| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:BPTF-PEBP1 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: BPTF-PEBP1 | FusionPDB ID: 10095 | FusionGDB2.0 ID: 10095 | Hgene | Tgene | Gene symbol | BPTF | PEBP1 | Gene ID | 2186 | 5037 |
| Gene name | bromodomain PHD finger transcription factor | phosphatidylethanolamine binding protein 1 | |
| Synonyms | FAC1|FALZ|NEDDFL|NURF301 | HCNP|HCNPpp|HEL-210|HEL-S-34|HEL-S-96|PBP|PEBP|PEBP-1|RKIP | |
| Cytomap | 17q24.2 | 12q24.23 | |
| Type of gene | protein-coding | protein-coding | |
| Description | nucleosome-remodeling factor subunit BPTFbromodomain and PHD domain transcription factorbromodomain and PHD finger-containing transcription factorfetal Alz-50 clone 1 proteinfetal Alz-50 reactive clone 1fetal Alzheimer antigennucleosome remodeling f | phosphatidylethanolamine-binding protein 1Raf kinase inhibitory proteinepididymis luminal protein 210epididymis secretory protein Li 34epididymis secretory protein Li 96hippocampal cholinergic neurostimulating peptideneuropolypeptide h3prostatic bi | |
| Modification date | 20200313 | 20200327 | |
| UniProtAcc | Q12830 | . | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000306378, ENST00000321892, ENST00000335221, ENST00000424123, ENST00000577770, | ENST00000542939, ENST00000261313, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 27 X 21 X 13=7371 | 6 X 5 X 4=120 |
| # samples | 42 | 7 | |
| ** MAII score | log2(42/7371*10)=-4.1333991254172 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(7/120*10)=-0.777607578663552 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: BPTF [Title/Abstract] AND PEBP1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | BPTF(65960520)-PEBP1(118575844), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | BPTF-PEBP1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. BPTF-PEBP1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. BPTF-PEBP1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. BPTF-PEBP1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | BPTF | GO:0000122 | negative regulation of transcription by RNA polymerase II | 10727212 |
| Hgene | BPTF | GO:0006338 | chromatin remodeling | 14609955 |
 Fusion gene breakpoints across BPTF (5'-gene) Fusion gene breakpoints across BPTF (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
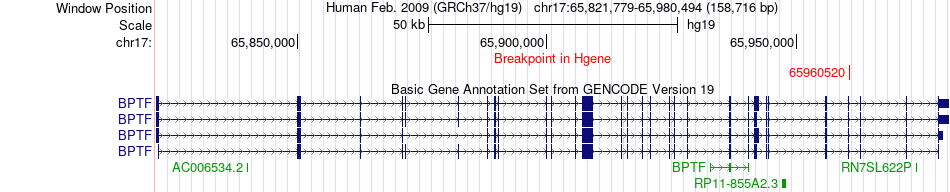 |
 Fusion gene breakpoints across PEBP1 (3'-gene) Fusion gene breakpoints across PEBP1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
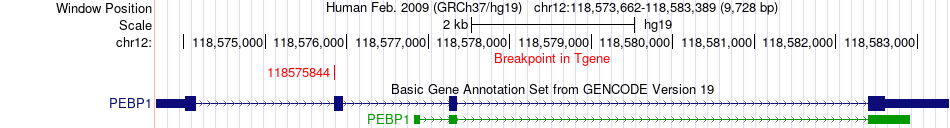 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-GM-A2D9-01A | BPTF | chr17 | 65960520 | + | PEBP1 | chr12 | 118575844 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000335221 | BPTF | chr17 | 65960520 | + | ENST00000261313 | PEBP1 | chr12 | 118575844 | + | 9674 | 8464 | 61 | 8892 | 2943 |
