| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:CD74-NRG1 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: CD74-NRG1 | FusionPDB ID: 14642 | FusionGDB2.0 ID: 14642 | Hgene | Tgene | Gene symbol | CD74 | NRG1 | Gene ID | 972 | 3084 |
| Gene name | CD74 molecule | neuregulin 1 | |
| Synonyms | DHLAG|HLADG|II|Ia-GAMMA|p33 | ARIA|GGF|GGF2|HGL|HRG|HRG1|HRGA|MST131|MSTP131|NDF|NRG1-IT2|SMDF | |
| Cytomap | 5q33.1 | 8p12 | |
| Type of gene | protein-coding | protein-coding | |
| Description | HLA class II histocompatibility antigen gamma chainCD74 antigen (invariant polypeptide of major histocompatibility complex, class II antigen-associated)CD74 molecule, major histocompatibility complex, class II invariant chainHLA-DR antigens-associated | pro-neuregulin-1, membrane-bound isoformacetylcholine receptor-inducing activityglial growth factor 2heregulin, alpha (45kD, ERBB2 p185-activator)neu differentiation factorpro-NRG1sensory and motor neuron derived factor | |
| Modification date | 20200313 | 20200320 | |
| UniProtAcc | P04233 | Q02297 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000377795, ENST00000009530, ENST00000353334, ENST00000524315, | ENST00000287842, ENST00000287845, ENST00000338921, ENST00000341377, ENST00000356819, ENST00000405005, ENST00000519301, ENST00000520407, ENST00000520502, ENST00000521670, ENST00000523079, ENST00000539990, ENST00000523681, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 43 X 51 X 20=43860 | 25 X 17 X 14=5950 |
| # samples | 59 | 27 | |
| ** MAII score | log2(59/43860*10)=-6.21604704731175 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(27/5950*10)=-4.46185835603184 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: CD74 [Title/Abstract] AND NRG1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | |||
| Anticipated loss of major functional domain due to fusion event. | CD74-NRG1 seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. CD74-NRG1 seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. CD74-NRG1 seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CD74 | GO:0001516 | prostaglandin biosynthetic process | 12782713 |
| Hgene | CD74 | GO:0001934 | positive regulation of protein phosphorylation | 24942581 |
| Hgene | CD74 | GO:0002792 | negative regulation of peptide secretion | 19849849 |
| Hgene | CD74 | GO:0033674 | positive regulation of kinase activity | 24942581 |
| Hgene | CD74 | GO:0043066 | negative regulation of apoptotic process | 12782713 |
| Hgene | CD74 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling | 24942581 |
| Hgene | CD74 | GO:0043410 | positive regulation of MAPK cascade | 24942581 |
| Hgene | CD74 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator | 17045821 |
| Hgene | CD74 | GO:0045657 | positive regulation of monocyte differentiation | 24942581 |
| Hgene | CD74 | GO:0045893 | positive regulation of transcription, DNA-templated | 24942581 |
| Hgene | CD74 | GO:0046598 | positive regulation of viral entry into host cell | 24942581 |
| Hgene | CD74 | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | 17045821 |
| Hgene | CD74 | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 17045821|24942581 |
| Tgene | NRG1 | GO:0003222 | ventricular trabecula myocardium morphogenesis | 17336907 |
| Tgene | NRG1 | GO:0031334 | positive regulation of protein complex assembly | 10559227 |
| Tgene | NRG1 | GO:0038127 | ERBB signaling pathway | 11389077 |
| Tgene | NRG1 | GO:0038129 | ERBB3 signaling pathway | 27353365 |
| Tgene | NRG1 | GO:0045892 | negative regulation of transcription, DNA-templated | 15073182 |
| Tgene | NRG1 | GO:0051048 | negative regulation of secretion | 10559227 |
| Tgene | NRG1 | GO:0060379 | cardiac muscle cell myoblast differentiation | 17336907 |
| Tgene | NRG1 | GO:0060956 | endocardial cell differentiation | 17336907 |
 Fusion gene breakpoints across CD74 (5'-gene) Fusion gene breakpoints across CD74 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 Fusion gene breakpoints across NRG1 (3'-gene) Fusion gene breakpoints across NRG1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerKB3 | . | . | CD74 | chr5 | 149782125 | - | NRG1 | chr8 | 32585466 | + |
| ChimerKB3 | . | . | CD74 | chr5 | 149784242 | - | NRG1 | chr8 | 32585466 | + |
| ChimerKB3 | . | . | CD74 | chr5 | 149784242 | - | NRG1 | chr8 | 32600192 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000353334 | CD74 | chr5 | 149782125 | - | ENST00000519301 | NRG1 | chr8 | 32585466 | + | 2307 | 868 | 180 | 2303 | 707 |
| ENST00000353334 | CD74 | chr5 | 149782125 | - | ENST00000520407 | NRG1 | chr8 | 32585466 | + | 1548 | 868 | 180 | 1091 | 303 |
| ENST00000353334 | CD74 | chr5 | 149782125 | - | ENST00000523079 | NRG1 | chr8 | 32585466 | + | 2208 | 868 | 180 | 1628 | 482 |
| ENST00000353334 | CD74 | chr5 | 149782125 | - | ENST00000338921 | NRG1 | chr8 | 32585466 | + | 2941 | 868 | 180 | 2312 | 710 |
| ENST00000353334 | CD74 | chr5 | 149782125 | - | ENST00000356819 | NRG1 | chr8 | 32585466 | + | 2932 | 868 | 180 | 2303 | 707 |
| ENST00000353334 | CD74 | chr5 | 149782125 | - | ENST00000287845 | NRG1 | chr8 | 32585466 | + | 2932 | 868 | 180 | 2303 | 707 |
| ENST00000353334 | CD74 | chr5 | 149782125 | - | ENST00000341377 | NRG1 | chr8 | 32585466 | + | 2976 | 868 | 1109 | 2347 | 412 |
| ENST00000353334 | CD74 | chr5 | 149782125 | - | ENST00000287842 | NRG1 | chr8 | 32585466 | + | 2389 | 868 | 180 | 2279 | 699 |
| ENST00000353334 | CD74 | chr5 | 149782125 | - | ENST00000521670 | NRG1 | chr8 | 32585466 | + | 2027 | 868 | 180 | 1754 | 524 |
| ENST00000353334 | CD74 | chr5 | 149782125 | - | ENST00000405005 | NRG1 | chr8 | 32585466 | + | 2358 | 868 | 180 | 2288 | 702 |
| ENST00000353334 | CD74 | chr5 | 149782125 | - | ENST00000520502 | NRG1 | chr8 | 32585466 | + | 1531 | 868 | 180 | 1091 | 303 |
| ENST00000353334 | CD74 | chr5 | 149782125 | - | ENST00000539990 | NRG1 | chr8 | 32585466 | + | 2386 | 868 | 180 | 2279 | 699 |
| ENST00000009530 | CD74 | chr5 | 149782125 | - | ENST00000519301 | NRG1 | chr8 | 32585466 | + | 2321 | 882 | 2 | 2317 | 771 |
| ENST00000009530 | CD74 | chr5 | 149782125 | - | ENST00000520407 | NRG1 | chr8 | 32585466 | + | 1562 | 882 | 2 | 1105 | 367 |
| ENST00000009530 | CD74 | chr5 | 149782125 | - | ENST00000523079 | NRG1 | chr8 | 32585466 | + | 2222 | 882 | 2 | 1642 | 546 |
| ENST00000009530 | CD74 | chr5 | 149782125 | - | ENST00000338921 | NRG1 | chr8 | 32585466 | + | 2955 | 882 | 2 | 2326 | 774 |
| ENST00000009530 | CD74 | chr5 | 149782125 | - | ENST00000356819 | NRG1 | chr8 | 32585466 | + | 2946 | 882 | 2 | 2317 | 771 |
| ENST00000009530 | CD74 | chr5 | 149782125 | - | ENST00000287845 | NRG1 | chr8 | 32585466 | + | 2946 | 882 | 2 | 2317 | 771 |
| ENST00000009530 | CD74 | chr5 | 149782125 | - | ENST00000341377 | NRG1 | chr8 | 32585466 | + | 2990 | 882 | 1123 | 2361 | 412 |
| ENST00000009530 | CD74 | chr5 | 149782125 | - | ENST00000287842 | NRG1 | chr8 | 32585466 | + | 2403 | 882 | 2 | 2293 | 763 |
| ENST00000009530 | CD74 | chr5 | 149782125 | - | ENST00000521670 | NRG1 | chr8 | 32585466 | + | 2041 | 882 | 2 | 1768 | 588 |
| ENST00000009530 | CD74 | chr5 | 149782125 | - | ENST00000405005 | NRG1 | chr8 | 32585466 | + | 2372 | 882 | 2 | 2302 | 766 |
| ENST00000009530 | CD74 | chr5 | 149782125 | - | ENST00000520502 | NRG1 | chr8 | 32585466 | + | 1545 | 882 | 2 | 1105 | 367 |
| ENST00000009530 | CD74 | chr5 | 149782125 | - | ENST00000539990 | NRG1 | chr8 | 32585466 | + | 2400 | 882 | 2 | 2293 | 763 |
| ENST00000353334 | CD74 | chr5 | 149784242 | - | ENST00000519301 | NRG1 | chr8 | 32585466 | + | 2244 | 805 | 180 | 2240 | 686 |
| ENST00000353334 | CD74 | chr5 | 149784242 | - | ENST00000520407 | NRG1 | chr8 | 32585466 | + | 1485 | 805 | 180 | 1028 | 282 |
| ENST00000353334 | CD74 | chr5 | 149784242 | - | ENST00000523079 | NRG1 | chr8 | 32585466 | + | 2145 | 805 | 180 | 1565 | 461 |
| ENST00000353334 | CD74 | chr5 | 149784242 | - | ENST00000338921 | NRG1 | chr8 | 32585466 | + | 2878 | 805 | 180 | 2249 | 689 |
| ENST00000353334 | CD74 | chr5 | 149784242 | - | ENST00000356819 | NRG1 | chr8 | 32585466 | + | 2869 | 805 | 180 | 2240 | 686 |
| ENST00000353334 | CD74 | chr5 | 149784242 | - | ENST00000287845 | NRG1 | chr8 | 32585466 | + | 2869 | 805 | 180 | 2240 | 686 |
| ENST00000353334 | CD74 | chr5 | 149784242 | - | ENST00000341377 | NRG1 | chr8 | 32585466 | + | 2913 | 805 | 1046 | 2284 | 412 |
| ENST00000353334 | CD74 | chr5 | 149784242 | - | ENST00000287842 | NRG1 | chr8 | 32585466 | + | 2326 | 805 | 180 | 2216 | 678 |
| ENST00000353334 | CD74 | chr5 | 149784242 | - | ENST00000521670 | NRG1 | chr8 | 32585466 | + | 1964 | 805 | 180 | 1691 | 503 |
| ENST00000353334 | CD74 | chr5 | 149784242 | - | ENST00000405005 | NRG1 | chr8 | 32585466 | + | 2295 | 805 | 180 | 2225 | 681 |
