| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:UNC79-BRF1 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: UNC79-BRF1 | FusionPDB ID: 96863 | FusionGDB2.0 ID: 96863 | Hgene | Tgene | Gene symbol | UNC79 | BRF1 | Gene ID | 57578 | 2972 |
| Gene name | unc-79 homolog, NALCN channel complex subunit | BRF1 RNA polymerase III transcription initiation factor subunit | |
| Synonyms | KIAA1409 | BRF|BRF-1|CFDS|GTF3B|HEL-S-76p|TAF3B2|TAF3C|TAFIII90|TF3B90|TFIIIB90|hBRF | |
| Cytomap | 14q32.12 | 14q32.33 | |
| Type of gene | protein-coding | protein-coding | |
| Description | protein unc-79 homologunc-79 homolog | transcription factor IIIB 90 kDa subunitB - related factor 1BRF1 homolog, subunit of RNA polymerase III transcription initiation factor IIIBBRF1, RNA polymerase III transcription initiation factor 90 kDa subunitTATA box binding protein (TBP)-associate | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q9P2D8 | Q92994 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000256339, ENST00000393151, ENST00000553484, ENST00000555664, | ENST00000327359, ENST00000440513, ENST00000379932, ENST00000379937, ENST00000392557, ENST00000446501, ENST00000547530, ENST00000548421, ENST00000549044, ENST00000551787, ENST00000546474, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 3 X 3 X 2=18 | 9 X 10 X 5=450 |
| # samples | 3 | 10 | |
| ** MAII score | log2(3/18*10)=0.736965594166206 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(10/450*10)=-2.16992500144231 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: UNC79 [Title/Abstract] AND BRF1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | UNC79(94069764)-BRF1(105752713), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | UNC79-BRF1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. UNC79-BRF1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. UNC79-BRF1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. UNC79-BRF1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across UNC79 (5'-gene) Fusion gene breakpoints across UNC79 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
