| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:BRCA2-RXFP2 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: BRCA2-RXFP2 | FusionPDB ID: 10159 | FusionGDB2.0 ID: 10159 | Hgene | Tgene | Gene symbol | BRCA2 | RXFP2 | Gene ID | 675 | 122042 |
| Gene name | BRCA2 DNA repair associated | relaxin family peptide receptor 2 | |
| Synonyms | BRCC2|BROVCA2|FACD|FAD|FAD1|FANCD|FANCD1|GLM3|PNCA2|XRCC11 | GPR106|GREAT|INSL3R|LGR8|LGR8.1|RXFPR2 | |
| Cytomap | 13q13.1 | 13q13.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | breast cancer type 2 susceptibility proteinBRCA1/BRCA2-containing complex, subunit 2Fanconi anemia group D1 proteinbreast and ovarian cancer susceptibility gene, early onsetbreast and ovarian cancer susceptibility protein 2breast cancer 2 tumor suppr | relaxin receptor 2G protein coupled receptor affecting testicular descentG-protein coupled receptor 106leucine-rich repeat-containing G-protein coupled receptor 8relaxin/insulin like family peptide receptor 2 | |
| Modification date | 20200329 | 20200313 | |
| UniProtAcc | P51587 | . | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000380152, ENST00000544455, | ENST00000298386, ENST00000380314, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 3 X 3 X 1=9 | 4 X 4 X 2=32 |
| # samples | 3 | 4 | |
| ** MAII score | log2(3/9*10)=1.73696559416621 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(4/32*10)=0.321928094887362 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: BRCA2 [Title/Abstract] AND RXFP2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | BRCA2(32945237)-RXFP2(32332395), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | BRCA2-RXFP2 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. BRCA2-RXFP2 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | BRCA2 | GO:0000724 | double-strand break repair via homologous recombination | 20729832 |
| Hgene | BRCA2 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation | 15930293 |
| Hgene | BRCA2 | GO:0043966 | histone H3 acetylation | 9619837 |
| Hgene | BRCA2 | GO:0043967 | histone H4 acetylation | 9619837 |
| Hgene | BRCA2 | GO:0045893 | positive regulation of transcription, DNA-templated | 9126734 |
| Hgene | BRCA2 | GO:0070200 | establishment of protein localization to telomere | 21076401 |
 Fusion gene breakpoints across BRCA2 (5'-gene) Fusion gene breakpoints across BRCA2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
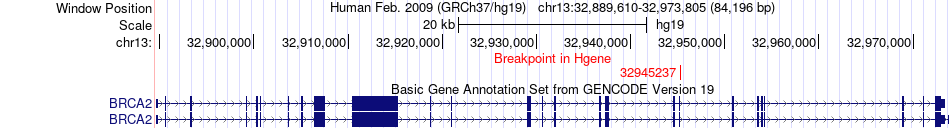 |
 Fusion gene breakpoints across RXFP2 (3'-gene) Fusion gene breakpoints across RXFP2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
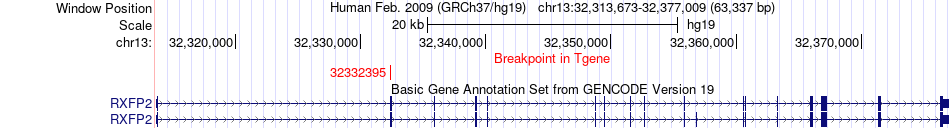 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-A2-A0ET-01A | BRCA2 | chr13 | 32945237 | + | RXFP2 | chr13 | 32332395 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000380152 | BRCA2 | chr13 | 32945237 | + | ENST00000380314 | RXFP2 | chr13 | 32332395 | + | 11431 | 8865 | 233 | 10963 | 3576 |
| ENST00000380152 | BRCA2 | chr13 | 32945237 | + | ENST00000298386 | RXFP2 | chr13 | 32332395 | + | 11503 | 8865 | 233 | 11035 | 3600 |
| ENST00000544455 | BRCA2 | chr13 | 32945237 | + | ENST00000380314 | RXFP2 | chr13 | 32332395 | + | 11425 | 8859 | 227 | 10957 | 3576 |
| ENST00000544455 | BRCA2 | chr13 | 32945237 | + | ENST00000298386 | RXFP2 | chr13 | 32332395 | + | 11497 | 8859 | 227 | 11029 | 3600 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000380152 | ENST00000380314 | BRCA2 | chr13 | 32945237 | + | RXFP2 | chr13 | 32332395 | + | 0.00020541 | 0.99979466 |
| ENST00000380152 | ENST00000298386 | BRCA2 | chr13 | 32945237 | + | RXFP2 | chr13 | 32332395 | + | 0.000146505 | 0.9998535 |
| ENST00000544455 | ENST00000380314 | BRCA2 | chr13 | 32945237 | + | RXFP2 | chr13 | 32332395 | + | 0.000205225 | 0.9997948 |