| ENST00000353334 | CD74 | chr5 | 149784242 | - | ENST00000520502 | NRG1 | chr8 | 32585466 | + | 1468 | 805 | 180 | 1028 | 282 |
| ENST00000353334 | CD74 | chr5 | 149784242 | - | ENST00000539990 | NRG1 | chr8 | 32585466 | + | 2323 | 805 | 180 | 2216 | 678 |
| ENST00000009530 | CD74 | chr5 | 149784242 | - | ENST00000519301 | NRG1 | chr8 | 32585466 | + | 2066 | 627 | 2 | 2062 | 686 |
| ENST00000009530 | CD74 | chr5 | 149784242 | - | ENST00000520407 | NRG1 | chr8 | 32585466 | + | 1307 | 627 | 2 | 850 | 282 |
| ENST00000009530 | CD74 | chr5 | 149784242 | - | ENST00000523079 | NRG1 | chr8 | 32585466 | + | 1967 | 627 | 2 | 1387 | 461 |
| ENST00000009530 | CD74 | chr5 | 149784242 | - | ENST00000338921 | NRG1 | chr8 | 32585466 | + | 2700 | 627 | 2 | 2071 | 689 |
| ENST00000009530 | CD74 | chr5 | 149784242 | - | ENST00000356819 | NRG1 | chr8 | 32585466 | + | 2691 | 627 | 2 | 2062 | 686 |
| ENST00000009530 | CD74 | chr5 | 149784242 | - | ENST00000287845 | NRG1 | chr8 | 32585466 | + | 2691 | 627 | 2 | 2062 | 686 |
| ENST00000009530 | CD74 | chr5 | 149784242 | - | ENST00000341377 | NRG1 | chr8 | 32585466 | + | 2735 | 627 | 868 | 2106 | 412 |
| ENST00000009530 | CD74 | chr5 | 149784242 | - | ENST00000287842 | NRG1 | chr8 | 32585466 | + | 2148 | 627 | 2 | 2038 | 678 |
| ENST00000009530 | CD74 | chr5 | 149784242 | - | ENST00000521670 | NRG1 | chr8 | 32585466 | + | 1786 | 627 | 2 | 1513 | 503 |
| ENST00000009530 | CD74 | chr5 | 149784242 | - | ENST00000405005 | NRG1 | chr8 | 32585466 | + | 2117 | 627 | 2 | 2047 | 681 |
| ENST00000009530 | CD74 | chr5 | 149784242 | - | ENST00000520502 | NRG1 | chr8 | 32585466 | + | 1290 | 627 | 2 | 850 | 282 |
| ENST00000009530 | CD74 | chr5 | 149784242 | - | ENST00000539990 | NRG1 | chr8 | 32585466 | + | 2145 | 627 | 2 | 2038 | 678 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >14642_14642_1_CD74-NRG1_CD74_chr5_149782125_ENST00000009530_NRG1_chr8_32585466_ENST00000287842_length(amino acids)=763AA_BP=293 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKVLTKCQEEVSHIPAVHPGSFRPKCDENGNYLPLQCYGSIGYCWCVFPNGTEVPNTRSRGHHN CSESLELEDPSSGLGVTKQDLGPATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKAEE LYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVERE AETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRD SPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAH DSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNPLAA -------------------------------------------------------------- >14642_14642_2_CD74-NRG1_CD74_chr5_149782125_ENST00000009530_NRG1_chr8_32585466_ENST00000287845_length(amino acids)=771AA_BP=293 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKVLTKCQEEVSHIPAVHPGSFRPKCDENGNYLPLQCYGSIGYCWCVFPNGTEVPNTRSRGHHN CSESLELEDPSSGLGVTKQDLGPATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKHLG IEFMEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVIS SEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHAR ETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFS SFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFL -------------------------------------------------------------- >14642_14642_3_CD74-NRG1_CD74_chr5_149782125_ENST00000009530_NRG1_chr8_32585466_ENST00000338921_length(amino acids)=774AA_BP=293 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKVLTKCQEEVSHIPAVHPGSFRPKCDENGNYLPLQCYGSIGYCWCVFPNGTEVPNTRSRGHHN CSESLELEDPSSGLGVTKQDLGPATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMKVQNQEK HSGEPFPEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKN VISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLR HARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQ QFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDT -------------------------------------------------------------- >14642_14642_4_CD74-NRG1_CD74_chr5_149782125_ENST00000009530_NRG1_chr8_32585466_ENST00000341377_length(amino acids)=412AA_BP= MASFYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVI SSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHA RETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQF SSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPF -------------------------------------------------------------- >14642_14642_5_CD74-NRG1_CD74_chr5_149782125_ENST00000009530_NRG1_chr8_32585466_ENST00000356819_length(amino acids)=771AA_BP=293 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKVLTKCQEEVSHIPAVHPGSFRPKCDENGNYLPLQCYGSIGYCWCVFPNGTEVPNTRSRGHHN CSESLELEDPSSGLGVTKQDLGPATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKHLG IEFMEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVIS SEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHAR ETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFS SFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFL -------------------------------------------------------------- >14642_14642_6_CD74-NRG1_CD74_chr5_149782125_ENST00000009530_NRG1_chr8_32585466_ENST00000405005_length(amino acids)=766AA_BP=293 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKVLTKCQEEVSHIPAVHPGSFRPKCDENGNYLPLQCYGSIGYCWCVFPNGTEVPNTRSRGHHN CSESLELEDPSSGLGVTKQDLGPATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMKVQNQEK AEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIV EREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDS YRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHN PAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNP -------------------------------------------------------------- >14642_14642_7_CD74-NRG1_CD74_chr5_149782125_ENST00000009530_NRG1_chr8_32585466_ENST00000519301_length(amino acids)=771AA_BP=293 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKVLTKCQEEVSHIPAVHPGSFRPKCDENGNYLPLQCYGSIGYCWCVFPNGTEVPNTRSRGHHN CSESLELEDPSSGLGVTKQDLGPATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKHLG IEFMEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVIS SEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHAR ETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFS SFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFL -------------------------------------------------------------- >14642_14642_8_CD74-NRG1_CD74_chr5_149782125_ENST00000009530_NRG1_chr8_32585466_ENST00000520407_length(amino acids)=367AA_BP=293 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKVLTKCQEEVSHIPAVHPGSFRPKCDENGNYLPLQCYGSIGYCWCVFPNGTEVPNTRSRGHHN CSESLELEDPSSGLGVTKQDLGPATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYSTST -------------------------------------------------------------- >14642_14642_9_CD74-NRG1_CD74_chr5_149782125_ENST00000009530_NRG1_chr8_32585466_ENST00000520502_length(amino acids)=367AA_BP=293 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKVLTKCQEEVSHIPAVHPGSFRPKCDENGNYLPLQCYGSIGYCWCVFPNGTEVPNTRSRGHHN CSESLELEDPSSGLGVTKQDLGPATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYSTST -------------------------------------------------------------- >14642_14642_10_CD74-NRG1_CD74_chr5_149782125_ENST00000009530_NRG1_chr8_32585466_ENST00000521670_length(amino acids)=588AA_BP=293 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKVLTKCQEEVSHIPAVHPGSFRPKCDENGNYLPLQCYGSIGYCWCVFPNGTEVPNTRSRGHHN CSESLELEDPSSGLGVTKQDLGPATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMKVQNQEK AEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIV EREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDS -------------------------------------------------------------- >14642_14642_11_CD74-NRG1_CD74_chr5_149782125_ENST00000009530_NRG1_chr8_32585466_ENST00000523079_length(amino acids)=546AA_BP=293 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKVLTKCQEEVSHIPAVHPGSFRPKCDENGNYLPLQCYGSIGYCWCVFPNGTEVPNTRSRGHHN CSESLELEDPSSGLGVTKQDLGPATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKAEE LYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVERE AETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRD -------------------------------------------------------------- >14642_14642_12_CD74-NRG1_CD74_chr5_149782125_ENST00000009530_NRG1_chr8_32585466_ENST00000539990_length(amino acids)=763AA_BP=293 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKVLTKCQEEVSHIPAVHPGSFRPKCDENGNYLPLQCYGSIGYCWCVFPNGTEVPNTRSRGHHN CSESLELEDPSSGLGVTKQDLGPATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKAEE LYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVERE AETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRD SPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAH DSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNPLAA -------------------------------------------------------------- >14642_14642_13_CD74-NRG1_CD74_chr5_149782125_ENST00000353334_NRG1_chr8_32585466_ENST00000287842_length(amino acids)=699AA_BP=229 