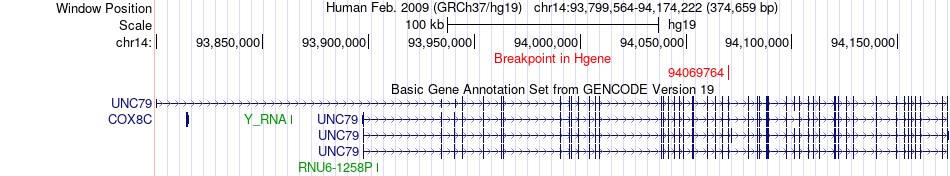 |
 Fusion gene breakpoints across BRF1 (3'-gene) Fusion gene breakpoints across BRF1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
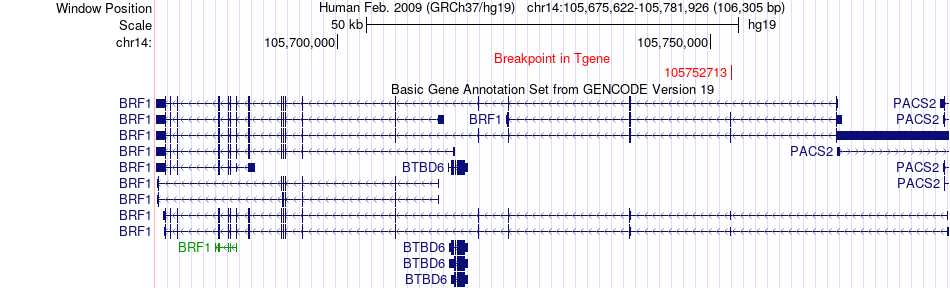 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LGG | TCGA-DU-A5TU-01A | UNC79 | chr14 | 94069764 | + | BRF1 | chr14 | 105752713 | - |
| ChimerDB4 | LGG | TCGA-DU-A5TU | UNC79 | chr14 | 94069764 | + | BRF1 | chr14 | 105752713 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000256339 | UNC79 | chr14 | 94069764 | + | ENST00000546474 | BRF1 | chr14 | 105752713 | - | 6993 | 3878 | 325 | 5727 | 1800 |
| ENST00000555664 | UNC79 | chr14 | 94069764 | + | ENST00000546474 | BRF1 | chr14 | 105752713 | - | 7562 | 4447 | 627 | 6296 | 1889 |
| ENST00000553484 | UNC79 | chr14 | 94069764 | + | ENST00000546474 | BRF1 | chr14 | 105752713 | - | 7023 | 3908 | 88 | 5757 | 1889 |
| ENST00000393151 | UNC79 | chr14 | 94069764 | + | ENST00000546474 | BRF1 | chr14 | 105752713 | - | 6869 | 3754 | 0 | 5603 | 1867 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000256339 | ENST00000546474 | UNC79 | chr14 | 94069764 | + | BRF1 | chr14 | 105752713 | - | 0.005968276 | 0.9940317 |
| ENST00000555664 | ENST00000546474 | UNC79 | chr14 | 94069764 | + | BRF1 | chr14 | 105752713 | - | 0.004972517 | 0.9950275 |
| ENST00000553484 | ENST00000546474 | UNC79 | chr14 | 94069764 | + | BRF1 | chr14 | 105752713 | - | 0.003298995 | 0.99670094 |
| ENST00000393151 | ENST00000546474 | UNC79 | chr14 | 94069764 | + | BRF1 | chr14 | 105752713 | - | 0.003011887 | 0.9969881 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >96863_96863_1_UNC79-BRF1_UNC79_chr14_94069764_ENST00000256339_BRF1_chr14_105752713_ENST00000546474_length(amino acids)=1800AA_BP=1185 MQEYHNRVLHNIYPVPSGTDIANTLKYFSQTLLSVLRDAPSERGPQSRDAQLSDYPSLDYQGLYVTLVTLLDLVPLLQHGQHDLGQSIFY TTTCLLPFLNDDILSTLPYTMISTLATFPPFLHKDIIEYLSTSFLPMAILGSSRREGVPAHVNLSASSMLMIAMQYTSNPVYHCQLLECL MKYKQEVWKDLLYVIAYGPSQVKPPAVQMLFHYWPNLKPPGAISEYRGLQYTAWNPIHCQHIECHNAINKPAVKMCIDPSLSVALGDKPP PLYLCEECSERIAGDHSEWLIDVLLPQAEISAICQKKNCSSHVRRAVVTCFSAGCCGRHGNRPVRYCKRCHSNHHSNEVGAAAETHLYQT