| ENST00000321892 | BPTF | chr17 | 65960520 | + | ENST00000261313 | PEBP1 | chr12 | 118575844 | + | 10103 | 8893 | 61 | 9321 | 3086 |
| ENST00000306378 | BPTF | chr17 | 65960520 | + | ENST00000261313 | PEBP1 | chr12 | 118575844 | + | 9724 | 8514 | 60 | 8942 | 2960 |
| ENST00000424123 | BPTF | chr17 | 65960520 | + | ENST00000261313 | PEBP1 | chr12 | 118575844 | + | 9196 | 7986 | 0 | 8414 | 2804 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000335221 | ENST00000261313 | BPTF | chr17 | 65960520 | + | PEBP1 | chr12 | 118575844 | + | 0.000472572 | 0.99952745 |
| ENST00000321892 | ENST00000261313 | BPTF | chr17 | 65960520 | + | PEBP1 | chr12 | 118575844 | + | 0.000431559 | 0.9995684 |
| ENST00000306378 | ENST00000261313 | BPTF | chr17 | 65960520 | + | PEBP1 | chr12 | 118575844 | + | 0.000401196 | 0.9995988 |
| ENST00000424123 | ENST00000261313 | BPTF | chr17 | 65960520 | + | PEBP1 | chr12 | 118575844 | + | 0.000431461 | 0.9995685 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >10095_10095_1_BPTF-PEBP1_BPTF_chr17_65960520_ENST00000306378_PEBP1_chr12_118575844_ENST00000261313_length(amino acids)=2960AA_BP=2811 MRGRRGRPPKQPAAPAAERCAPAPPPPPPPPTSGPIGGLRSRHRGSSRGRWAAAQAEVAPKTRLSSPRGGSSSRRKPPPPPPAPPSTSAP GRGGRGGGGGRTGGGGGGGHLARTTAARRAVNKVVYDDHESEEEEEEEDMVSEEEEEEDGDAEETQDSEDDEEDEMEEDDDDSDYPEEME DDDDDASYCTESSFRSHSTYSSTPGRRKPRVHRPRSPILEEKDIPPLEFPKSSEDLMVPNEHIMNVIAIYEVLRNFGTVLRLSPFRFEDF CAALVSQEQCTLMAEMHVVLLKAVLREEDTSNTTFGPADLKDSVNSTLYFIDGMTWPEVLRVYCESDKEYHHVLPYQEAEDYPYGPVENK IKVLQFLVDQFLTTNIAREELMSEGVIQYDDHCRVCHKLGDLLCCETCSAVYHLECVKPPLEEVPEDEWQCEVCVAHKVPGVTDCVAEIQ KNKPYIRHEPIGYDRSRRKYWFLNRRLIIEEDTENENEKKIWYYSTKVQLAELIDCLDKDYWEAELCKILEEMREEIHRHMDITEDLTNK ARGSNKSFLAAANEEILESIRAKKGDIDNVKSPEETEKDKNETENDSKDAEKNREEFEDQSLEKDSDDKTPDDDPEQGKSEVGDFKSEKS NGELSESPGAGKGASGSTRIITRLRNPDSKLSQLKSQQVAAAAHEANKLFKEGKEVLVVNSQGEISRLSTKKEVIMKGNINNYFKLGQEG KYRVYHNQYSTNSFALNKHQHREDHDKRRHLAHKFCLTPAGEFKWNGSVHGSKVLTISTLRLTITQLENNIPSSFLHPNWASHRANWIKA VQMCSKPREFALALAILECAVKPVVMLPIWRESLGHTRLHRMTSIEREEKEKVKKKEKKQEEEETMQQATWVKYTFPVKHQVWKQKGEEY RVTGYGGWSWISKTHVYRFVPKLPGNTNVNYRKSLEGTKNNMDENMDESDKRKCSRSPKKIKIEPDSEKDEVKGSDAAKGADQNEMDISK ITEKKDQDVKELLDSDSDKPCKEEPMEVDDDMKTESHVNCQESSQVDVVNVSEGFHLRTSYKKKTKSSKLDGLLERRIKQFTLEEKQRLE KIKLEGGIKGIGKTSTNSSKNLSESPVITKAKEGCQSDSMRQEQSPNANNDQPEDLIQGCSESDSSVLRMSDPSHTTNKLYPKDRVLDDV SIRSPETKCPKQNSIENDIEEKVSDLASRGQEPSKSKTKGNDFFIDDSKLASADDIGTLICKNKKPLIQEESDTIVSSSKSALHSSVPKS TNDRDATPLSRAMDFEGKLGCDSESNSTLENSSDTVSIQDSSEEDMIVQNSNESISEQFRTREQDVEVLEPLKCELVSGESTGNCEDRLP VKGTEANGKKPSQQKKLEERPVNKCSDQIKLKNTTDKKNNENRESEKKGQRTSTFQINGKDNKPKIYLKGECLKEISESRVVSGNVEPKV NNINKIIPENDIKSLTVKESAIRPFINGDVIMEDFNERNSSETKSHLLSSSDAEGNYRDSLETLPSTKESDSTQTTTPSASCPESNSVNQ VEDMEIETSEVKKVTSSPITSEEESNLSNDFIDENGLPINKNENVNGESKRKTVITEVTTMTSTVATESKTVIKVEKGDKQTVVSSTENC AKSTVTTTTTTVTKLSTPSTGGSVDIISVKEQSKTVVTTTVTDSLTTTGGTLVTSMTVSKEYSTRDKVKLMKFSRPKKTRSGTALPSYRK