| ENST00000544455 | ENST00000298386 | BRCA2 | chr13 | 32945237 | + | RXFP2 | chr13 | 32332395 | + | 0.000146572 | 0.9998534 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >10159_10159_1_BRCA2-RXFP2_BRCA2_chr13_32945237_ENST00000380152_RXFP2_chr13_32332395_ENST00000298386_length(amino acids)=3600AA_BP=2875 MPIGSKERPTFFEIFKTRCNKADLGPISLNWFEELSSEAPPYNSEPAEESEHKNNNYEPNLFKTPQRKPSYNQLASTPIIFKEQGLTLPL YQSPVKELDKFKLDLGRNVPNSRHKSLRTVKTKMDQADDVSCPLLNSCLSESPVVLQCTHVTPQRDKSVVCGSLFHTPKFVKGRQTPKHI SESLGAEVDPDMSWSSSLATPPTLSSTVLIVRNEEASETVFPHDTTANVKSYFSNHDESLKKNDRFIASVTDSENTNQREAASHGFGKTS GNSFKVNSCKDHIGKSMPNVLEDEVYETVVDTSEEDSFSLCFSKCRTKNLQKVRTSKTRKKIFHEANADECEKSKNQVKEKYSFVSEVEP NDTDPLDSNVANQKPFESGSDKISKEVVPSLACEWSQLTLSGLNGAQMEKIPLLHISSCDQNISEKDLLDTENKRKKDFLTSENSLPRIS SLPKSEKPLNEETVVNKRDEEQHLESHTDCILAVKQAISGTSPVASSFQGIKKSIFRIRESPKETFNASFSGHMTDPNFKKETEASESGL EIHTVCSQKEDSLCPNLIDNGSWPATTTQNSVALKNAGLISTLKKKTNKFIYAIHDETSYKGKKIPKDQKSELINCSAQFEANAFEAPLT FANADSGLLHSSVKRSCSQNDSEEPTLSLTSSFGTILRKCSRNETCSNNTVISQDLDYKEAKCNKEKLQLFITPEADSLSCLQEGQCEND PKSKKVSDIKEEVLAAACHPVQHSKVEYSDTDFQSQKSLLYDHENASTLILTPTSKDVLSNLVMISRGKESYKMSDKLKGNNYESDVELT KNIPMEKNQDVCALNENYKNVELLPPEKYMRVASPSRKVQFNQNTNLRVIQKNQEETTSISKITVNPDSEELFSDNENNFVFQVANERNN LALGNTKELHETDLTCVNEPIFKNSTMVLYGDTGDKQATQVSIKKDLVYVLAEENKNSVKQHIKMTLGQDLKSDISLNIDKIPEKNNDYM NKWAGLLGPISNHSFGGSFRTASNKEIKLSEHNIKKSKMFFKDIEEQYPTSLACVEIVNTLALDNQKKLSKPQSINTVSAHLQSSVVVSD CKNSHITPQMLFSKQDFNSNHNLTPSQKAEITELSTILEESGSQFEFTQFRKPSYILQKSTFEVPENQMTILKTTSEECRDADLHVIMNA PSIGQVDSSKQFEGTVEIKRKFAGLLKNDCNKSASGYLTDENEVGFRGFYSAHGTKLNVSTEALQKAVKLFSDIENISEETSAEVHPISL SSSKCHDSVVSMFKIENHNDKTVSEKNNKCQLILQNNIEMTTGTFVEEITENYKRNTENEDNKYTAASRNSHNLEFDGSDSSKNDTVCIH KDETDLLFTDQHNICLKLSGQFMKEGNTQIKEDLSDLTFLEVAKAQEACHGNTSNKEQLTATKTEQNIKDFETSDTFFQTASGKNISVAK ESFNKIVNFFDQKPEELHNFSLNSELHSDIRKNKMDILSYEETDIVKHKILKESVPVGTGNQLVTFQGQPERDEKIKEPTLLGFHTASGK KVKIAKESLDKVKNLFDEKEQGTSEITSFSHQWAKTLKYREACKDLELACETIEITAAPKCKEMQNSLNNDKNLVSIETVVPPKLLSDNL CRQTENLKTSKSIFLKVKVHENVEKETAKSPATCYTNQSPYSVIENSALAFYTSCSRKTSVSQTSLLEAKKWLREGIFDGQPERINTADY VGNYLYENNSNSTIAENDKNHLSEKQDTYLSNSSMSNSYSYHSDEVYNDSGYLSKNKLDSGIEPVLKNVEDQKNTSFSKVISNVKDANAY PQTVNEDICVEELVTSSSPCKNKNAAIKLSISNSNNFEVGPPAFRIASGKIVCVSHETIKKVKDIFTDSFSKVIKENNENKSKICQTKIM AGCYEALDDSEDILHNSLDNDECSTHSHKVFADIQSEEILQHNQNMSGLEKVSKISPCDVSLETSDICKCSIGKLHKSVSSANTCGIFST ASGKSVQVSDASLQNARQVFSEIEDSTKQVFSKVLFKSNEHSDQLTREENTAIRTPEHLISQKGFSYNVVNSSAFSGFSTASGKQVSILE SSLHKVKGVLEEFDLIRTEHSLHYSPTSRQNVSKILPRVDKRNPEHCVNSEMEKTCSKEFKLSNNLNVEGGSSENNHSIKVSPYLSQFQQ DKQQLVLGTKVSLVENIHVLGKEQASPKNVKMEIGKTETFSDVPVKTNIEVCSTYSKDSENYFETEAVEIAKAFMEDDELTDSKLPSHAT HSLFTCPENEEMVLSNSRIGKRRGEPLILVGEPSIKRNLLNEFDRIIENQEKSLKASKSTPDGTIKDRRLFMHHVSLEPITCVPFRTTKE RQEIQNPNFTAPGQEFLSKSHLYEHLTLEKSSSNLAVSGHPFYQVSATRNEKMRHLITTGRPTKVFVPPFKTKSHFHRVEQCVRNINLEE NRQKQNIDGHGSDDSKNKINDNEIHQFNKNNSNQAVAVTFTKCEEEPLDLITSLQNARDIQDMRIKKKQRQRVFPQPGSLYLAKTSTLPR ISLKAAVGGQVPSACSHKQLYTYGVSKHCIKINSKNAESFQFHTEDYFGKESLWTGKGIQLADGGWLIPSNDGKAGKEEFYRALCDTPGV DPKLISRIWVYNHYRWIIWKLAAMECAFPKEFANRCLSPERVLLQLKYRYDTEIDRSRRSAIKKIMERDDTAAKTLVLCVSDIISLSANI SETSSNKTSSADTQKVAIIELTDGWYAVKAQLDPPLLAVLKNGRLTVGQKIILHGAELVGSPDACTPLEAPESLMLKISANSTRPARWYT KLGFFPDPRPFPLPLSSLFSDGGNVGCVDVIIQRAYPIQWMEKTSSGLYIFRNEREEEKEAAKYVEAQQKRLEALFTKIQEEFEEHEDFA LTQGSMITPSCQKGYFPCGNLTKCLPRAFHCDGKDDCGNGADEENCGDTSGWATIFGTVHGNANSVALTQECFLKQYPQCCDCKETELEC VNGDLKSVPMISNNVTLLSLKKNKIHSLPDKVFIKYTKLKKIFLQHNCIRHISRKAFFGLCNLQILYLNHNCITTLRPGIFKDLHQLTWL ILDDNPITRISQRLFTGLNSLFFLSMVNNYLEALPKQMCAQMPQLNWVDLEGNRIKYLTNSTFLSCDSLTVLFLPRNQIGFVPEKTFSSL KNLGELDLSSNTITELSPHLFKDLKLLQKLNLSSNPLMYLHKNQFESLKQLQSLDLERIEIPNINTRMFQPMKNLSHIYFKNFRYCSYAP HVRICMPLTDGISSFEDLLANNILRIFVWVIAFITCFGNLFVIGMRSFIKAENTTHAMSIKILCCADCLMGVYLFFVGIFDIKYRGQYQK YALLWMESVQCRLMGFLAMLSTEVSVLLLTYLTLEKFLVIVFPFSNIRPGKRQTSVILICIWMAGFLIAVIPFWNKDYFGNFYGKNGVCF PLYYDQTEDIGSKGYSLGIFLGVNLLAFLIIVFSYITMFCSIQKTALQTTEVRNCFGREVAVANRFFFIVFSDAICWIPVFVVKILSLFR VEIPDTMTSWIVIFFLPVNSALNPILYTLTTNFFKDKLKQLLHKHQRKSIFKIKKKSLSTSIVWIEDSSSLKLGVLNKITLGDSIMKPVS -------------------------------------------------------------- >10159_10159_2_BRCA2-RXFP2_BRCA2_chr13_32945237_ENST00000380152_RXFP2_chr13_32332395_ENST00000380314_length(amino acids)=3576AA_BP=2875 MPIGSKERPTFFEIFKTRCNKADLGPISLNWFEELSSEAPPYNSEPAEESEHKNNNYEPNLFKTPQRKPSYNQLASTPIIFKEQGLTLPL