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKESLELEDPSSGLGVTKQDLGPATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRY LCKCPNEFTGDRCQNYVMASFYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNP PPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGR LNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLV TPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSS -------------------------------------------------------------- >14642_14642_14_CD74-NRG1_CD74_chr5_149782125_ENST00000353334_NRG1_chr8_32585466_ENST00000287845_length(amino acids)=707AA_BP=229 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKESLELEDPSSGLGVTKQDLGPATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRY LCKCPNEFTGDRCQNYVMASFYKHLGIEFMEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIA NGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSS PTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFME EERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVD -------------------------------------------------------------- >14642_14642_15_CD74-NRG1_CD74_chr5_149782125_ENST00000353334_NRG1_chr8_32585466_ENST00000338921_length(amino acids)=710AA_BP=229 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKESLELEDPSSGLGVTKQDLGPATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRY LCKCQPGFTGARCTENVPMKVQNQEKHSGEPFPEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMM NIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSR HSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSP FMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRL -------------------------------------------------------------- >14642_14642_16_CD74-NRG1_CD74_chr5_149782125_ENST00000353334_NRG1_chr8_32585466_ENST00000341377_length(amino acids)=412AA_BP= MASFYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVI SSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHA RETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQF SSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPF -------------------------------------------------------------- >14642_14642_17_CD74-NRG1_CD74_chr5_149782125_ENST00000353334_NRG1_chr8_32585466_ENST00000356819_length(amino acids)=707AA_BP=229 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKESLELEDPSSGLGVTKQDLGPATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRY LCKCPNEFTGDRCQNYVMASFYKHLGIEFMEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIA NGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSS PTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFME EERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVD -------------------------------------------------------------- >14642_14642_18_CD74-NRG1_CD74_chr5_149782125_ENST00000353334_NRG1_chr8_32585466_ENST00000405005_length(amino acids)=702AA_BP=229 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKESLELEDPSSGLGVTKQDLGPATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRY LCKCQPGFTGARCTENVPMKVQNQEKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHH PNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGP RGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPL LLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSS -------------------------------------------------------------- >14642_14642_19_CD74-NRG1_CD74_chr5_149782125_ENST00000353334_NRG1_chr8_32585466_ENST00000519301_length(amino acids)=707AA_BP=229 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKESLELEDPSSGLGVTKQDLGPATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRY LCKCPNEFTGDRCQNYVMASFYKHLGIEFMEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIA NGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSS PTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFME EERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVD -------------------------------------------------------------- >14642_14642_20_CD74-NRG1_CD74_chr5_149782125_ENST00000353334_NRG1_chr8_32585466_ENST00000520407_length(amino acids)=303AA_BP=229 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKESLELEDPSSGLGVTKQDLGPATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRY -------------------------------------------------------------- >14642_14642_21_CD74-NRG1_CD74_chr5_149782125_ENST00000353334_NRG1_chr8_32585466_ENST00000520502_length(amino acids)=303AA_BP=229 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKESLELEDPSSGLGVTKQDLGPATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRY -------------------------------------------------------------- >14642_14642_22_CD74-NRG1_CD74_chr5_149782125_ENST00000353334_NRG1_chr8_32585466_ENST00000521670_length(amino acids)=524AA_BP=229 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKESLELEDPSSGLGVTKQDLGPATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRY LCKCQPGFTGARCTENVPMKVQNQEKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHH PNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGP -------------------------------------------------------------- >14642_14642_23_CD74-NRG1_CD74_chr5_149782125_ENST00000353334_NRG1_chr8_32585466_ENST00000523079_length(amino acids)=482AA_BP=229 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKESLELEDPSSGLGVTKQDLGPATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRY LCKCPNEFTGDRCQNYVMASFYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNP PPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGR -------------------------------------------------------------- >14642_14642_24_CD74-NRG1_CD74_chr5_149782125_ENST00000353334_NRG1_chr8_32585466_ENST00000539990_length(amino acids)=699AA_BP=229 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKESLELEDPSSGLGVTKQDLGPATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRY LCKCPNEFTGDRCQNYVMASFYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNP PPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGR LNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLV TPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSS -------------------------------------------------------------- >14642_14642_25_CD74-NRG1_CD74_chr5_149784242_ENST00000009530_NRG1_chr8_32585466_ENST00000287842_length(amino acids)=678AA_BP=208 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASF YKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEH IVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETP DSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFH HNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQ -------------------------------------------------------------- >14642_14642_26_CD74-NRG1_CD74_chr5_149784242_ENST00000009530_NRG1_chr8_32585466_ENST00000287845_length(amino acids)=686AA_BP=208 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASF YKHLGIEFMEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVS KNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSF LRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHH PQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGE -------------------------------------------------------------- >14642_14642_27_CD74-NRG1_CD74_chr5_149784242_ENST00000009530_NRG1_chr8_32585466_ENST00000338921_length(amino acids)=689AA_BP=208 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMKV QNQEKHSGEPFPEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQ YVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPREC NSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKF DHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDER -------------------------------------------------------------- >14642_14642_28_CD74-NRG1_CD74_chr5_149784242_ENST00000009530_NRG1_chr8_32585466_ENST00000341377_length(amino acids)=412AA_BP= MASFYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVI SSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHA RETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQF SSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPF -------------------------------------------------------------- >14642_14642_29_CD74-NRG1_CD74_chr5_149784242_ENST00000009530_NRG1_chr8_32585466_ENST00000356819_length(amino acids)=686AA_BP=208 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASF YKHLGIEFMEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVS KNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSF LRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHH PQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGE -------------------------------------------------------------- >14642_14642_30_CD74-NRG1_CD74_chr5_149784242_ENST00000009530_NRG1_chr8_32585466_ENST00000405005_length(amino acids)=681AA_BP=208 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMKV QNQEKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVIS SEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHAR ETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFS SFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFL -------------------------------------------------------------- >14642_14642_31_CD74-NRG1_CD74_chr5_149784242_ENST00000009530_NRG1_chr8_32585466_ENST00000519301_length(amino acids)=686AA_BP=208 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASF YKHLGIEFMEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVS KNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSF LRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHH PQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGE -------------------------------------------------------------- >14642_14642_32_CD74-NRG1_CD74_chr5_149784242_ENST00000009530_NRG1_chr8_32585466_ENST00000520407_length(amino acids)=282AA_BP=208 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASF -------------------------------------------------------------- >14642_14642_33_CD74-NRG1_CD74_chr5_149784242_ENST00000009530_NRG1_chr8_32585466_ENST00000520502_length(amino acids)=282AA_BP=208 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASF -------------------------------------------------------------- >14642_14642_34_CD74-NRG1_CD74_chr5_149784242_ENST00000009530_NRG1_chr8_32585466_ENST00000521670_length(amino acids)=503AA_BP=208 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMKV QNQEKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVIS SEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHAR -------------------------------------------------------------- >14642_14642_35_CD74-NRG1_CD74_chr5_149784242_ENST00000009530_NRG1_chr8_32585466_ENST00000523079_length(amino acids)=461AA_BP=208 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASF YKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEH IVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETP -------------------------------------------------------------- >14642_14642_36_CD74-NRG1_CD74_chr5_149784242_ENST00000009530_NRG1_chr8_32585466_ENST00000539990_length(amino acids)=678AA_BP=208 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASF YKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEH IVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETP DSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFH HNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQ -------------------------------------------------------------- >14642_14642_37_CD74-NRG1_CD74_chr5_149784242_ENST00000353334_NRG1_chr8_32585466_ENST00000287842_length(amino acids)=678AA_BP=208 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASF YKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEH IVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETP DSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFH HNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQ -------------------------------------------------------------- >14642_14642_38_CD74-NRG1_CD74_chr5_149784242_ENST00000353334_NRG1_chr8_32585466_ENST00000287845_length(amino acids)=686AA_BP=208 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASF YKHLGIEFMEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVS KNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSF LRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHH PQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGE -------------------------------------------------------------- >14642_14642_39_CD74-NRG1_CD74_chr5_149784242_ENST00000353334_NRG1_chr8_32585466_ENST00000338921_length(amino acids)=689AA_BP=208 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMKV QNQEKHSGEPFPEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQ YVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPREC NSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKF DHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDER -------------------------------------------------------------- >14642_14642_40_CD74-NRG1_CD74_chr5_149784242_ENST00000353334_NRG1_chr8_32585466_ENST00000341377_length(amino acids)=412AA_BP= MASFYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVI SSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHA RETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQF SSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPF -------------------------------------------------------------- >14642_14642_41_CD74-NRG1_CD74_chr5_149784242_ENST00000353334_NRG1_chr8_32585466_ENST00000356819_length(amino acids)=686AA_BP=208 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASF YKHLGIEFMEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVS KNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSF LRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHH PQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGE -------------------------------------------------------------- >14642_14642_42_CD74-NRG1_CD74_chr5_149784242_ENST00000353334_NRG1_chr8_32585466_ENST00000405005_length(amino acids)=681AA_BP=208 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMKV QNQEKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVIS SEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHAR ETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFS SFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFL -------------------------------------------------------------- >14642_14642_43_CD74-NRG1_CD74_chr5_149784242_ENST00000353334_NRG1_chr8_32585466_ENST00000519301_length(amino acids)=686AA_BP=208 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASF YKHLGIEFMEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVS KNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSF LRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHH PQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGE -------------------------------------------------------------- >14642_14642_44_CD74-NRG1_CD74_chr5_149784242_ENST00000353334_NRG1_chr8_32585466_ENST00000520407_length(amino acids)=282AA_BP=208 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASF -------------------------------------------------------------- >14642_14642_45_CD74-NRG1_CD74_chr5_149784242_ENST00000353334_NRG1_chr8_32585466_ENST00000520502_length(amino acids)=282AA_BP=208 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASF -------------------------------------------------------------- >14642_14642_46_CD74-NRG1_CD74_chr5_149784242_ENST00000353334_NRG1_chr8_32585466_ENST00000521670_length(amino acids)=503AA_BP=208 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMKV QNQEKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVIS SEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHAR -------------------------------------------------------------- >14642_14642_47_CD74-NRG1_CD74_chr5_149784242_ENST00000353334_NRG1_chr8_32585466_ENST00000523079_length(amino acids)=461AA_BP=208 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASF YKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEH IVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETP -------------------------------------------------------------- >14642_14642_48_CD74-NRG1_CD74_chr5_149784242_ENST00000353334_NRG1_chr8_32585466_ENST00000539990_length(amino acids)=678AA_BP=208 MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALYTGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQL ENLRMKLPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNADPLKVYPPLKGSFPENLRHLKNTMETIDWKV FESWMHHWLLFEMSRHSLEQKPTDAPPKATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASF YKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEH IVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETP DSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFH HNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQ -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr5:/chr8:) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CD74 | NRG1 |
