| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:CCNT1-HSPD1 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: CCNT1-HSPD1 | FusionPDB ID: 14191 | FusionGDB2.0 ID: 14191 | Hgene | Tgene | Gene symbol | CCNT1 | HSPD1 | Gene ID | 904 | 3329 |
| Gene name | cyclin T1 | heat shock protein family D (Hsp60) member 1 | |
| Synonyms | CCNT|CYCT1|HIVE1 | CPN60|GROEL|HLD4|HSP-60|HSP60|HSP65|HuCHA60|SPG13 | |
| Cytomap | 12q13.11-q13.12 | 2q33.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | cyclin-T1CDK9-associated C-type proteinMLLT10/CCNT1 fusioncyclin C-related proteinhuman immunodeficiency virus type 1 (HIV-1) expression (elevated) 1 | 60 kDa heat shock protein, mitochondrial60 kDa chaperoninP60 lymphocyte proteinchaperonin 60epididymis secretory sperm binding proteinheat shock 60kDa protein 1 (chaperonin)heat shock protein 65mitochondrial matrix protein P1short heat shock prote | |
| Modification date | 20200313 | 20200315 | |
| UniProtAcc | O60563 | P10809 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000261900, | ENST00000544407, ENST00000345042, ENST00000388968, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 6 X 8 X 4=192 | 14 X 10 X 6=840 |
| # samples | 7 | 16 | |
| ** MAII score | log2(7/192*10)=-1.45567948377619 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(16/840*10)=-2.39231742277876 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: CCNT1 [Title/Abstract] AND HSPD1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | CCNT1(49099550)-HSPD1(198353971), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | CCNT1-HSPD1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. CCNT1-HSPD1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CCNT1 | GO:0045944 | positive regulation of transcription by RNA polymerase II | 15563843 |
| Tgene | HSPD1 | GO:0002368 | B cell cytokine production | 16148103 |
| Tgene | HSPD1 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway | 16148103 |
| Tgene | HSPD1 | GO:0002842 | positive regulation of T cell mediated immune response to tumor cell | 10663613 |
| Tgene | HSPD1 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process | 17823127 |
| Tgene | HSPD1 | GO:0006986 | response to unfolded protein | 11050098 |
| Tgene | HSPD1 | GO:0032727 | positive regulation of interferon-alpha production | 17164250 |
| Tgene | HSPD1 | GO:0032729 | positive regulation of interferon-gamma production | 17164250 |
| Tgene | HSPD1 | GO:0032733 | positive regulation of interleukin-10 production | 16148103 |
| Tgene | HSPD1 | GO:0032735 | positive regulation of interleukin-12 production | 17164250 |
| Tgene | HSPD1 | GO:0032755 | positive regulation of interleukin-6 production | 16148103 |
| Tgene | HSPD1 | GO:0042026 | protein refolding | 11050098 |
| Tgene | HSPD1 | GO:0042100 | B cell proliferation | 16148103 |
| Tgene | HSPD1 | GO:0042110 | T cell activation | 15371451|17164250|18256040 |
| Tgene | HSPD1 | GO:0042113 | B cell activation | 16148103 |
| Tgene | HSPD1 | GO:0043032 | positive regulation of macrophage activation | 17164250 |
| Tgene | HSPD1 | GO:0044406 | adhesion of symbiont to host | 20633027 |
| Tgene | HSPD1 | GO:0048291 | isotype switching to IgG isotypes | 16148103 |
| Tgene | HSPD1 | GO:0050870 | positive regulation of T cell activation | 16148103|17164250 |
 Fusion gene breakpoints across CCNT1 (5'-gene) Fusion gene breakpoints across CCNT1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
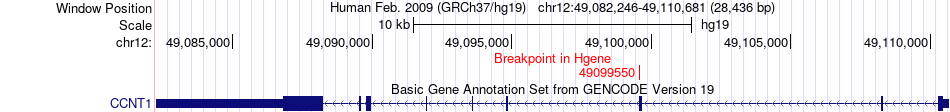 |
 Fusion gene breakpoints across HSPD1 (3'-gene) Fusion gene breakpoints across HSPD1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
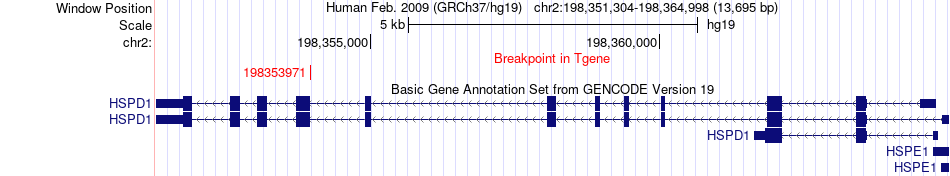 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-IN-A7NT | CCNT1 | chr12 | 49099550 | - | HSPD1 | chr2 | 198353971 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000261900 | CCNT1 | chr12 | 49099550 | - | ENST00000388968 | HSPD1 | chr2 | 198353971 | - | 1813 | 595 | 223 | 1347 | 374 |
| ENST00000261900 | CCNT1 | chr12 | 49099550 | - | ENST00000345042 | HSPD1 | chr2 | 198353971 | - | 1808 | 595 | 223 | 1347 | 374 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000261900 | ENST00000388968 | CCNT1 | chr12 | 49099550 | - | HSPD1 | chr2 | 198353971 | - | 0.000711489 | 0.99928844 |