YQSPVKELDKFKLDLGRNVPNSRHKSLRTVKTKMDQADDVSCPLLNSCLSESPVVLQCTHVTPQRDKSVVCGSLFHTPKFVKGRQTPKHI SESLGAEVDPDMSWSSSLATPPTLSSTVLIVRNEEASETVFPHDTTANVKSYFSNHDESLKKNDRFIASVTDSENTNQREAASHGFGKTS GNSFKVNSCKDHIGKSMPNVLEDEVYETVVDTSEEDSFSLCFSKCRTKNLQKVRTSKTRKKIFHEANADECEKSKNQVKEKYSFVSEVEP NDTDPLDSNVANQKPFESGSDKISKEVVPSLACEWSQLTLSGLNGAQMEKIPLLHISSCDQNISEKDLLDTENKRKKDFLTSENSLPRIS SLPKSEKPLNEETVVNKRDEEQHLESHTDCILAVKQAISGTSPVASSFQGIKKSIFRIRESPKETFNASFSGHMTDPNFKKETEASESGL EIHTVCSQKEDSLCPNLIDNGSWPATTTQNSVALKNAGLISTLKKKTNKFIYAIHDETSYKGKKIPKDQKSELINCSAQFEANAFEAPLT FANADSGLLHSSVKRSCSQNDSEEPTLSLTSSFGTILRKCSRNETCSNNTVISQDLDYKEAKCNKEKLQLFITPEADSLSCLQEGQCEND PKSKKVSDIKEEVLAAACHPVQHSKVEYSDTDFQSQKSLLYDHENASTLILTPTSKDVLSNLVMISRGKESYKMSDKLKGNNYESDVELT KNIPMEKNQDVCALNENYKNVELLPPEKYMRVASPSRKVQFNQNTNLRVIQKNQEETTSISKITVNPDSEELFSDNENNFVFQVANERNN LALGNTKELHETDLTCVNEPIFKNSTMVLYGDTGDKQATQVSIKKDLVYVLAEENKNSVKQHIKMTLGQDLKSDISLNIDKIPEKNNDYM NKWAGLLGPISNHSFGGSFRTASNKEIKLSEHNIKKSKMFFKDIEEQYPTSLACVEIVNTLALDNQKKLSKPQSINTVSAHLQSSVVVSD CKNSHITPQMLFSKQDFNSNHNLTPSQKAEITELSTILEESGSQFEFTQFRKPSYILQKSTFEVPENQMTILKTTSEECRDADLHVIMNA PSIGQVDSSKQFEGTVEIKRKFAGLLKNDCNKSASGYLTDENEVGFRGFYSAHGTKLNVSTEALQKAVKLFSDIENISEETSAEVHPISL SSSKCHDSVVSMFKIENHNDKTVSEKNNKCQLILQNNIEMTTGTFVEEITENYKRNTENEDNKYTAASRNSHNLEFDGSDSSKNDTVCIH KDETDLLFTDQHNICLKLSGQFMKEGNTQIKEDLSDLTFLEVAKAQEACHGNTSNKEQLTATKTEQNIKDFETSDTFFQTASGKNISVAK ESFNKIVNFFDQKPEELHNFSLNSELHSDIRKNKMDILSYEETDIVKHKILKESVPVGTGNQLVTFQGQPERDEKIKEPTLLGFHTASGK KVKIAKESLDKVKNLFDEKEQGTSEITSFSHQWAKTLKYREACKDLELACETIEITAAPKCKEMQNSLNNDKNLVSIETVVPPKLLSDNL CRQTENLKTSKSIFLKVKVHENVEKETAKSPATCYTNQSPYSVIENSALAFYTSCSRKTSVSQTSLLEAKKWLREGIFDGQPERINTADY VGNYLYENNSNSTIAENDKNHLSEKQDTYLSNSSMSNSYSYHSDEVYNDSGYLSKNKLDSGIEPVLKNVEDQKNTSFSKVISNVKDANAY PQTVNEDICVEELVTSSSPCKNKNAAIKLSISNSNNFEVGPPAFRIASGKIVCVSHETIKKVKDIFTDSFSKVIKENNENKSKICQTKIM AGCYEALDDSEDILHNSLDNDECSTHSHKVFADIQSEEILQHNQNMSGLEKVSKISPCDVSLETSDICKCSIGKLHKSVSSANTCGIFST ASGKSVQVSDASLQNARQVFSEIEDSTKQVFSKVLFKSNEHSDQLTREENTAIRTPEHLISQKGFSYNVVNSSAFSGFSTASGKQVSILE SSLHKVKGVLEEFDLIRTEHSLHYSPTSRQNVSKILPRVDKRNPEHCVNSEMEKTCSKEFKLSNNLNVEGGSSENNHSIKVSPYLSQFQQ DKQQLVLGTKVSLVENIHVLGKEQASPKNVKMEIGKTETFSDVPVKTNIEVCSTYSKDSENYFETEAVEIAKAFMEDDELTDSKLPSHAT HSLFTCPENEEMVLSNSRIGKRRGEPLILVGEPSIKRNLLNEFDRIIENQEKSLKASKSTPDGTIKDRRLFMHHVSLEPITCVPFRTTKE RQEIQNPNFTAPGQEFLSKSHLYEHLTLEKSSSNLAVSGHPFYQVSATRNEKMRHLITTGRPTKVFVPPFKTKSHFHRVEQCVRNINLEE NRQKQNIDGHGSDDSKNKINDNEIHQFNKNNSNQAVAVTFTKCEEEPLDLITSLQNARDIQDMRIKKKQRQRVFPQPGSLYLAKTSTLPR ISLKAAVGGQVPSACSHKQLYTYGVSKHCIKINSKNAESFQFHTEDYFGKESLWTGKGIQLADGGWLIPSNDGKAGKEEFYRALCDTPGV DPKLISRIWVYNHYRWIIWKLAAMECAFPKEFANRCLSPERVLLQLKYRYDTEIDRSRRSAIKKIMERDDTAAKTLVLCVSDIISLSANI SETSSNKTSSADTQKVAIIELTDGWYAVKAQLDPPLLAVLKNGRLTVGQKIILHGAELVGSPDACTPLEAPESLMLKISANSTRPARWYT KLGFFPDPRPFPLPLSSLFSDGGNVGCVDVIIQRAYPIQWMEKTSSGLYIFRNEREEEKEAAKYVEAQQKRLEALFTKIQEEFEEHEDFA LTQGSMITPSCQKGYFPCGNLTKCLPRAFHCDGKDDCGNGADEENCGDTSGWATIFGTVHGNANSVALTQECFLKQYPQCCDCKETELEC VNGDLKSVPMISNNVTLLSLKKNKIHSLPDKVFIKYTKLKKIFLQHNCIRHISRKAFFGLCNLQILYLNHNCITTLRPGIFKDLHQLTWL ILDDNPITRISQRLFTGLNSLFFLSMVNNYLEALPKQMCAQMPQLNWVDLEGNRIKYLTNSTFLSCDSLTVLDLSSNTITELSPHLFKDL KLLQKLNLSSNPLMYLHKNQFESLKQLQSLDLERIEIPNINTRMFQPMKNLSHIYFKNFRYCSYAPHVRICMPLTDGISSFEDLLANNIL RIFVWVIAFITCFGNLFVIGMRSFIKAENTTHAMSIKILCCADCLMGVYLFFVGIFDIKYRGQYQKYALLWMESVQCRLMGFLAMLSTEV SVLLLTYLTLEKFLVIVFPFSNIRPGKRQTSVILICIWMAGFLIAVIPFWNKDYFGNFYGKNGVCFPLYYDQTEDIGSKGYSLGIFLGVN LLAFLIIVFSYITMFCSIQKTALQTTEVRNCFGREVAVANRFFFIVFSDAICWIPVFVVKILSLFRVEIPDTMTSWIVIFFLPVNSALNP -------------------------------------------------------------- >10159_10159_3_BRCA2-RXFP2_BRCA2_chr13_32945237_ENST00000544455_RXFP2_chr13_32332395_ENST00000298386_length(amino acids)=3600AA_BP=2875 MPIGSKERPTFFEIFKTRCNKADLGPISLNWFEELSSEAPPYNSEPAEESEHKNNNYEPNLFKTPQRKPSYNQLASTPIIFKEQGLTLPL YQSPVKELDKFKLDLGRNVPNSRHKSLRTVKTKMDQADDVSCPLLNSCLSESPVVLQCTHVTPQRDKSVVCGSLFHTPKFVKGRQTPKHI SESLGAEVDPDMSWSSSLATPPTLSSTVLIVRNEEASETVFPHDTTANVKSYFSNHDESLKKNDRFIASVTDSENTNQREAASHGFGKTS