| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:CCDC104-NDRG1 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: CCDC104-NDRG1 | FusionPDB ID: 13566 | FusionGDB2.0 ID: 13566 | Hgene | Tgene | Gene symbol | CCDC104 | NDRG1 | Gene ID | 112942 | 10397 |
| Gene name | cilia and flagella associated protein 36 | N-myc downstream regulated 1 | |
| Synonyms | BARTL1|CCDC104 | CAP43|CMT4D|DRG-1|DRG1|GC4|HMSNL|NDR1|NMSL|PROXY1|RIT42|RTP|TARG1|TDD5 | |
| Cytomap | 2p16.1 | 8q24.22 | |
| Type of gene | protein-coding | protein-coding | |
| Description | cilia- and flagella-associated protein 36coiled-coil domain containing 104coiled-coil domain-containing protein 104 | protein NDRG1N-myc downstream-regulated gene 1 proteindifferentiation-related gene 1 proteinnickel-specific induction protein Cap43protein regulated by oxygen-1reducing agents and tunicamycin-responsive protein | |
| Modification date | 20200313 | 20200328 | |
| UniProtAcc | . | Q92597 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000339012, ENST00000349456, ENST00000403007, ENST00000406691, ENST00000407816, ENST00000490934, | ENST00000522476, ENST00000414097, ENST00000518066, ENST00000518176, ENST00000521414, ENST00000537882, ENST00000323851, ENST00000354944, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 3 X 3 X 3=27 | 24 X 17 X 10=4080 |
| # samples | 4 | 26 | |
| ** MAII score | log2(4/27*10)=0.567040592723894 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(26/4080*10)=-3.9719856238304 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: CCDC104 [Title/Abstract] AND NDRG1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | CCDC104(55750958)-NDRG1(134276895), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | CCDC104-NDRG1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. CCDC104-NDRG1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across CCDC104 (5'-gene) Fusion gene breakpoints across CCDC104 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
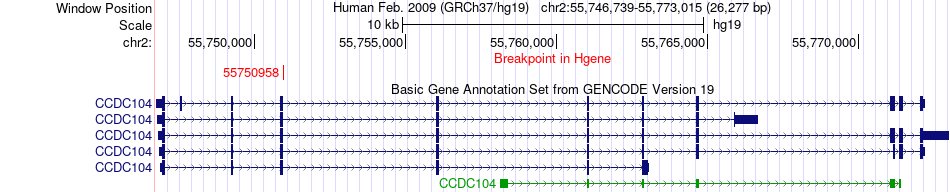 |
 Fusion gene breakpoints across NDRG1 (3'-gene) Fusion gene breakpoints across NDRG1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
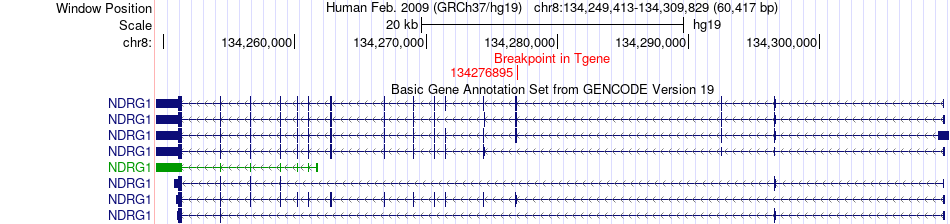 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | Non-Cancer | TCGA-DO-A1JZ-11A | CCDC104 | chr2 | 55750958 | + | NDRG1 | chr8 | 134276895 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000339012 | CCDC104 | chr2 | 55750958 | + | ENST00000323851 | NDRG1 | chr8 | 134276895 | - | 3348 | 555 | 180 | 1640 | 486 |