| FUNCTION: Plays a critical role in MHC class II antigen processing by stabilizing peptide-free class II alpha/beta heterodimers in a complex soon after their synthesis and directing transport of the complex from the endoplasmic reticulum to the endosomal/lysosomal system where the antigen processing and binding of antigenic peptides to MHC class II takes place. Serves as cell surface receptor for the cytokine MIF.; FUNCTION: [Class-II-associated invariant chain peptide]: Binds to the peptide-binding site of MHC class II alpha/beta heterodimers forming an alpha-beta-CLIP complex, thereby preventing the loading of antigenic peptides to the MHC class II complex until its release by HLA-DM in the endosome. {ECO:0000269|PubMed:1448172}.; FUNCTION: [Isoform p41]: Stabilizes the conformation of mature CTSL by binding to its active site and serving as a chaperone to help maintain a pool of mature enzyme in endocytic compartments and extracellular space of antigen-presenting cells (APCs). Has antiviral activity by stymieing the endosomal entry of Ebola virus and coronaviruses, including SARS-CoV-2 (PubMed:32855215). Disrupts cathepsin-mediated Ebola virus glycoprotein processing, which prevents viral fusion and entry. This antiviral activity is specific to p41 isoform (PubMed:32855215). {ECO:0000250|UniProtKB:P04441, ECO:0000269|PubMed:32855215}. | FUNCTION: Direct ligand for ERBB3 and ERBB4 tyrosine kinase receptors. Concomitantly recruits ERBB1 and ERBB2 coreceptors, resulting in ligand-stimulated tyrosine phosphorylation and activation of the ERBB receptors. The multiple isoforms perform diverse functions such as inducing growth and differentiation of epithelial, glial, neuronal, and skeletal muscle cells; inducing expression of acetylcholine receptor in synaptic vesicles during the formation of the neuromuscular junction; stimulating lobuloalveolar budding and milk production in the mammary gland and inducing differentiation of mammary tumor cells; stimulating Schwann cell proliferation; implication in the development of the myocardium such as trabeculation of the developing heart. Isoform 10 may play a role in motor and sensory neuron development. Binds to ERBB4 (PubMed:10867024, PubMed:7902537). Binds to ERBB3 (PubMed:20682778). Acts as a ligand for integrins and binds (via EGF domain) to integrins ITGAV:ITGB3 or ITGA6:ITGB4. Its binding to integrins and subsequent ternary complex formation with integrins and ERRB3 are essential for NRG1-ERBB signaling. Induces the phosphorylation and activation of MAPK3/ERK1, MAPK1/ERK2 and AKT1 (PubMed:20682778). Ligand-dependent ERBB4 endocytosis is essential for the NRG1-mediated activation of these kinases in neurons (By similarity). {ECO:0000250|UniProtKB:P43322, ECO:0000269|PubMed:10867024, ECO:0000269|PubMed:1348215, ECO:0000269|PubMed:20682778, ECO:0000269|PubMed:7902537}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
Top |
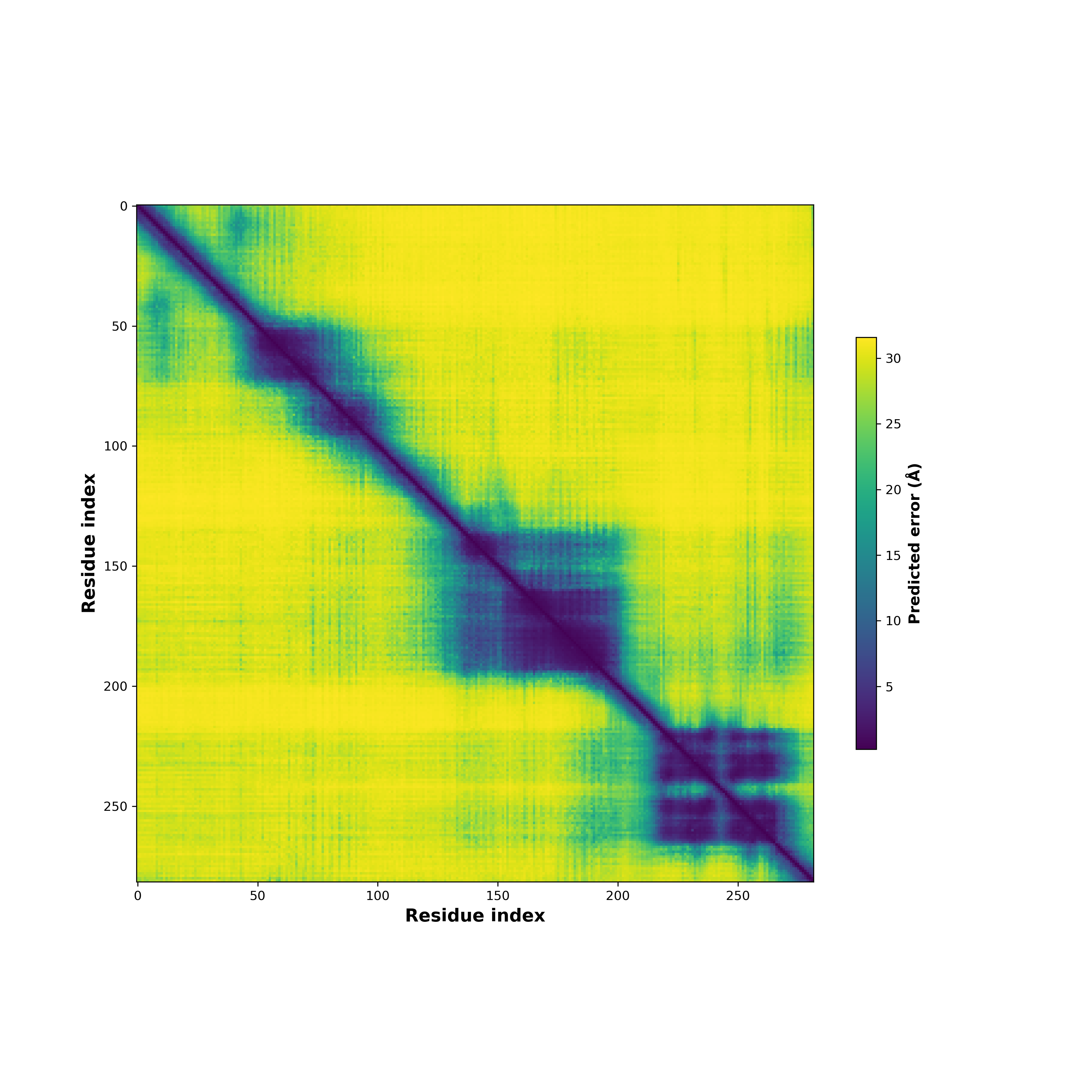
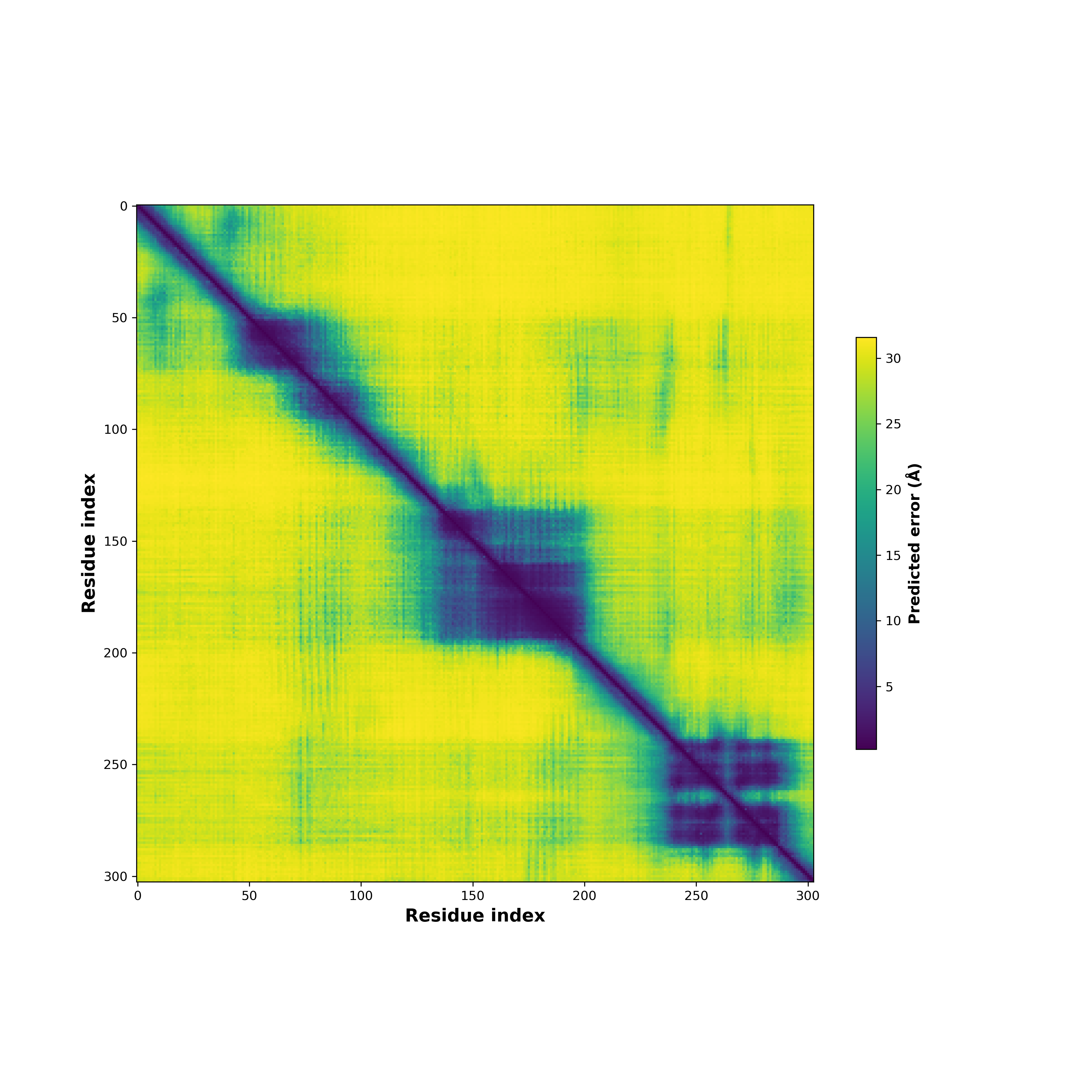
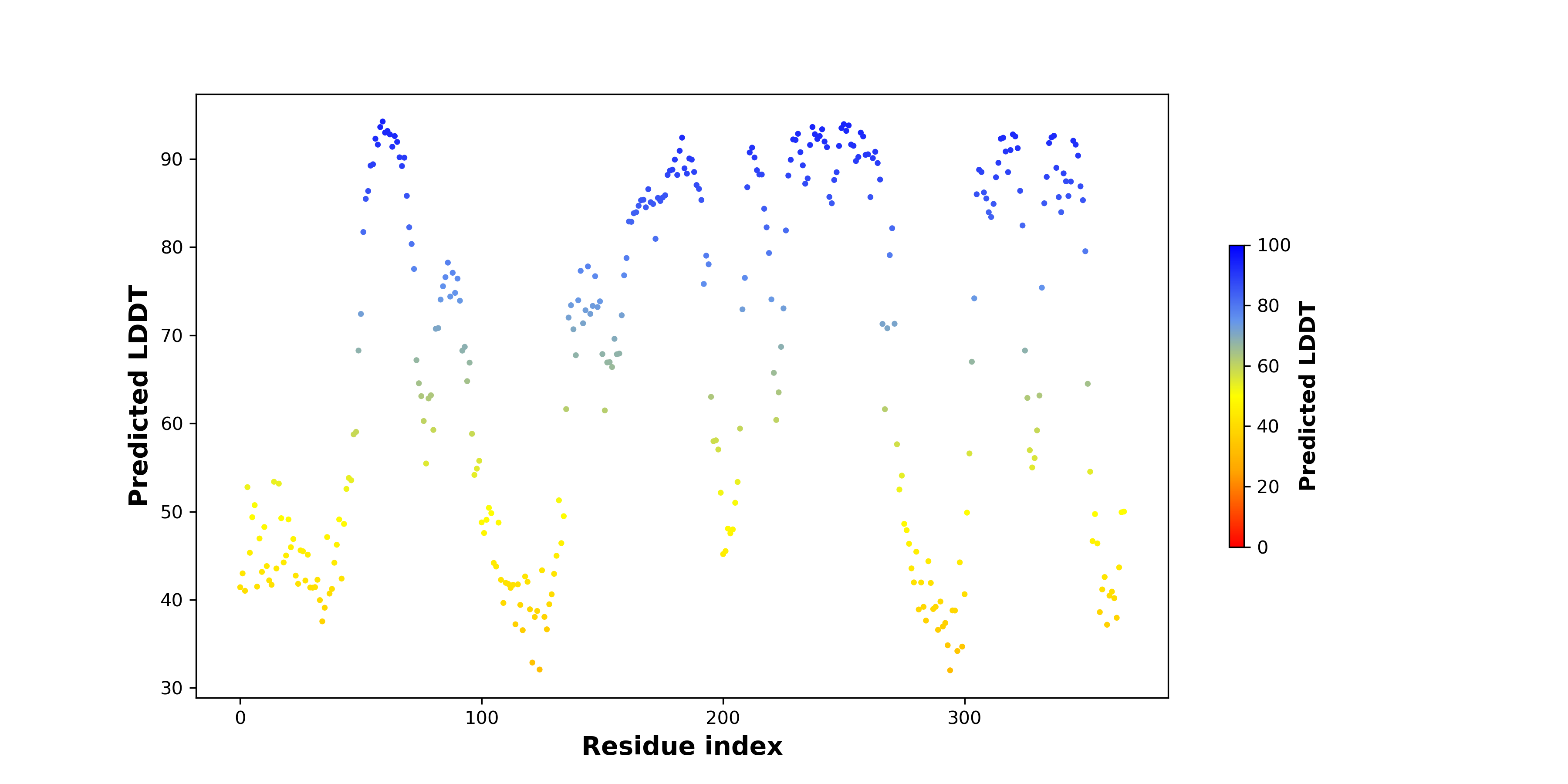
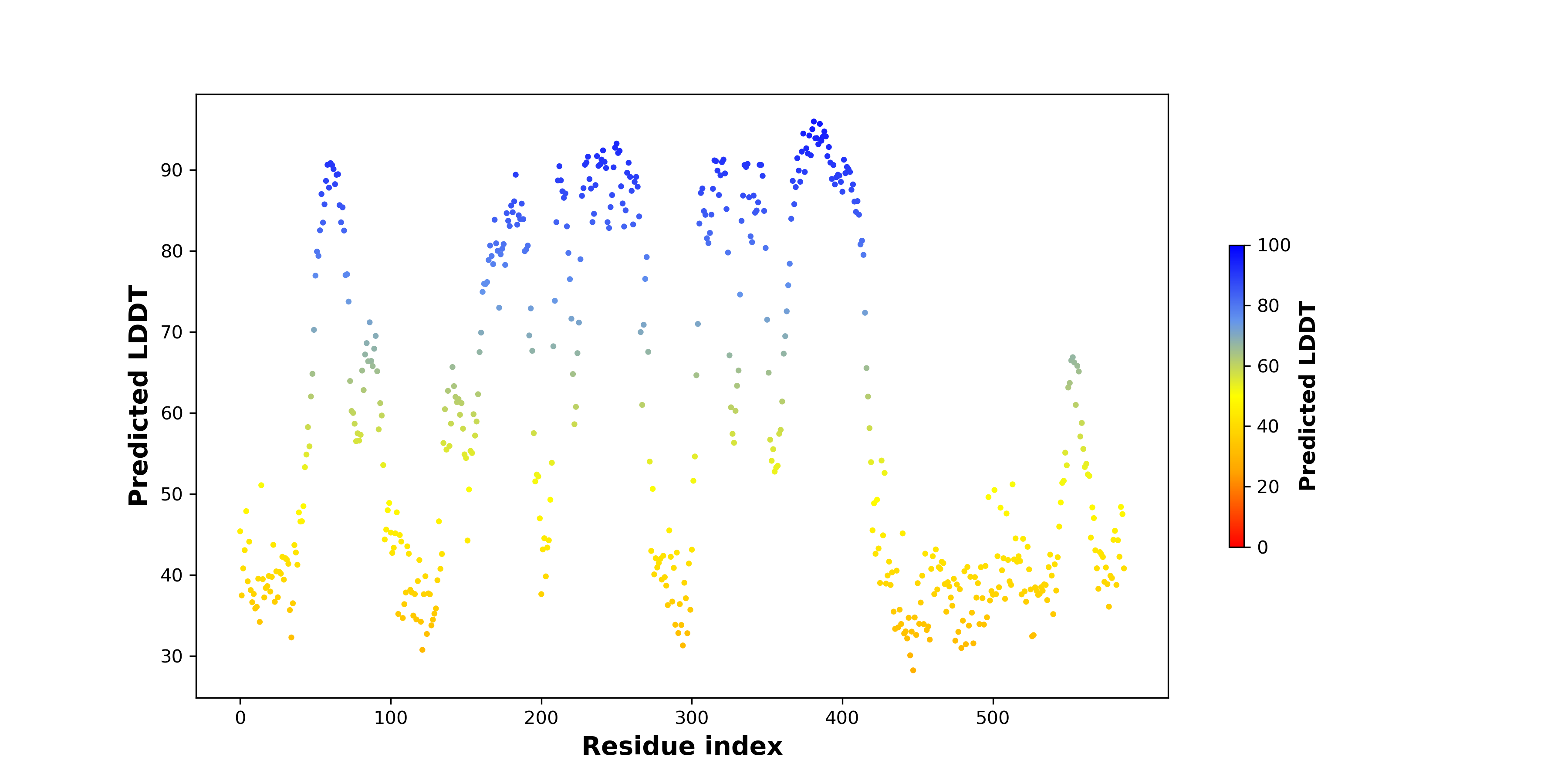
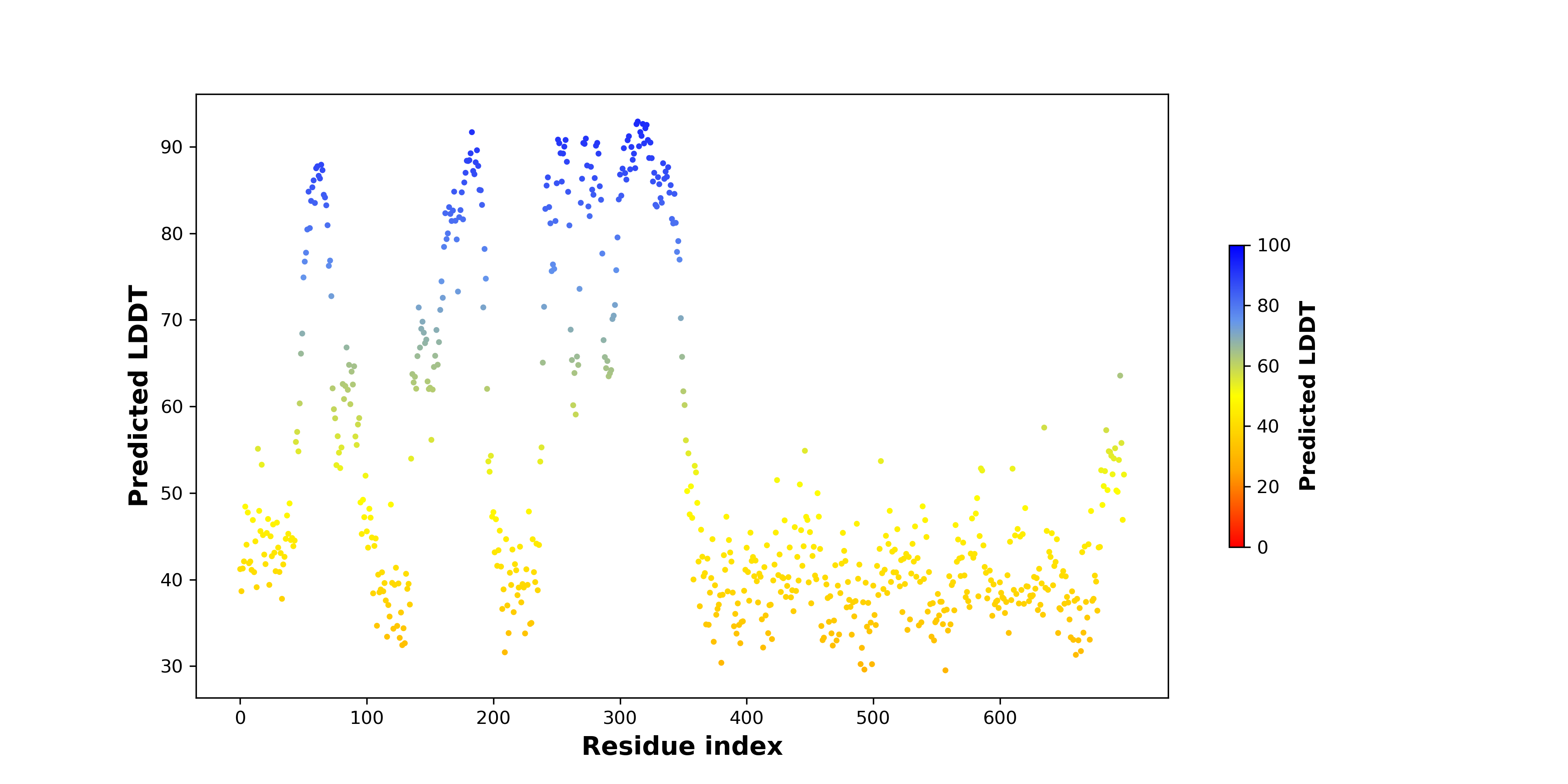
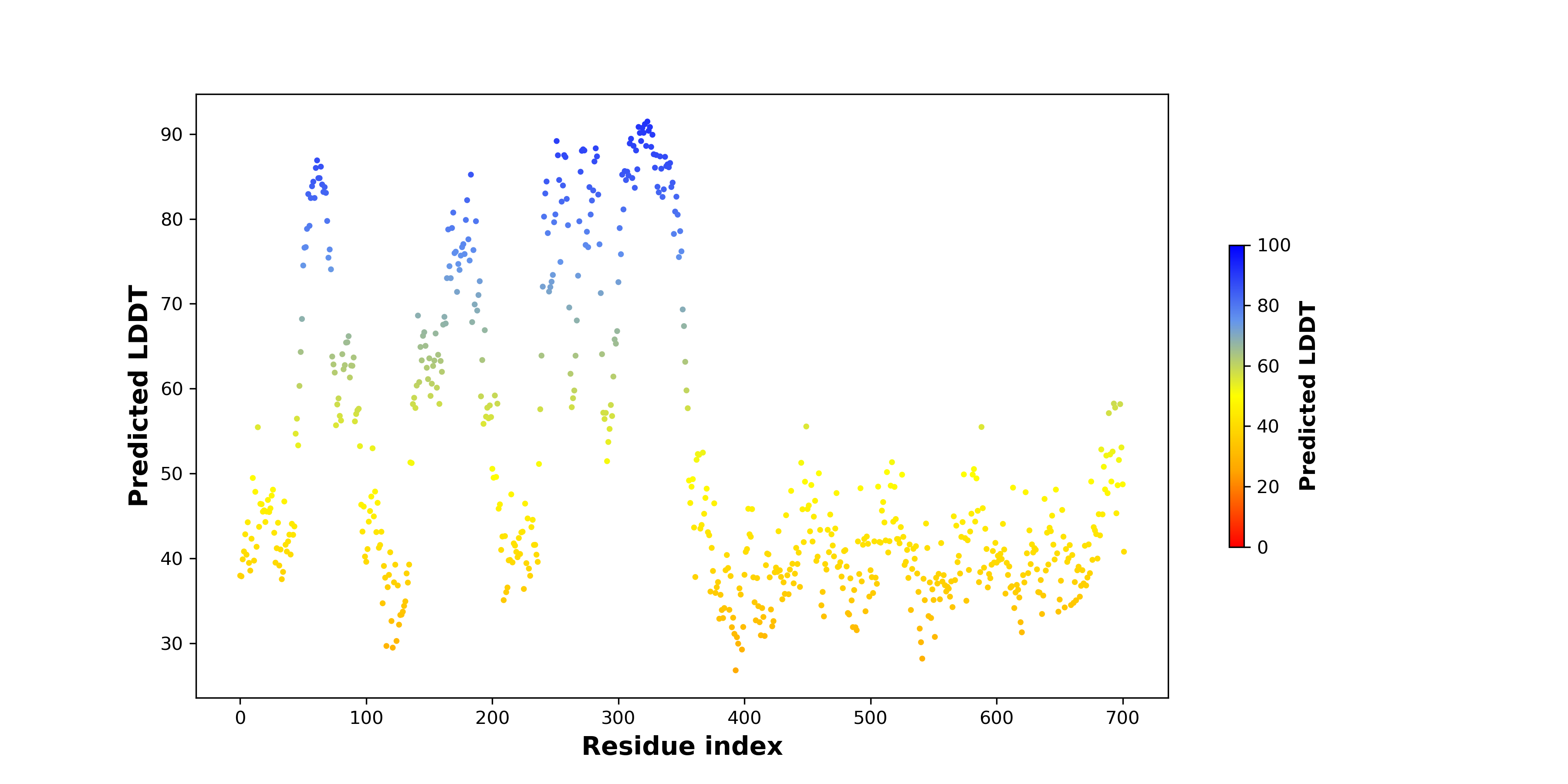
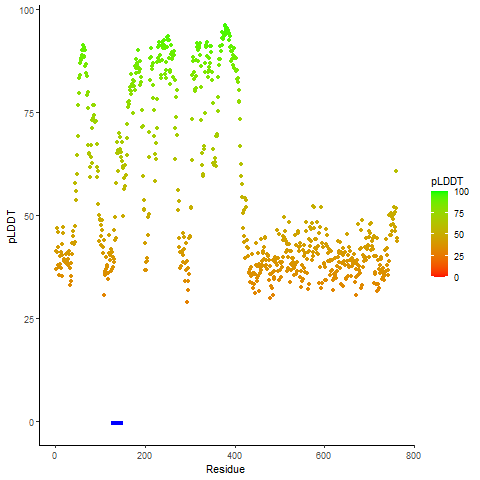
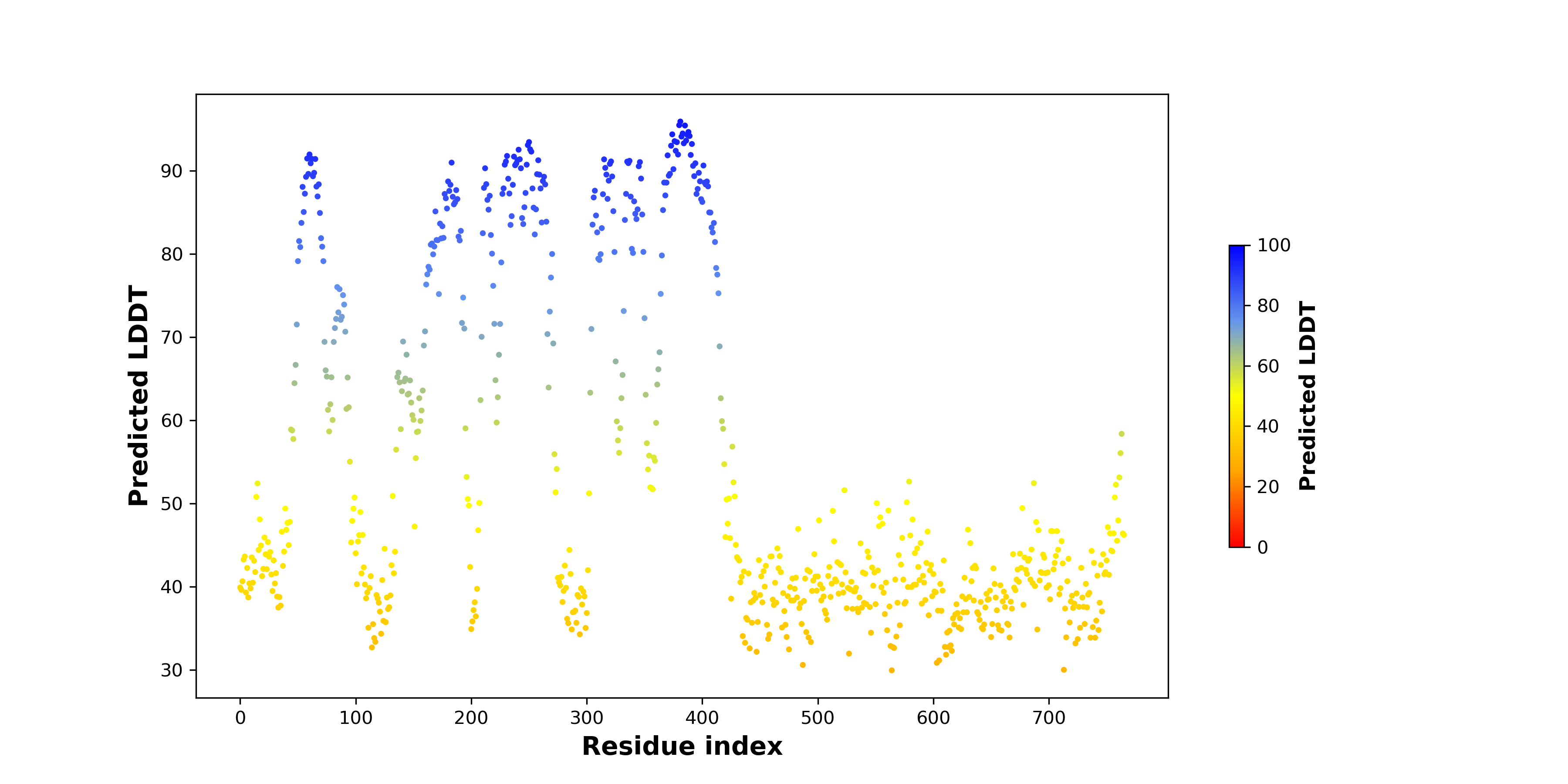
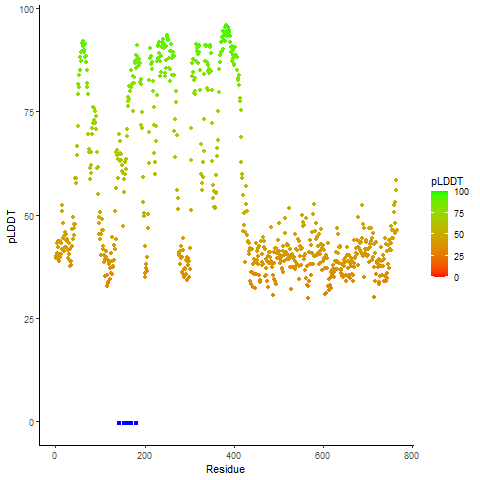
Fusion Protein Structures |
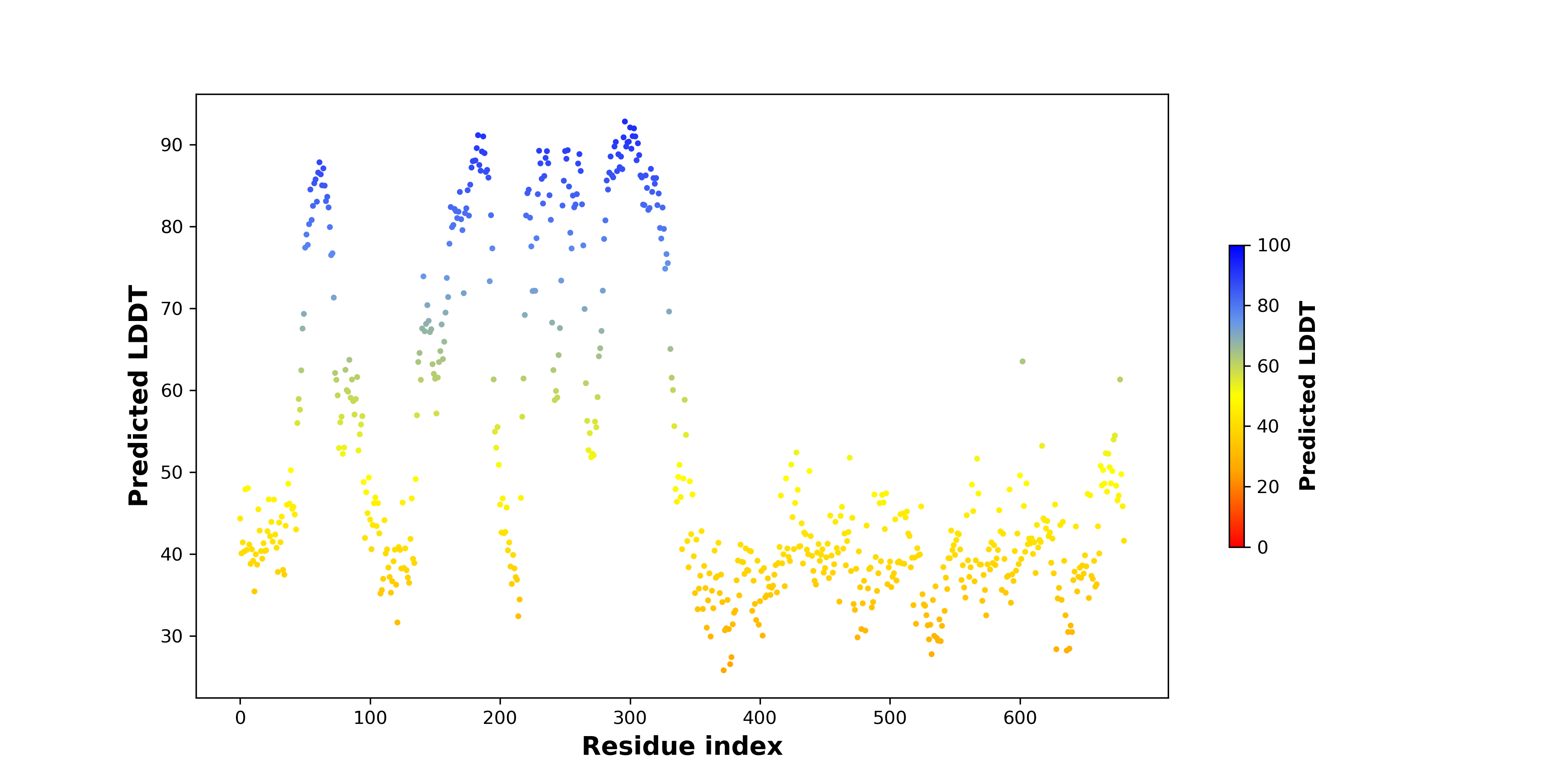
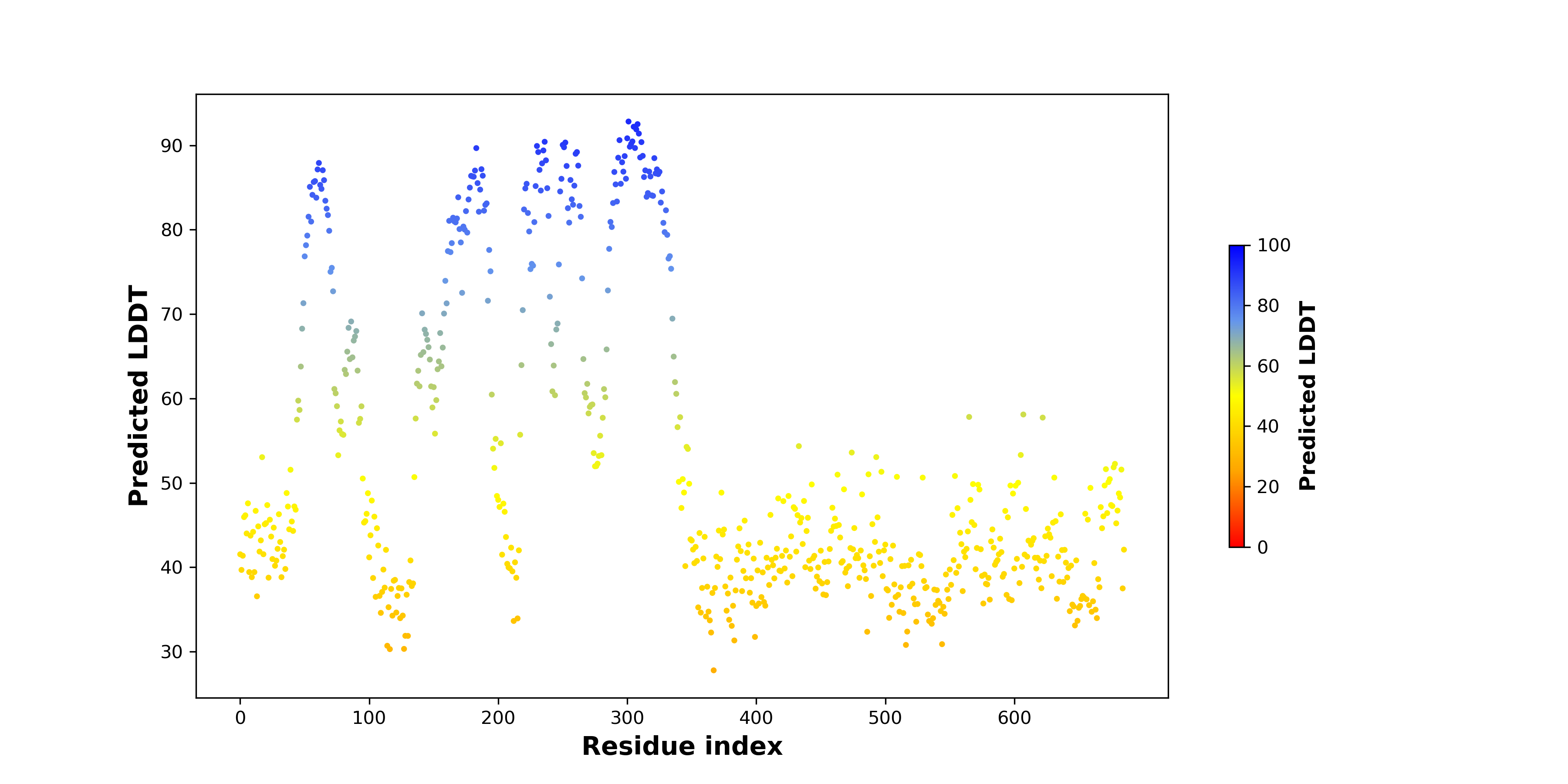
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
| PDB file (52) >>>52.pdbFusion protein BP residue: 208 CIF file (52) >>>52.cif | CD74 | chr5 | 149784242 | - | NRG1 | chr8 | 32585466 | + | MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALY TGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQLENLRMKLPKP PKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNAD PLKVYPPLKGSFPENLRHLKNTMETIDWKVFESWMHHWLLFEMSRHSLEQ KPTDAPPKATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYL | 282 |
| 3D view using mol* of 52 (AA BP:208) | ||||||||||
 | ||||||||||
| PDB file (64) >>>64.pdbFusion protein BP residue: 229 CIF file (64) >>>64.cif | CD74 | chr5 | 149782125 | - | NRG1 | chr8 | 32585466 | + | MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALY TGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQLENLRMKLPKP PKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNAD PLKVYPPLKGSFPENLRHLKNTMETIDWKVFESWMHHWLLFEMSRHSLEQ KPTDAPPKESLELEDPSSGLGVTKQDLGPATSTSTTGTSHLVKCAEKEKT FCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYSTSTPFLS | 303 |
| 3D view using mol* of 64 (AA BP:229) | ||||||||||
 | ||||||||||
| PDB file (121) >>>121.pdbFusion protein BP residue: 293 CIF file (121) >>>121.cif | CD74 | chr5 | 149782125 | - | NRG1 | chr8 | 32585466 | + | MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALY TGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQLENLRMKLPKP PKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNAD PLKVYPPLKGSFPENLRHLKNTMETIDWKVFESWMHHWLLFEMSRHSLEQ KPTDAPPKVLTKCQEEVSHIPAVHPGSFRPKCDENGNYLPLQCYGSIGYC WCVFPNGTEVPNTRSRGHHNCSESLELEDPSSGLGVTKQDLGPATSTSTT GTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNY | 367 |
| 3D view using mol* of 121 (AA BP:293) | ||||||||||
 | ||||||||||
| PDB file (175) >>>175.pdbFusion protein BP residue: CIF file (175) >>>175.cif | CD74 | chr5 | 149782125 | - | NRG1 | chr8 | 32585466 | + | MASFYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRL RQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREA ETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVEN SRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAM TTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPL LLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETT QEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETE DERVGEDTPFLGIQNPLAASLEATPAFRLADSRTNPAGRFSTQEEIQARL | 412 |
| 3D view using mol* of 175 (AA BP:) | ||||||||||
 | ||||||||||
| PDB file (233) >>>233.pdbFusion protein BP residue: 208 CIF file (233) >>>233.cif | CD74 | chr5 | 149784242 | - | NRG1 | chr8 | 32585466 | + | MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALY TGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQLENLRMKLPKP PKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNAD PLKVYPPLKGSFPENLRHLKNTMETIDWKVFESWMHHWLLFEMSRHSLEQ KPTDAPPKATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYL CKCPNEFTGDRCQNYVMASFYKAEELYQKRVLTITGICIALLVVGIMCVV AYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQY VSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTES ILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETP | 461 |
| 3D view using mol* of 233 (AA BP:208) | ||||||||||
 | ||||||||||
| PDB file (269) >>>269.pdbFusion protein BP residue: 229 CIF file (269) >>>269.cif | CD74 | chr5 | 149782125 | - | NRG1 | chr8 | 32585466 | + | MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALY TGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQLENLRMKLPKP PKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNAD PLKVYPPLKGSFPENLRHLKNTMETIDWKVFESWMHHWLLFEMSRHSLEQ KPTDAPPKESLELEDPSSGLGVTKQDLGPATSTSTTGTSHLVKCAEKEKT FCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKAEELYQK RVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMN IANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTA HHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGR | 482 |
| 3D view using mol* of 269 (AA BP:229) | ||||||||||
 | ||||||||||
| PDB file (308) >>>308.pdbFusion protein BP residue: 208 CIF file (308) >>>308.cif | CD74 | chr5 | 149784242 | - | NRG1 | chr8 | 32585466 | + | MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALY TGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQLENLRMKLPKP PKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNAD PLKVYPPLKGSFPENLRHLKNTMETIDWKVFESWMHHWLLFEMSRHSLEQ KPTDAPPKATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYL CKCQPGFTGARCTENVPMKVQNQEKAEELYQKRVLTITGICIALLVVGIM CVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLV NQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGH TESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHAR ETPDSYRDSPHSERHNLIAELRRNKAHRSKCMQIQLSATHLRSSSIPHLG | 503 |
| 3D view using mol* of 308 (AA BP:208) | ||||||||||
 | ||||||||||
| PDB file (350) >>>350.pdbFusion protein BP residue: 229 CIF file (350) >>>350.cif | CD74 | chr5 | 149782125 | - | NRG1 | chr8 | 32585466 | + | MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALY TGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQLENLRMKLPKP PKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNAD PLKVYPPLKGSFPENLRHLKNTMETIDWKVFESWMHHWLLFEMSRHSLEQ KPTDAPPKESLELEDPSSGLGVTKQDLGPATSTSTTGTSHLVKCAEKEKT FCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMKVQNQEKAEEL YQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNN MMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYT STAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGP RGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERHNLIAELRRNKAHRS | 524 |
| 3D view using mol* of 350 (AA BP:229) | ||||||||||
 | ||||||||||
| PDB file (384) >>>384.pdbFusion protein BP residue: 293 CIF file (384) >>>384.cif | CD74 | chr5 | 149782125 | - | NRG1 | chr8 | 32585466 | + | MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALY TGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQLENLRMKLPKP PKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNAD PLKVYPPLKGSFPENLRHLKNTMETIDWKVFESWMHHWLLFEMSRHSLEQ KPTDAPPKVLTKCQEEVSHIPAVHPGSFRPKCDENGNYLPLQCYGSIGYC WCVFPNGTEVPNTRSRGHHNCSESLELEDPSSGLGVTKQDLGPATSTSTT GTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNY VMASFYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDR LRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVERE AETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVE | 546 |
| 3D view using mol* of 384 (AA BP:293) | ||||||||||