SPPPINTRECGAEELVCAVEAVISLLKEAEFHAEQREHELNRRRQLGLSSSHHSLDNADFDNKDDDKHDQRLLSQFGIWFLVSLCTPSEN TPTESLARLVAMVFQWFHSTAYMMDDEVGSLVEKLKPQFVTKWLKTVCDVRFDVMVMCLLPKPMEFARVGGYWDKSCSTVTQLKEGLNRI LCLIPYNVINQSVWECIMPEWLEAIRTEVPDNQLKEFREVLSKMFDIELCPLPFSMEEMFGFISCRFTGYPSSVQEQALLWLHVLSELDI MVPLQLLISMFSDGVNSVKELANQRKSRVSELAGNLASRRVSVASDPGRRVQHNMLSPFHSPFQSPFRSPLRSPFRSPFKNFGHPGGRTI DFDCEDDEMNLNCFILMFDLLLKQMELQDDGITMGLEHSLSKDIISIINNVFQAPWGGSHTCQKDEKAIECNLCQSSILCYQLACELLER LAPKEESRLVEPTDSLEDSLLSSRPEFIIGPEGEEEENPASKHGENPGNCTEPVEHAAVKNDTERKFCYQQLPVTLRLIYTIFQEMAKFE EPDILFNMLNCLKILCLHGECLYIARKDHPQFLAYIQDHMLIASLWRVVKSEFSQLSSLAVPLLLHALSLPHGADIFWTIINGNFNSKDW KMRFEAVEKVAVICRFLDIHSVTKNHLLKYSLAHAFCCFLTAVEDVNPAVATRAGLLLDTIKRPALQGLCLCLDFQFDTVVKDRPTILSK LLLLHFLKQDIPALSWEFFVNRFETLSLEAQLHLDCNKEFPFPTTITAVRTNVANLSDAALWKIKRARFARNRQKSVRSLRDSVKGPVES KRALSLPETLTSKIRAGKTPTLGGGFHVNLGKESRAQTLQNGRRHIHHLGNQLQLNQHCLDTAFNFFKMAVSRHLTRGRKMAHVIAACLY LVCRTEGTPHMLLDLSDLLQVNVYVLGKTFLLLARELCINAPAIDPCLYIPRFAHLLEFGEKNHEVSMTALRLLQRMKRDWMHTGRRPSG LCGAALLVAARMHDFRRTVKEVISVVKVCESTLRKRLTEFEDTPTSQLTIDEFMKIDLEEECDPPSYTAGQRKLRMKQLEQVLSKKLEEV EGEISSYQDAIEIELENSRPKAKGGLASLAKDGSTEDTASSLCGEEDTEDEELEAAASHLNKDLYRELLGGAPGSSEAAGSPEWGGRPPA LGSLLDPLPTAASLGISDSIRECISSQSSDPKDASGDGELDLSGIDDLEIDRYILNESEARVKAELWMRENAEYLREQREKEARIAKEKE LGIYKEHKPKKSCKRREPIQASTAREAIEKMLEQKKISSKINYSVLRGLSSAGGGSPHREDAQPEHSASARKLSRRRTPASRSGADPVTS VGKRLRPLVSTQPAKKVATGEALLPSSPTLGAEPARPQAVLVESGPVSYHADEEADEEEPDEEDGEPCVSALQMMGSNDYGCDGDEDDGY -------------------------------------------------------------- >96863_96863_2_UNC79-BRF1_UNC79_chr14_94069764_ENST00000393151_BRF1_chr14_105752713_ENST00000546474_length(amino acids)=1867AA_BP=1252 MSTKAEQFASKIRYLQEYHNRVLHNIYPVPSGTDIANTLKYFSQTLLSILSRTGKKENQDASNLTVPMTMCLFPVPFPLTPSLRPQVSSI NPTVTRSLLYSVLRDAPSERGPQSRDAQLSDYPSLDYQGLYVTLVTLLDLVPLLQHGQHDLGQSIFYTTTCLLPFLNDDILSTLPYTMIS TLATFPPFLHKDIIEYLSTSFLPMAILGSSRREGVPAHVNLSASSMLMIAMQYTSNPVYHCQLLECLMKYKQEVWKDLLYVIAYGPSQVK PPAVQMLFHYWPNLKPPGAISEYRGLQYTAWNPIHCQHIECHNAINKPAVKMCIDPSLSVALGDKPPPLYLCEECSERIAGDHSEWLIDV