FVTKSSKKSIFVLPNDDLKKLARKGGIREVPYFNYNAKPALDIWPYPSPRPTFGITWRYRLQTVKSLAGVSLMLRLLWASLRWDDMAAKA PPGGGTTRTETSETEITTTEIIKRRDVGPYGIRSEYCIRKIICPIGVPETPKETPTPQRKGLRSSALRPKRPETPKQTGPVIIETWVAEE ELELWEIRAFAERVEKEKAQAVEQQAKKRLEQQKPTVIATSTTSPTSSTTSTISPAQKVMVAPISGSVTTGTKMVLTTKVGSPATVTFQQ NKNFHQTFATWVKQGQSNSGVVQVQQKVLGIIPSSTGTSQQTFTSFQPRTATVTIRPNTSGSGGTTSNSQVITGPQIRPGMTVIRTPLQQ STLGKAIIRTPVMVQPGAPQQVMTQIIRGQPVSTAVSAPNTVSSTPGQKSLTSATSTSNIQSSASQPPRPQQGQVKLTMAQLTQLTQGHG GNQGLTVVIQGQGQTTGQLQLIPQGVTVLPGPGQQLMQAAMPNGTVQRFLFTPLATTATTASTTTTTVSTTAAGTGEQRQSKLSPQMQVH QDKTLPPAQSSSVGPAEAQPQTAQPSAQPQPQTQPQSPAQPEVQTQPEVQTQTTVSSHVPSEAQPTHAQSSKPQVAAQSQPQSNVQGQSP VRVQSPSQTRIRPSTPSQLSPGQQSQVQTTTSQPIPIQPHTSLQIPSQGQPQSQPQVQSSTQTLSSGQTLNQVTVSSPSRPQLQIQQPQP QVIAVPQLQQQVQVLSQIQSQVVAQIQAQQSGVPQQIKLQLPIQIQQSSAVQTHQIQNVVTVQAASVQEQLQRVQQLRDQQQKKKQQQIE IKREHTLQASNQSEIIQKQVVMKHNAVIEHLKQKKSMTPAEREENQRMIVCNQVMKYILDKIDKEEKQAAKKRKREESVEQKRSKQNATK LSALLFKHKEQLRAEILKKRALLDKDLQIEVQEELKRDLKIKKEKDLMQLAQATAVAAPCPPVTPAPPAPPAPPPSPPPPPAVQHTGLLS TPTLPAASQKRKREEEKDSSSKSKKKKMISTTSKETKKDTKLYCICKTPYDESKFYIGCDRCQNWYHGRCVGILQSEAELIDEYVCPQCQ STEDAMTVLTPLTEKDYEGLKRVLRSLQVKNRPTSISWDGLDSGKLYTLVLTDPDAPSRKDPKYREWHHFLVVNMKGNDISSGTVLSDYV -------------------------------------------------------------- >10095_10095_2_BPTF-PEBP1_BPTF_chr17_65960520_ENST00000321892_PEBP1_chr12_118575844_ENST00000261313_length(amino acids)=3086AA_BP=2937 MRGRRGRPPKQPAAPAAERCAPAPPPPPPPPTSGPIGGLRSRHRGSSRGRWAAAQAEVAPKTRLSSPRGGSSSRRKPPPPPPAPPSTSAP GRGGRGGGGGRTGGGGGGGHLARTTAARRAVNKVVYDDHESEEEEEEEDMVSEEEEEEDGDAEETQDSEDDEEDEMEEDDDDSDYPEEME DDDDDASYCTESSFRSHSTYSSTPGRRKPRVHRPRSPILEEKDIPPLEFPKSSEDLMVPNEHIMNVIAIYEVLRNFGTVLRLSPFRFEDF CAALVSQEQCTLMAEMHVVLLKAVLREEDTSNTTFGPADLKDSVNSTLYFIDGMTWPEVLRVYCESDKEYHHVLPYQEAEDYPYGPVENK IKVLQFLVDQFLTTNIAREELMSEGVIQYDDHCRVCHKLGDLLCCETCSAVYHLECVKPPLEEVPEDEWQCEVCVAHKVPGVTDCVAEIQ KNKPYIRHEPIGYDRSRRKYWFLNRRLIIEEDTENENEKKIWYYSTKVQLAELIDCLDKDYWEAELCKILEEMREEIHRHMDITEDLTNK ARGSNKSFLAAANEEILESIRAKKGDIDNVKSPEETEKDKNETENDSKDAEKNREEFEDQSLEKDSDDKTPDDDPEQGKSEEPTEVGDKG NSVSANLGDNTTNATSEETSPSEGRSPVGCLSETPDSSNMAEKKVASELPQDVPEEPNKTCESSNTSATTTSIQPNLENSNSSSELNSSQ SESAKAADDPENGERESHTPVSIQEEIVGDFKSEKSNGELSESPGAGKGASGSTRIITRLRNPDSKLSQLKSQQVAAAAHEANKLFKEGK EVLVVNSQGEISRLSTKKEVIMKGNINNYFKLGQEGKYRVYHNQYSTNSFALNKHQHREDHDKRRHLAHKFCLTPAGEFKWNGSVHGSKV LTISTLRLTITQLENNIPSSFLHPNWASHRANWIKAVQMCSKPREFALALAILECAVKPVVMLPIWRESLGHTRLHRMTSIEREEKEKVK KKEKKQEEEETMQQATWVKYTFPVKHQVWKQKGEEYRVTGYGGWSWISKTHVYRFVPKLPGNTNVNYRKSLEGTKNNMDENMDESDKRKC SRSPKKIKIEPDSEKDEVKGSDAAKGADQNEMDISKITEKKDQDVKELLDSDSDKPCKEEPMEVDDDMKTESHVNCQESSQVDVVNVSEG FHLRTSYKKKTKSSKLDGLLERRIKQFTLEEKQRLEKIKLEGGIKGIGKTSTNSSKNLSESPVITKAKEGCQSDSMRQEQSPNANNDQPE DLIQGCSESDSSVLRMSDPSHTTNKLYPKDRVLDDVSIRSPETKCPKQNSIENDIEEKVSDLASRGQEPSKSKTKGNDFFIDDSKLASAD DIGTLICKNKKPLIQEESDTIVSSSKSALHSSVPKSTNDRDATPLSRAMDFEGKLGCDSESNSTLENSSDTVSIQDSSEEDMIVQNSNES