 | ||||||||||
| PDB file (451) >>>451.pdbFusion protein BP residue: 293 CIF file (451) >>>451.cif | CD74 | chr5 | 149782125 | - | NRG1 | chr8 | 32585466 | + | MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALY TGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQLENLRMKLPKP PKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNAD PLKVYPPLKGSFPENLRHLKNTMETIDWKVFESWMHHWLLFEMSRHSLEQ KPTDAPPKVLTKCQEEVSHIPAVHPGSFRPKCDENGNYLPLQCYGSIGYC WCVFPNGTEVPNTRSRGHHNCSESLELEDPSSGLGVTKQDLGPATSTSTT GTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTEN VPMKVQNQEKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKL HDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIV EREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMS SVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERH | 588 |
| 3D view using mol* of 451 (AA BP:293) | ||||||||||
 | ||||||||||
| PDB file (548) >>>548.pdbFusion protein BP residue: 208 CIF file (548) >>>548.cif | CD74 | chr5 | 149784242 | - | NRG1 | chr8 | 32585466 | + | MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALY TGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQLENLRMKLPKP PKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNAD PLKVYPPLKGSFPENLRHLKNTMETIDWKVFESWMHHWLLFEMSRHSLEQ KPTDAPPKATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYL CKCPNEFTGDRCQNYVMASFYKAEELYQKRVLTITGICIALLVVGIMCVV AYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQY VSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTES ILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETP DSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVS MPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSL PASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEV DSNTSSQSSNSESETEDERVGEDTPFLGIQNPLAASLEATPAFRLADSRT | 678 |
| 3D view using mol* of 548 (AA BP:208) | ||||||||||
 | ||||||||||
| PDB file (549) >>>549.pdbFusion protein BP residue: 208 CIF file (549) >>>549.cif | CD74 | chr5 | 149784242 | - | NRG1 | chr8 | 32585466 | + | MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALY TGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQLENLRMKLPKP PKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNAD PLKVYPPLKGSFPENLRHLKNTMETIDWKVFESWMHHWLLFEMSRHSLEQ KPTDAPPKATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYL CKCQPGFTGARCTENVPMKVQNQEKAEELYQKRVLTITGICIALLVVGIM CVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLV NQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGH TESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHAR ETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSM TVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDS NSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANR LEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNPLAASLEATPAFRLAD | 681 |
| 3D view using mol* of 549 (AA BP:208) | ||||||||||
 | ||||||||||
| PDB file (559) >>>559.pdbFusion protein BP residue: 208 CIF file (559) >>>559.cif | CD74 | chr5 | 149784242 | - | NRG1 | chr8 | 32585466 | + | MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALY TGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQLENLRMKLPKP PKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNAD PLKVYPPLKGSFPENLRHLKNTMETIDWKVFESWMHHWLLFEMSRHSLEQ KPTDAPPKATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYL CKCPNEFTGDRCQNYVMASFYKHLGIEFMEAEELYQKRVLTITGICIALL VVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPE NVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHS WSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSF LRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSP PVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHN PAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNG HIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNPLAASLEATPA | 686 |
| 3D view using mol* of 559 (AA BP:208) | ||||||||||
 | ||||||||||
| PDB file (563) >>>563.pdbFusion protein BP residue: 208 CIF file (563) >>>563.cif | CD74 | chr5 | 149784242 | - | NRG1 | chr8 | 32585466 | + | MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALY TGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQLENLRMKLPKP PKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNAD PLKVYPPLKGSFPENLRHLKNTMETIDWKVFESWMHHWLLFEMSRHSLEQ KPTDAPPKATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYL CKCQPGFTGARCTENVPMKVQNQEKHSGEPFPEAEELYQKRVLTITGICI ALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNP PPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTP SHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPREC NSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSE MSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSF HHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTK PNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNPLAASLEA | 689 |
| 3D view using mol* of 563 (AA BP:208) | ||||||||||
 | ||||||||||
| PDB file (568) >>>568.pdbFusion protein BP residue: 229 CIF file (568) >>>568.cif | CD74 | chr5 | 149782125 | - | NRG1 | chr8 | 32585466 | + | MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALY TGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQLENLRMKLPKP PKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNAD PLKVYPPLKGSFPENLRHLKNTMETIDWKVFESWMHHWLLFEMSRHSLEQ KPTDAPPKESLELEDPSSGLGVTKQDLGPATSTSTTGTSHLVKCAEKEKT FCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKAEELYQK RVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMN IANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTA HHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGR LNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHT PSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKF DHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKL ANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGI | 699 |
| 3D view using mol* of 568 (AA BP:229) | ||||||||||
 | ||||||||||
| PDB file (575) >>>575.pdbFusion protein BP residue: 229 CIF file (575) >>>575.cif | CD74 | chr5 | 149782125 | - | NRG1 | chr8 | 32585466 | + | MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALY TGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQLENLRMKLPKP PKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNAD PLKVYPPLKGSFPENLRHLKNTMETIDWKVFESWMHHWLLFEMSRHSLEQ KPTDAPPKESLELEDPSSGLGVTKQDLGPATSTSTTGTSHLVKCAEKEKT FCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMKVQNQEKAEEL YQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNN MMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYT STAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGP RGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVD FHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLRE KKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPV KKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPF LGIQNPLAASLEATPAFRLADSRTNPAGRFSTQEEIQARLSSVIANQDPI | 702 |
| 3D view using mol* of 575 (AA BP:229) | ||||||||||
 | ||||||||||
| PDB file (580) >>>580.pdbFusion protein BP residue: 229 CIF file (580) >>>580.cif | CD74 | chr5 | 149782125 | - | NRG1 | chr8 | 32585466 | + | MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALY TGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQLENLRMKLPKP PKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNAD PLKVYPPLKGSFPENLRHLKNTMETIDWKVFESWMHHWLLFEMSRHSLEQ KPTDAPPKESLELEDPSSGLGVTKQDLGPATSTSTTGTSHLVKCAEKEKT FCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKHLGIEFM EAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLR SERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFS TSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSS PTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPAR MSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTP PRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEP AQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVG EDTPFLGIQNPLAASLEATPAFRLADSRTNPAGRFSTQEEIQARLSSVIA | 707 |
| 3D view using mol* of 580 (AA BP:229) | ||||||||||
 | ||||||||||
| PDB file (584) >>>584.pdbFusion protein BP residue: 229 CIF file (584) >>>584.cif | CD74 | chr5 | 149782125 | - | NRG1 | chr8 | 32585466 | + | MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALY TGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQLENLRMKLPKP PKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNAD PLKVYPPLKGSFPENLRHLKNTMETIDWKVFESWMHHWLLFEMSRHSLEQ KPTDAPPKESLELEDPSSGLGVTKQDLGPATSTSTTGTSHLVKCAEKEKT FCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMKVQNQEKHSGE PFPEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQ SLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAET SFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSR HSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTT PARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLL VTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQE YEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDE RVGEDTPFLGIQNPLAASLEATPAFRLADSRTNPAGRFSTQEEIQARLSS | 710 |