GNSFKVNSCKDHIGKSMPNVLEDEVYETVVDTSEEDSFSLCFSKCRTKNLQKVRTSKTRKKIFHEANADECEKSKNQVKEKYSFVSEVEP NDTDPLDSNVANQKPFESGSDKISKEVVPSLACEWSQLTLSGLNGAQMEKIPLLHISSCDQNISEKDLLDTENKRKKDFLTSENSLPRIS SLPKSEKPLNEETVVNKRDEEQHLESHTDCILAVKQAISGTSPVASSFQGIKKSIFRIRESPKETFNASFSGHMTDPNFKKETEASESGL EIHTVCSQKEDSLCPNLIDNGSWPATTTQNSVALKNAGLISTLKKKTNKFIYAIHDETSYKGKKIPKDQKSELINCSAQFEANAFEAPLT FANADSGLLHSSVKRSCSQNDSEEPTLSLTSSFGTILRKCSRNETCSNNTVISQDLDYKEAKCNKEKLQLFITPEADSLSCLQEGQCEND PKSKKVSDIKEEVLAAACHPVQHSKVEYSDTDFQSQKSLLYDHENASTLILTPTSKDVLSNLVMISRGKESYKMSDKLKGNNYESDVELT KNIPMEKNQDVCALNENYKNVELLPPEKYMRVASPSRKVQFNQNTNLRVIQKNQEETTSISKITVNPDSEELFSDNENNFVFQVANERNN LALGNTKELHETDLTCVNEPIFKNSTMVLYGDTGDKQATQVSIKKDLVYVLAEENKNSVKQHIKMTLGQDLKSDISLNIDKIPEKNNDYM NKWAGLLGPISNHSFGGSFRTASNKEIKLSEHNIKKSKMFFKDIEEQYPTSLACVEIVNTLALDNQKKLSKPQSINTVSAHLQSSVVVSD CKNSHITPQMLFSKQDFNSNHNLTPSQKAEITELSTILEESGSQFEFTQFRKPSYILQKSTFEVPENQMTILKTTSEECRDADLHVIMNA PSIGQVDSSKQFEGTVEIKRKFAGLLKNDCNKSASGYLTDENEVGFRGFYSAHGTKLNVSTEALQKAVKLFSDIENISEETSAEVHPISL SSSKCHDSVVSMFKIENHNDKTVSEKNNKCQLILQNNIEMTTGTFVEEITENYKRNTENEDNKYTAASRNSHNLEFDGSDSSKNDTVCIH KDETDLLFTDQHNICLKLSGQFMKEGNTQIKEDLSDLTFLEVAKAQEACHGNTSNKEQLTATKTEQNIKDFETSDTFFQTASGKNISVAK ESFNKIVNFFDQKPEELHNFSLNSELHSDIRKNKMDILSYEETDIVKHKILKESVPVGTGNQLVTFQGQPERDEKIKEPTLLGFHTASGK KVKIAKESLDKVKNLFDEKEQGTSEITSFSHQWAKTLKYREACKDLELACETIEITAAPKCKEMQNSLNNDKNLVSIETVVPPKLLSDNL CRQTENLKTSKSIFLKVKVHENVEKETAKSPATCYTNQSPYSVIENSALAFYTSCSRKTSVSQTSLLEAKKWLREGIFDGQPERINTADY VGNYLYENNSNSTIAENDKNHLSEKQDTYLSNSSMSNSYSYHSDEVYNDSGYLSKNKLDSGIEPVLKNVEDQKNTSFSKVISNVKDANAY PQTVNEDICVEELVTSSSPCKNKNAAIKLSISNSNNFEVGPPAFRIASGKIVCVSHETIKKVKDIFTDSFSKVIKENNENKSKICQTKIM AGCYEALDDSEDILHNSLDNDECSTHSHKVFADIQSEEILQHNQNMSGLEKVSKISPCDVSLETSDICKCSIGKLHKSVSSANTCGIFST ASGKSVQVSDASLQNARQVFSEIEDSTKQVFSKVLFKSNEHSDQLTREENTAIRTPEHLISQKGFSYNVVNSSAFSGFSTASGKQVSILE SSLHKVKGVLEEFDLIRTEHSLHYSPTSRQNVSKILPRVDKRNPEHCVNSEMEKTCSKEFKLSNNLNVEGGSSENNHSIKVSPYLSQFQQ DKQQLVLGTKVSLVENIHVLGKEQASPKNVKMEIGKTETFSDVPVKTNIEVCSTYSKDSENYFETEAVEIAKAFMEDDELTDSKLPSHAT HSLFTCPENEEMVLSNSRIGKRRGEPLILVGEPSIKRNLLNEFDRIIENQEKSLKASKSTPDGTIKDRRLFMHHVSLEPITCVPFRTTKE RQEIQNPNFTAPGQEFLSKSHLYEHLTLEKSSSNLAVSGHPFYQVSATRNEKMRHLITTGRPTKVFVPPFKTKSHFHRVEQCVRNINLEE NRQKQNIDGHGSDDSKNKINDNEIHQFNKNNSNQAVAVTFTKCEEEPLDLITSLQNARDIQDMRIKKKQRQRVFPQPGSLYLAKTSTLPR ISLKAAVGGQVPSACSHKQLYTYGVSKHCIKINSKNAESFQFHTEDYFGKESLWTGKGIQLADGGWLIPSNDGKAGKEEFYRALCDTPGV DPKLISRIWVYNHYRWIIWKLAAMECAFPKEFANRCLSPERVLLQLKYRYDTEIDRSRRSAIKKIMERDDTAAKTLVLCVSDIISLSANI SETSSNKTSSADTQKVAIIELTDGWYAVKAQLDPPLLAVLKNGRLTVGQKIILHGAELVGSPDACTPLEAPESLMLKISANSTRPARWYT KLGFFPDPRPFPLPLSSLFSDGGNVGCVDVIIQRAYPIQWMEKTSSGLYIFRNEREEEKEAAKYVEAQQKRLEALFTKIQEEFEEHEDFA LTQGSMITPSCQKGYFPCGNLTKCLPRAFHCDGKDDCGNGADEENCGDTSGWATIFGTVHGNANSVALTQECFLKQYPQCCDCKETELEC VNGDLKSVPMISNNVTLLSLKKNKIHSLPDKVFIKYTKLKKIFLQHNCIRHISRKAFFGLCNLQILYLNHNCITTLRPGIFKDLHQLTWL ILDDNPITRISQRLFTGLNSLFFLSMVNNYLEALPKQMCAQMPQLNWVDLEGNRIKYLTNSTFLSCDSLTVLFLPRNQIGFVPEKTFSSL KNLGELDLSSNTITELSPHLFKDLKLLQKLNLSSNPLMYLHKNQFESLKQLQSLDLERIEIPNINTRMFQPMKNLSHIYFKNFRYCSYAP HVRICMPLTDGISSFEDLLANNILRIFVWVIAFITCFGNLFVIGMRSFIKAENTTHAMSIKILCCADCLMGVYLFFVGIFDIKYRGQYQK YALLWMESVQCRLMGFLAMLSTEVSVLLLTYLTLEKFLVIVFPFSNIRPGKRQTSVILICIWMAGFLIAVIPFWNKDYFGNFYGKNGVCF PLYYDQTEDIGSKGYSLGIFLGVNLLAFLIIVFSYITMFCSIQKTALQTTEVRNCFGREVAVANRFFFIVFSDAICWIPVFVVKILSLFR VEIPDTMTSWIVIFFLPVNSALNPILYTLTTNFFKDKLKQLLHKHQRKSIFKIKKKSLSTSIVWIEDSSSLKLGVLNKITLGDSIMKPVS -------------------------------------------------------------- >10159_10159_4_BRCA2-RXFP2_BRCA2_chr13_32945237_ENST00000544455_RXFP2_chr13_32332395_ENST00000380314_length(amino acids)=3576AA_BP=2875 MPIGSKERPTFFEIFKTRCNKADLGPISLNWFEELSSEAPPYNSEPAEESEHKNNNYEPNLFKTPQRKPSYNQLASTPIIFKEQGLTLPL YQSPVKELDKFKLDLGRNVPNSRHKSLRTVKTKMDQADDVSCPLLNSCLSESPVVLQCTHVTPQRDKSVVCGSLFHTPKFVKGRQTPKHI SESLGAEVDPDMSWSSSLATPPTLSSTVLIVRNEEASETVFPHDTTANVKSYFSNHDESLKKNDRFIASVTDSENTNQREAASHGFGKTS GNSFKVNSCKDHIGKSMPNVLEDEVYETVVDTSEEDSFSLCFSKCRTKNLQKVRTSKTRKKIFHEANADECEKSKNQVKEKYSFVSEVEP NDTDPLDSNVANQKPFESGSDKISKEVVPSLACEWSQLTLSGLNGAQMEKIPLLHISSCDQNISEKDLLDTENKRKKDFLTSENSLPRIS