ISEQFRTREQDVEVLEPLKCELVSGESTGNCEDRLPVKGTEANGKKPSQQKKLEERPVNKCSDQIKLKNTTDKKNNENRESEKKGQRTST FQINGKDNKPKIYLKGECLKEISESRVVSGNVEPKVNNINKIIPENDIKSLTVKESAIRPFINGDVIMEDFNERNSSETKSHLLSSSDAE GNYRDSLETLPSTKESDSTQTTTPSASCPESNSVNQVEDMEIETSEVKKVTSSPITSEEESNLSNDFIDENGLPINKNENVNGESKRKTV ITEVTTMTSTVATESKTVIKVEKGDKQTVVSSTENCAKSTVTTTTTTVTKLSTPSTGGSVDIISVKEQSKTVVTTTVTDSLTTTGGTLVT SMTVSKEYSTRDKVKLMKFSRPKKTRSGTALPSYRKFVTKSSKKSIFVLPNDDLKKLARKGGIREVPYFNYNAKPALDIWPYPSPRPTFG ITWRYRLQTVKSLAGVSLMLRLLWASLRWDDMAAKAPPGGGTTRTETSETEITTTEIIKRRDVGPYGIRSEYCIRKIICPIGVPETPKET PTPQRKGLRSSALRPKRPETPKQTGPVIIETWVAEEELELWEIRAFAERVEKEKAQAVEQQAKKRLEQQKPTVIATSTTSPTSSTTSTIS PAQKVMVAPISGSVTTGTKMVLTTKVGSPATVTFQQNKNFHQTFATWVKQGQSNSGVVQVQQKVLGIIPSSTGTSQQTFTSFQPRTATVT IRPNTSGSGGTTSNSQVITGPQIRPGMTVIRTPLQQSTLGKAIIRTPVMVQPGAPQQVMTQIIRGQPVSTAVSAPNTVSSTPGQKSLTSA TSTSNIQSSASQPPRPQQGQVKLTMAQLTQLTQGHGGNQGLTVVIQGQGQTTGQLQLIPQGVTVLPGPGQQLMQAAMPNGTVQRFLFTPL ATTATTASTTTTTVSTTAAGTGEQRQSKLSPQMQVHQDKTLPPAQSSSVGPAEAQPQTAQPSAQPQPQTQPQSPAQPEVQTQPEVQTQTT VSSHVPSEAQPTHAQSSKPQVAAQSQPQSNVQGQSPVRVQSPSQTRIRPSTPSQLSPGQQSQVQTTTSQPIPIQPHTSLQIPSQGQPQSQ PQVQSSTQTLSSGQTLNQVTVSSPSRPQLQIQQPQPQVIAVPQLQQQVQVLSQIQSQVVAQIQAQQSGVPQQIKLQLPIQIQQSSAVQTH QIQNVVTVQAASVQEQLQRVQQLRDQQQKKKQQQIEIKREHTLQASNQSEIIQKQVVMKHNAVIEHLKQKKSMTPAEREENQRMIVCNQV MKYILDKIDKEEKQAAKKRKREESVEQKRSKQNATKLSALLFKHKEQLRAEILKKRALLDKDLQIEVQEELKRDLKIKKEKDLMQLAQAT AVAAPCPPVTPAPPAPPAPPPSPPPPPAVQHTGLLSTPTLPAASQKRKREEEKDSSSKSKKKKMISTTSKETKKDTKLYCICKTPYDESK FYIGCDRCQNWYHGRCVGILQSEAELIDEYVCPQCQSTEDAMTVLTPLTEKDYEGLKRVLRSLQVKNRPTSISWDGLDSGKLYTLVLTDP DAPSRKDPKYREWHHFLVVNMKGNDISSGTVLSDYVGSGPPKGTGLHRYVWLVYEQDRPLKCDEPILSNRSGDHRGKFKVASFRKKYELR -------------------------------------------------------------- >10095_10095_3_BPTF-PEBP1_BPTF_chr17_65960520_ENST00000335221_PEBP1_chr12_118575844_ENST00000261313_length(amino acids)=2943AA_BP=2794 MRGRRGRPPKQPAAPAAERCAPAPPPPPPPPTSGPIGGLRSRHRGSSRGRWAAAQAEVAPKTRLSSPRGGSSSRRKPPPPPPAPPSTSAP GRGGRGGGGGRTGGGGGGGHLARTTAARRAVNKVVYDDHESEEEEEEEDMVSEEEEEEDGDAEETQDSEDDEEDEMEEDDDDSDYPEEME DDDDDASYCTESSFRSHSTYSSTPGRRKPRVHRPRSPILEEKDIPPLEFPKSSEDLMVPNEHIMNVIAIYEVLRNFGTVLRLSPFRFEDF CAALVSQEQCTLMAEMHVVLLKAVLREEDTSNTTFGPADLKDSVNSTLYFIDGMTWPEVLRVYCESDKEYHHVLPYQEAEDYPYGPVENK IKVLQFLVDQFLTTNIAREELMSEGVIQYDDHCRVCHKLGDLLCCETCSAVYHLECVKPPLEEVPEDEWQCEVCVAHKVPGVTDCVAEIQ KNKPYIRHEPIGYDRSRRKYWFLNRRLIIEEDTENENEKKIWYYSTKVQLAELIDCLDKDYWEAELCKILEEMREEIHRHMDITEDLTNK ARGSNKSFLAAANEEILESIRAKKGDIDNVKSPEETEKDKNETENDSKDAEKNREEFEDQSLEKDSDDKTPDDDPEQGKSEEPTEVGDKG NSVSANLGDNTTNATSEETSPSEGRSPVGCLSETPDSSNMAEKKVASELPQDVPEEPNKTCESSNTSATTTSIQPNLENSNSSSELNSSQ SESAKAADDPENGERESHTPVSIQEEIVGDFKSEKSNGELSESPGAGKGASGSTRIITRLRNPDSKLSQLKSQQVAAAAHEANKLFKEGK EVLVVNSQGEISRLSTKKEVIMKGNINNYFKLGQEGKYRVYHNQYSTNSFALNKHQHREDHDKRRHLAHKFCLTPAGEFKWNGSVHGSKV LTISTLRLTITQLENNIPSSFLHPNWASHRANWIKAVQMCSKPREFALALAILECAVKPVVMLPIWRESLGHTRLHRMTSIEREEKEKVK KKEKKQEEEETMQQATWVKYTFPVKHQVWKQKGEEYRVTGYGGWSWISKTHVYRFVPKLPGNTNVNYRKSLEGTKNNMDENMDESDKRKC