LLPQAEISAICQKKNCSSHVRRAVVTCFSAGCCGRHGNRPVRYCKRCHSNHHSNEVGAAAETHLYQTSPPPINTRECGAEELVCAVEAVI SLLKEAEFHAEQREHELNRRRQLGLSSSHHSLDNADFDNKDDDKHDQRLLSQFGIWFLVSLCTPSENTPTESLARLVAMVFQWFHSTAYM MDDEVGSLVEKLKPQFVTKWLKTVCDVRFDVMVMCLLPKPMEFARVGGYWDKSCSTVTQLKEGLNRILCLIPYNVINQSVWECIMPEWLE AIRTEVPDNQLKEFREVLSKMFDIELCPLPFSMEEMFGFISCRFTGYPSSVQEQALLWLHVLSELDIMVPLQLLISMFSDGVNSVKELAN QRKSRVSELAGNLASRRVSVASDPGRRVQHNMLSPFHSPFQSPFRSPLRSPFRSPFKNFGHPGGRTIDFDCEDDEMNLNCFILMFDLLLK QMELQDDGITMGLEHSLSKDIISIINNVFQAPWGGSHTCQKDEKAIECNLCQSSILCYQLACELLERLAPKEESRLVEPTDSLEDSLLSS RPEFIIGPEGEEEENPASKHGENPGNCTEPVEHAAVKNDTERKFCYQQLPVTLRLIYTIFQEMAKFEEPDILFNMLNCLKILCLHGECLY IARKDHPQFLAYIQDHMLIASLWRVVKSEFSQLSSLAVPLLLHALSLPHGADIFWTIINGNFNSKDWKMRFEAVEKVAVICRFLDIHSVT KNHLLKYSLAHAFCCFLTAVEDVNPAVATRAGLLLDTIKRPALQGLCLCLDFQFDTVVKDRPTILSKLLLLHFLKQDIPALSWEFFVNRF ETLSLEAQLHLDCNKEFPFPTTITAVRTNVANLSDAALWKIKRARFARNRQKSVRSLRDSVKGPVESKRALSLPETLTSKIRAGKTPTLG GGFHVNLGKESRAQTLQNGRRHIHHLGNQLQLNQHCLDTAFNFFKMAVSRHLTRGRKMAHVIAACLYLVCRTEGTPHMLLDLSDLLQVNV YVLGKTFLLLARELCINAPAIDPCLYIPRFAHLLEFGEKNHEVSMTALRLLQRMKRDWMHTGRRPSGLCGAALLVAARMHDFRRTVKEVI SVVKVCESTLRKRLTEFEDTPTSQLTIDEFMKIDLEEECDPPSYTAGQRKLRMKQLEQVLSKKLEEVEGEISSYQDAIEIELENSRPKAK GGLASLAKDGSTEDTASSLCGEEDTEDEELEAAASHLNKDLYRELLGGAPGSSEAAGSPEWGGRPPALGSLLDPLPTAASLGISDSIREC ISSQSSDPKDASGDGELDLSGIDDLEIDRYILNESEARVKAELWMRENAEYLREQREKEARIAKEKELGIYKEHKPKKSCKRREPIQAST AREAIEKMLEQKKISSKINYSVLRGLSSAGGGSPHREDAQPEHSASARKLSRRRTPASRSGADPVTSVGKRLRPLVSTQPAKKVATGEAL -------------------------------------------------------------- >96863_96863_3_UNC79-BRF1_UNC79_chr14_94069764_ENST00000553484_BRF1_chr14_105752713_ENST00000546474_length(amino acids)=1889AA_BP=1274 MAERGSSHDTDFGAKPFQLDSTMSTKAEQFASKIRYLQEYHNRVLHNIYPVPSGTDIANTLKYFSQTLLSILSRTGKKENQDASNLTVPM TMCLFPVPFPLTPSLRPQVSSINPTVTRSLLYSVLRDAPSERGPQSRDAQLSDYPSLDYQGLYVTLVTLLDLVPLLQHGQHDLGQSIFYT TTCLLPFLNDDILSTLPYTMISTLATFPPFLHKDIIEYLSTSFLPMAILGSSRREGVPAHVNLSASSMLMIAMQYTSNPVYHCQLLECLM KYKQEVWKDLLYVIAYGPSQVKPPAVQMLFHYWPNLKPPGAISEYRGLQYTAWNPIHCQHIECHNAINKPAVKMCIDPSLSVALGDKPPP LYLCEECSERIAGDHSEWLIDVLLPQAEISAICQKKNCSSHVRRAVVTCFSAGCCGRHGNRPVRYCKRCHSNHHSNEVGAAAETHLYQTS