SLPKSEKPLNEETVVNKRDEEQHLESHTDCILAVKQAISGTSPVASSFQGIKKSIFRIRESPKETFNASFSGHMTDPNFKKETEASESGL EIHTVCSQKEDSLCPNLIDNGSWPATTTQNSVALKNAGLISTLKKKTNKFIYAIHDETSYKGKKIPKDQKSELINCSAQFEANAFEAPLT FANADSGLLHSSVKRSCSQNDSEEPTLSLTSSFGTILRKCSRNETCSNNTVISQDLDYKEAKCNKEKLQLFITPEADSLSCLQEGQCEND PKSKKVSDIKEEVLAAACHPVQHSKVEYSDTDFQSQKSLLYDHENASTLILTPTSKDVLSNLVMISRGKESYKMSDKLKGNNYESDVELT KNIPMEKNQDVCALNENYKNVELLPPEKYMRVASPSRKVQFNQNTNLRVIQKNQEETTSISKITVNPDSEELFSDNENNFVFQVANERNN LALGNTKELHETDLTCVNEPIFKNSTMVLYGDTGDKQATQVSIKKDLVYVLAEENKNSVKQHIKMTLGQDLKSDISLNIDKIPEKNNDYM NKWAGLLGPISNHSFGGSFRTASNKEIKLSEHNIKKSKMFFKDIEEQYPTSLACVEIVNTLALDNQKKLSKPQSINTVSAHLQSSVVVSD CKNSHITPQMLFSKQDFNSNHNLTPSQKAEITELSTILEESGSQFEFTQFRKPSYILQKSTFEVPENQMTILKTTSEECRDADLHVIMNA PSIGQVDSSKQFEGTVEIKRKFAGLLKNDCNKSASGYLTDENEVGFRGFYSAHGTKLNVSTEALQKAVKLFSDIENISEETSAEVHPISL SSSKCHDSVVSMFKIENHNDKTVSEKNNKCQLILQNNIEMTTGTFVEEITENYKRNTENEDNKYTAASRNSHNLEFDGSDSSKNDTVCIH KDETDLLFTDQHNICLKLSGQFMKEGNTQIKEDLSDLTFLEVAKAQEACHGNTSNKEQLTATKTEQNIKDFETSDTFFQTASGKNISVAK ESFNKIVNFFDQKPEELHNFSLNSELHSDIRKNKMDILSYEETDIVKHKILKESVPVGTGNQLVTFQGQPERDEKIKEPTLLGFHTASGK KVKIAKESLDKVKNLFDEKEQGTSEITSFSHQWAKTLKYREACKDLELACETIEITAAPKCKEMQNSLNNDKNLVSIETVVPPKLLSDNL CRQTENLKTSKSIFLKVKVHENVEKETAKSPATCYTNQSPYSVIENSALAFYTSCSRKTSVSQTSLLEAKKWLREGIFDGQPERINTADY VGNYLYENNSNSTIAENDKNHLSEKQDTYLSNSSMSNSYSYHSDEVYNDSGYLSKNKLDSGIEPVLKNVEDQKNTSFSKVISNVKDANAY PQTVNEDICVEELVTSSSPCKNKNAAIKLSISNSNNFEVGPPAFRIASGKIVCVSHETIKKVKDIFTDSFSKVIKENNENKSKICQTKIM AGCYEALDDSEDILHNSLDNDECSTHSHKVFADIQSEEILQHNQNMSGLEKVSKISPCDVSLETSDICKCSIGKLHKSVSSANTCGIFST ASGKSVQVSDASLQNARQVFSEIEDSTKQVFSKVLFKSNEHSDQLTREENTAIRTPEHLISQKGFSYNVVNSSAFSGFSTASGKQVSILE SSLHKVKGVLEEFDLIRTEHSLHYSPTSRQNVSKILPRVDKRNPEHCVNSEMEKTCSKEFKLSNNLNVEGGSSENNHSIKVSPYLSQFQQ DKQQLVLGTKVSLVENIHVLGKEQASPKNVKMEIGKTETFSDVPVKTNIEVCSTYSKDSENYFETEAVEIAKAFMEDDELTDSKLPSHAT HSLFTCPENEEMVLSNSRIGKRRGEPLILVGEPSIKRNLLNEFDRIIENQEKSLKASKSTPDGTIKDRRLFMHHVSLEPITCVPFRTTKE RQEIQNPNFTAPGQEFLSKSHLYEHLTLEKSSSNLAVSGHPFYQVSATRNEKMRHLITTGRPTKVFVPPFKTKSHFHRVEQCVRNINLEE NRQKQNIDGHGSDDSKNKINDNEIHQFNKNNSNQAVAVTFTKCEEEPLDLITSLQNARDIQDMRIKKKQRQRVFPQPGSLYLAKTSTLPR ISLKAAVGGQVPSACSHKQLYTYGVSKHCIKINSKNAESFQFHTEDYFGKESLWTGKGIQLADGGWLIPSNDGKAGKEEFYRALCDTPGV DPKLISRIWVYNHYRWIIWKLAAMECAFPKEFANRCLSPERVLLQLKYRYDTEIDRSRRSAIKKIMERDDTAAKTLVLCVSDIISLSANI SETSSNKTSSADTQKVAIIELTDGWYAVKAQLDPPLLAVLKNGRLTVGQKIILHGAELVGSPDACTPLEAPESLMLKISANSTRPARWYT KLGFFPDPRPFPLPLSSLFSDGGNVGCVDVIIQRAYPIQWMEKTSSGLYIFRNEREEEKEAAKYVEAQQKRLEALFTKIQEEFEEHEDFA LTQGSMITPSCQKGYFPCGNLTKCLPRAFHCDGKDDCGNGADEENCGDTSGWATIFGTVHGNANSVALTQECFLKQYPQCCDCKETELEC VNGDLKSVPMISNNVTLLSLKKNKIHSLPDKVFIKYTKLKKIFLQHNCIRHISRKAFFGLCNLQILYLNHNCITTLRPGIFKDLHQLTWL ILDDNPITRISQRLFTGLNSLFFLSMVNNYLEALPKQMCAQMPQLNWVDLEGNRIKYLTNSTFLSCDSLTVLDLSSNTITELSPHLFKDL KLLQKLNLSSNPLMYLHKNQFESLKQLQSLDLERIEIPNINTRMFQPMKNLSHIYFKNFRYCSYAPHVRICMPLTDGISSFEDLLANNIL RIFVWVIAFITCFGNLFVIGMRSFIKAENTTHAMSIKILCCADCLMGVYLFFVGIFDIKYRGQYQKYALLWMESVQCRLMGFLAMLSTEV SVLLLTYLTLEKFLVIVFPFSNIRPGKRQTSVILICIWMAGFLIAVIPFWNKDYFGNFYGKNGVCFPLYYDQTEDIGSKGYSLGIFLGVN LLAFLIIVFSYITMFCSIQKTALQTTEVRNCFGREVAVANRFFFIVFSDAICWIPVFVVKILSLFRVEIPDTMTSWIVIFFLPVNSALNP -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr13:32945237/chr13:32332395) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| BRCA2 | . |
| FUNCTION: Involved in double-strand break repair and/or homologous recombination. Binds RAD51 and potentiates recombinational DNA repair by promoting assembly of RAD51 onto single-stranded DNA (ssDNA). Acts by targeting RAD51 to ssDNA over double-stranded DNA, enabling RAD51 to displace replication protein-A (RPA) from ssDNA and stabilizing RAD51-ssDNA filaments by blocking ATP hydrolysis. Part of a PALB2-scaffolded HR complex containing RAD51C and which is thought to play a role in DNA repair by HR. May participate in S phase checkpoint activation. Binds selectively to ssDNA, and to ssDNA in tailed duplexes and replication fork structures. May play a role in the extension step after strand invasion at replication-dependent DNA double-strand breaks; together with PALB2 