SRSPKKIKIEPDSEKDEVKGSDAAKGADQNEMDISKITEKKDQDVKELLDSDSDKPCKEEPMEVDDDMKTESHVNCQESSQVDVVNVSEG FHLRTSYKKKTKSSKLDGLLERRIKQFTLEEKQRLEKIKLEGGIKGIGKTSTNSSKNLSESPVITKAKEGCQSDSMRQEQSPNANNDQPE DLIQGCSESDSSVLRMSDPSHTTNKLYPKDRVLDDVSIRSPETKCPKQNSIENDIEEKVSDLASRGQEPSKSKTKGNDFFIDDSKLASAD DIGTLICKNKKPLIQEESDTIVSSSKSALHSSVPKSTNDRDATPLSRAMDFEGKLGCDSESNSTLENSSDTVSIQDSSEEDMIVQNSNES ISEQFRTREQDVEVLEPLKCELVSGESTGNCEDRLPVKGTEANGKKPSQQKKLEERPVNKCSDQIKLKNTTDKKNNENRESEKKGQRTST FQINGKDNKPKIYLKGECLKEISESRVVSGNVEPKVNNINKIIPENDIKSLTVKESAIRPFINGDVIMEDFNERNSSETKSHLLSSSDAE GNYRDSLETLPSTKESDSTQTTTPSASCPESNSVNQVEDMEIETSEVKKVTSSPITSEEESNLSNDFIDENGLPINKNENVNGESKRKTV ITEVTTMTSTVATESKTVIKVEKGDKQTVVSSTENCAKSTVTTTTTTVTKLSTPSTGGSVDIISVKEQSKTVVTTTVTDSLTTTGGTLVT SMTVSKEYSTRDKVKLMKFSRPKKTRSGTALPSYRKFVTKSSKKSIFVLPNDDLKKLARKGGIREVPYFNYNAKPALDIWPYPSPRPTFG ITWRYRLQTVKSLAGVSLMLRLLWASLRWDDMAAKAPPGGGTTRTETSETEITTTEIIKRRDVGPYGIRSEYCIRKIICPIGVPETPKET PTPQRKGLRSSALRPKRPETPKQTGPVIIETWVAEEELELWEIRAFAERVEKEKAQAVEQQAKKRLEQQKPTVIATSTTSPTSSTTSTIS PAQKVMVAPISGSVTTGTKMVLTTKVGSPATVTFQQNKNFHQTFATWVKQGQSNSGVVQVQQKVLGIIPSSTGTSQQTFTSFQPRTATVT IRPNTSGSGGTTSNSQVITGPQIRPGMTVIRTPLQQSTLGKAIIRTPVMVQPGAPQQVMTQIIRGQPVSTAVSAPNTVSSTPGQKSLTSA TSTSNIQSSASQPPRPQQGQVKLTMAQLTQLTQGHGGNQGLTVVIQGQGQTTGQLQLIPQGVTVLPGPGQQLMQAAMPNGTVQRFLFTPL ATTATTASTTTTTVSTTAAGTGEQRQSKLSPQMQVHQDKTLPPAQSSSVGPAEAQPQTAQPSAQPQPQTQPQSPAQPEVQTQPEVQTQTT VSSHVPSEAQPTHAQSSKPQVAAQSQPQSNVQGQSPVRVQSPSQTRIRPSTPSQLSPGQQSQVQTTTSQPIPIQPHTSLQIPSQGQPQSQ PQVVMKHNAVIEHLKQKKSMTPAEREENQRMIVCNQVMKYILDKIDKEEKQAAKKRKREESVEQKRSKQNATKLSALLFKHKEQLRAEIL KKRALLDKDLQIEVQEELKRDLKIKKEKDLMQLAQATAVAAPCPPVTPAPPAPPAPPPSPPPPPAVQHTGLLSTPTLPAASQKRKREEEK DSSSKSKKKKMISTTSKETKKDTKLYCICKTPYDESKFYIGCDRCQNWYHGRCVGILQSEAELIDEYVCPQCQSTEDAMTVLTPLTEKDY EGLKRVLRSLQVKNRPTSISWDGLDSGKLYTLVLTDPDAPSRKDPKYREWHHFLVVNMKGNDISSGTVLSDYVGSGPPKGTGLHRYVWLV -------------------------------------------------------------- >10095_10095_4_BPTF-PEBP1_BPTF_chr17_65960520_ENST00000424123_PEBP1_chr12_118575844_ENST00000261313_length(amino acids)=2804AA_BP=2655 MVSEEEEEEDGDAEETQDSEDDEEDEMEEDDDDSDYPEEMEDDDDDASYCTESSFRSHSTYSSTPGRRKPRVHRPRSPILEEKDIPPLEF PKSSEDLMVPNEHIMNVIAIYEVLRNFGTVLRLSPFRFEDFCAALVSQEQCTLMAEMHVVLLKAVLREEDTSNTTFGPADLKDSVNSTLY FIDGMTWPEVLRVYCESDKEYHHVLPYQEAEDYPYGPVENKIKVLQFLVDQFLTTNIAREELMSEGVIQYDDHCRVCHKLGDLLCCETCS AVYHLECVKPPLEEVPEDEWQCEVCVAHKVPGVTDCVAEIQKNKPYIRHEPIGYDRSRRKYWFLNRRLIIEEDTENENEKKIWYYSTKVQ LAELIDCLDKDYWEAELCKILEEMREEIHRHMDITEDLTNKARGSNKSFLAAANEEILESIRAKKGDIDNVKSPEETEKDKNETENDSKD AEKNREEFEDQSLEKDSDDKTPDDDPEQGKSEEPTEVGDKGNSVSANLGDNTTNATSEETSPSEGRSPVGCLSETPDSSNMAEKKVASEL PQDVPEEPNKTCESSNTSATTTSIQPNLENSNSSSELNSSQSESAKAADDPENGERESHTPVSIQEEIVGDFKSEKSNGELSESPGAGKG ASGSTRIITRLRNPDSKLSQLKSQQVAAAAHEANKLFKEGKEVLVVNSQGEISRLSTKKEVIMKGNINNYFKLGQEGKYRVYHNQYSTNS FALNKHQHREDHDKRRHLAHKFCLTPAGEFKWNGSVHGSKVLTISTLRLTITQLENNIPSSFLHPNWASHRANWIKAVQMCSKPREFALA