| 3D view using mol* of 584 (AA BP:229) | ||||||||||
 | ||||||||||
| PDB file (624) >>>624.pdbFusion protein BP residue: 293 CIF file (624) >>>624.cif | CD74 | chr5 | 149782125 | - | NRG1 | chr8 | 32585466 | + | MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALY TGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQLENLRMKLPKP PKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNAD PLKVYPPLKGSFPENLRHLKNTMETIDWKVFESWMHHWLLFEMSRHSLEQ KPTDAPPKVLTKCQEEVSHIPAVHPGSFRPKCDENGNYLPLQCYGSIGYC WCVFPNGTEVPNTRSRGHHNCSESLELEDPSSGLGVTKQDLGPATSTSTT GTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNY VMASFYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDR LRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVERE AETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVE NSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSA MTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERP LLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYET TQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESET EDERVGEDTPFLGIQNPLAASLEATPAFRLADSRTNPAGRFSTQEEIQAR | 763 |
| 3D view using mol* of 624 (AA BP:293) | ||||||||||
 | ||||||||||
| PDB file (626) >>>626.pdbFusion protein BP residue: 293 CIF file (626) >>>626.cif | CD74 | chr5 | 149782125 | - | NRG1 | chr8 | 32585466 | + | MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALY TGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQLENLRMKLPKP PKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNAD PLKVYPPLKGSFPENLRHLKNTMETIDWKVFESWMHHWLLFEMSRHSLEQ KPTDAPPKVLTKCQEEVSHIPAVHPGSFRPKCDENGNYLPLQCYGSIGYC WCVFPNGTEVPNTRSRGHHNCSESLELEDPSSGLGVTKQDLGPATSTSTT GTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTEN VPMKVQNQEKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKL HDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIV EREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMS SVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERY VSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEE ERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEE YETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSE SETEDERVGEDTPFLGIQNPLAASLEATPAFRLADSRTNPAGRFSTQEEI | 766 |
| 3D view using mol* of 626 (AA BP:293) | ||||||||||
 | ||||||||||
| PDB file (629) >>>629.pdbFusion protein BP residue: 293 CIF file (629) >>>629.cif | CD74 | chr5 | 149782125 | - | NRG1 | chr8 | 32585466 | + | MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALY TGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQLENLRMKLPKP PKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNAD PLKVYPPLKGSFPENLRHLKNTMETIDWKVFESWMHHWLLFEMSRHSLEQ KPTDAPPKVLTKCQEEVSHIPAVHPGSFRPKCDENGNYLPLQCYGSIGYC WCVFPNGTEVPNTRSRGHHNCSESLELEDPSSGLGVTKQDLGPATSTSTT GTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNY VMASFYKHLGIEFMEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKK QRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVIS SEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHS VIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSP HSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVS PFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRI VEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQ SSNSESETEDERVGEDTPFLGIQNPLAASLEATPAFRLADSRTNPAGRFS | 771 |
| 3D view using mol* of 629 (AA BP:293) | ||||||||||
 | ||||||||||
| PDB file (631) >>>631.pdbFusion protein BP residue: 293 CIF file (631) >>>631.cif | CD74 | chr5 | 149782125 | - | NRG1 | chr8 | 32585466 | + | MHRRRSRSCREDQKPVMDDQRDLISNNEQLPMLGRRPGAPESKCSRGALY TGFSILVTLLLAGQATTAYFLYQQQGRLDKLTVTSQNLQLENLRMKLPKP PKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTEDHVMHLLQNAD PLKVYPPLKGSFPENLRHLKNTMETIDWKVFESWMHHWLLFEMSRHSLEQ KPTDAPPKVLTKCQEEVSHIPAVHPGSFRPKCDENGNYLPLQCYGSIGYC WCVFPNGTEVPNTRSRGHHNCSESLELEDPSSGLGVTKQDLGPATSTSTT GTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTEN VPMKVQNQEKHSGEPFPEAEELYQKRVLTITGICIALLVVGIMCVVAYCK TKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKN VISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSE SHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYR DSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSM AVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASP LRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNT SSQSSNSESETEDERVGEDTPFLGIQNPLAASLEATPAFRLADSRTNPAG | 774 |
| 3D view using mol* of 631 (AA BP:293) | ||||||||||
 | ||||||||||
Top |
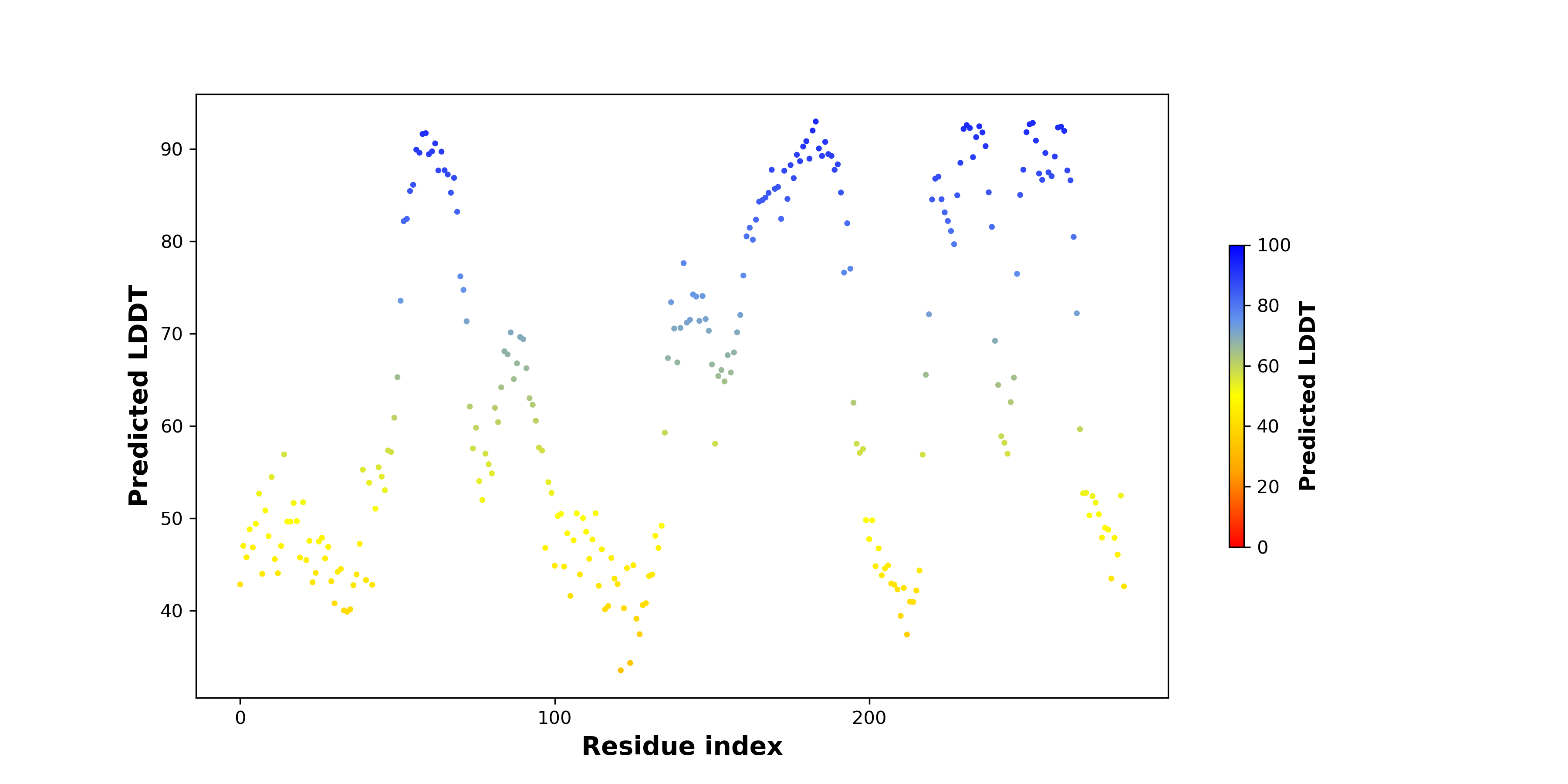
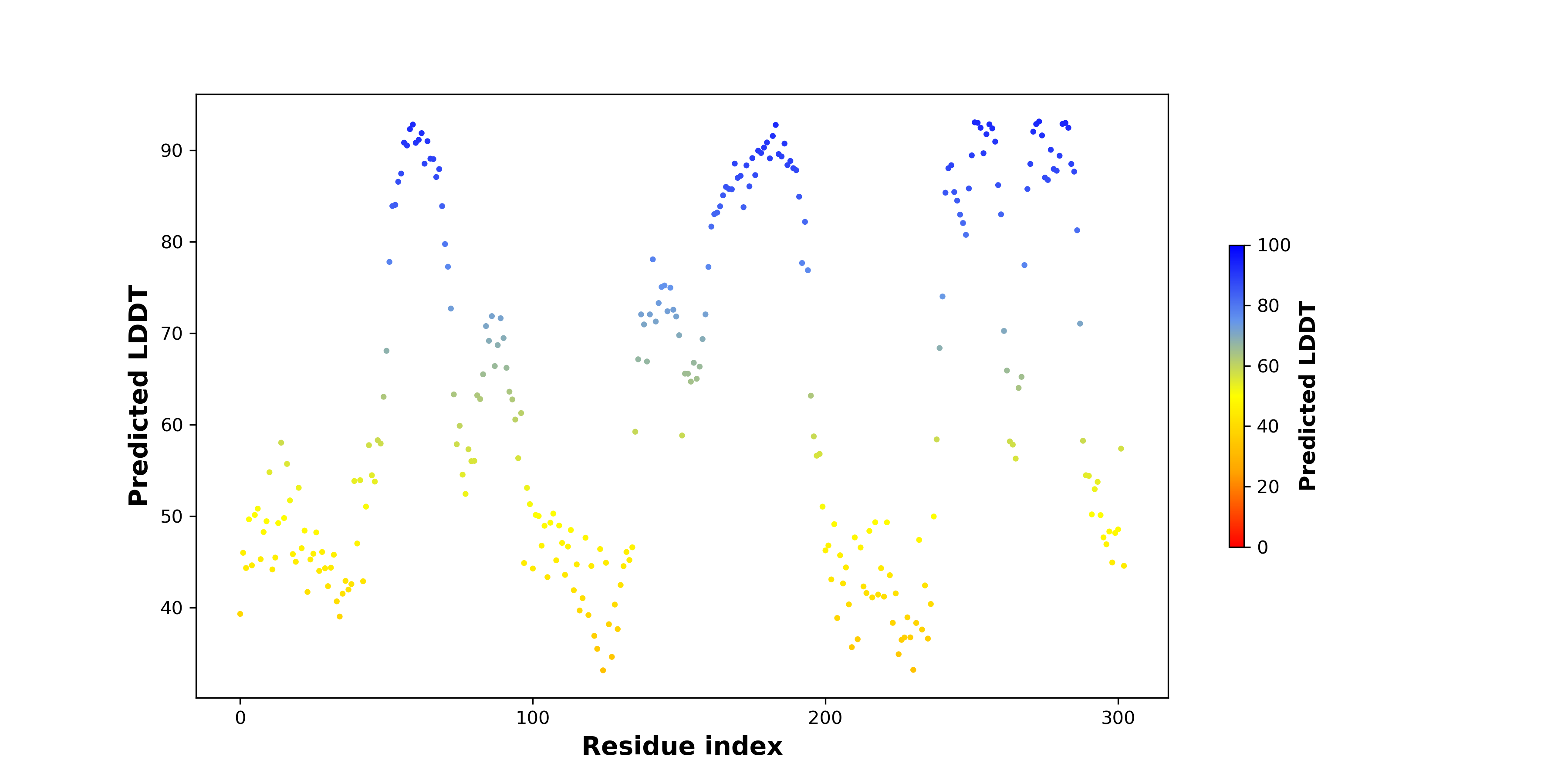
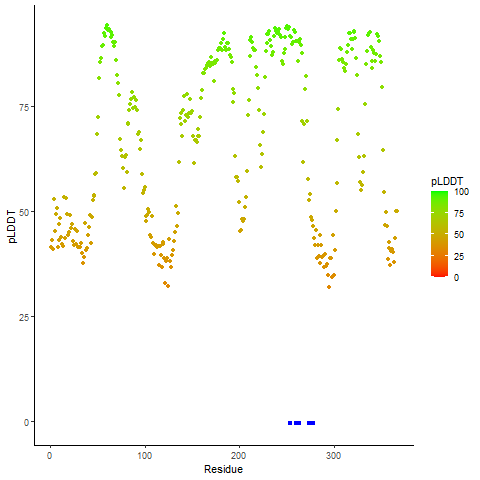
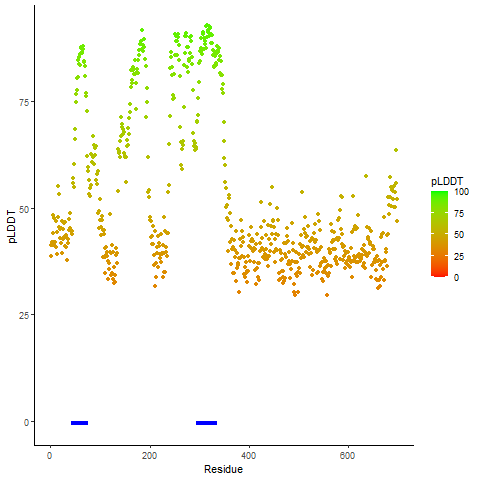
pLDDT score distribution |
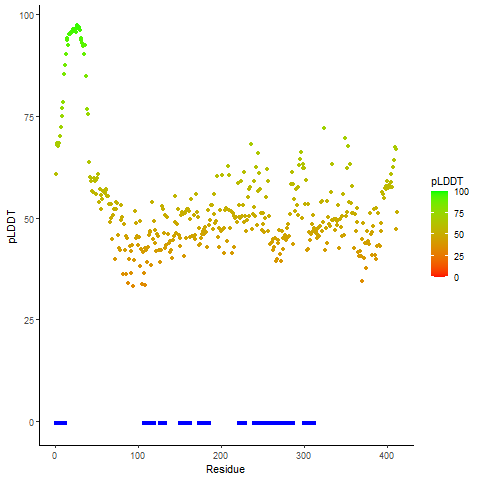
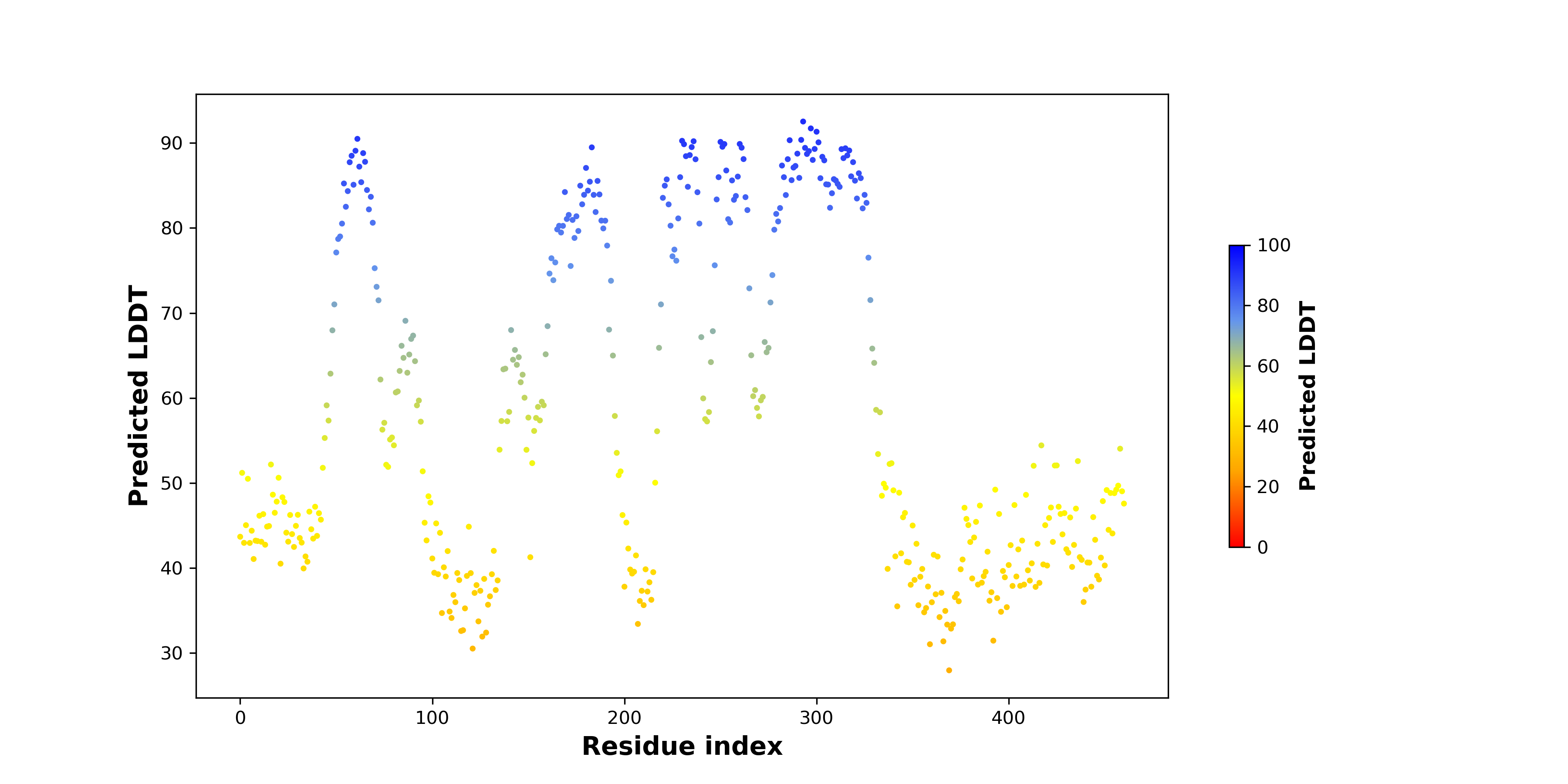
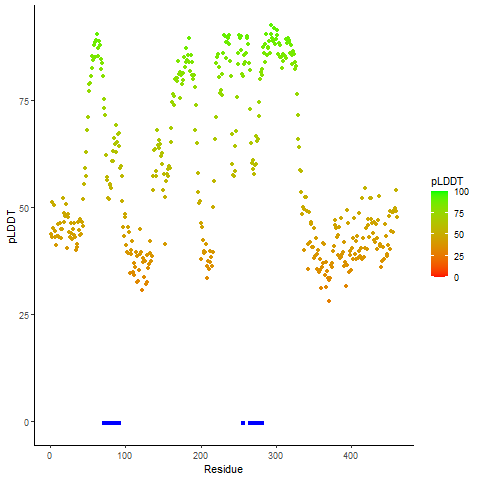
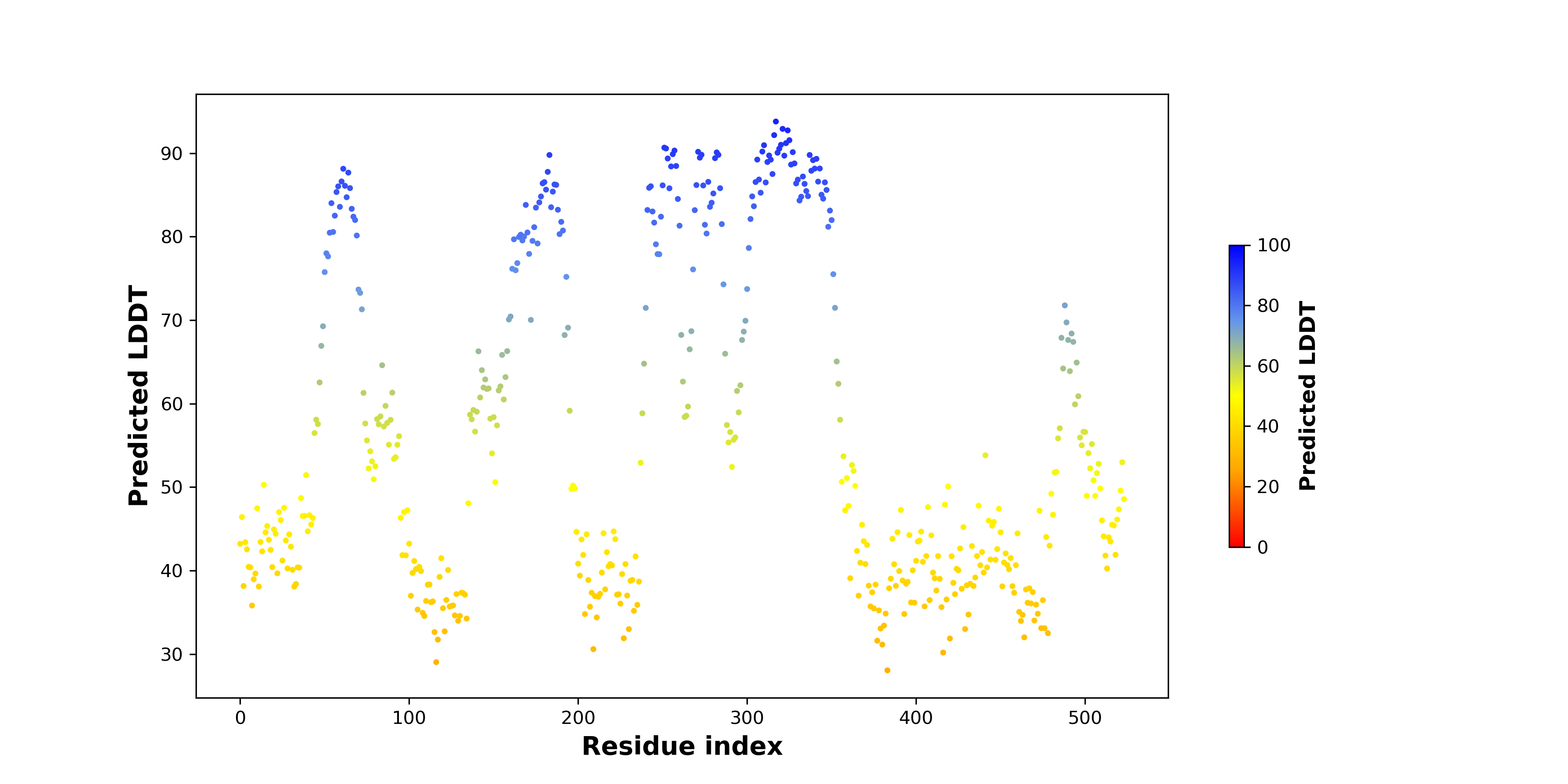
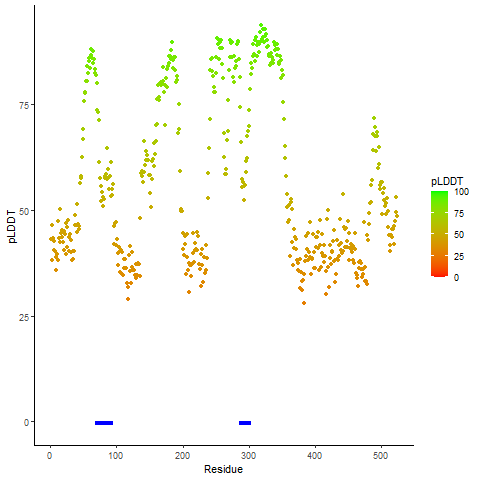
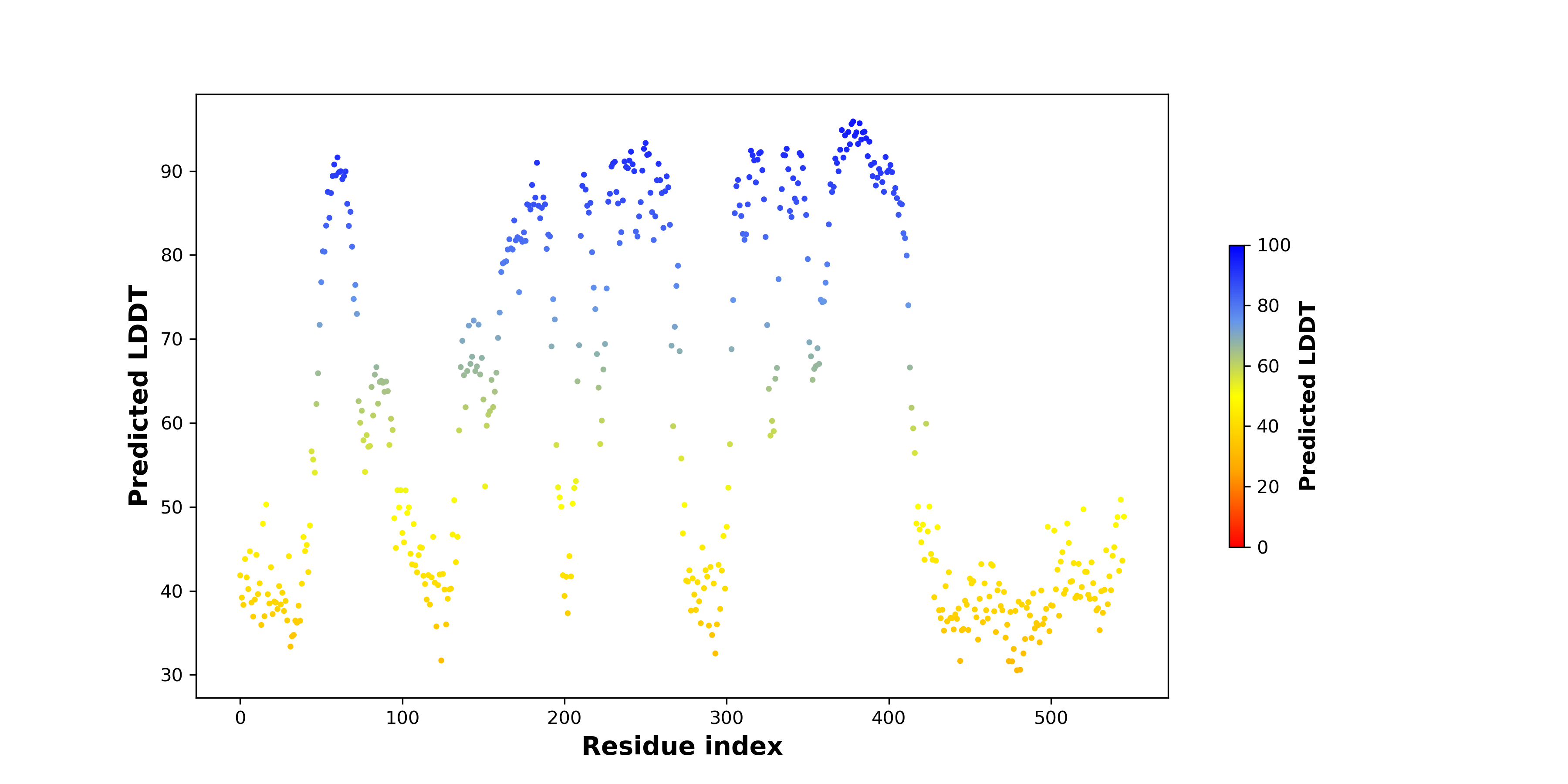
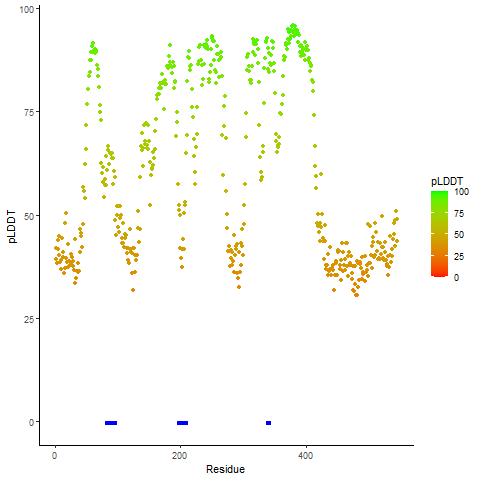
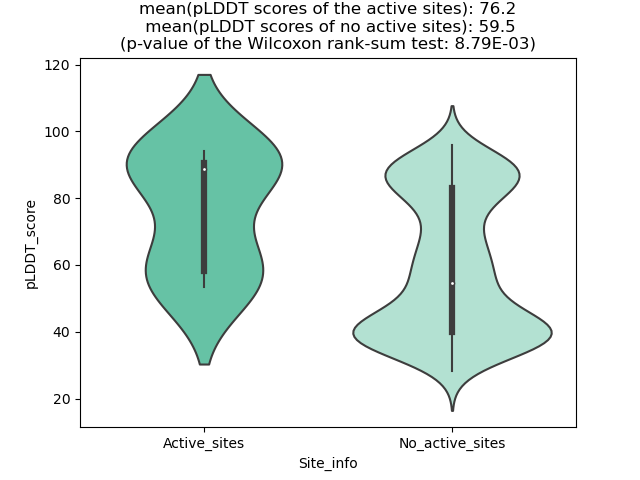
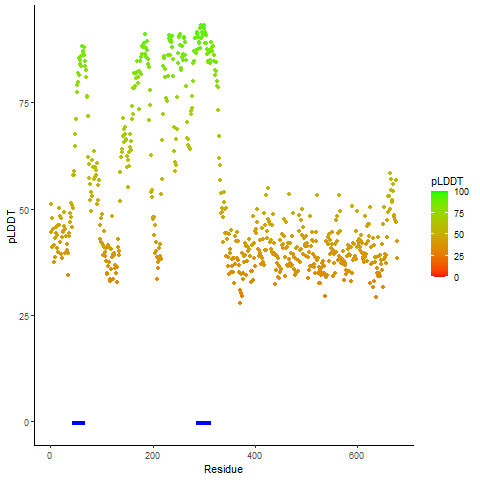
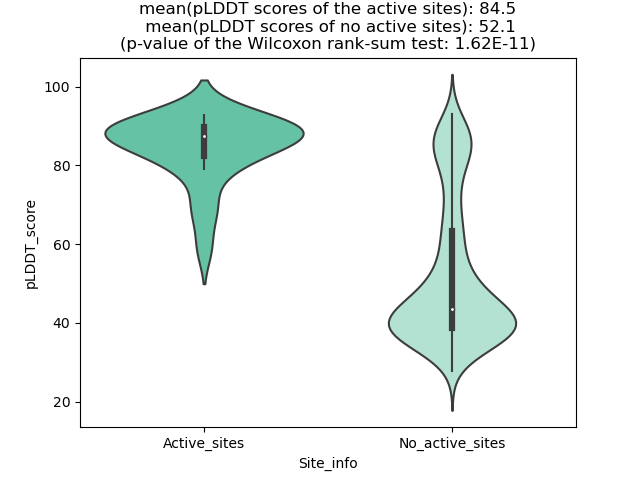
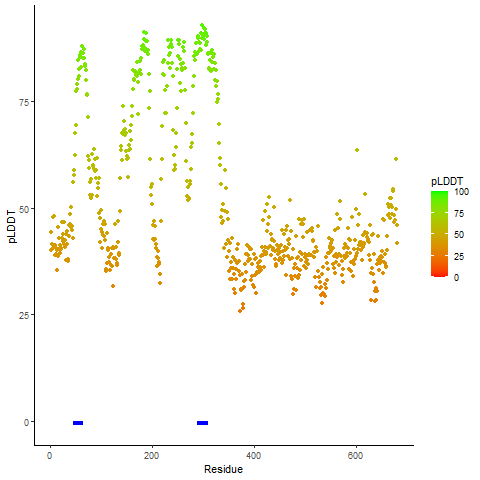
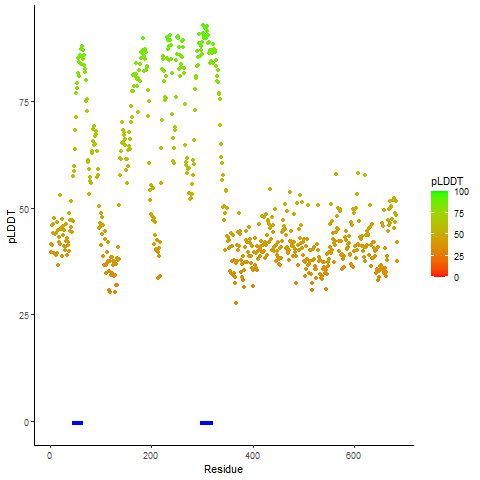
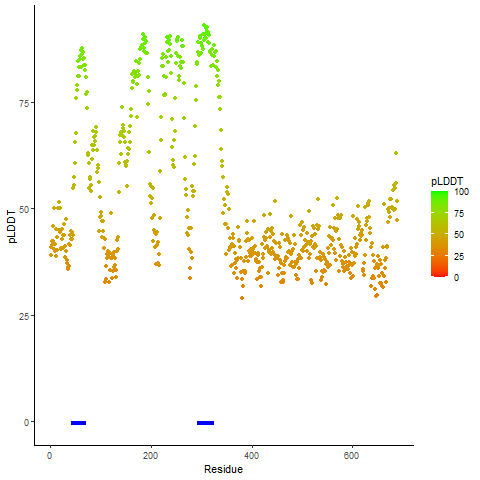
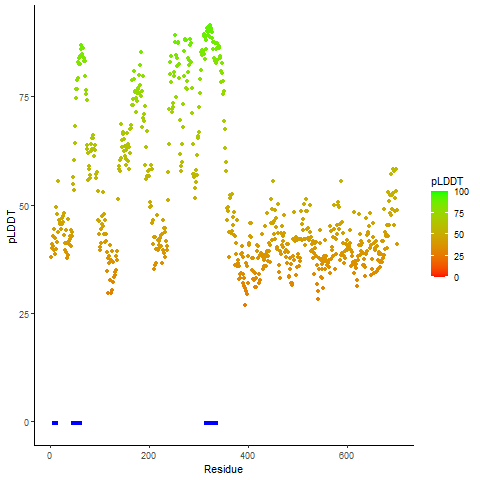
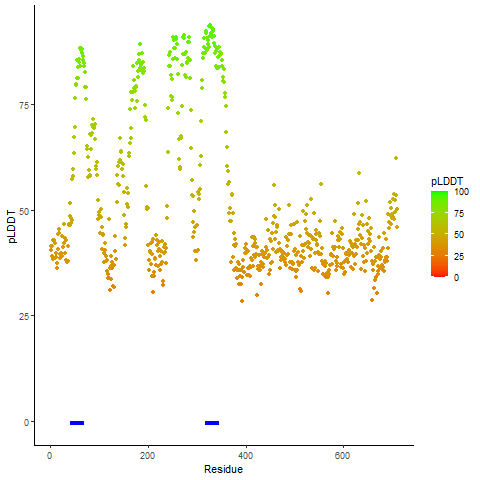
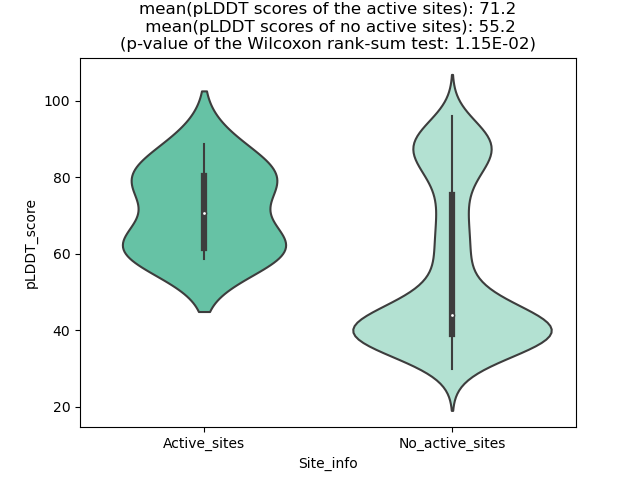
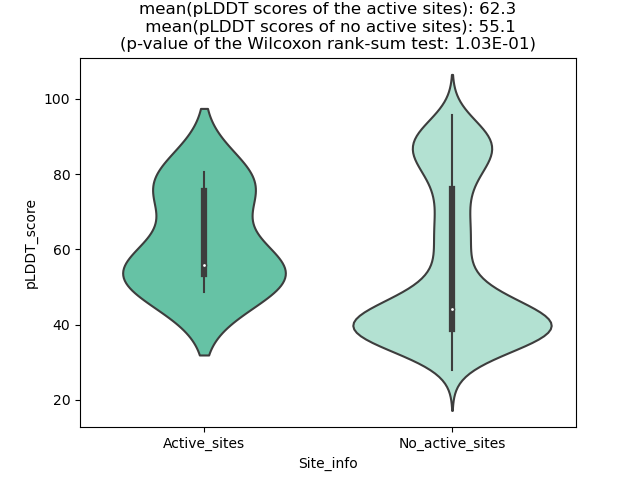
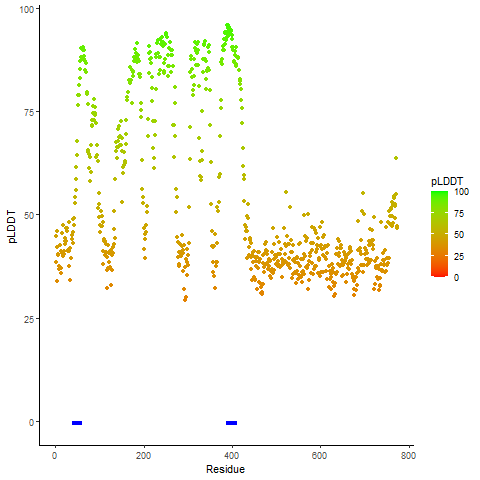
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
CD74_pLDDT.png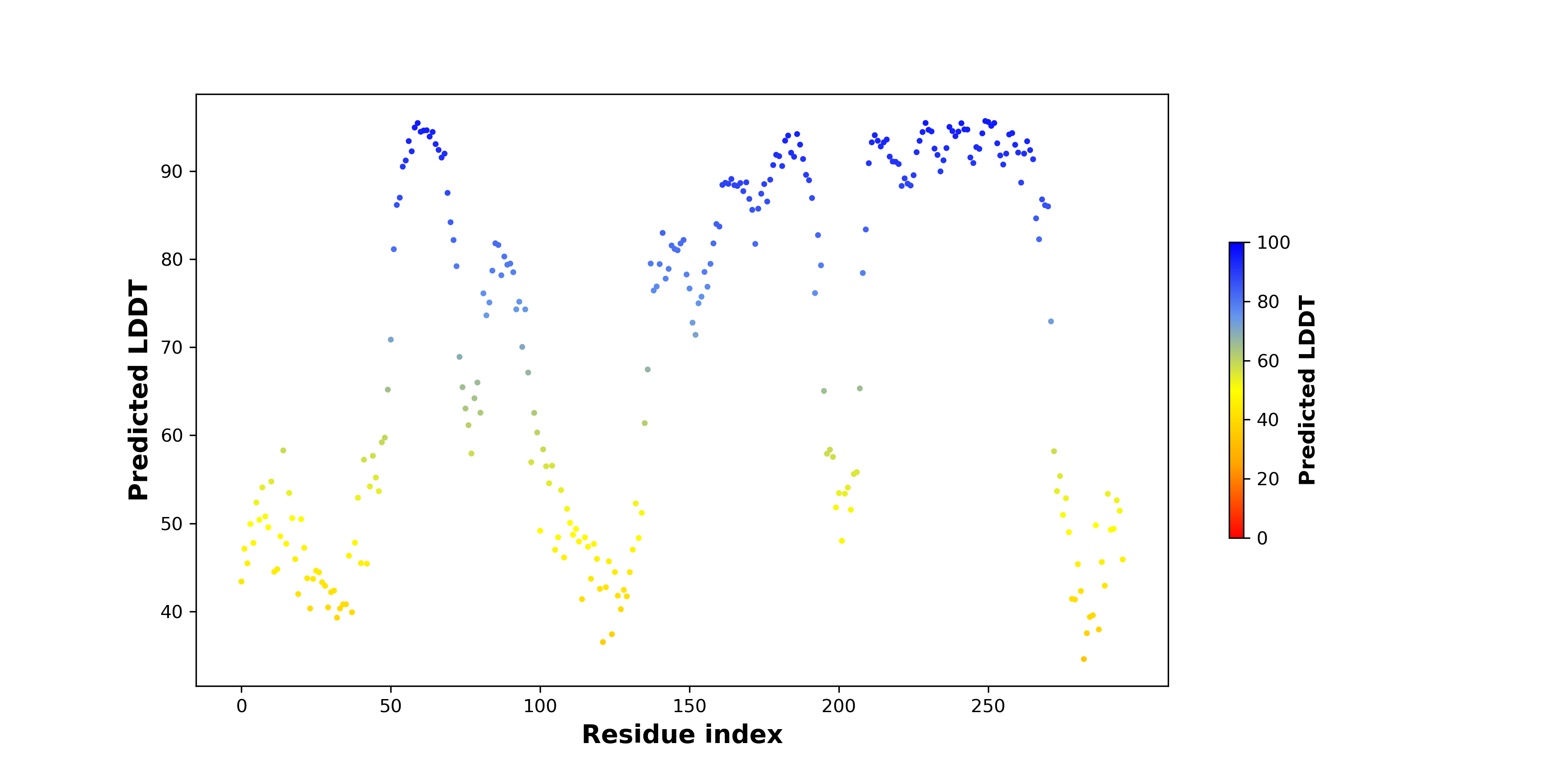 |
NRG1_pLDDT.png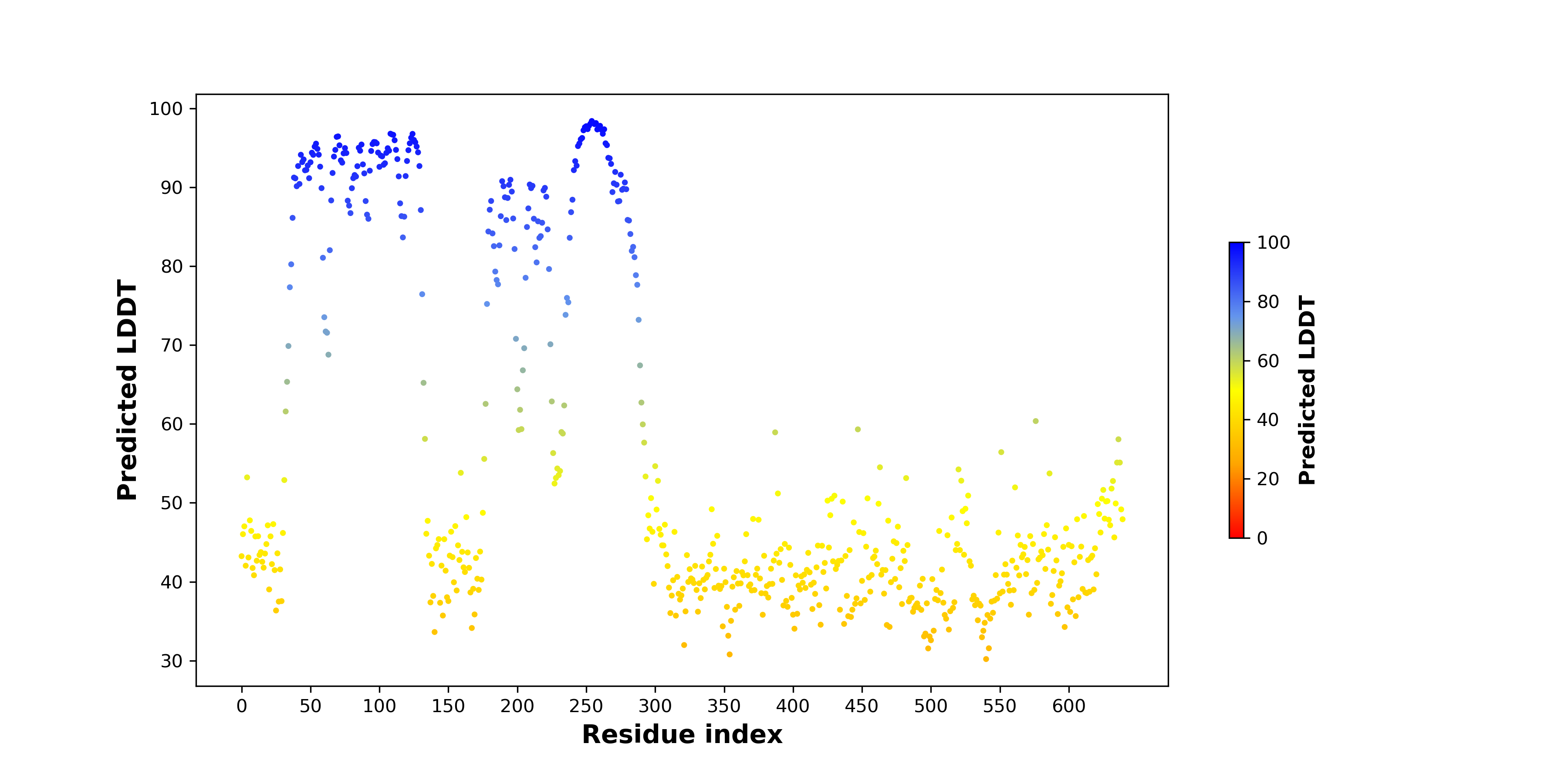 |
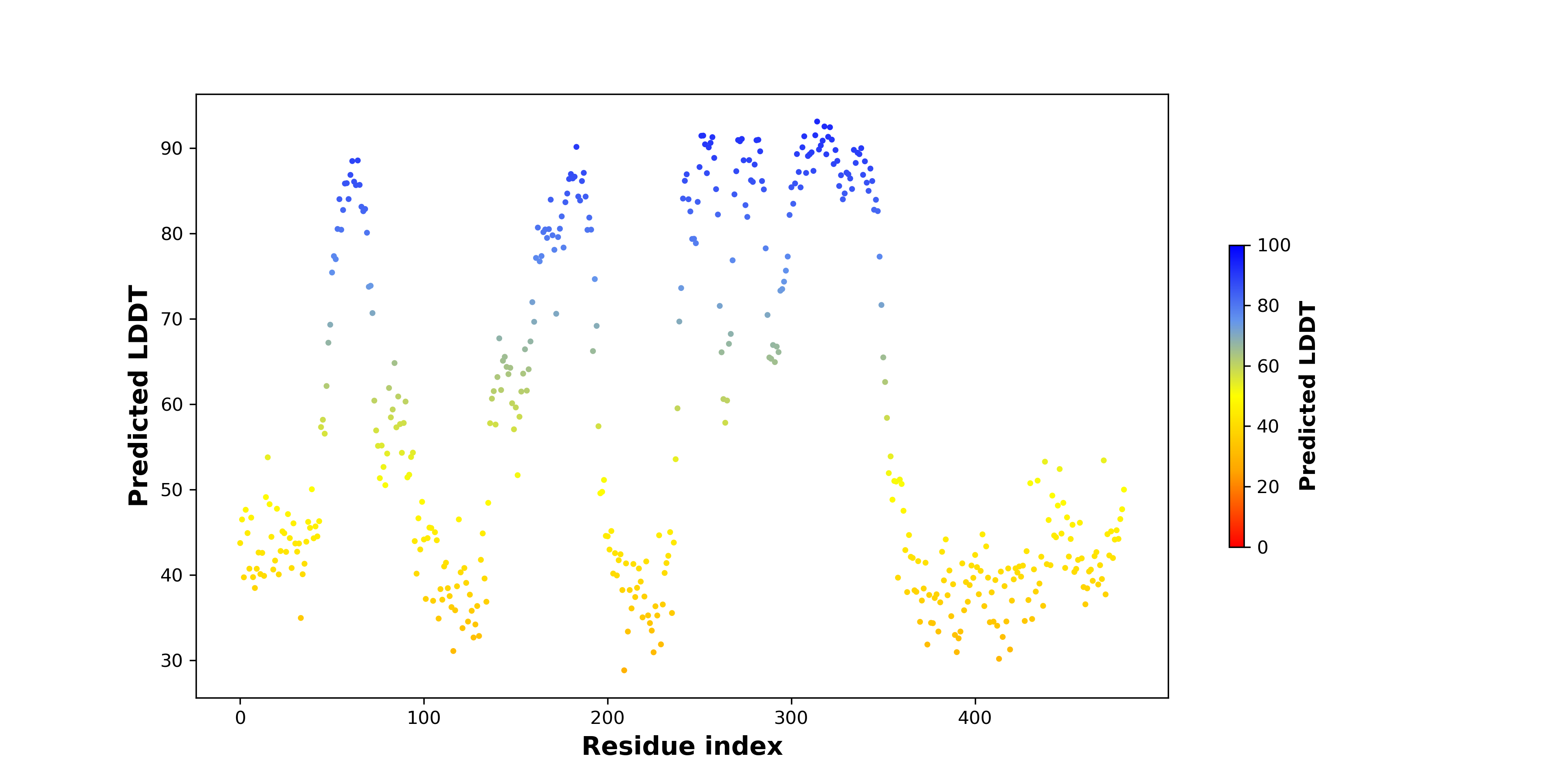
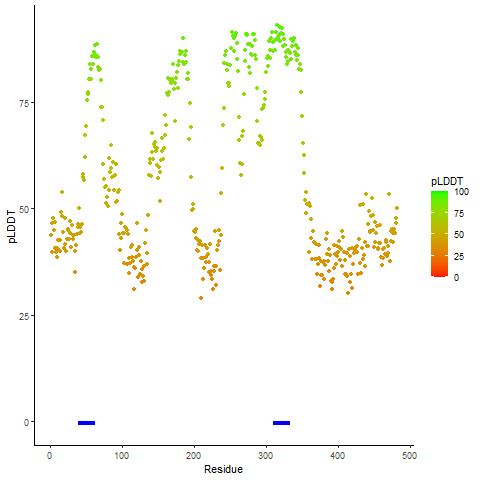
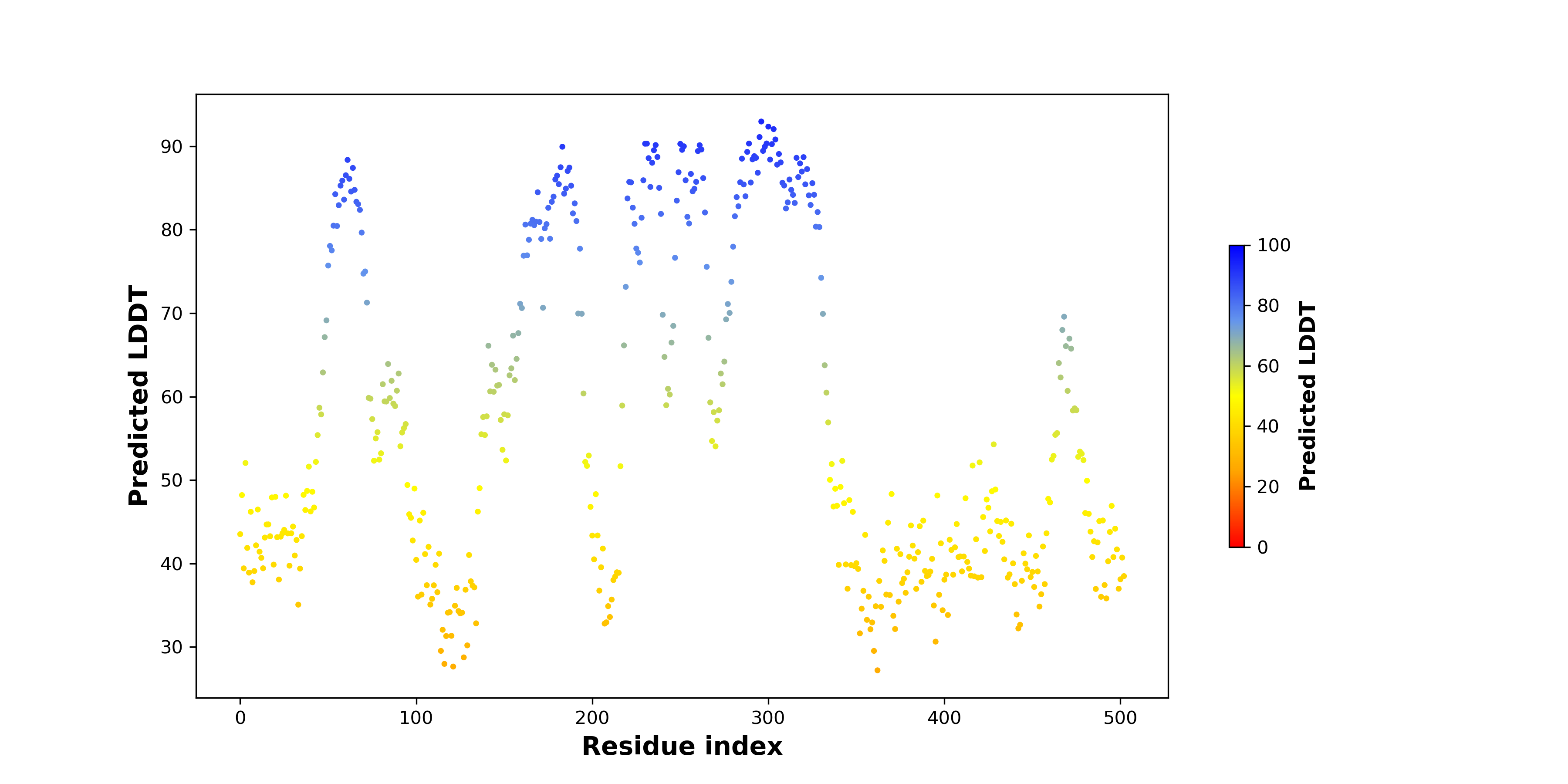
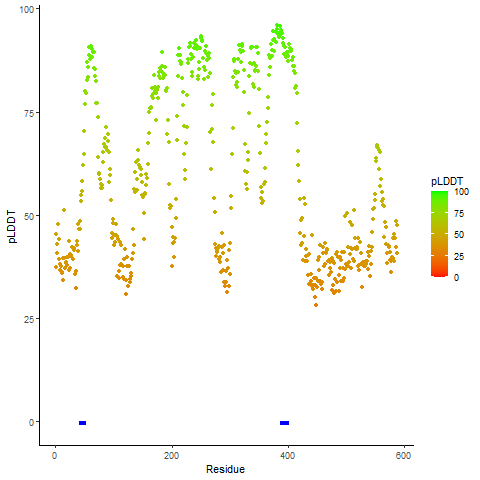
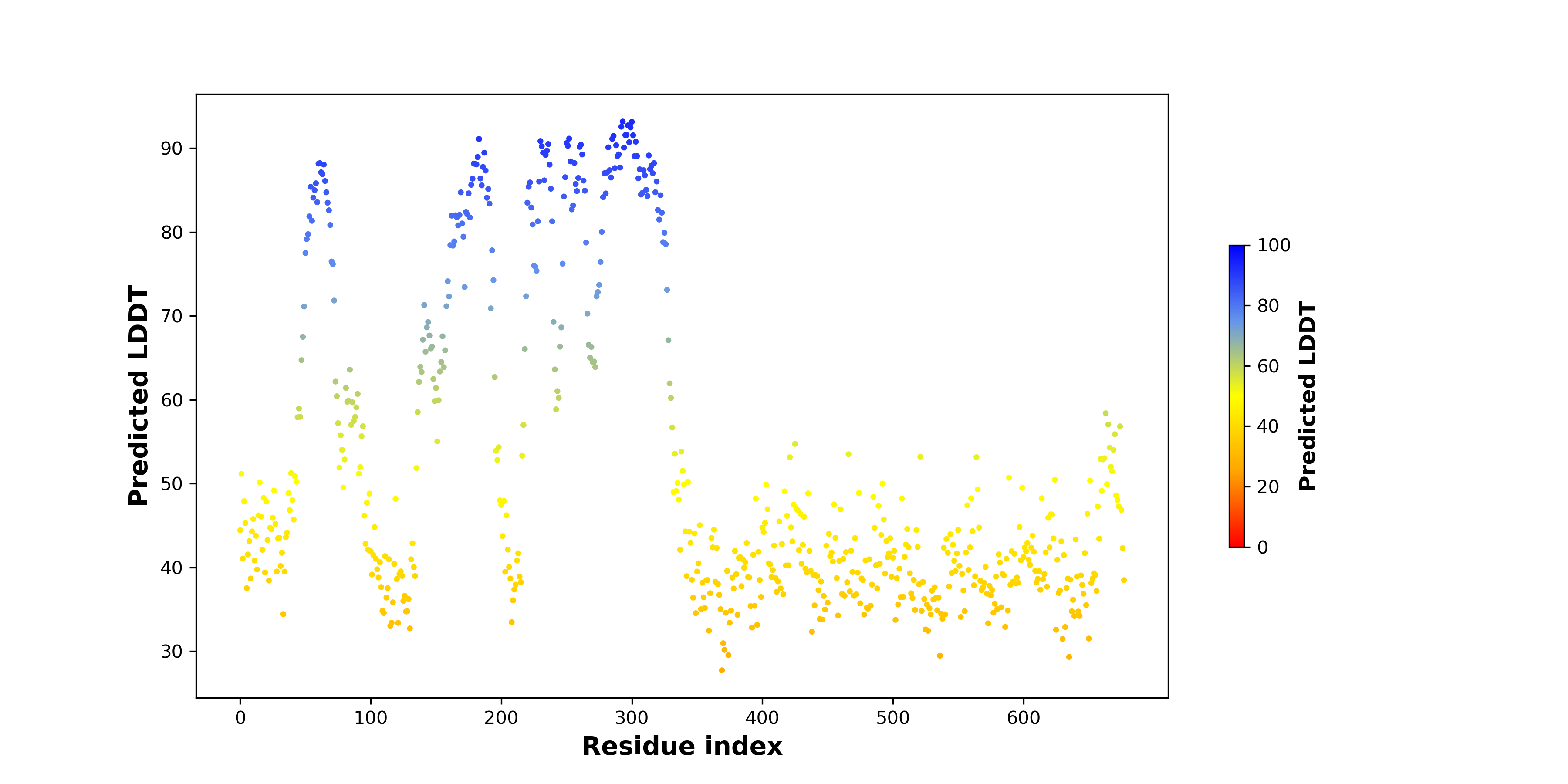
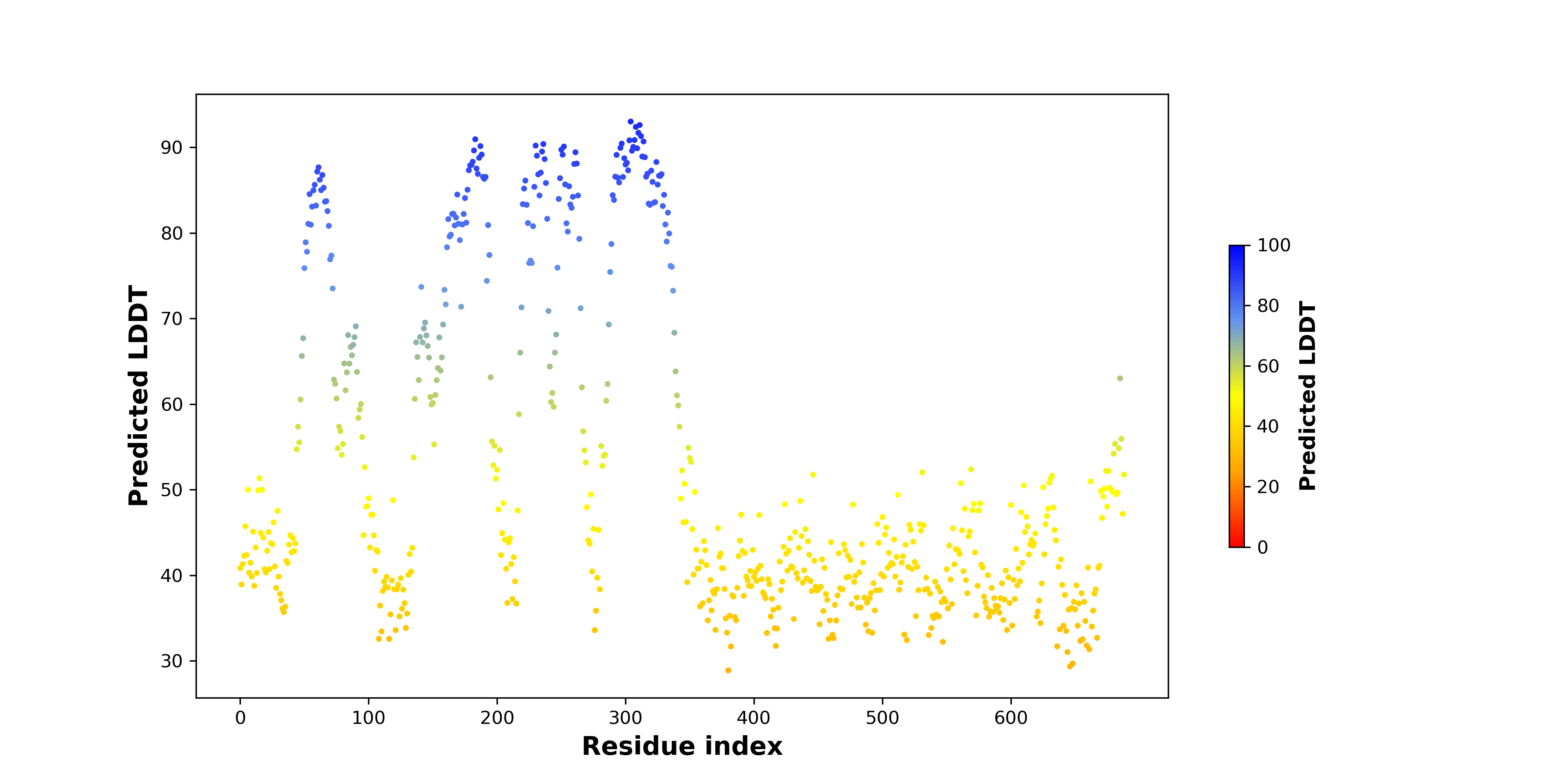
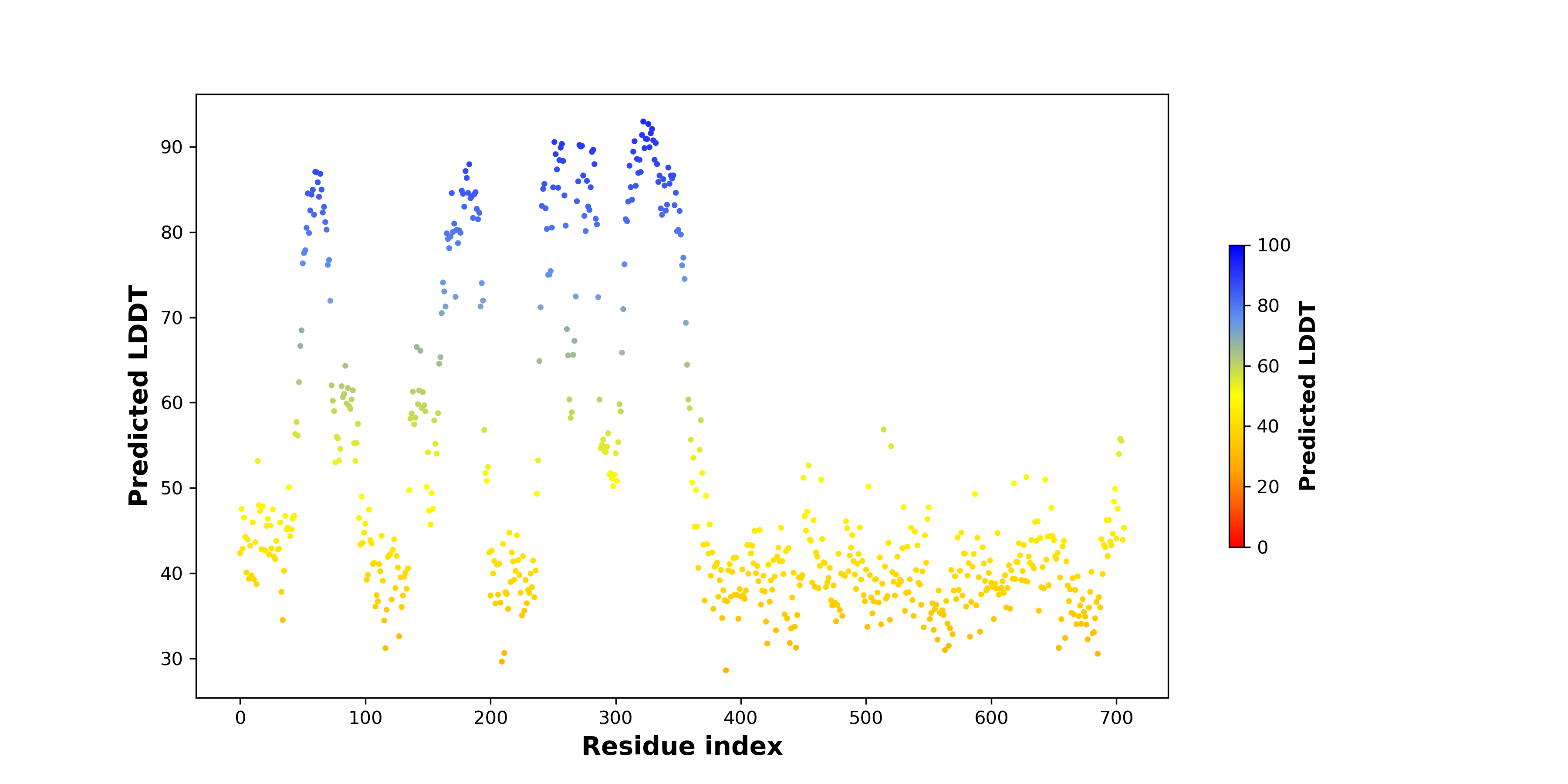
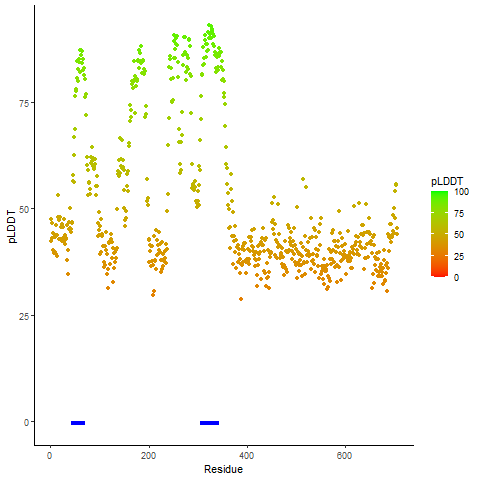
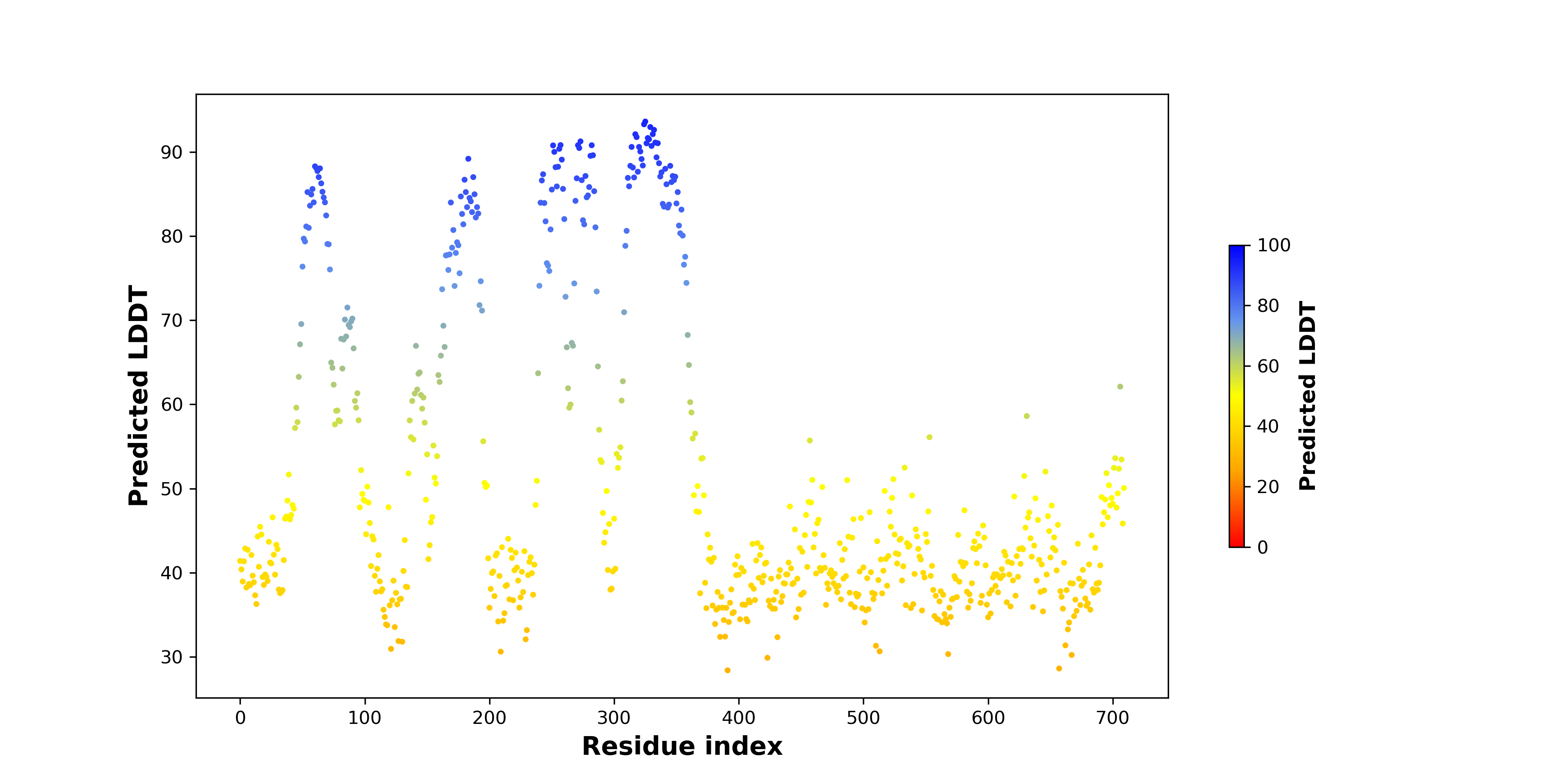
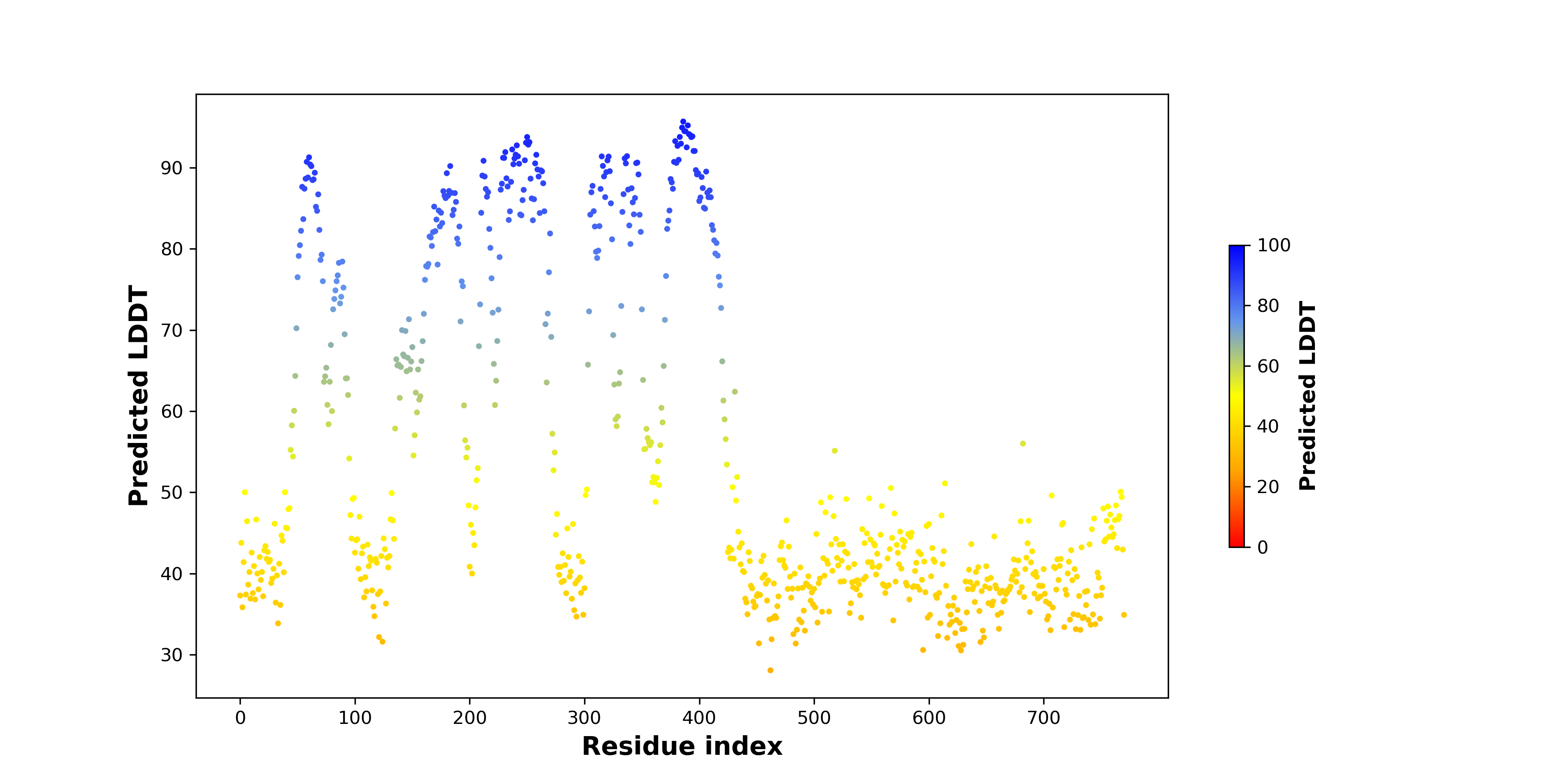
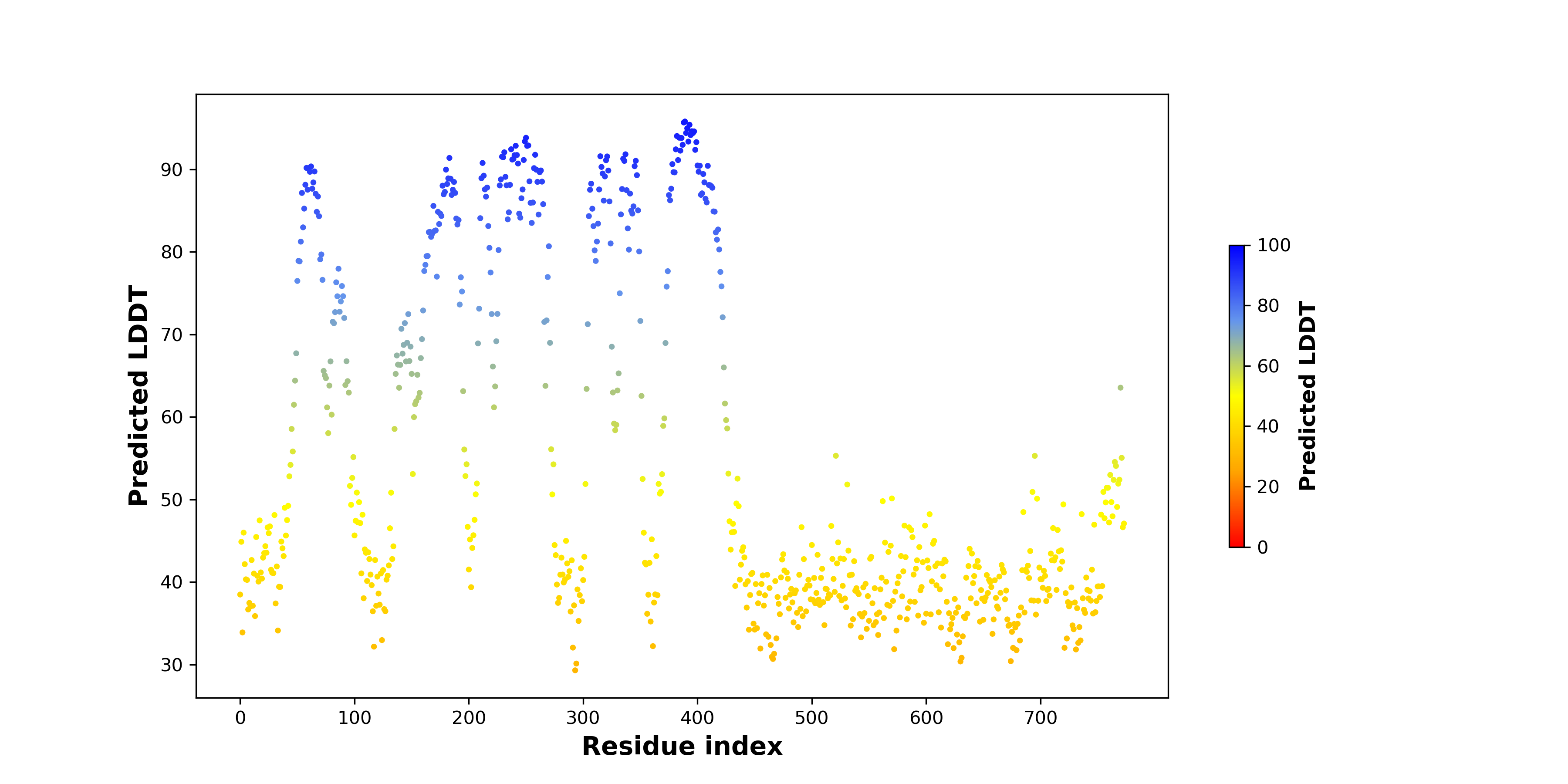
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
Top |
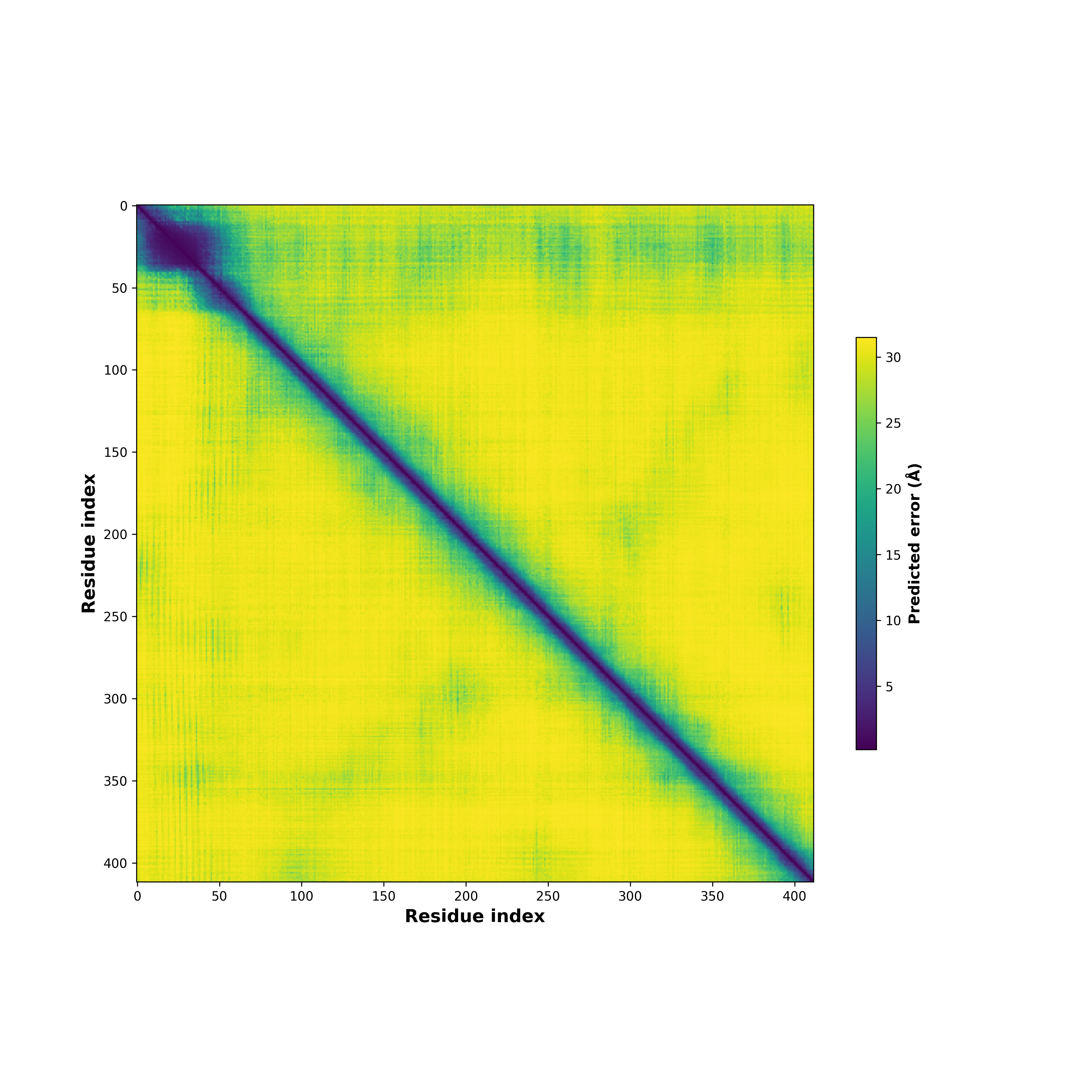
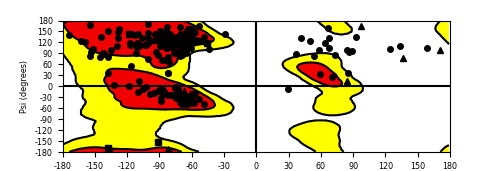
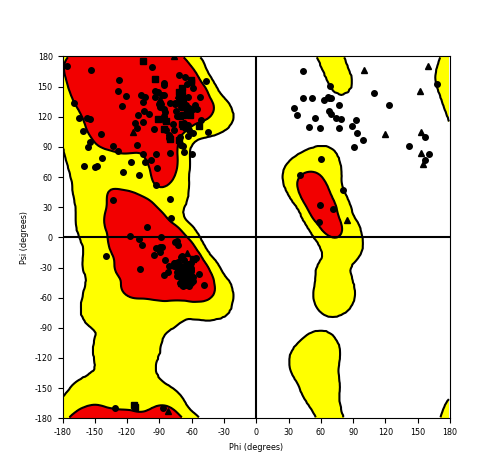
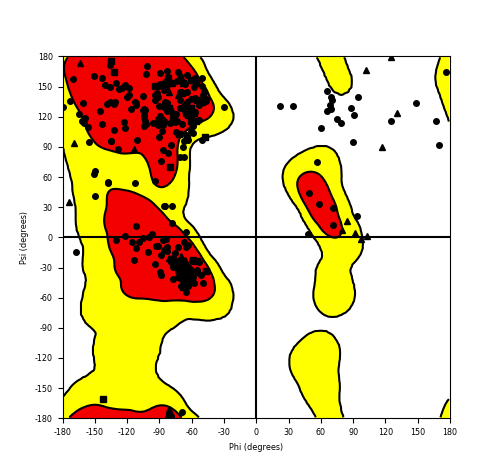
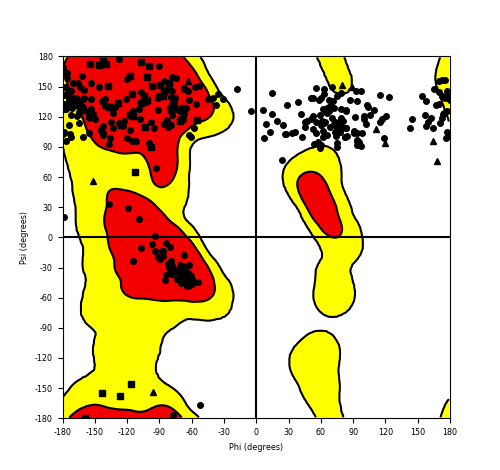
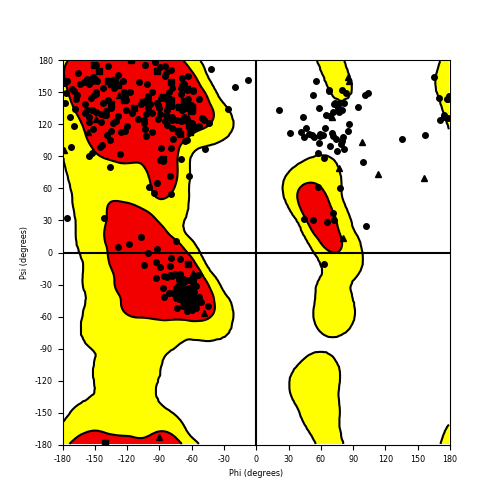
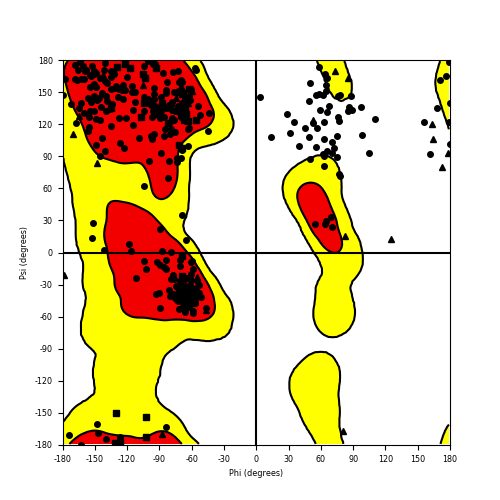
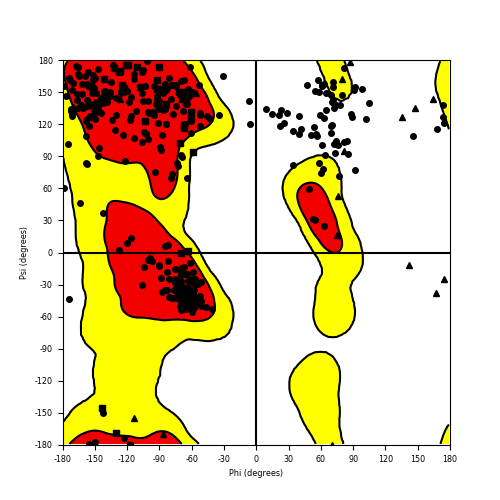
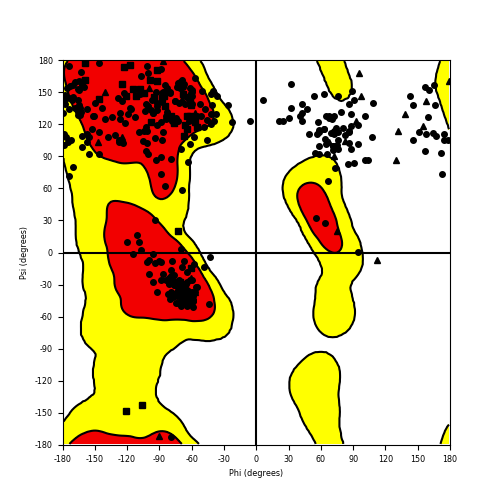
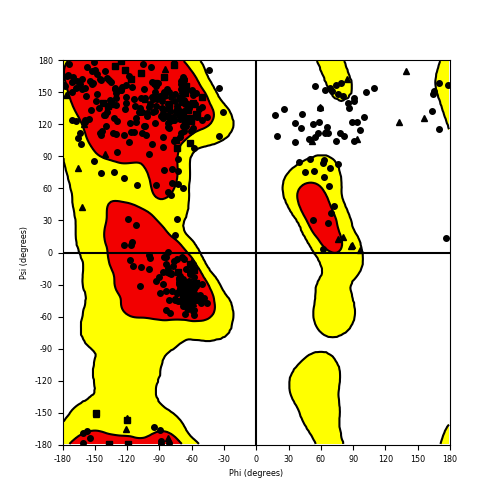
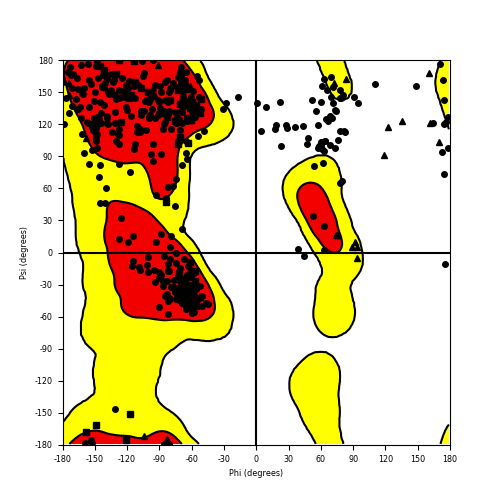
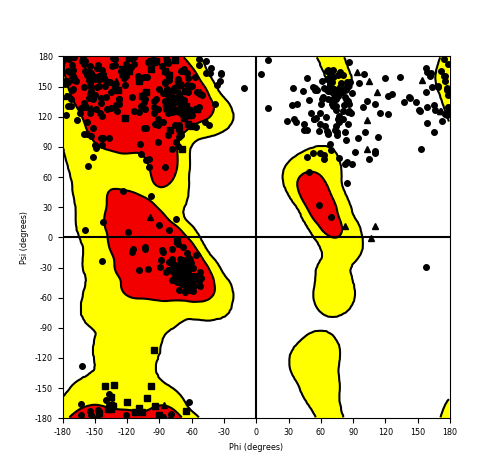
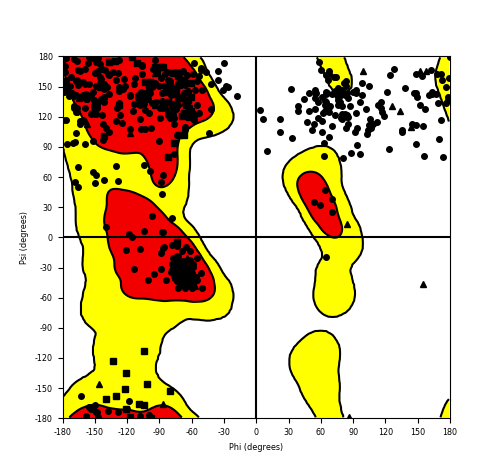
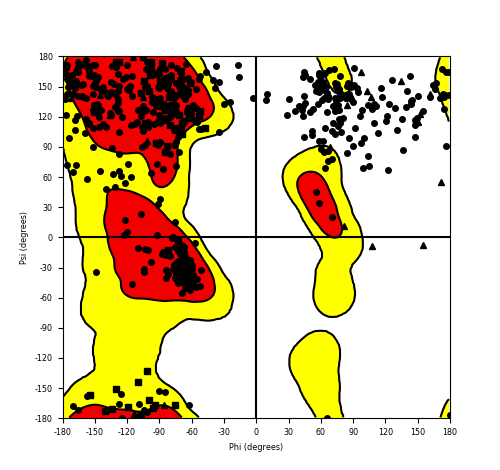
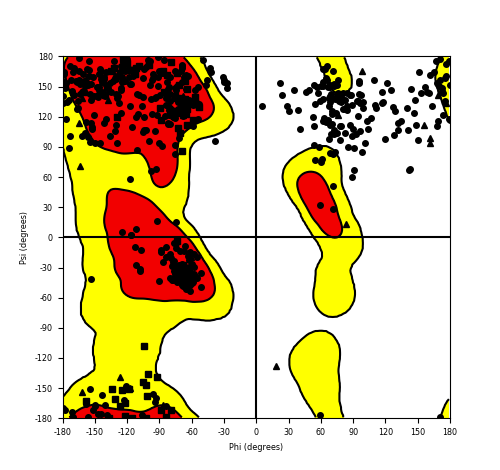
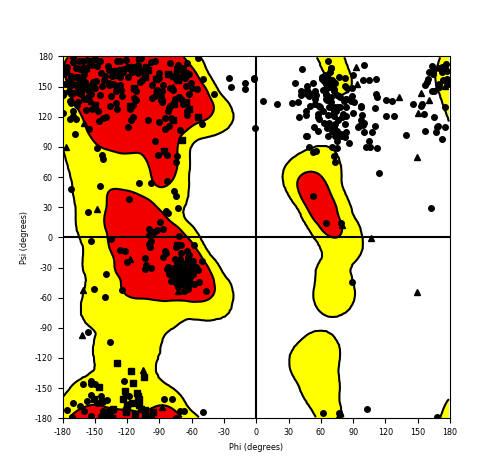
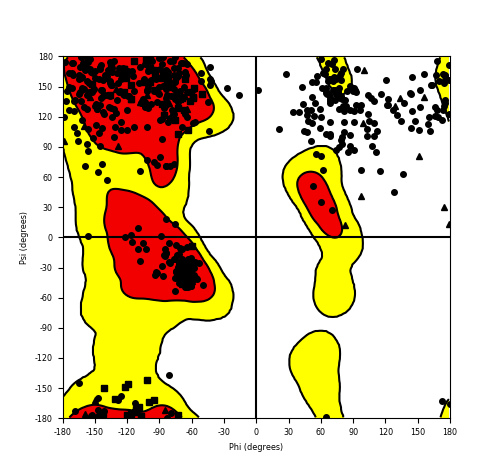
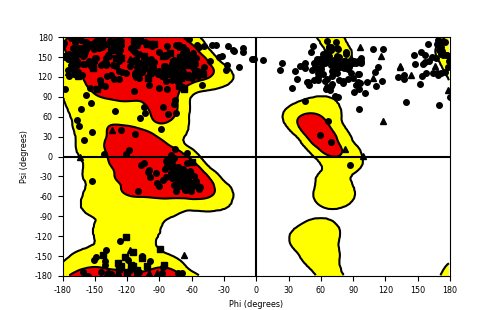
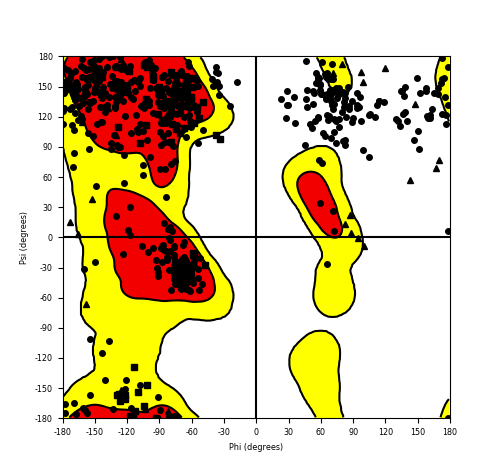
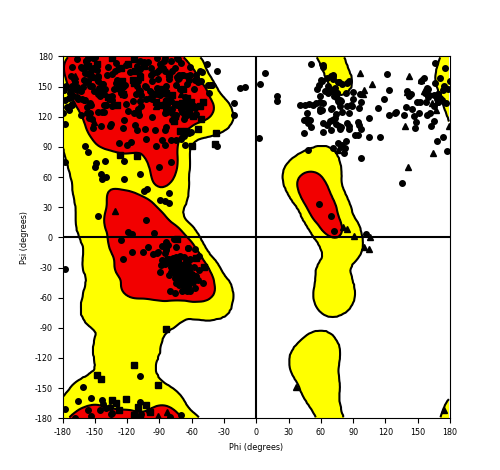
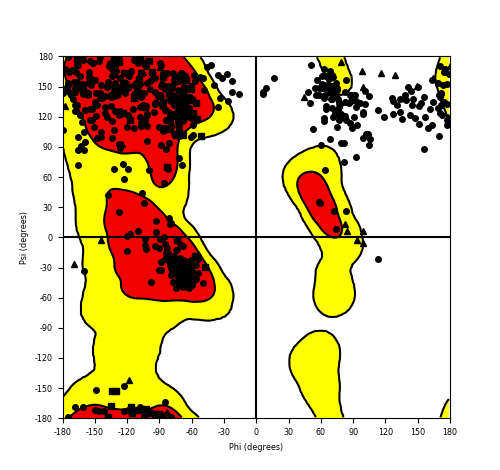
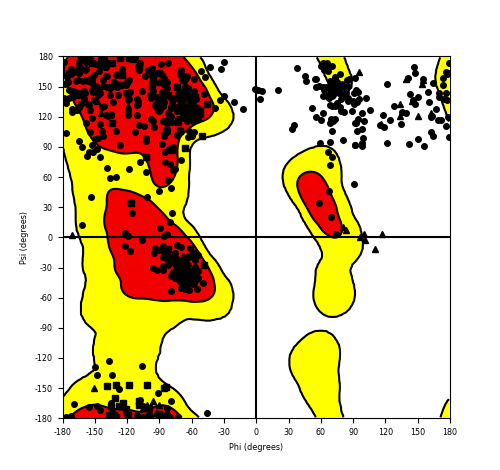
Ramachandran Plot of Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
Top |
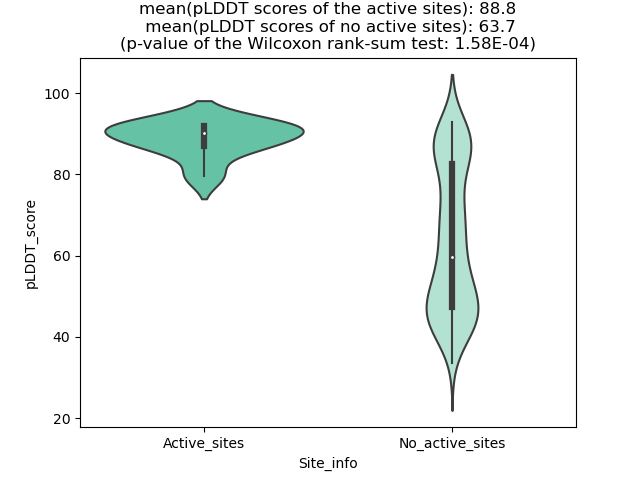
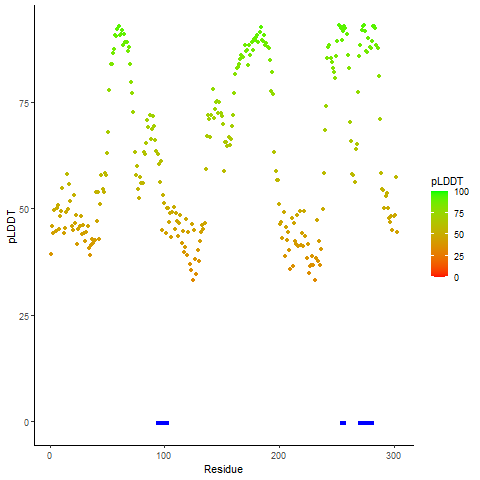
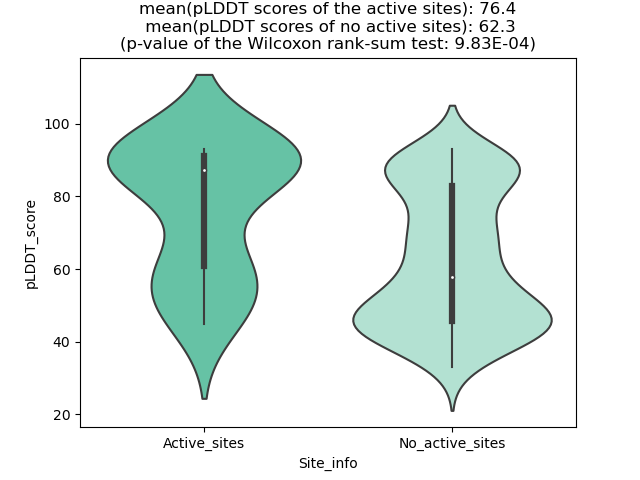
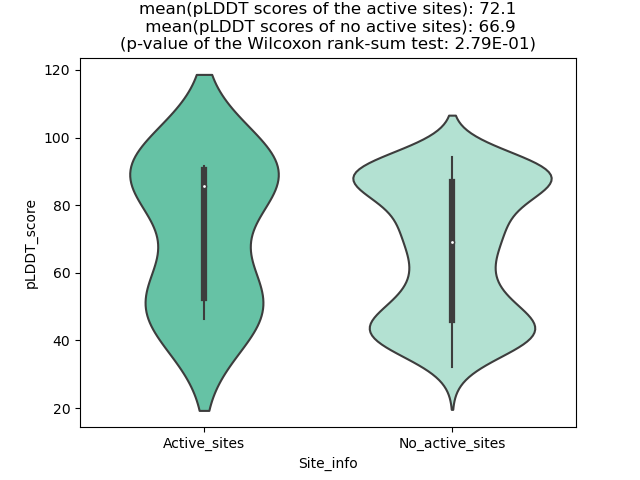
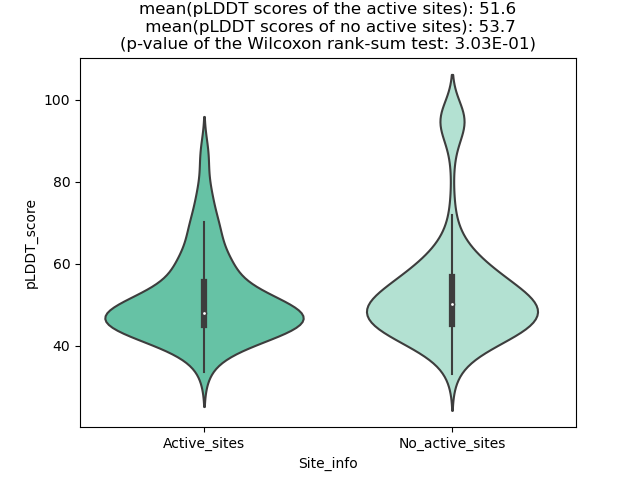
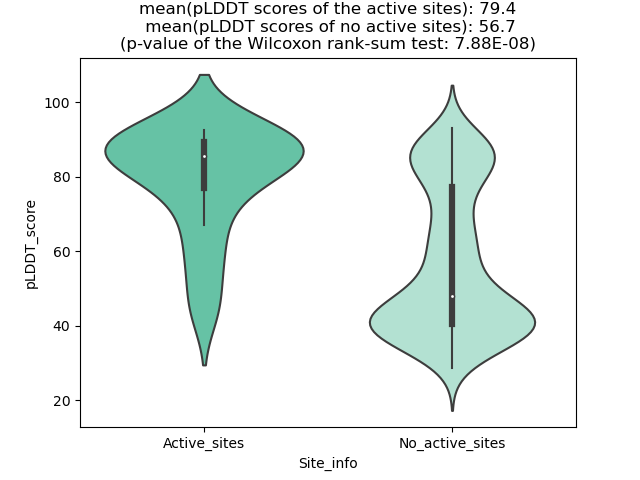
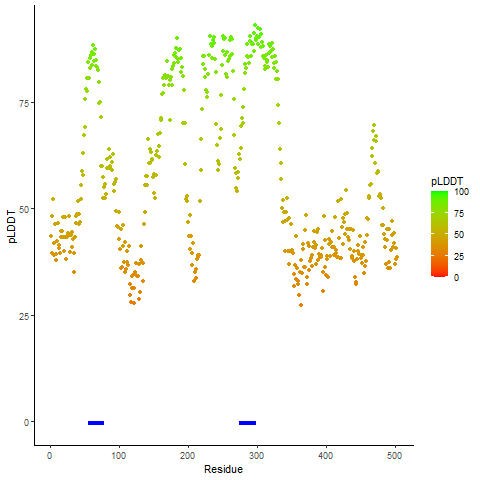
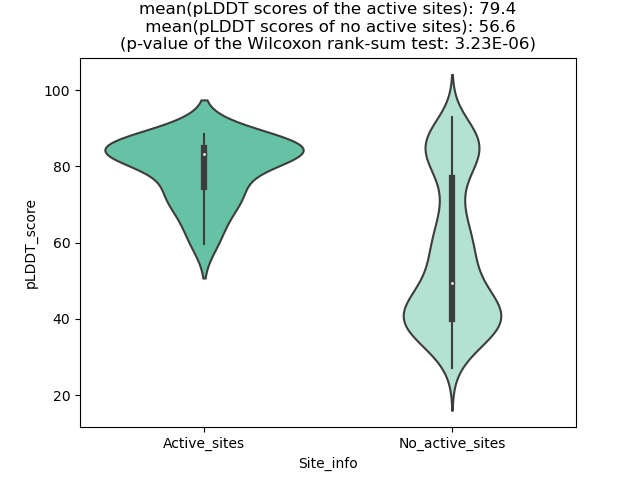
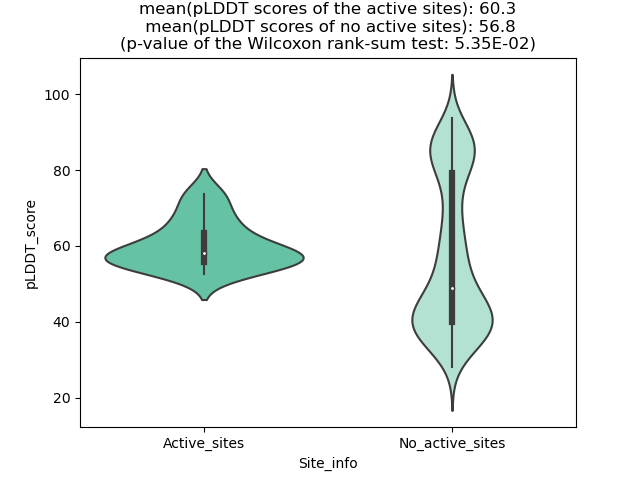
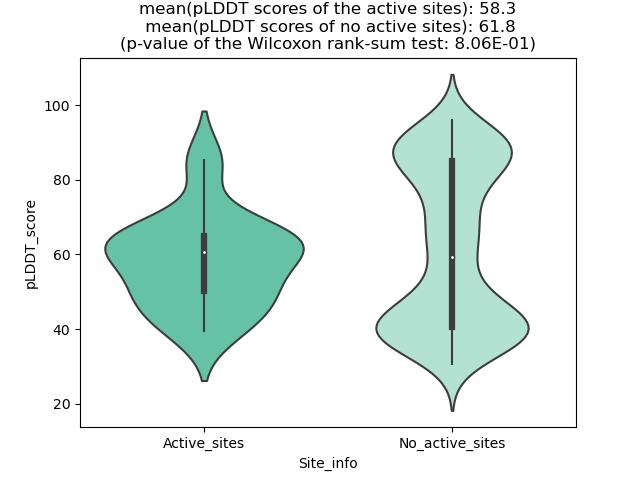
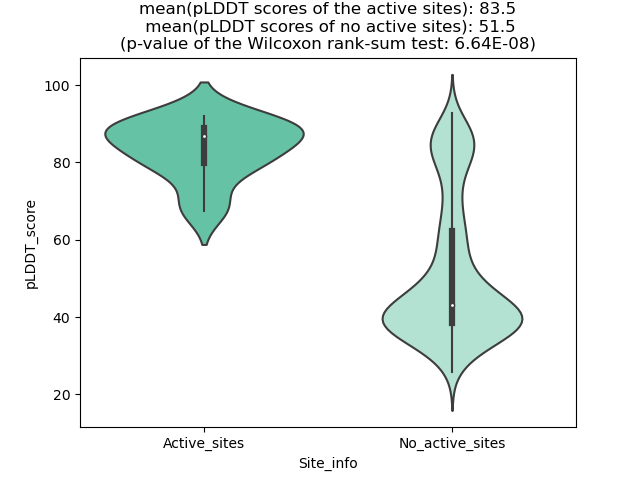
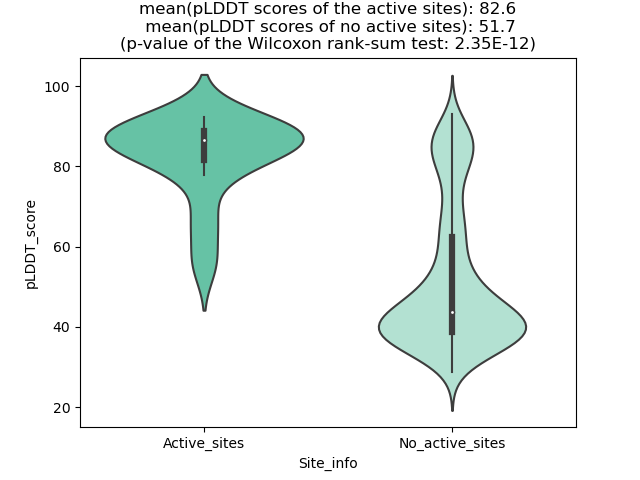
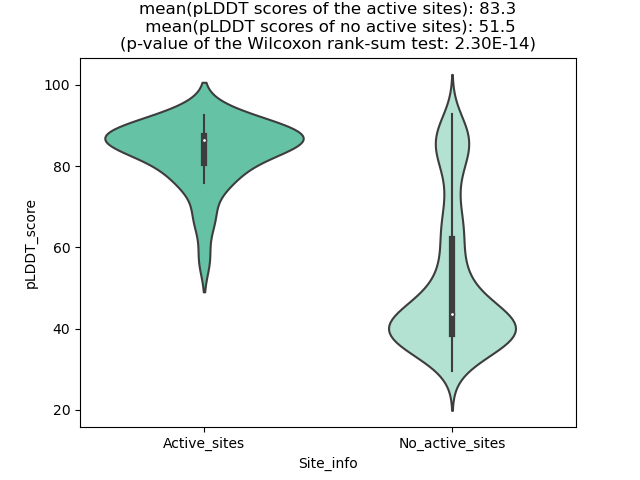
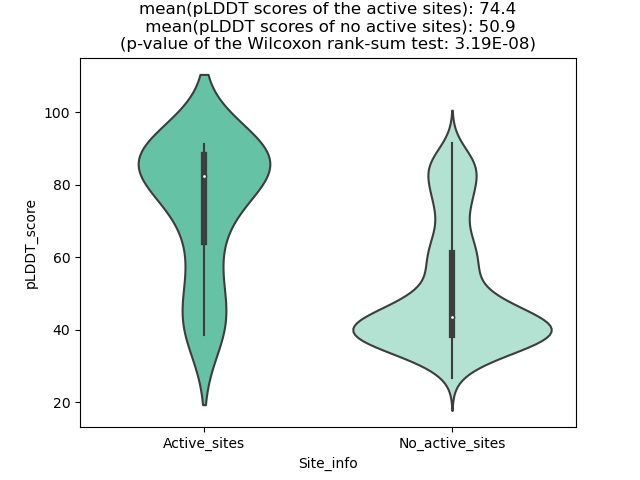
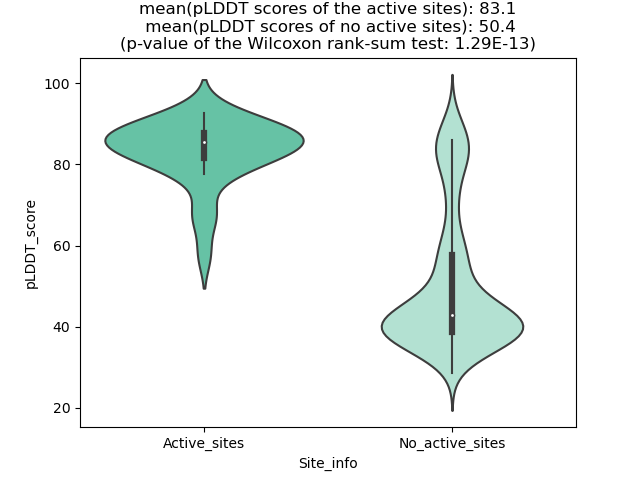
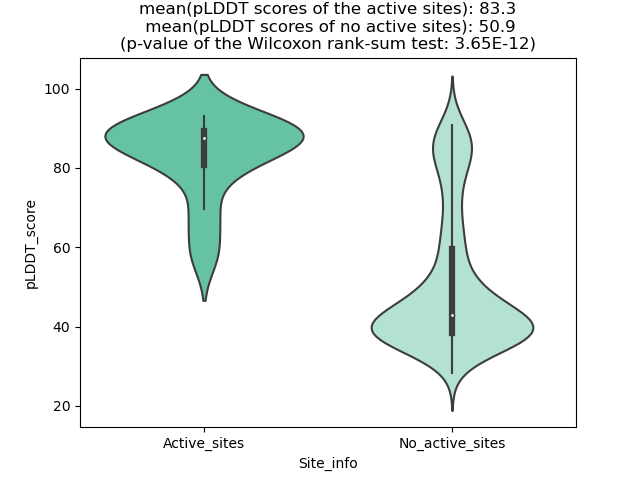
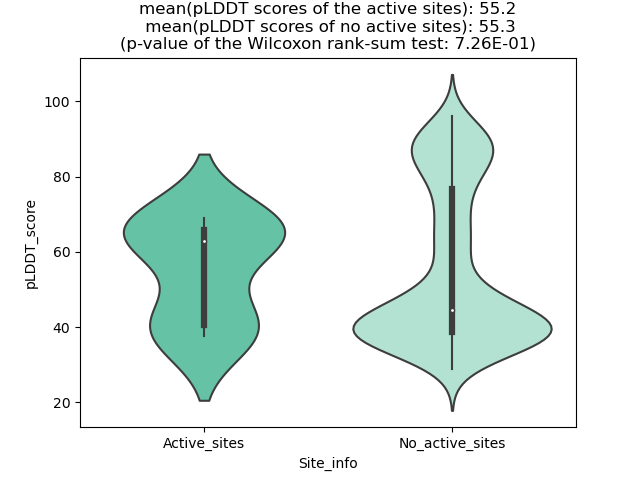
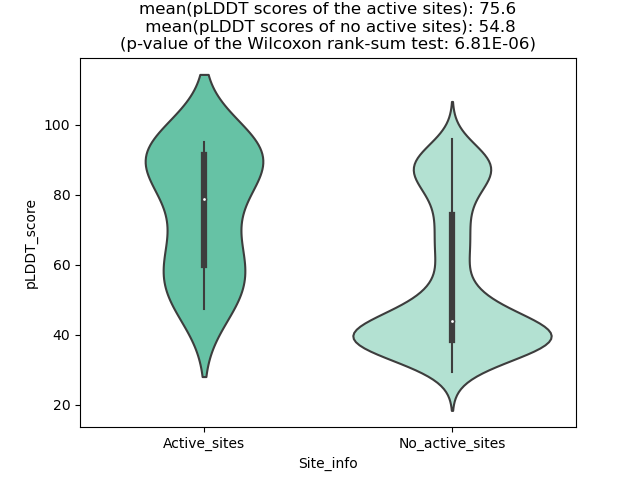
Potential Active Site Information |
 The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. |
| Fusion AA seq ID in FusionPDB | Site score | Size | D score | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
| 52 | 0.555 | 29 | 0.503 | 63.455 | 0.724 | 0.512 | 0.733 | 0.128 | 0.952 | 0.135 | 1.096 | Chain A: 222,223,228,231,234,235,236,237 |
| 64 | 0.833 | 69 | 0.835 | 282.632 | 0.743 | 0.61 | 0.782 | 0.271 | 0.958 | 0.282 | 1.454 | Chain A: 94,95,97,98,99,100,102,255,257,271,272,27 3,274,275,276,277,280,281 |
| 121 | 0.69 | 40 | 0.68 | 70.315 | 0.6 | 0.556 | 0.786 | 0.368 | 0.761 | 0.484 | 1.928 | Chain A: 254,260,261,262,263,274,275,277,278 |
