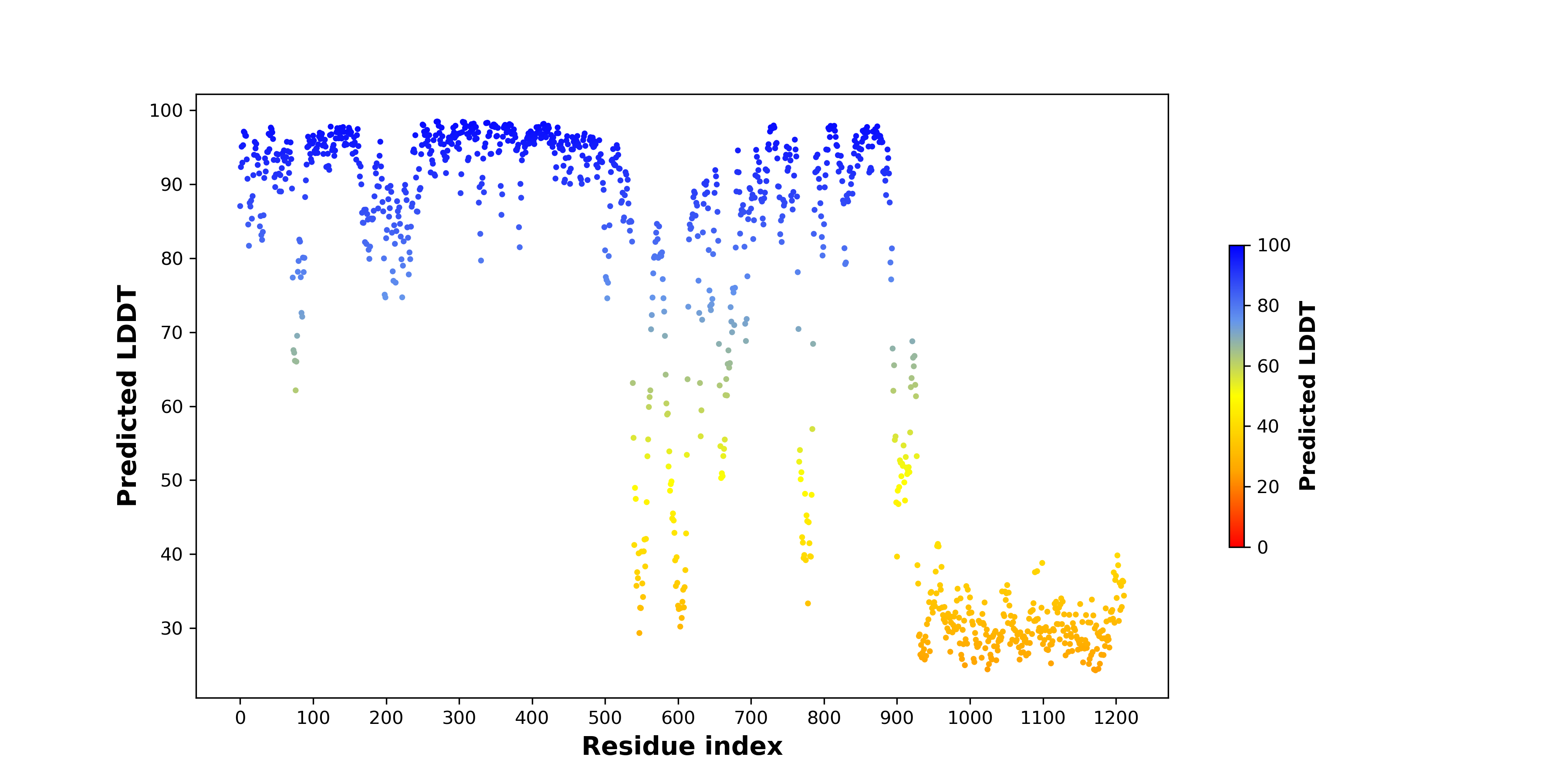
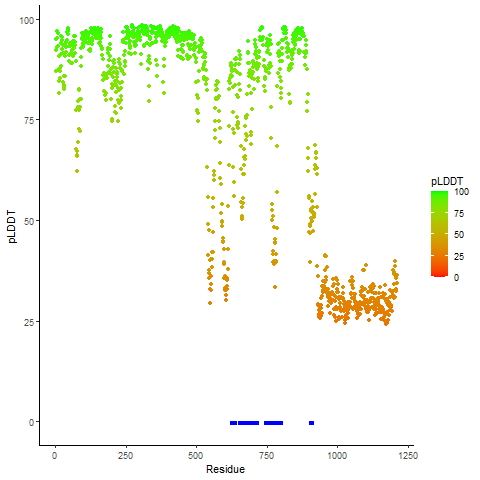
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
B4GALT5_pLDDT.png |
ERBB4_pLDDT.png |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
Top |
Ramachandran Plot of Fusion Protein Structure |
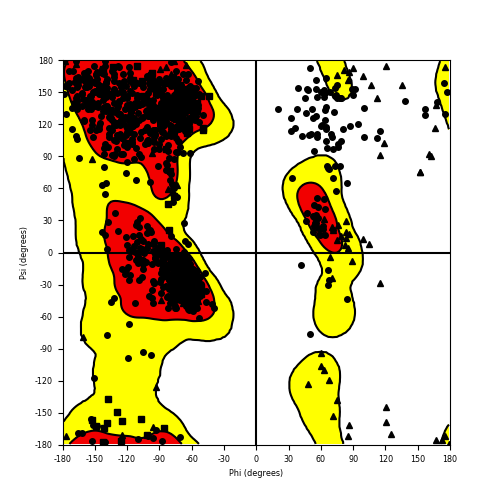
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| Fusion AA seq ID in FusionPDB and their Ramachandran plots |
| B4GALT5_ERBB4_847.png |
 |
| B4GALT5_ERBB4_851.png |
 |
| B4GALT5_ERBB4_857.png |
 |
Top |
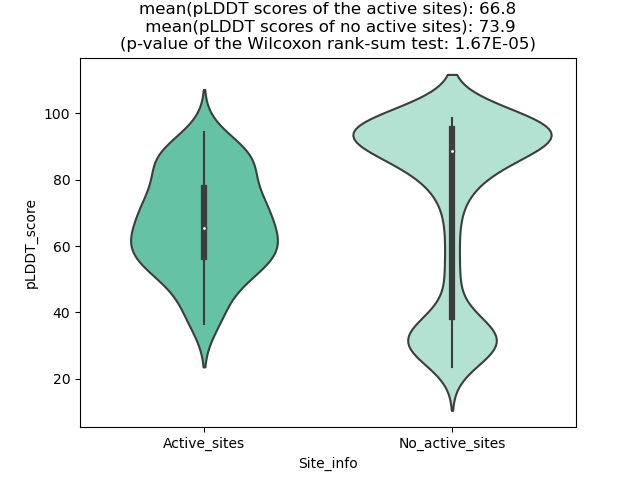
Potential Active Site Information |
 The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. |
| Fusion AA seq ID in FusionPDB | Site score | Size | D score | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
| 847 | 1.031 | 718 | 1.055 | 1785.315 | 0.552 | 0.729 | 0.944 | 0.668 | 0.988 | 0.677 | 1.112 | Chain A: 196,198,199,244,247,248,593,594,595,596,5 97,598,599,600,601,602,603,604,605,606,607,608,609 ,610,611,613,614,615,629,630,631,632,633,634,636,6 53,655,657,661,665,668,669,670,671,672,674,675,676 ,677,678,679,680,681,685,686,687,688,689,698,700,7 07,709,710,737,741,743,744,746,747,749,751,752,754 ,764,765,766,767,768,769,770,771,772,773,774,775,7 76,778,779,780,781,782,783,784,785,786,787,788,789 ,790,791,800,801,816,823,824,826,827,830,924,927,9 28,929,1014,1015,1016,1017,1018,1019,1020,1021,102 2,1150,1151,1152,1153 |
| 851 | 1.033 | 306 | 1.004 | 786.499 | 0.46 | 0.747 | 1.024 | 0.588 | 1.178 | 0.499 | 0.861 | Chain A: 582,583,585,586,587,588,589,600,601,602,6 03,604,605,622,623,624,631,647,648,651,654,655,657 ,658,659,661,662,664,665,667,668,669,670,676,677,6 78,679,727,731,733,734,736,756,757,758,759,760,761 ,762,763,766,768,769,770,771,772,773,774,775,776,7 77,789,791 |