LAILECAVKPVVMLPIWRESLGHTRLHRMTSIEREEKEKVKKKEKKQEEEETMQQATWVKYTFPVKHQVWKQKGEEYRVTGYGGWSWISK THVYRFVPKLPGNTNVNYRKSLEGTKNNMDENMDESDKRKCSRSPKKIKIEPDSEKDEVKGSDAAKGADQNEMDISKITEKKDQDVKELL DSDSDKPCKEEPMEVDDDMKTESHVNCQESSQVDVVNVSEGFHLRTSYKKKTKSSKLDGLLERRIKQFTLEEKQRLEKIKLEGGIKGIGK TSTNSSKNLSESPVITKAKEGCQSDSMRQEQSPNANNDQPEDLIQGCSESDSSVLRMSDPSHTTNKLYPKDRVLDDVSIRSPETKCPKQN SIENDIEEKVSDLASRGQEPSKSKTKGNDFFIDDSKLASADDIGTLICKNKKPLIQEESDTIVSSSKSALHSSVPKSTNDRDATPLSRAM DFEGKLGCDSESNSTLENSSDTVSIQDSSEEDMIVQNSNESISEQFRTREQDVEVLEPLKCELVSGESTGNCEDRLPVKGTEANGKKPSQ QKKLEERPVNKCSDQIKLKNTTDKKNNENRESEKKGQRTSTFQINGKDNKPKIYLKGECLKEISESRVVSGNVEPKVNNINKIIPENDIK SLTVKESAIRPFINGDVIMEDFNERNSSETKSHLLSSSDAEGNYRDSLETLPSTKESDSTQTTTPSASCPESNSVNQVEDMEIETSEVKK VTSSPITSEEESNLSNDFIDENGLPINKNENVNGESKRKTVITEVTTMTSTVATESKTVIKVEKGDKQTVVSSTENCAKSTVTTTTTTVT KLSTPSTGGSVDIISVKEQSKTVVTTTVTDSLTTTGGTLVTSMTVSKEYSTRDKVKLMKFSRPKKTRSGTALPSYRKFVTKSSKKSIFVL PNDDLKKLARKGGIREVPYFNYNAKPALDIWPYPSPRPTFGITWRYRLQTVKSLAGVSLMLRLLWASLRWDDMAAKAPPGGGTTRTETSE TEITTTEIIKRRDVGPYGIRSEYCIRKIICPIGVPETPKETPTPQRKGLRSSALRPKRPETPKQTGPVIIETWVAEEELELWEIRAFAER VEKEKAQAVEQQAKKRLEQQKPTVIATSTTSPTSSTTSTISPAQKVMVAPISGSVTTGTKMVLTTKVGSPATVTFQQNKNFHQTFATWVK QGQSNSGVVQVQQKVLGIIPSSTGTSQQTFTSFQPRTATVTIRPNTSGSGGTTSNSQVITGPQIRPGMTVIRTPLQQSTLGKAIIRTPVM VQPGAPQQVMTQIIRGQPVSTAVSAPNTVSSTPGQKSLTSATSTSNIQSSASQPPRPQQGQVKLTMAQLTQLTQGHGGNQGLTVVIQGQG QTTGQLQLIPQGVTVLPGPGQQLMQAAMPNGTVQRFLFTPLATTATTASTTTTTVSTTAAGTGEQRQSKLSPQMQVHQDKTLPPAQSSSV GPAEAQPQTAQPSAQPQPQTQPQSPAQPEVQTQPEVQTQTTVSSHVPSEAQPTHAQSSKPQVAAQSQPQSNVQGQSPVRVQSPSQTRIRP STPSQLSPGQQSQVQTTTSQPIPIQPHTSLQIPSQGQPQSQPQVVMKHNAVIEHLKQKKSMTPAEREENQRMIVCNQVMKYILDKIDKEE KQAAKKRKREESVEQKRSKQNATKLSALLFKHKEQLRAEILKKRALLDKDLQIEVQEELKRDLKIKKEKDLMQLAQATAVAAPCPPVTPA PPAPPAPPPSPPPPPAVQHTGLLSTPTLPAASQKRKREEEKDSSSKSKKKKMISTTSKETKKDTKLYCICKTPYDESKFYIGCDRCQNWY HGRCVGILQSEAELIDEYVCPQCQSTEDAMTVLTPLTEKDYEGLKRVLRSLQVKNRPTSISWDGLDSGKLYTLVLTDPDAPSRKDPKYRE WHHFLVVNMKGNDISSGTVLSDYVGSGPPKGTGLHRYVWLVYEQDRPLKCDEPILSNRSGDHRGKFKVASFRKKYELRAPVAGTCYQAEW -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr17:65960520/chr12:118575844) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| BPTF | . |
| FUNCTION: Histone-binding component of NURF (nucleosome-remodeling factor), a complex which catalyzes ATP-dependent nucleosome sliding and facilitates transcription of chromatin. Specifically recognizes H3 tails trimethylated on 'Lys-4' (H3K4me3), which mark transcription start sites of virtually all active genes. May also regulate transcription through direct binding to DNA or transcription factors. | FUNCTION: Might normally function as a transcriptional repressor. EWS-fusion-proteins (EFPS) may play a role in the tumorigenic process. They may disturb gene expression by mimicking, or interfering with the normal function of CTD-POLII within the transcription initiation complex. They may also contribute to an aberrant activation of the fusion protein target genes. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000306378 | + | 25 | 28 | 2022_2050 | 2818.0 | 2921.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000306378 | + | 25 | 28 | 2706_2732 | 2818.0 | 2921.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000306378 | + | 25 | 28 | 574_604 | 2818.0 | 2921.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000306378 | + | 25 | 28 | 978_1007 | 2818.0 | 2921.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000321892 | + | 27 | 30 | 2022_2050 | 2944.0 | 3047.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000321892 | + | 27 | 30 | 2706_2732 | 2944.0 | 3047.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000321892 | + | 27 | 30 | 574_604 | 2944.0 | 3047.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000321892 | + | 27 | 30 | 978_1007 | 2944.0 | 3047.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000335221 | + | 27 | 30 | 2022_2050 | 2801.0 | 2904.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000335221 | + | 27 | 30 | 2706_2732 | 2801.0 | 2904.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000335221 | + | 27 | 30 | 574_604 | 2801.0 | 2904.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000335221 | + | 27 | 30 | 978_1007 | 2801.0 | 2904.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000306378 | + | 25 | 28 | 143_180 | 2818.0 | 2921.0 | Compositional bias | Note=Glu-rich |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000306378 | + | 25 | 28 | 149_185 | 2818.0 | 2921.0 | Compositional bias | Note=Asp-rich |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000306378 | + | 25 | 28 | 1709_1803 | 2818.0 | 2921.0 | Compositional bias | Note=Thr-rich |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000306378 | + | 25 | 28 | 2338_2361 | 2818.0 | 2921.0 | Compositional bias | Note=Thr-rich |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000306378 | + | 25 | 28 | 2795_2817 | 2818.0 | 2921.0 | Compositional bias | Note=Pro-rich |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000321892 | + | 27 | 30 | 143_180 | 2944.0 | 3047.0 | Compositional bias | Note=Glu-rich |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000321892 | + | 27 | 30 | 149_185 | 2944.0 | 3047.0 | Compositional bias | Note=Asp-rich |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000321892 | + | 27 | 30 | 1709_1803 | 2944.0 | 3047.0 | Compositional bias | Note=Thr-rich |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000321892 | + | 27 | 30 | 2338_2361 | 2944.0 | 3047.0 | Compositional bias | Note=Thr-rich |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000321892 | + | 27 | 30 | 2795_2817 | 2944.0 | 3047.0 | Compositional bias | Note=Pro-rich |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000321892 | + | 27 | 30 | 2848_2853 | 2944.0 | 