| ENST00000339012 | CCDC104 | chr2 | 55750958 | + | ENST00000354944 | NDRG1 | chr8 | 134276895 | - | 3137 | 555 | 180 | 1430 | 416 |
| ENST00000406691 | CCDC104 | chr2 | 55750958 | + | ENST00000323851 | NDRG1 | chr8 | 134276895 | - | 3231 | 438 | 138 | 1523 | 461 |
| ENST00000406691 | CCDC104 | chr2 | 55750958 | + | ENST00000354944 | NDRG1 | chr8 | 134276895 | - | 3020 | 438 | 138 | 1313 | 391 |
| ENST00000349456 | CCDC104 | chr2 | 55750958 | + | ENST00000323851 | NDRG1 | chr8 | 134276895 | - | 3223 | 430 | 130 | 1515 | 461 |
| ENST00000349456 | CCDC104 | chr2 | 55750958 | + | ENST00000354944 | NDRG1 | chr8 | 134276895 | - | 3012 | 430 | 130 | 1305 | 391 |
| ENST00000407816 | CCDC104 | chr2 | 55750958 | + | ENST00000323851 | NDRG1 | chr8 | 134276895 | - | 3187 | 394 | 94 | 1479 | 461 |
| ENST00000407816 | CCDC104 | chr2 | 55750958 | + | ENST00000354944 | NDRG1 | chr8 | 134276895 | - | 2976 | 394 | 94 | 1269 | 391 |
| ENST00000403007 | CCDC104 | chr2 | 55750958 | + | ENST00000323851 | NDRG1 | chr8 | 134276895 | - | 3146 | 353 | 53 | 1438 | 461 |
| ENST00000403007 | CCDC104 | chr2 | 55750958 | + | ENST00000354944 | NDRG1 | chr8 | 134276895 | - | 2935 | 353 | 53 | 1228 | 391 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000339012 | ENST00000323851 | CCDC104 | chr2 | 55750958 | + | NDRG1 | chr8 | 134276895 | - | 0.004019482 | 0.9959805 |
| ENST00000339012 | ENST00000354944 | CCDC104 | chr2 | 55750958 | + | NDRG1 | chr8 | 134276895 | - | 0.004329093 | 0.9956709 |
| ENST00000406691 | ENST00000323851 | CCDC104 | chr2 | 55750958 | + | NDRG1 | chr8 | 134276895 | - | 0.004946064 | 0.9950539 |
| ENST00000406691 | ENST00000354944 | CCDC104 | chr2 | 55750958 | + | NDRG1 | chr8 | 134276895 | - | 0.007260216 | 0.99273974 |
| ENST00000349456 | ENST00000323851 | CCDC104 | chr2 | 55750958 | + | NDRG1 | chr8 | 134276895 | - | 0.004896717 | 0.99510324 |
| ENST00000349456 | ENST00000354944 | CCDC104 | chr2 | 55750958 | + | NDRG1 | chr8 | 134276895 | - | 0.007156894 | 0.99284315 |
| ENST00000407816 | ENST00000323851 | CCDC104 | chr2 | 55750958 | + | NDRG1 | chr8 | 134276895 | - | 0.00499837 | 0.9950016 |
| ENST00000407816 | ENST00000354944 | CCDC104 | chr2 | 55750958 | + | NDRG1 | chr8 | 134276895 | - | 0.00721328 | 0.9927867 |
| ENST00000403007 | ENST00000323851 | CCDC104 | chr2 | 55750958 | + | NDRG1 | chr8 | 134276895 | - | 0.004715209 | 0.99528486 |