is involved in both POLH localization at collapsed replication forks and DNA polymerization activity. In concert with NPM1, regulates centrosome duplication. Interacts with the TREX-2 complex (transcription and export complex 2) subunits PCID2 and SEM1, and is required to prevent R-loop-associated DNA damage and thus transcription-associated genomic instability. Silencing of BRCA2 promotes R-loop accumulation at actively transcribed genes in replicating and non-replicating cells, suggesting that BRCA2 mediates the control of R-loop associated genomic instability, independently of its known role in homologous recombination (PubMed:24896180). {ECO:0000269|PubMed:15115758, ECO:0000269|PubMed:15199141, ECO:0000269|PubMed:15671039, ECO:0000269|PubMed:18317453, ECO:0000269|PubMed:20729832, ECO:0000269|PubMed:20729858, ECO:0000269|PubMed:20729859, ECO:0000269|PubMed:21084279, ECO:0000269|PubMed:21719596, ECO:0000269|PubMed:24485656, ECO:0000269|PubMed:24896180}. | FUNCTION: Might normally function as a transcriptional repressor. EWS-fusion-proteins (EFPS) may play a role in the tumorigenic process. They may disturb gene expression by mimicking, or interfering with the normal function of CTD-POLII within the transcription initiation complex. They may also contribute to an aberrant activation of the fusion protein target genes. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | BRCA2 | chr13:32945237 | chr13:32332395 | ENST00000380152 | + | 20 | 27 | 1410_1595 | 2877.3333333333335 | 3419.0 | Region | Note=Required for stimulation of POLH DNA polymerization activity |
| Hgene | BRCA2 | chr13:32945237 | chr13:32332395 | ENST00000544455 | + | 20 | 28 | 1410_1595 | 2877.3333333333335 | 3376.3333333333335 | Region | Note=Required for stimulation of POLH DNA polymerization activity |
| Hgene | BRCA2 | chr13:32945237 | chr13:32332395 | ENST00000380152 | + | 20 | 27 | 1002_1036 | 2877.3333333333335 | 3419.0 | Repeat | Note=BRCA2 1 |
| Hgene | BRCA2 | chr13:32945237 | chr13:32332395 | ENST00000380152 | + | 20 | 27 | 1212_1246 | 2877.3333333333335 | 3419.0 | Repeat | Note=BRCA2 2 |
| Hgene | BRCA2 | chr13:32945237 | chr13:32332395 | ENST00000380152 | + | 20 | 27 | 1421_1455 | 2877.3333333333335 | 3419.0 | Repeat | Note=BRCA2 3 |
| Hgene | BRCA2 | chr13:32945237 | chr13:32332395 | ENST00000380152 | + | 20 | 27 | 1517_1551 | 2877.3333333333335 | 3419.0 | Repeat | Note=BRCA2 4 |
| Hgene | BRCA2 | chr13:32945237 | chr13:32332395 | ENST00000380152 | + | 20 | 27 | 1664_1698 | 2877.3333333333335 | 3419.0 | Repeat | Note=BRCA2 5 |
| Hgene | BRCA2 | chr13:32945237 | chr13:32332395 | ENST00000380152 | + | 20 | 27 | 1837_1871 | 2877.3333333333335 | 3419.0 | Repeat | Note=BRCA2 6 |
| Hgene | BRCA2 | chr13:32945237 | chr13:32332395 | ENST00000380152 | + | 20 | 27 | 1971_2005 | 2877.3333333333335 | 3419.0 | Repeat | Note=BRCA2 7 |
| Hgene | BRCA2 | chr13:32945237 | chr13:32332395 | ENST00000380152 | + | 20 | 27 | 2051_2085 | 2877.3333333333335 | 3419.0 | Repeat | Note=BRCA2 8 |
| Hgene | BRCA2 | chr13:32945237 | chr13:32332395 | ENST00000544455 | + | 20 | 28 | 1002_1036 | 2877.3333333333335 | 3376.3333333333335 | Repeat | Note=BRCA2 1 |
| Hgene | BRCA2 | chr13:32945237 | chr13:32332395 | ENST00000544455 | + | 20 | 28 | 1212_1246 | 2877.3333333333335 | 3376.3333333333335 | Repeat | Note=BRCA2 2 |
| Hgene | BRCA2 | chr13:32945237 | chr13:32332395 | ENST00000544455 | + | 20 | 28 | 1421_1455 | 2877.3333333333335 | 3376.3333333333335 | Repeat | Note=BRCA2 3 |
| Hgene | BRCA2 | chr13:32945237 | chr13:32332395 | ENST00000544455 | + | 20 | 28 | 1517_1551 | 2877.3333333333335 | 3376.3333333333335 | Repeat | Note=BRCA2 4 |
| Hgene | BRCA2 | chr13:32945237 | chr13:32332395 | ENST00000544455 | + | 20 | 28 | 1664_1698 | 2877.3333333333335 | 3376.3333333333335 | Repeat | Note=BRCA2 5 |
| Hgene | BRCA2 | chr13:32945237 | chr13:32332395 | ENST00000544455 | + | 20 | 28 | 1837_1871 | 2877.3333333333335 | 3376.3333333333335 | Repeat | Note=BRCA2 6 |