PPPINTRECGAEELVCAVEAVISLLKEAEFHAEQREHELNRRRQLGLSSSHHSLDNADFDNKDDDKHDQRLLSQFGIWFLVSLCTPSENT PTESLARLVAMVFQWFHSTAYMMDDEVGSLVEKLKPQFVTKWLKTVCDVRFDVMVMCLLPKPMEFARVGGYWDKSCSTVTQLKEGLNRIL CLIPYNVINQSVWECIMPEWLEAIRTEVPDNQLKEFREVLSKMFDIELCPLPFSMEEMFGFISCRFTGYPSSVQEQALLWLHVLSELDIM VPLQLLISMFSDGVNSVKELANQRKSRVSELAGNLASRRVSVASDPGRRVQHNMLSPFHSPFQSPFRSPLRSPFRSPFKNFGHPGGRTID FDCEDDEMNLNCFILMFDLLLKQMELQDDGITMGLEHSLSKDIISIINNVFQAPWGGSHTCQKDEKAIECNLCQSSILCYQLACELLERL APKEESRLVEPTDSLEDSLLSSRPEFIIGPEGEEEENPASKHGENPGNCTEPVEHAAVKNDTERKFCYQQLPVTLRLIYTIFQEMAKFEE PDILFNMLNCLKILCLHGECLYIARKDHPQFLAYIQDHMLIASLWRVVKSEFSQLSSLAVPLLLHALSLPHGADIFWTIINGNFNSKDWK MRFEAVEKVAVICRFLDIHSVTKNHLLKYSLAHAFCCFLTAVEDVNPAVATRAGLLLDTIKRPALQGLCLCLDFQFDTVVKDRPTILSKL LLLHFLKQDIPALSWEFFVNRFETLSLEAQLHLDCNKEFPFPTTITAVRTNVANLSDAALWKIKRARFARNRQKSVRSLRDSVKGPVESK RALSLPETLTSKIRAGKTPTLGGGFHVNLGKESRAQTLQNGRRHIHHLGNQLQLNQHCLDTAFNFFKMAVSRHLTRGRKMAHVIAACLYL VCRTEGTPHMLLDLSDLLQVNVYVLGKTFLLLARELCINAPAIDPCLYIPRFAHLLEFGEKNHEVSMTALRLLQRMKRDWMHTGRRPSGL CGAALLVAARMHDFRRTVKEVISVVKVCESTLRKRLTEFEDTPTSQLTIDEFMKIDLEEECDPPSYTAGQRKLRMKQLEQVLSKKLEEVE GEISSYQDAIEIELENSRPKAKGGLASLAKDGSTEDTASSLCGEEDTEDEELEAAASHLNKDLYRELLGGAPGSSEAAGSPEWGGRPPAL GSLLDPLPTAASLGISDSIRECISSQSSDPKDASGDGELDLSGIDDLEIDRYILNESEARVKAELWMRENAEYLREQREKEARIAKEKEL GIYKEHKPKKSCKRREPIQASTAREAIEKMLEQKKISSKINYSVLRGLSSAGGGSPHREDAQPEHSASARKLSRRRTPASRSGADPVTSV -------------------------------------------------------------- >96863_96863_4_UNC79-BRF1_UNC79_chr14_94069764_ENST00000555664_BRF1_chr14_105752713_ENST00000546474_length(amino acids)=1889AA_BP=1274 MAERGSSHDTDFGAKPFQLDSTMSTKAEQFASKIRYLQEYHNRVLHNIYPVPSGTDIANTLKYFSQTLLSILSRTGKKENQDASNLTVPM TMCLFPVPFPLTPSLRPQVSSINPTVTRSLLYSVLRDAPSERGPQSRDAQLSDYPSLDYQGLYVTLVTLLDLVPLLQHGQHDLGQSIFYT TTCLLPFLNDDILSTLPYTMISTLATFPPFLHKDIIEYLSTSFLPMAILGSSRREGVPAHVNLSASSMLMIAMQYTSNPVYHCQLLECLM KYKQEVWKDLLYVIAYGPSQVKPPAVQMLFHYWPNLKPPGAISEYRGLQYTAWNPIHCQHIECHNAINKPAVKMCIDPSLSVALGDKPPP LYLCEECSERIAGDHSEWLIDVLLPQAEISAICQKKNCSSHVRRAVVTCFSAGCCGRHGNRPVRYCKRCHSNHHSNEVGAAAETHLYQTS PPPINTRECGAEELVCAVEAVISLLKEAEFHAEQREHELNRRRQLGLSSSHHSLDNADFDNKDDDKHDQRLLSQFGIWFLVSLCTPSENT