| ENST00000403007 | ENST00000354944 | CCDC104 | chr2 | 55750958 | + | NDRG1 | chr8 | 134276895 | - | 0.006953044 | 0.99304694 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >13566_13566_1_CCDC104-NDRG1_CCDC104_chr2_55750958_ENST00000339012_NDRG1_chr8_134276895_ENST00000323851_length(amino acids)=486AA_BP=124 MPLWGGMAAEEEDEVEWVVESIAGFLRGPDWSIPILDFVEQKCEVNCKGGHVITPGSPEPVILVACVPLVFDDEEESKLTYTEIHQEYKE LVEKLLEGYLKEIGINEDQFQEACTSPLAKTHTSQEQDIETLHGSVHVTLCGTPKGNRPVILTYHDIGMNHKTCYNPLFNYEDMQEITQH FAVCHVDAPGQQDGAASFPAGYMYPSMDQLAEMLPGVLQQFGLKSIIGMGTGAGAYILTRFALNNPEMVEGLVLINVNPCAEGWMDWAAS KISGWTQALPDMVVSHLFGKEEMQSNVEVVHTYRQHIVNDMNPGNLHLFINAYNSRRDLEIERPMPGTHTVTLQCPALLVVGDSSPAVDA VVECNSKLDPTKTTLLKMADCGGLPQISQPAKLAEAFKYFVQGMGYMPSASMTRLMRSRTASGSSVTSLDGTRSRSHTSEGTRSRSHTSE -------------------------------------------------------------- >13566_13566_2_CCDC104-NDRG1_CCDC104_chr2_55750958_ENST00000339012_NDRG1_chr8_134276895_ENST00000354944_length(amino acids)=416AA_BP=124 MPLWGGMAAEEEDEVEWVVESIAGFLRGPDWSIPILDFVEQKCEVNCKGGHVITPGSPEPVILVACVPLVFDDEEESKLTYTEIHQEYKE LVEKLLEGYLKEIGINEDQFQEACTSPLAKTHTSQEQDIETLHGSVHVTLCGTPKGNRPVILTYHDIGMNHKTCAYILTRFALNNPEMVE GLVLINVNPCAEGWMDWAASKISGWTQALPDMVVSHLFGKEEMQSNVEVVHTYRQHIVNDMNPGNLHLFINAYNSRRDLEIERPMPGTHT VTLQCPALLVVGDSSPAVDAVVECNSKLDPTKTTLLKMADCGGLPQISQPAKLAEAFKYFVQGMGYMPSASMTRLMRSRTASGSSVTSLD -------------------------------------------------------------- >13566_13566_3_CCDC104-NDRG1_CCDC104_chr2_55750958_ENST00000349456_NDRG1_chr8_134276895_ENST00000323851_length(amino acids)=461AA_BP=99 MPLWGGMAAEEEDEVEWVVESIAGFLRGPDWSIPILDFVEQKCEVFDDEEESKLTYTEIHQEYKELVEKLLEGYLKEIGINEDQFQEACT SPLAKTHTSQEQDIETLHGSVHVTLCGTPKGNRPVILTYHDIGMNHKTCYNPLFNYEDMQEITQHFAVCHVDAPGQQDGAASFPAGYMYP SMDQLAEMLPGVLQQFGLKSIIGMGTGAGAYILTRFALNNPEMVEGLVLINVNPCAEGWMDWAASKISGWTQALPDMVVSHLFGKEEMQS NVEVVHTYRQHIVNDMNPGNLHLFINAYNSRRDLEIERPMPGTHTVTLQCPALLVVGDSSPAVDAVVECNSKLDPTKTTLLKMADCGGLP QISQPAKLAEAFKYFVQGMGYMPSASMTRLMRSRTASGSSVTSLDGTRSRSHTSEGTRSRSHTSEGTRSRSHTSEGAHLDITPNSGAAGN -------------------------------------------------------------- >13566_13566_4_CCDC104-NDRG1_CCDC104_chr2_55750958_ENST00000349456_NDRG1_chr8_134276895_ENST00000354944_length(amino acids)=391AA_BP=99 MPLWGGMAAEEEDEVEWVVESIAGFLRGPDWSIPILDFVEQKCEVFDDEEESKLTYTEIHQEYKELVEKLLEGYLKEIGINEDQFQEACT SPLAKTHTSQEQDIETLHGSVHVTLCGTPKGNRPVILTYHDIGMNHKTCAYILTRFALNNPEMVEGLVLINVNPCAEGWMDWAASKISGW TQALPDMVVSHLFGKEEMQSNVEVVHTYRQHIVNDMNPGNLHLFINAYNSRRDLEIERPMPGTHTVTLQCPALLVVGDSSPAVDAVVECN SKLDPTKTTLLKMADCGGLPQISQPAKLAEAFKYFVQGMGYMPSASMTRLMRSRTASGSSVTSLDGTRSRSHTSEGTRSRSHTSEGTRSR -------------------------------------------------------------- >13566_13566_5_CCDC104-NDRG1_CCDC104_chr2_55750958_ENST00000403007_NDRG1_chr8_134276895_ENST00000323851_length(amino