| ENST00000261900 | ENST00000345042 | CCNT1 | chr12 | 49099550 | - | HSPD1 | chr2 | 198353971 | - | 0.000743295 | 0.9992567 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >14191_14191_1_CCNT1-HSPD1_CCNT1_chr12_49099550_ENST00000261900_HSPD1_chr2_198353971_ENST00000345042_length(amino acids)=374AA_BP=124 MEGERKNNNKRWYFTREQLENSPSRRFGVDPDKELSYRQQAANLLQDMGQRLNVSQLTINTAIVYMHRFYMIQSFTQFPGNSVAPAALFL AAKVEEQPKKLEHVIKVAHTCLHPQESLPDTRSEVFGEEGLTLNLEDVQPHDLGKVGEVIVTKDDAMLLKGKGDKAQIEKRIQEIIEQLD VTTSEYEKEKLNERLAKLSDGVAVLKVGGTSDVEVNEKKDRVTDALNATRAAVEEGIVLGGGCALLRCIPALDSLTPANEDQKIGIEIIK RTLKIPAMTIAKNAGVEGSLIVEKIMQSSSEVGYDAMAGDFVNMVEKGIIDPTKVVRTALLDAAGVASLLTTAEVVVTEIPKEEKDPGMG -------------------------------------------------------------- >14191_14191_2_CCNT1-HSPD1_CCNT1_chr12_49099550_ENST00000261900_HSPD1_chr2_198353971_ENST00000388968_length(amino acids)=374AA_BP=124 MEGERKNNNKRWYFTREQLENSPSRRFGVDPDKELSYRQQAANLLQDMGQRLNVSQLTINTAIVYMHRFYMIQSFTQFPGNSVAPAALFL AAKVEEQPKKLEHVIKVAHTCLHPQESLPDTRSEVFGEEGLTLNLEDVQPHDLGKVGEVIVTKDDAMLLKGKGDKAQIEKRIQEIIEQLD VTTSEYEKEKLNERLAKLSDGVAVLKVGGTSDVEVNEKKDRVTDALNATRAAVEEGIVLGGGCALLRCIPALDSLTPANEDQKIGIEIIK RTLKIPAMTIAKNAGVEGSLIVEKIMQSSSEVGYDAMAGDFVNMVEKGIIDPTKVVRTALLDAAGVASLLTTAEVVVTEIPKEEKDPGMG -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr12:49099550/chr2:198353971) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CCNT1 | HSPD1 |
| FUNCTION: Regulatory subunit of the cyclin-dependent kinase pair (CDK9/cyclin-T1) complex, also called positive transcription elongation factor B (P-TEFb), which is proposed to facilitate the transition from abortive to productive elongation by phosphorylating the CTD (C-terminal domain) of the large subunit of RNA polymerase II (RNA Pol II). {ECO:0000269|PubMed:16109376, ECO:0000269|PubMed:16109377}.; FUNCTION: (Microbial infection) In case of HIV or SIV infections, binds to the transactivation domain of the viral nuclear transcriptional activator, Tat, thereby increasing Tat's affinity for the transactivating response RNA element (TAR RNA). Serves as an essential cofactor for Tat, by promoting RNA Pol II activation, allowing transcription of viral genes. {ECO:0000269|PubMed:10329125, ECO:0000269|PubMed:10329126}. | FUNCTION: Chaperonin implicated in mitochondrial protein import and macromolecular assembly. Together with Hsp10, facilitates the correct folding of imported proteins. May also prevent misfolding and promote the refolding and proper assembly of unfolded polypeptides generated under stress conditions in the mitochondrial matrix (PubMed:1346131, PubMed:11422376). The functional units of these chaperonins consist of heptameric rings of the large subunit Hsp60, which function as a back-to-back double ring. In a cyclic reaction, Hsp60 ring complexes bind one unfolded substrate protein per ring, followed by the binding of ATP and association with 2 heptameric rings of the co-chaperonin Hsp10. This leads to sequestration of the substrate protein in the inner cavity of Hsp60 where, for a certain period of time, it can fold undisturbed by other cell components. Synchronous hydrolysis of ATP in all Hsp60 subunits results in the dissociation of the chaperonin rings and the release of ADP and the folded substrate protein (Probable). {ECO:0000269|PubMed:11422376, ECO:0000269|PubMed:1346131, ECO:0000305|PubMed:25918392}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CCNT1 | chr12:49099550 | chr2:198353971 | ENST00000261900 | - | 3 | 9 | 384_425 | 124.0 | 727.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CCNT1 | chr12:49099550 | chr2:198353971 | ENST00000261900 | - | 3 | 9 | 506_530 | 124.0 | 727.0 | Compositional bias | Note=His-rich |
| Hgene | CCNT1 | chr12:49099550 | chr2:198353971 | ENST00000261900 | - | 3 | 9 | 548_629 | 124.0 | 727.0 | Compositional bias | Note=Ser-rich |
| Hgene | CCNT1 | chr12:49099550 | chr2:198353971 | ENST00000261900 | - | 3 | 9 | 708_725 | 124.0 | 727.0 | Compositional bias | Note=Pro-rich |
| Tgene | HSPD1 | chr12:49099550 | chr2:198353971 | ENST00000345042 | 7 | 12 | 111_115 | 323.0 | 574.0 | Nucleotide binding | ATP | |
| Tgene | HSPD1 | chr12:49099550 | chr2:198353971 | ENST00000388968 | 7 | 12 | 111_115 | 323.0 | 574.0 | Nucleotide binding | ATP |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| CCNT1 | |
| HSPD1 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to CCNT1-HSPD1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to CCNT1-HSPD1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