| Hgene | BRCA2 | chr13:32945237 | chr13:32332395 | ENST00000544455 | + | 20 | 28 | 1971_2005 | 2877.3333333333335 | 3376.3333333333335 | Repeat | Note=BRCA2 7 |
| Hgene | BRCA2 | chr13:32945237 | chr13:32332395 | ENST00000544455 | + | 20 | 28 | 2051_2085 | 2877.3333333333335 | 3376.3333333333335 | Repeat | Note=BRCA2 8 |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000298386 | 0 | 18 | 44_81 | 31.333333333333332 | 755.0 | Domain | LDL-receptor class A | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000380314 | 0 | 17 | 44_81 | 31.333333333333332 | 731.0 | Domain | LDL-receptor class A | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000298386 | 0 | 18 | 138_159 | 31.333333333333332 | 755.0 | Repeat | Note=LRR 1 | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000298386 | 0 | 18 | 162_183 | 31.333333333333332 | 755.0 | Repeat | Note=LRR 2 | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000298386 | 0 | 18 | 186_207 | 31.333333333333332 | 755.0 | Repeat | Note=LRR 3 | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000298386 | 0 | 18 | 210_231 | 31.333333333333332 | 755.0 | Repeat | Note=LRR 4 | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000298386 | 0 | 18 | 234_255 | 31.333333333333332 | 755.0 | Repeat | Note=LRR 5 | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000298386 | 0 | 18 | 258_279 | 31.333333333333332 | 755.0 | Repeat | Note=LRR 6 | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000298386 | 0 | 18 | 282_303 | 31.333333333333332 | 755.0 | Repeat | Note=LRR 7 | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000298386 | 0 | 18 | 306_327 | 31.333333333333332 | 755.0 | Repeat | Note=LRR 8 | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000298386 | 0 | 18 | 330_351 | 31.333333333333332 | 755.0 | Repeat | Note=LRR 9 | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000298386 | 0 | 18 | 354_375 | 31.333333333333332 | 755.0 | Repeat | Note=LRR 10 | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000380314 | 0 | 17 | 138_159 | 31.333333333333332 | 731.0 | Repeat | Note=LRR 1 | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000380314 | 0 | 17 | 162_183 | 31.333333333333332 | 731.0 | Repeat | Note=LRR 2 | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000380314 | 0 | 17 | 186_207 | 31.333333333333332 | 731.0 | Repeat | Note=LRR 3 | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000380314 | 0 | 17 | 210_231 | 31.333333333333332 | 731.0 | Repeat | Note=LRR 4 | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000380314 | 0 | 17 | 234_255 | 31.333333333333332 | 731.0 | Repeat | Note=LRR 5 | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000380314 | 0 | 17 | 258_279 | 31.333333333333332 | 731.0 | Repeat | Note=LRR 6 | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000380314 | 0 | 17 | 282_303 | 31.333333333333332 | 731.0 | Repeat | Note=LRR 7 | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000380314 | 0 | 17 | 306_327 | 31.333333333333332 | 731.0 | Repeat | Note=LRR 8 | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000380314 | 0 | 17 | 330_351 | 31.333333333333332 | 731.0 | Repeat | Note=LRR 9 | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000380314 | 0 | 17 | 354_375 | 31.333333333333332 | 731.0 | Repeat | Note=LRR 10 | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000298386 | 0 | 18 | 438_455 | 31.333333333333332 | 755.0 | Topological domain | Cytoplasmic | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000298386 | 0 | 18 | 477_495 | 31.333333333333332 | 755.0 | Topological domain | Extracellular | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000298386 | 0 | 18 | 519_537 | 31.333333333333332 | 755.0 | Topological domain | Cytoplasmic | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000298386 | 0 | 18 | 559_592 | 31.333333333333332 | 755.0 | Topological domain | Extracellular | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000298386 | 0 | 18 | 614_639 | 31.333333333333332 | 755.0 | Topological domain | Cytoplasmic | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000298386 | 0 | 18 | 661_670 | 31.333333333333332 | 755.0 | Topological domain | Extracellular | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000298386 | 0 | 18 | 692_754 | 31.333333333333332 | 755.0 | Topological domain | Cytoplasmic | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000380314 | 0 | 17 | 438_455 | 31.333333333333332 | 731.0 | Topological domain | Cytoplasmic | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000380314 | 0 | 17 | 477_495 | 31.333333333333332 | 731.0 | Topological domain | Extracellular | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000380314 | 0 | 17 | 519_537 | 31.333333333333332 | 731.0 | Topological domain | Cytoplasmic | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000380314 | 0 | 17 | 559_592 | 31.333333333333332 | 731.0 | Topological domain | Extracellular | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000380314 | 0 | 17 | 614_639 | 31.333333333333332 | 731.0 | Topological domain | Cytoplasmic | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000380314 | 0 | 17 | 661_670 | 31.333333333333332 | 731.0 | Topological domain | Extracellular | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000380314 | 0 | 17 | 692_754 | 31.333333333333332 | 731.0 | Topological domain | Cytoplasmic | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000298386 | 0 | 18 | 417_437 | 31.333333333333332 | 755.0 | Transmembrane | Helical%3B Name%3D1 | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000298386 | 0 | 18 | 456_476 | 31.333333333333332 | 755.0 | Transmembrane | Helical%3B Name%3D2 | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000298386 | 0 | 18 | 496_518 | 31.333333333333332 | 755.0 | Transmembrane | Helical%3B Name%3D3 | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000298386 | 0 | 18 | 538_558 | 31.333333333333332 | 755.0 | Transmembrane | Helical%3B Name%3D4 | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000298386 | 0 | 18 | 593_613 | 31.333333333333332 | 755.0 | Transmembrane | Helical%3B Name%3D5 | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000298386 | 0 | 18 | 640_660 | 31.333333333333332 | 755.0 | Transmembrane | Helical%3B Name%3D6 | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000298386 | 0 | 18 | 671_691 | 31.333333333333332 | 755.0 | Transmembrane | Helical%3B Name%3D7 | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000380314 | 0 | 17 | 417_437 | 31.333333333333332 | 731.0 | Transmembrane | Helical%3B Name%3D1 | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000380314 | 0 | 17 | 456_476 | 31.333333333333332 | 731.0 | Transmembrane | Helical%3B Name%3D2 | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000380314 | 0 | 17 | 496_518 | 31.333333333333332 | 731.0 | Transmembrane | Helical%3B Name%3D3 | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000380314 | 0 | 17 | 538_558 | 31.333333333333332 | 731.0 | Transmembrane | Helical%3B Name%3D4 | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000380314 | 0 | 17 | 593_613 | 31.333333333333332 | 731.0 | Transmembrane | Helical%3B Name%3D5 | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000380314 | 0 | 17 | 640_660 | 31.333333333333332 | 731.0 | Transmembrane | Helical%3B Name%3D6 | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000380314 | 0 | 17 | 671_691 | 31.333333333333332 | 731.0 | Transmembrane | Helical%3B Name%3D7 |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000298386 | 0 | 18 | 1_416 | 31.333333333333332 | 755.0 | Topological domain | Extracellular | |
| Tgene | RXFP2 | chr13:32945237 | chr13:32332395 | ENST00000380314 | 0 | 17 | 1_416 | 31.333333333333332 | 731.0 | Topological domain | Extracellular |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| BRCA2 | |
| RXFP2 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to BRCA2-RXFP2 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to BRCA2-RXFP2 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