acids)=461AA_BP=99 MPLWGGMAAEEEDEVEWVVESIAGFLRGPDWSIPILDFVEQKCEVFDDEEESKLTYTEIHQEYKELVEKLLEGYLKEIGINEDQFQEACT SPLAKTHTSQEQDIETLHGSVHVTLCGTPKGNRPVILTYHDIGMNHKTCYNPLFNYEDMQEITQHFAVCHVDAPGQQDGAASFPAGYMYP SMDQLAEMLPGVLQQFGLKSIIGMGTGAGAYILTRFALNNPEMVEGLVLINVNPCAEGWMDWAASKISGWTQALPDMVVSHLFGKEEMQS NVEVVHTYRQHIVNDMNPGNLHLFINAYNSRRDLEIERPMPGTHTVTLQCPALLVVGDSSPAVDAVVECNSKLDPTKTTLLKMADCGGLP QISQPAKLAEAFKYFVQGMGYMPSASMTRLMRSRTASGSSVTSLDGTRSRSHTSEGTRSRSHTSEGTRSRSHTSEGAHLDITPNSGAAGN -------------------------------------------------------------- >13566_13566_6_CCDC104-NDRG1_CCDC104_chr2_55750958_ENST00000403007_NDRG1_chr8_134276895_ENST00000354944_length(amino acids)=391AA_BP=99 MPLWGGMAAEEEDEVEWVVESIAGFLRGPDWSIPILDFVEQKCEVFDDEEESKLTYTEIHQEYKELVEKLLEGYLKEIGINEDQFQEACT SPLAKTHTSQEQDIETLHGSVHVTLCGTPKGNRPVILTYHDIGMNHKTCAYILTRFALNNPEMVEGLVLINVNPCAEGWMDWAASKISGW TQALPDMVVSHLFGKEEMQSNVEVVHTYRQHIVNDMNPGNLHLFINAYNSRRDLEIERPMPGTHTVTLQCPALLVVGDSSPAVDAVVECN SKLDPTKTTLLKMADCGGLPQISQPAKLAEAFKYFVQGMGYMPSASMTRLMRSRTASGSSVTSLDGTRSRSHTSEGTRSRSHTSEGTRSR -------------------------------------------------------------- >13566_13566_7_CCDC104-NDRG1_CCDC104_chr2_55750958_ENST00000406691_NDRG1_chr8_134276895_ENST00000323851_length(amino acids)=461AA_BP=99 MPLWGGMAAEEEDEVEWVVESIAGFLRGPDWSIPILDFVEQKCEVFDDEEESKLTYTEIHQEYKELVEKLLEGYLKEIGINEDQFQEACT SPLAKTHTSQEQDIETLHGSVHVTLCGTPKGNRPVILTYHDIGMNHKTCYNPLFNYEDMQEITQHFAVCHVDAPGQQDGAASFPAGYMYP SMDQLAEMLPGVLQQFGLKSIIGMGTGAGAYILTRFALNNPEMVEGLVLINVNPCAEGWMDWAASKISGWTQALPDMVVSHLFGKEEMQS NVEVVHTYRQHIVNDMNPGNLHLFINAYNSRRDLEIERPMPGTHTVTLQCPALLVVGDSSPAVDAVVECNSKLDPTKTTLLKMADCGGLP QISQPAKLAEAFKYFVQGMGYMPSASMTRLMRSRTASGSSVTSLDGTRSRSHTSEGTRSRSHTSEGTRSRSHTSEGAHLDITPNSGAAGN -------------------------------------------------------------- >13566_13566_8_CCDC104-NDRG1_CCDC104_chr2_55750958_ENST00000406691_NDRG1_chr8_134276895_ENST00000354944_length(amino acids)=391AA_BP=99 MPLWGGMAAEEEDEVEWVVESIAGFLRGPDWSIPILDFVEQKCEVFDDEEESKLTYTEIHQEYKELVEKLLEGYLKEIGINEDQFQEACT SPLAKTHTSQEQDIETLHGSVHVTLCGTPKGNRPVILTYHDIGMNHKTCAYILTRFALNNPEMVEGLVLINVNPCAEGWMDWAASKISGW TQALPDMVVSHLFGKEEMQSNVEVVHTYRQHIVNDMNPGNLHLFINAYNSRRDLEIERPMPGTHTVTLQCPALLVVGDSSPAVDAVVECN SKLDPTKTTLLKMADCGGLPQISQPAKLAEAFKYFVQGMGYMPSASMTRLMRSRTASGSSVTSLDGTRSRSHTSEGTRSRSHTSEGTRSR -------------------------------------------------------------- >13566_13566_9_CCDC104-NDRG1_CCDC104_chr2_55750958_ENST00000407816_NDRG1_chr8_134276895_ENST00000323851_length(amino acids)=461AA_BP=99 