PTESLARLVAMVFQWFHSTAYMMDDEVGSLVEKLKPQFVTKWLKTVCDVRFDVMVMCLLPKPMEFARVGGYWDKSCSTVTQLKEGLNRIL CLIPYNVINQSVWECIMPEWLEAIRTEVPDNQLKEFREVLSKMFDIELCPLPFSMEEMFGFISCRFTGYPSSVQEQALLWLHVLSELDIM VPLQLLISMFSDGVNSVKELANQRKSRVSELAGNLASRRVSVASDPGRRVQHNMLSPFHSPFQSPFRSPLRSPFRSPFKNFGHPGGRTID FDCEDDEMNLNCFILMFDLLLKQMELQDDGITMGLEHSLSKDIISIINNVFQAPWGGSHTCQKDEKAIECNLCQSSILCYQLACELLERL APKEESRLVEPTDSLEDSLLSSRPEFIIGPEGEEEENPASKHGENPGNCTEPVEHAAVKNDTERKFCYQQLPVTLRLIYTIFQEMAKFEE PDILFNMLNCLKILCLHGECLYIARKDHPQFLAYIQDHMLIASLWRVVKSEFSQLSSLAVPLLLHALSLPHGADIFWTIINGNFNSKDWK MRFEAVEKVAVICRFLDIHSVTKNHLLKYSLAHAFCCFLTAVEDVNPAVATRAGLLLDTIKRPALQGLCLCLDFQFDTVVKDRPTILSKL LLLHFLKQDIPALSWEFFVNRFETLSLEAQLHLDCNKEFPFPTTITAVRTNVANLSDAALWKIKRARFARNRQKSVRSLRDSVKGPVESK RALSLPETLTSKIRAGKTPTLGGGFHVNLGKESRAQTLQNGRRHIHHLGNQLQLNQHCLDTAFNFFKMAVSRHLTRGRKMAHVIAACLYL VCRTEGTPHMLLDLSDLLQVNVYVLGKTFLLLARELCINAPAIDPCLYIPRFAHLLEFGEKNHEVSMTALRLLQRMKRDWMHTGRRPSGL CGAALLVAARMHDFRRTVKEVISVVKVCESTLRKRLTEFEDTPTSQLTIDEFMKIDLEEECDPPSYTAGQRKLRMKQLEQVLSKKLEEVE GEISSYQDAIEIELENSRPKAKGGLASLAKDGSTEDTASSLCGEEDTEDEELEAAASHLNKDLYRELLGGAPGSSEAAGSPEWGGRPPAL GSLLDPLPTAASLGISDSIRECISSQSSDPKDASGDGELDLSGIDDLEIDRYILNESEARVKAELWMRENAEYLREQREKEARIAKEKEL GIYKEHKPKKSCKRREPIQASTAREAIEKMLEQKKISSKINYSVLRGLSSAGGGSPHREDAQPEHSASARKLSRRRTPASRSGADPVTSV -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr14:94069764/chr14:105752713) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| UNC79 | BRF1 |
| FUNCTION: Component of the NALCN sodium channel complex, a cation channel activated either by neuropeptides substance P or neurotensin that controls neuronal excitability. {ECO:0000250}. | FUNCTION: General activator of RNA polymerase which utilizes different TFIIIB complexes at structurally distinct promoters. The isoform 1 is involved in the transcription of tRNA, adenovirus VA1, 7SL and 5S RNA. Isoform 2 is required for transcription of the U6 promoter. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | BRF1 | chr14:94069764 | chr14:105752713 | ENST00000327359 | 0 | 18 | 185_269 | 0 | 563.0 | Repeat | Note=2 | |