| 857 | 1.085 | 335 | 1.121 | 989.555 | 0.531 | 0.779 | 1.017 | 1.327 | 0.879 | 1.509 | 0.962 | Chain A: 626,628,629,630,631,632,633,634,636,638,6 53,655,657,658,661,665,668,669,671,672,675,676,685 ,698,700,702,703,705,706,707,710,711,714,715,746,7 47,751,752,754,764,765,766,767,768,769,770,779,780 ,781,782,783,784,785,786,787,791,799,801,905,906,9 07,910,911 |
Top |
Potentially Interacting Small Molecules through Virtual Screening |
 The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. |
| Fusion AA seq ID in FusionPDB | ZINC ID | DrugBank ID | Drug name | Docking score | Glide gscore |
Top |
 Drug information from DrugBank of the top 20 interacting small molecules. Drug information from DrugBank of the top 20 interacting small molecules. |
| ZINC ID | DrugBank ID | Drug name | Drug type | SMILES | Drug group |
Top |
Biochemical Features of Small Molecules |
 ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) |
| ZINC ID | mol_MW | dipole | SASA | FOSA | FISA | PISA | WPSA | volume | donorHB | accptHB | IP | Human Oral Absorption | Percent Human Oral Absorption | Rule Of Five | Rule Of Three |
Top |
Drug Toxicity Information |
 Toxicity information of individual drugs using eToxPred Toxicity information of individual drugs using eToxPred |
| ZINC ID | Smile | Surface Accessibility | Toxicity |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| B4GALT5 | ELAVL1, HLA-DPA1, NXPH2, IL20RB, GINM1, TNF, LIPH, IL27RA, FCGRT, SLC39A9, KIAA1467, IL13RA2, SYT12, KERA, GABRG2, CRP, HSPA13, MGAT4C, AVP, TPCN2, TRIM25, SH2D3C, HNRNPH1, TMEM59, NRSN1, C16orf92, TMX1, TMPRSS13, OPTC, IGF2R, CCL3, CSTL1, GPR114, CSF2, HLA-DQA1, CTLA4, PMCH, B3GNT7, EPO, CORT, IFNE, CHST8, ERP27, ADIPOQ, IDS, VTI1A, B4GALT6, PCDH20, OMA1, PGLYRP3, ATG9A, NAPA, EDDM3A, HS3ST1, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| B4GALT5 |  |