MPLWGGMAAEEEDEVEWVVESIAGFLRGPDWSIPILDFVEQKCEVFDDEEESKLTYTEIHQEYKELVEKLLEGYLKEIGINEDQFQEACT SPLAKTHTSQEQDIETLHGSVHVTLCGTPKGNRPVILTYHDIGMNHKTCYNPLFNYEDMQEITQHFAVCHVDAPGQQDGAASFPAGYMYP SMDQLAEMLPGVLQQFGLKSIIGMGTGAGAYILTRFALNNPEMVEGLVLINVNPCAEGWMDWAASKISGWTQALPDMVVSHLFGKEEMQS NVEVVHTYRQHIVNDMNPGNLHLFINAYNSRRDLEIERPMPGTHTVTLQCPALLVVGDSSPAVDAVVECNSKLDPTKTTLLKMADCGGLP QISQPAKLAEAFKYFVQGMGYMPSASMTRLMRSRTASGSSVTSLDGTRSRSHTSEGTRSRSHTSEGTRSRSHTSEGAHLDITPNSGAAGN -------------------------------------------------------------- >13566_13566_10_CCDC104-NDRG1_CCDC104_chr2_55750958_ENST00000407816_NDRG1_chr8_134276895_ENST00000354944_length(amino acids)=391AA_BP=99 MPLWGGMAAEEEDEVEWVVESIAGFLRGPDWSIPILDFVEQKCEVFDDEEESKLTYTEIHQEYKELVEKLLEGYLKEIGINEDQFQEACT SPLAKTHTSQEQDIETLHGSVHVTLCGTPKGNRPVILTYHDIGMNHKTCAYILTRFALNNPEMVEGLVLINVNPCAEGWMDWAASKISGW TQALPDMVVSHLFGKEEMQSNVEVVHTYRQHIVNDMNPGNLHLFINAYNSRRDLEIERPMPGTHTVTLQCPALLVVGDSSPAVDAVVECN SKLDPTKTTLLKMADCGGLPQISQPAKLAEAFKYFVQGMGYMPSASMTRLMRSRTASGSSVTSLDGTRSRSHTSEGTRSRSHTSEGTRSR -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr2:55750958/chr8:134276895) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | NDRG1 |
| FUNCTION: Might normally function as a transcriptional repressor. EWS-fusion-proteins (EFPS) may play a role in the tumorigenic process. They may disturb gene expression by mimicking, or interfering with the normal function of CTD-POLII within the transcription initiation complex. They may also contribute to an aberrant activation of the fusion protein target genes. | FUNCTION: Stress-responsive protein involved in hormone responses, cell growth, and differentiation. Acts as a tumor suppressor in many cell types. Necessary but not sufficient for p53/TP53-mediated caspase activation and apoptosis. Has a role in cell trafficking, notably of the Schwann cell, and is necessary for the maintenance and development of the peripheral nerve myelin sheath. Required for vesicular recycling of CDH1 and TF. May also function in lipid trafficking. Protects cells from spindle disruption damage. Functions in p53/TP53-dependent mitotic spindle checkpoint. Regulates microtubule dynamics and maintains euploidy. {ECO:0000269|PubMed:15247272, ECO:0000269|PubMed:15377670, ECO:0000269|PubMed:17786215, ECO:0000269|PubMed:9766676}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | NDRG1 | chr2:55750958 | chr8:134276895 | ENST00000323851 | 2 | 16 | 339_368 | 33.0 | 395.0 | Region | Note=3 X 10 AA tandem repeats of G-T-R-S-R-S-H-T-S-E | |