| Tgene | BRF1 | chr14:94069764 | chr14:105752713 | ENST00000327359 | 0 | 18 | 91_172 | 0 | 563.0 | Repeat | Note=1 | |
| Tgene | BRF1 | chr14:94069764 | chr14:105752713 | ENST00000379937 | 0 | 17 | 185_269 | 0 | 651.0 | Repeat | Note=2 | |
| Tgene | BRF1 | chr14:94069764 | chr14:105752713 | ENST00000379937 | 0 | 17 | 91_172 | 0 | 651.0 | Repeat | Note=1 | |
| Tgene | BRF1 | chr14:94069764 | chr14:105752713 | ENST00000392557 | 0 | 14 | 185_269 | 0 | 474.0 | Repeat | Note=2 | |
| Tgene | BRF1 | chr14:94069764 | chr14:105752713 | ENST00000392557 | 0 | 14 | 91_172 | 0 | 474.0 | Repeat | Note=1 | |
| Tgene | BRF1 | chr14:94069764 | chr14:105752713 | ENST00000440513 | 0 | 18 | 185_269 | 0 | 585.0 | Repeat | Note=2 | |
| Tgene | BRF1 | chr14:94069764 | chr14:105752713 | ENST00000440513 | 0 | 18 | 91_172 | 0 | 585.0 | Repeat | Note=1 | |
| Tgene | BRF1 | chr14:94069764 | chr14:105752713 | ENST00000446501 | 0 | 13 | 185_269 | 0 | 440.0 | Repeat | Note=2 | |
| Tgene | BRF1 | chr14:94069764 | chr14:105752713 | ENST00000446501 | 0 | 13 | 91_172 | 0 | 440.0 | Repeat | Note=1 | |
| Tgene | BRF1 | chr14:94069764 | chr14:105752713 | ENST00000546474 | 0 | 18 | 185_269 | 61.333333333333336 | 678.0 | Repeat | Note=2 | |
| Tgene | BRF1 | chr14:94069764 | chr14:105752713 | ENST00000546474 | 0 | 18 | 91_172 | 61.333333333333336 | 678.0 | Repeat | Note=1 | |
| Tgene | BRF1 | chr14:94069764 | chr14:105752713 | ENST00000548421 | 0 | 4 | 185_269 | 61.333333333333336 | 209.0 | Repeat | Note=2 | |
| Tgene | BRF1 | chr14:94069764 | chr14:105752713 | ENST00000548421 | 0 | 4 | 91_172 | 61.333333333333336 | 209.0 | Repeat | Note=1 | |
| Tgene | BRF1 | chr14:94069764 | chr14:105752713 | ENST00000327359 | 0 | 18 | 2_33 | 0 | 563.0 | Zinc finger | TFIIB-type | |
| Tgene | BRF1 | chr14:94069764 | chr14:105752713 | ENST00000379937 | 0 | 17 | 2_33 | 0 | 651.0 | Zinc finger | TFIIB-type | |
| Tgene | BRF1 | chr14:94069764 | chr14:105752713 | ENST00000392557 | 0 | 14 | 2_33 | 0 | 474.0 | Zinc finger | TFIIB-type | |
| Tgene | BRF1 | chr14:94069764 | chr14:105752713 | ENST00000440513 | 0 | 18 | 2_33 | 0 | 585.0 | Zinc finger | TFIIB-type | |