| ERBB4 |  |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to B4GALT5-ERBB4 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to B4GALT5-ERBB4 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | B4GALT5 | C0023893 | Liver Cirrhosis, Experimental | 1 | CTD_human |
| Hgene | B4GALT5 | C0087031 | Juvenile-Onset Still Disease | 1 | CTD_human |
| Hgene | B4GALT5 | C3495559 | Juvenile arthritis | 1 | CTD_human |
| Hgene | B4GALT5 | C3714758 | Juvenile psoriatic arthritis | 1 | CTD_human |
| Hgene | B4GALT5 | C4552091 | Polyarthritis, Juvenile, Rheumatoid Factor Negative | 1 | CTD_human |
| Hgene | B4GALT5 | C4704862 | Polyarthritis, Juvenile, Rheumatoid Factor Positive | 1 | CTD_human |
| Tgene | ERBB4 | C0005586 | Bipolar Disorder | 5 | PSYGENET |
| Tgene | ERBB4 | C0036341 | Schizophrenia | 4 | PSYGENET |
| Tgene | ERBB4 | C0004238 | Atrial Fibrillation | 2 | CTD_human |
| Tgene | ERBB4 | C0235480 | Paroxysmal atrial fibrillation | 2 | CTD_human |
| Tgene | ERBB4 | C2585653 | Persistent atrial fibrillation | 2 | CTD_human |
| Tgene | ERBB4 | C3468561 | familial atrial fibrillation | 2 | CTD_human |
| Tgene | ERBB4 | C0002736 | Amyotrophic Lateral Sclerosis | 1 | ORPHANET |
| Tgene | ERBB4 | C0007114 | Malignant neoplasm of skin | 1 | CTD_human |
| Tgene | ERBB4 | C0016978 | gallbladder neoplasm | 1 | CTD_human |
| Tgene | ERBB4 | C0025202 | melanoma | 1 | CGI;CTD_human |
| Tgene | ERBB4 | C0037286 | Skin Neoplasms | 1 | CTD_human |
| Tgene | ERBB4 | C0153452 | Malignant neoplasm of gallbladder | 1 | CTD_human |
| Tgene | ERBB4 | C3715155 | AMYOTROPHIC LATERAL SCLEROSIS 19 | 1 | CTD_human;GENOMICS_ENGLAND;UNIPROT |