| Tgene | NDRG1 | chr2:55750958 | chr8:134276895 | ENST00000414097 | 2 | 16 | 339_368 | 33.0 | 395.0 | Region | Note=3 X 10 AA tandem repeats of G-T-R-S-R-S-H-T-S-E | |
| Tgene | NDRG1 | chr2:55750958 | chr8:134276895 | ENST00000522476 | 0 | 14 | 339_368 | 0 | 329.0 | Region | Note=3 X 10 AA tandem repeats of G-T-R-S-R-S-H-T-S-E | |
| Tgene | NDRG1 | chr2:55750958 | chr8:134276895 | ENST00000537882 | 0 | 15 | 339_368 | 0 | 314.0 | Region | Note=3 X 10 AA tandem repeats of G-T-R-S-R-S-H-T-S-E | |
| Tgene | NDRG1 | chr2:55750958 | chr8:134276895 | ENST00000323851 | 2 | 16 | 339_348 | 33.0 | 395.0 | Repeat | Note=1 | |
| Tgene | NDRG1 | chr2:55750958 | chr8:134276895 | ENST00000323851 | 2 | 16 | 349_358 | 33.0 | 395.0 | Repeat | Note=2 | |
| Tgene | NDRG1 | chr2:55750958 | chr8:134276895 | ENST00000323851 | 2 | 16 | 359_368 | 33.0 | 395.0 | Repeat | Note=3 | |
| Tgene | NDRG1 | chr2:55750958 | chr8:134276895 | ENST00000414097 | 2 | 16 | 339_348 | 33.0 | 395.0 | Repeat | Note=1 | |
| Tgene | NDRG1 | chr2:55750958 | chr8:134276895 | ENST00000414097 | 2 | 16 | 349_358 | 33.0 | 395.0 | Repeat | Note=2 | |
| Tgene | NDRG1 | chr2:55750958 | chr8:134276895 | ENST00000414097 | 2 | 16 | 359_368 | 33.0 | 395.0 | Repeat | Note=3 | |
| Tgene | NDRG1 | chr2:55750958 | chr8:134276895 | ENST00000522476 | 0 | 14 | 339_348 | 0 | 329.0 | Repeat | Note=1 | |
| Tgene | NDRG1 | chr2:55750958 | chr8:134276895 | ENST00000522476 | 0 | 14 | 349_358 | 0 | 329.0 | Repeat | Note=2 | |
| Tgene | NDRG1 | chr2:55750958 | chr8:134276895 | ENST00000522476 | 0 | 14 | 359_368 | 0 | 329.0 | Repeat | Note=3 | |
| Tgene | NDRG1 | chr2:55750958 | chr8:134276895 | ENST00000537882 | 0 | 15 | 339_348 | 0 | 314.0 | Repeat | Note=1 | |
| Tgene | NDRG1 | chr2:55750958 | chr8:134276895 | ENST00000537882 | 0 | 15 | 349_358 | 0 | 314.0 | Repeat | Note=2 | |
| Tgene | NDRG1 | chr2:55750958 | chr8:134276895 | ENST00000537882 | 0 | 15 | 359_368 | 0 | 314.0 | Repeat | Note=3 |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CCDC104 | chr2:55750958 | chr8:134276895 | ENST00000339012 | + | 4 | 11 | 150_187 | 119.0 | 368.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CCDC104 | chr2:55750958 | chr8:134276895 | ENST00000349456 | + | 3 | 10 | 150_187 | 94.0 | 343.0 | Coiled coil | Ontology_term=ECO:0000255 |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| NDRG1 | ARL4D, PHYHIP, S100B, HSP90AA1, HSPA5, VCP, CANX, PSMC3, PSMC2, PSMD2, XRCC6, RUVBL2, ILF3, SEC23A, COPB2, CLTC, AP2M1, AP1M2, MME, CTNNB1, ACTG1, KIF5B, PPP2R2A, TLE3, HSD17B4, CNDP2, DARS, DLST, ACSL3, FASN, MAOA, LDHA, PKM, RPN2, TARS, SHMT2, SLC25A6, ATP1A1, EEF2, EEF1G, EIF2S3, EIF3E, PABPC1, RPS3, RPS6, RPL24, RPL3, RPS16, RPS8, RPS20, RPS9, RPL4, RPS26, NCL, HNRNPF, HNRNPU, HNRNPH1, DDX1, DDX5, UPF1, EWSR1, CDH1, ANXA5, CLNS1A, IGBP1, SPAG9, MLH1, ACOT7, DDX39B, GSR, DPP3, EIF4H, FTO, KYNU, MVD, NAMPT, PEPD, PGD, BCL2L11, HSPA4, CUL2, TAF9, SGK1, SUZ12, EED, NDRG2, DHX15, G6PD, GART, HIP1R, CCDC22, CLIC4, NAA50, RTCB, SH2D4A, THUMPD1, TIMM13, TSG101, USP14, XPO1, NIF3L1, NTMT1, TARDBP, UFM1, TMEM17, ERBB3, FXYD1, PDE4DIP, S1PR2, FAM60A, CDK15, TRIM25, HNRNPL, MYC, SKP2, RABAC1, GSK3B, NR4A1, YAP1, RTN1, Apoa1, Apoa2, Rabac1, Arl6ip1, APOA1, APOA2, RAB4A, ZBTB38, EZR, TGOLN2, LAMP1, PPM1F, HRAS, NRAS, KRAS, RFFL, DYNC1I2, CUL4A, CTBP2, ORF3a, PRKCB, RAB5A, SH2D3C, M, Rnf183, HULC, ARF6, C11orf52, CAV1, LAMTOR1, LYN, MARCKS, OCLN, RAB11A, RAB2A, RAB35, RAB9A, RHOB, STX4, STX6, STX7, ZFPL1, HTRA4, NAA40, ST6GALNAC6, OR2A4, HOXC5, RPL35A, FXYD3, SRRT, PPP2R2B, NAA11, YWHAH, GOT1, STX17, WIF1, GOLGA2, ATP1B1, COL10A1, CERS3, OPTN, SSBP2, ATP1B3, C2orf73, NTNG1, FGF12, OSGEP, CCR1, TIMMDC1, CEP19, MBNL1, CRCP, PINK1, NRSN1, CCDC53, SSUH2, DNAJB6, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| CCDC104 | |
| NDRG1 | 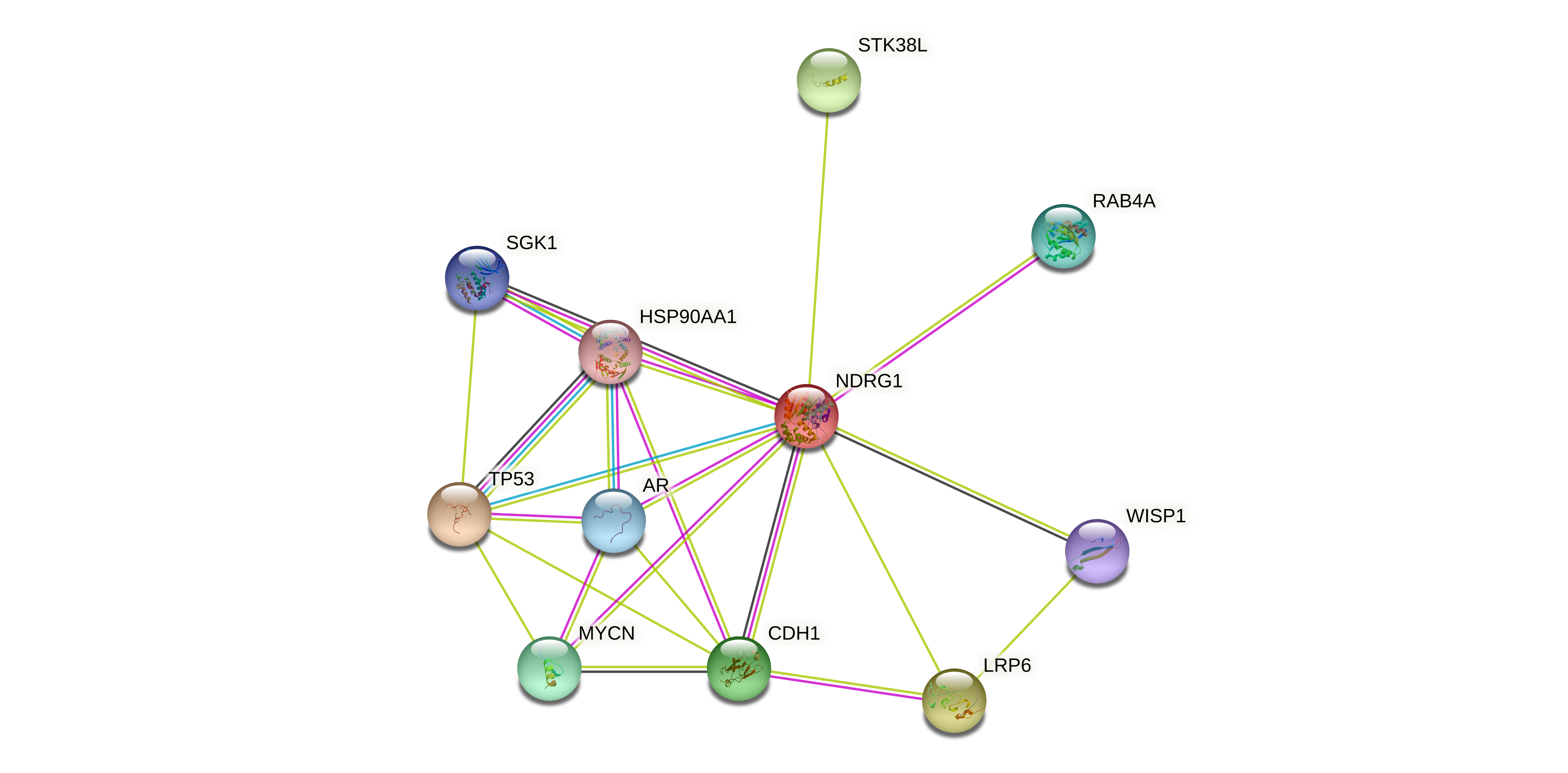 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to CCDC104-NDRG1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to CCDC104-NDRG1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Tgene | NDRG1 | C0027627 | Neoplasm Metastasis | 2 | CTD_human |
| Tgene | NDRG1 | C1832334 | CHARCOT-MARIE-TOOTH DISEASE, TYPE 4D | 2 | CTD_human;GENOMICS_ENGLAND;ORPHANET |
| Tgene | NDRG1 | C0004114 | Astrocytoma | 1 | CTD_human |
| Tgene | NDRG1 | C0006142 | Malignant neoplasm of breast | 1 | CTD_human |
| Tgene | NDRG1 | C0007131 | Non-Small Cell Lung Carcinoma | 1 | CTD_human |
| Tgene | NDRG1 | C0007134 | Renal Cell Carcinoma | 1 | CTD_human |
| Tgene | NDRG1 | C0017636 | Glioblastoma | 1 | CTD_human |
| Tgene | NDRG1 | C0022665 | Kidney Neoplasm | 1 | CTD_human |
| Tgene | NDRG1 | C0023893 | Liver Cirrhosis, Experimental | 1 | CTD_human |
| Tgene | NDRG1 | C0023903 | Liver neoplasms | 1 | CTD_human |
| Tgene | NDRG1 | C0025202 | melanoma | 1 | CTD_human |
| Tgene | NDRG1 | C0026640 | Mouth Neoplasms | 1 | CTD_human |
| Tgene | NDRG1 | C0029295 | Oropharyngeal Neoplasms | 1 | CTD_human |
| Tgene | NDRG1 | C0033578 | Prostatic Neoplasms | 1 | CTD_human |
| Tgene | NDRG1 | C0149925 | Small cell carcinoma of lung | 1 | CTD_human |