3047.0 | Compositional bias | Note=Poly-Lys |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000335221 | + | 27 | 30 | 143_180 | 2801.0 | 2904.0 | Compositional bias | Note=Glu-rich |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000335221 | + | 27 | 30 | 149_185 | 2801.0 | 2904.0 | Compositional bias | Note=Asp-rich |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000335221 | + | 27 | 30 | 1709_1803 | 2801.0 | 2904.0 | Compositional bias | Note=Thr-rich |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000335221 | + | 27 | 30 | 2338_2361 | 2801.0 | 2904.0 | Compositional bias | Note=Thr-rich |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000306378 | + | 25 | 28 | 240_300 | 2818.0 | 2921.0 | Domain | DDT |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000321892 | + | 27 | 30 | 240_300 | 2944.0 | 3047.0 | Domain | DDT |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000335221 | + | 27 | 30 | 240_300 | 2801.0 | 2904.0 | Domain | DDT |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000306378 | + | 25 | 28 | 390_437 | 2818.0 | 2921.0 | Zinc finger | PHD-type 1 |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000321892 | + | 27 | 30 | 2867_2918 | 2944.0 | 3047.0 | Zinc finger | PHD-type 2 |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000321892 | + | 27 | 30 | 390_437 | 2944.0 | 3047.0 | Zinc finger | PHD-type 1 |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000335221 | + | 27 | 30 | 390_437 | 2801.0 | 2904.0 | Zinc finger | PHD-type 1 |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000306378 | + | 25 | 28 | 2848_2853 | 2818.0 | 2921.0 | Compositional bias | Note=Poly-Lys |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000335221 | + | 27 | 30 | 2795_2817 | 2801.0 | 2904.0 | Compositional bias | Note=Pro-rich |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000335221 | + | 27 | 30 | 2848_2853 | 2801.0 | 2904.0 | Compositional bias | Note=Poly-Lys |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000306378 | + | 25 | 28 | 2944_3014 | 2818.0 | 2921.0 | Domain | Bromo |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000321892 | + | 27 | 30 | 2944_3014 | 2944.0 | 3047.0 | Domain | Bromo |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000335221 | + | 27 | 30 | 2944_3014 | 2801.0 | 2904.0 | Domain | Bromo |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000306378 | + | 25 | 28 | 2867_2918 | 2818.0 | 2921.0 | Zinc finger | PHD-type 2 |
| Hgene | BPTF | chr17:65960520 | chr12:118575844 | ENST00000335221 | + | 27 | 30 | 2867_2918 | 2801.0 | 2904.0 | Zinc finger | PHD-type 2 |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| BPTF | |
| PEBP1 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to BPTF-PEBP1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to BPTF-PEBP1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