| Tgene | BRF1 | chr14:94069764 | chr14:105752713 | ENST00000446501 | 0 | 13 | 2_33 | 0 | 440.0 | Zinc finger | TFIIB-type |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | UNC79 | chr14:94069764 | chr14:105752713 | ENST00000256339 | + | 26 | 50 | 2223_2243 | 1074.3333333333333 | 2459.0 | Transmembrane | Helical |
| Hgene | UNC79 | chr14:94069764 | chr14:105752713 | ENST00000256339 | + | 26 | 50 | 2466_2486 | 1074.3333333333333 | 2459.0 | Transmembrane | Helical |
| Hgene | UNC79 | chr14:94069764 | chr14:105752713 | ENST00000393151 | + | 26 | 50 | 2223_2243 | 1251.3333333333333 | 2636.0 | Transmembrane | Helical |
| Hgene | UNC79 | chr14:94069764 | chr14:105752713 | ENST00000393151 | + | 26 | 50 | 2466_2486 | 1251.3333333333333 | 2636.0 | Transmembrane | Helical |
| Hgene | UNC79 | chr14:94069764 | chr14:105752713 | ENST00000553484 | + | 26 | 51 | 2223_2243 | 1251.3333333333333 | 2658.0 | Transmembrane | Helical |
| Hgene | UNC79 | chr14:94069764 | chr14:105752713 | ENST00000553484 | + | 26 | 51 | 2466_2486 | 1251.3333333333333 | 2658.0 | Transmembrane | Helical |
| Tgene | BRF1 | chr14:94069764 | chr14:105752713 | ENST00000546474 | 0 | 18 | 2_33 | 61.333333333333336 | 678.0 | Zinc finger | TFIIB-type | |
| Tgene | BRF1 | chr14:94069764 | chr14:105752713 | ENST00000548421 | 0 | 4 | 2_33 | 61.333333333333336 | 209.0 | Zinc finger | TFIIB-type |
Top |
Fusion Protein Structures |
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
Top |
pLDDT score distribution |
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
UNC79_pLDDT.png |
BRF1_pLDDT.png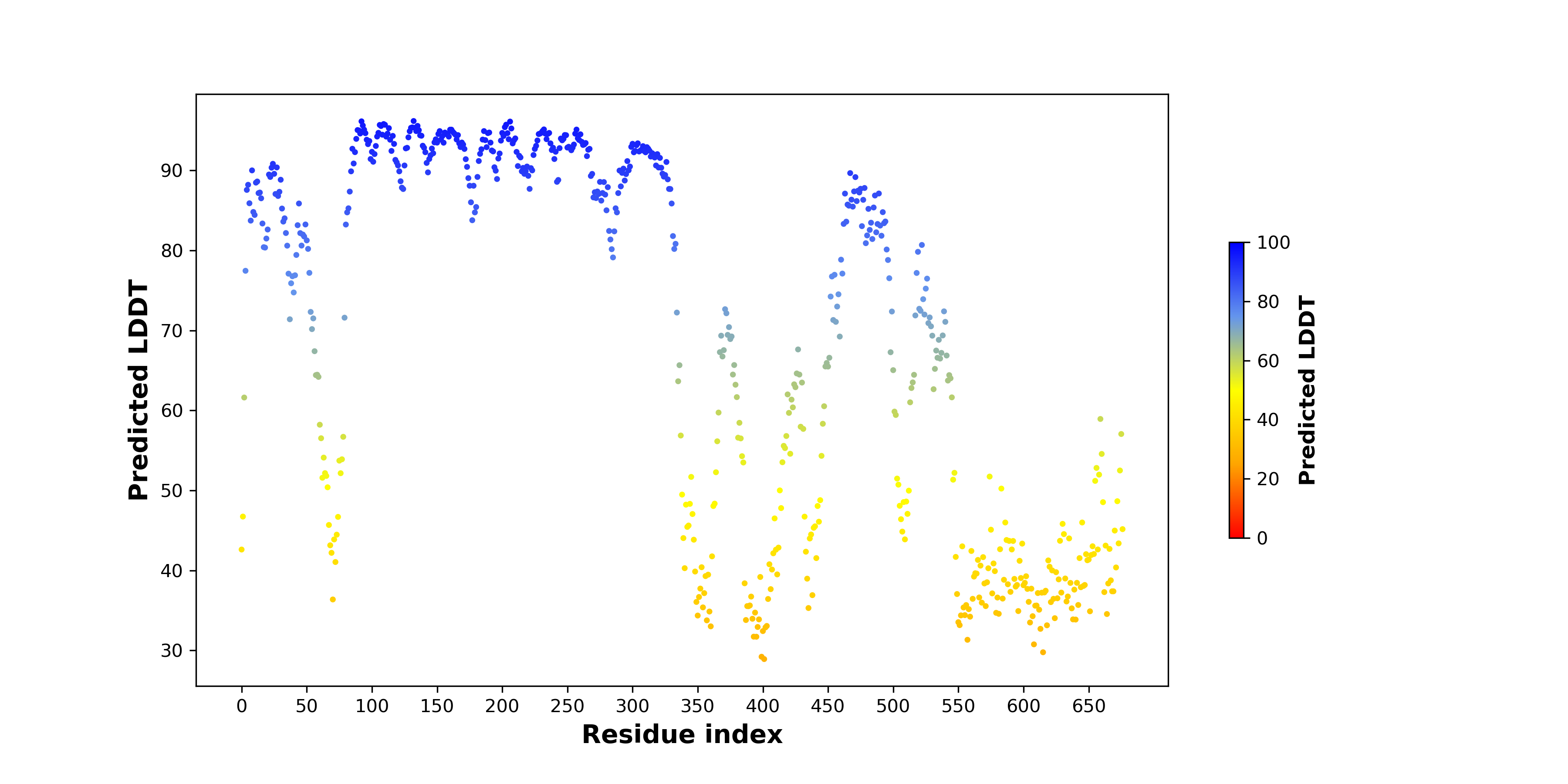 |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
Top |
Ramachandran Plot of Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| Fusion AA seq ID in FusionPDB and their Ramachandran plots |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| UNC79 | |
| BRF1 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to UNC79-BRF1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to UNC79-BRF1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