| Tgene | NDRG1 | C0153381 | Malignant neoplasm of mouth | 1 | CTD_human |
| Tgene | NDRG1 | C0205768 | Subependymal Giant Cell Astrocytoma | 1 | CTD_human |
| Tgene | NDRG1 | C0206658 | Smooth Muscle Tumor | 1 | CTD_human |
| Tgene | NDRG1 | C0206734 | Hemangioblastoma | 1 | CTD_human |
| Tgene | NDRG1 | C0279702 | Conventional (Clear Cell) Renal Cell Carcinoma | 1 | CTD_human |
| Tgene | NDRG1 | C0280783 | Juvenile Pilocytic Astrocytoma | 1 | CTD_human |
| Tgene | NDRG1 | C0280785 | Diffuse Astrocytoma | 1 | CTD_human |
| Tgene | NDRG1 | C0334579 | Anaplastic astrocytoma | 1 | CTD_human |
| Tgene | NDRG1 | C0334580 | Protoplasmic astrocytoma | 1 | CTD_human |
| Tgene | NDRG1 | C0334581 | Gemistocytic astrocytoma | 1 | CTD_human |
| Tgene | NDRG1 | C0334582 | Fibrillary Astrocytoma | 1 | CTD_human |
| Tgene | NDRG1 | C0334583 | Pilocytic Astrocytoma | 1 | CTD_human |
| Tgene | NDRG1 | C0334588 | Giant Cell Glioblastoma | 1 | CTD_human |
| Tgene | NDRG1 | C0338070 | Childhood Cerebral Astrocytoma | 1 | CTD_human |
| Tgene | NDRG1 | C0345904 | Malignant neoplasm of liver | 1 | CTD_human |
| Tgene | NDRG1 | C0376358 | Malignant neoplasm of prostate | 1 | CTD_human |
| Tgene | NDRG1 | C0547065 | Mixed oligoastrocytoma | 1 | CTD_human |
| Tgene | NDRG1 | C0678222 | Breast Carcinoma | 1 | CTD_human |
| Tgene | NDRG1 | C0740457 | Malignant neoplasm of kidney | 1 | CTD_human |
| Tgene | NDRG1 | C0750935 | Cerebral Astrocytoma | 1 | CTD_human |
| Tgene | NDRG1 | C0750936 | Intracranial Astrocytoma | 1 | CTD_human |
| Tgene | NDRG1 | C0751692 | Multiple Hemangioblastomas | 1 | CTD_human |
| Tgene | NDRG1 | C0887833 | Carcinoma, Pancreatic Ductal | 1 | CTD_human |
| Tgene | NDRG1 | C1168401 | Squamous cell carcinoma of the head and neck | 1 | CTD_human |
| Tgene | NDRG1 | C1257931 | Mammary Neoplasms, Human | 1 | CTD_human |
| Tgene | NDRG1 | C1266042 | Chromophobe Renal Cell Carcinoma | 1 | CTD_human |
| Tgene | NDRG1 | C1266043 | Sarcomatoid Renal Cell Carcinoma | 1 | CTD_human |
| Tgene | NDRG1 | C1266044 | Collecting Duct Carcinoma of the Kidney | 1 | CTD_human |
| Tgene | NDRG1 | C1306837 | Papillary Renal Cell Carcinoma | 1 | CTD_human |
| Tgene | NDRG1 | C1458155 | Mammary Neoplasms | 1 | CTD_human |
| Tgene | NDRG1 | C1621958 | Glioblastoma Multiforme | 1 | CTD_human |
| Tgene | NDRG1 | C1704230 | Grade I Astrocytoma | 1 | CTD_human |
| Tgene | NDRG1 | C2349952 | Oropharyngeal Carcinoma | 1 | CTD_human |
| Tgene | NDRG1 | C4704874 | Mammary Carcinoma, Human | 1 | CTD_human |