| 175 | 1.022735 | 658 | 1.026981 | 1574.027 | 0.50038 | 0.731992 | 0.948644 | 0.57119 | 1.080412 | 0.528678 | 0.891494 | Chain A: 1,2,3,4,5,6,7,8,9,10,11,12,107,108,109,11 0,111,112,113,114,115,117,119,127,129,132,151,154, 159,160,162,173,175,176,177,178,179,180,181,182,18 4,185,186,221,222,223,224,226,227,229,239,240,241, 243,244,245,246,247,248,249,250,251,252,253,254,25 5,256,257,258,259,260,261,262,263,264,265,266,267, 270,271,273,276,277,278,279,283,284,287,300,302,30 3,305,306,308,311,312 |
| 233 | 1.018 | 125 | 1.089 | 351.575 | 0.637 | 0.626 | 0.755 | 0.648 | 0.695 | 0.932 | 1.334 | Chain A: 71,74,75,77,78,81,82,84,85,88,89,92,256,2 65,266,267,270,271,274,277,278,281,282 |
| 269 | 1.091 | 119 | 1.212 | 289.149 | 0.61 | 0.627 | 0.867 | 2.043 | 0.32 | 6.391 | 0.699 | Chain A: 42,43,45,46,49,50,52,53,56,57,60,312,313, 316,317,319,320,321,323,324,325,327,328,329,331 |
| 308 | 0.915 | 65 | 0.991 | 201.341 | 0.717 | 0.592 | 0.734 | 1.298 | 0.374 | 3.468 | 0.541 | Chain A: 57,60,61,64,67,68,71,72,75,276,277,279,28 0,283,284,287,288,291,292,295 |
| 350 | 0.931 | 80 | 0.978 | 332.024 | 0.707 | 0.617 | 0.789 | 0.745 | 0.714 | 1.044 | 1.196 | Chain A: 71,74,75,78,81,82,84,85,86,88,89,90,91,92 ,288,290,291,293,294,297,298,300,301 |
| 384 | 0.852 | 77 | 0.824 | 210.259 | 0.694 | 0.611 | 0.817 | 0.306 | 1.129 | 0.272 | 0.466 | Chain A: 83,86,87,88,89,90,91,92,93,94,96,198,199, 201,202,204,205,206,207,209,340,341 |
| 451 | 0.784 | 41 | 0.77 | 119.707 | 0.539 | 0.683 | 1.037 | 0.906 | 0.741 | 1.222 | 0.293 | Chain A: 44,45,46,49,390,391,394,395,398 |
| 548 | 1.09 | 111 | 1.205 | 313.159 | 0.613 | 0.637 | 0.878 | 2.136 | 0.36 | 5.926 | 0.427 | Chain A: 46,49,50,52,53,56,57,60,61,64,288,291,292 ,295,296,298,299,302,303,304,306,307,310 |
| 549 | 0.991 | 58 | 1.088 | 117.992 | 0.58 | 0.677 | 0.939 | 3.116 | 0.104 | 29.858 | 12.515 | Chain A: 49,50,52,53,56,57,60,291,294,295,298,301, 302,305,306 |
| 563 | 1.092324 | 122 | 1.214693 | 336.483 | 0.605178 | 0.625694 | 0.838988 | 2.235645 | 0.308969 | 7.235825 | 0.576871 | Chain A: 45,46,49,50,52,53,56,57,60,61,64,67,295,2 98,299,302,303,306,307,309,310,313,314,317,318,321 |
| 568 | 1.071 | 156 | 1.178 | 425.32 | 0.603 | 0.627 | 0.841 | 1.764 | 0.423 | 4.169 | 0.468 | Chain A: 46,49,50,52,53,56,57,60,61,63,64,67,70,71 ,298,301,302,305,306,309,312,313,316,317,319,320,3 23,324,327,328,331 |
| 575 | 1.07 | 145 | 1.174 | 304.927 | 0.57 | 0.631 | 0.873 | 1.916 | 0.443 | 4.324 | 0.675 | Chain A: 7,8,9,10,11,46,49,50,52,53,56,57,60,315,3 16,319,320,322,323,326,327,330,331,334,335 |
| 580 | 1.061 | 143 | 1.178 | 335.111 | 0.645 | 0.596 | 0.774 | 1.781 | 0.368 | 4.842 | 0.591 | Chain A: 46,49,50,52,53,56,57,60,61,63,64,67,310,3 13,314,317,320,321,324,325,327,328,331,332,335,336 ,339 |
| 584 | 1.106 | 122 | 1.232 | 329.966 | 0.597 | 0.636 | 0.833 | 2.2 | 0.276 | 7.967 | 0.68 | Chain A: 45,46,49,50,52,53,56,57,60,61,63,64,320,3 23,324,327,328,330,331,334,335,338,339,342 |
| 624 | 0.708 | 35 | 0.728 | 83.006 | 0.748 | 0.55 | 0.696 | 1.152 | 0.486 | 2.374 | 1.208 | Chain A: 128,130,131,134,140,141,143,144,145,147 |
| 626 | 0.687 | 22 | 0.69 | 106.673 | 0.784 | 0.622 | 0.771 | 1.159 | 0.379 | 3.058 | 0.491 | Chain A: 143,155,156,157,158,162,165,166,169,181 |
| 629 | 0.685 | 26 | 0.676 | 76.832 | 0.671 | 0.62 | 0.858 | 1.153 | 0.54 | 2.136 | 1.4 | Chain A: 191,194,195,198,358,359,362,363 |
| 631 | 0.878395 | 59 | 0.933615 | 234.612 | 0.717703 | 0.604311 | 0.757114 | 1.325526 | 0.463188 | 2.861743 | 0.478688 | Chain A: 42,43,44,45,46,48,49,50,52,53,56,392,395, 398,399,400,402,403,406 |
Top |
Potentially Interacting Small Molecules through Virtual Screening |
 The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. |
| Fusion AA seq ID in FusionPDB | ZINC ID | DrugBank ID | Drug name | Docking score | Glide gscore |
| 52 | ZINC000003927870 | DB01073 | Fludarabine | -6.22965 | -6.32165 |
| 52 | ZINC000000388081 | DB05381 | Histamine | -5.60082 | -6.05582 |
| 52 | ZINC000003830813 | DB01077 | Etidronic acid | -5.69071 | -5.93681 |
| 52 | ZINC000000001795 | DB00627 | Niacin | -5.92129 | -5.93089 |
| 52 | ZINC000001531009 | DB00884 | Risedronic acid | -5.60444 | -5.79294 |
| 52 | ZINC000000001706 | DB13882 | Methyl nicotinate | -5.75745 | -5.75745 |
| 52 | ZINC000006467621 | DB11115 | Ensulizole | -5.18023 | -5.74683 |
| 52 | ZINC000000000882 | DB00173 | Adenine | -5.71092 | -5.73162 |
| 52 | ZINC000003831475 | DB14089 | Dimercaptosuccinic acid | -5.729 | -5.729 |
| 52 | ZINC000003803652 | DB00399 | Zoledronic acid | -5.09 | -5.6342 |
| 52 | ZINC000008015016 | DB14078 | Medronic acid | -5.29749 | -5.61109 |
| 52 | ZINC000003875368 | DB00698 | Nitrofurantoin | -4.31958 | -5.60068 |
| 52 | ZINC000013585233 | DB00360 | Sapropterin | -5.57229 | -5.59199 |
| 52 | ZINC000001035331 | DB00811 | Ribavirin | -5.56876 | -5.56876 |
| 52 | ZINC000038212689 | DB00544 | Fluorouracil | -5.3501 | -5.5501 |
| 52 | ZINC000003872277 | DB01119 | Diazoxide | -5.52148 | -5.52928 |
| 52 | ZINC000008101109 | DB00529 | Foscarnet | -5.48238 | -5.48878 |
| 52 | ZINC000002015035 | DB06262 | Droxidopa | -5.4699 | -5.4699 |
| 52 | ZINC000004640636 | DB00550 | Propylthiouracil | -5.40905 | -5.44765 |
| 52 | ZINC000003831477 | DB14089 | Dimercaptosuccinic acid | -5.40519 | -5.40519 |
Top |
 Drug information from DrugBank of the top 20 interacting small molecules. Drug information from DrugBank of the top 20 interacting small molecules. |
| ZINC ID | DrugBank ID | Drug name | Drug type | SMILES | Drug group |
| ZINC000003927870 | DB01073 | Fludarabine | Small molecule | NC1=NC(F)=NC2=C1N=CN2[C@@H]1O[C@H](CO)[C@@H](O)[C@@H]1O | Approved |
| ZINC000000388081 | DB05381 | Histamine | Small molecule | NCCC1=CNC=N1 | Approved|Investigational |
| ZINC000003830813 | DB01077 | Etidronic acid | Small molecule | CC(O)(P(O)(O)=O)P(O)(O)=O | Approved |
| ZINC000000001795 | DB00627 | Niacin | Small molecule | OC(=O)C1=CN=CC=C1 | Approved|Investigational|Nutraceutical |
| ZINC000001531009 | DB00884 | Risedronic acid | Small molecule | OC(CC1=CN=CC=C1)(P(O)(O)=O)P(O)(O)=O | Approved|Investigational |
| ZINC000000001706 | DB13882 | Methyl nicotinate | Small molecule | COC(=O)C1=CN=CC=C1 | Approved |
| ZINC000006467621 | DB11115 | Ensulizole | Small molecule | OS(=O)(=O)C1=CC2=C(C=C1)N=C(N2)C1=CC=CC=C1 | Approved |
| ZINC000000000882 | DB00173 | Adenine | Small molecule | NC1=C2NC=NC2=NC=N1 | Approved|Nutraceutical |
| ZINC000003803652 | DB00399 | Zoledronic acid | Small molecule | OC(CN1C=CN=C1)(P(O)(O)=O)P(O)(O)=O | Approved |
| ZINC000008015016 | DB14078 | Medronic acid | Small molecule | OP(O)(=O)CP(O)(O)=O | Approved|Investigational |
| ZINC000001035331 | DB00811 | Ribavirin | Small molecule | NC(=O)C1=NN(C=N1)[C@@H]1O[C@H](CO)[C@@H](O)[C@H]1O | Approved |
| ZINC000038212689 | DB00544 | Fluorouracil | Small molecule | FC1=CNC(=O)NC1=O | Approved |
| ZINC000003872277 | DB01119 | Diazoxide | Small molecule | CC1=NS(=O)(=O)C2=C(N1)C=CC(Cl)=C2 | Approved |
| ZINC000008101109 | DB00529 | Foscarnet | Small molecule | OC(=O)P(O)(O)=O | Approved |
| ZINC000002015035 | DB06262 | Droxidopa | Small molecule | N[C@@H]([C@H](O)C1=CC(O)=C(O)C=C1)C(O)=O | Approved|Investigational |
| ZINC000004640636 | DB00550 | Propylthiouracil | Small molecule | CCCC1=CC(=O)NC(=S)N1 | Approved|Investigational |
| ZINC000003831477 | DB14089 | Dimercaptosuccinic acid | Small molecule | OC(=O)C(S)C(S)C(O)=O | Approved|Experimental |
Top |
Biochemical Features of Small Molecules |
 ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) |
| ZINC ID | mol_MW | dipole | SASA | FOSA | FISA | PISA | WPSA | volume | donorHB | accptHB | IP | Human Oral Absorption | Percent Human Oral Absorption | Rule Of Five | Rule Of Three |
| ZINC000003927870 | 365.214 | 10.011 | 558.493 | 73.58 | 358.734 | 76.063 | 50.116 | 953.163 | 6 | 14.1 | 8.598 | 1 | 0 | 2 | 1 |
| ZINC000003927870 | 365.214 | 6.236 | 564.927 | 82.608 | 356.666 | 68.682 | 56.971 | 956.451 | 6 | 14.1 | 8.73 | 1 | 0 | 2 | 1 |
| ZINC000000388081 | 111.146 | 4.788 | 315.231 | 92.229 | 118.535 | 104.468 | 0 | 466.776 | 3 | 2.5 | 9.295 | 2 | 65.253 | 0 | 0 |
| ZINC000000388081 | 111.146 | 5.535 | 313.532 | 90.818 | 116.688 | 106.026 | 0 | 465.559 | 3 | 3 | 9.188 | 2 | 64.917 | 0 | 0 |
| ZINC000000388081 | 111.146 | 4.794 | 316.569 | 91.628 | 116.549 | 108.393 | 0 | 470.468 | 3 | 2.5 | 9.292 | 2 | 65.823 | 0 | 0 |
| ZINC000003830813 | 206.029 | 8.234 | 343.926 | 58.866 | 278.645 | 0 | 6.414 | 541.497 | 1 | 6.75 | 9.379 | 1 | 20.728 | 0 | 1 |
| ZINC000003830813 | 206.029 | 8.446 | 343.742 | 59.548 | 277.922 | 0 | 6.271 | 541.502 | 1 | 6.75 | 9.388 | 1 | 20.88 | 0 | 1 |
| ZINC000003830813 | 206.029 | 7.127 | 340.461 | 65.078 | 269.49 | 0 | 5.893 | 540.584 | 1 | 6.75 | 9.311 | 1 | 22.658 | 0 | 1 |
| ZINC000000001795 | 123.111 | 4.24 | 302.099 | 0 | 134.143 | 167.956 | 0 | 449.108 | 1 | 3.5 | 10.657 | 2 | 68.969 | 0 | 0 |
| ZINC000000001795 | 123.111 | 4.501 | 300.772 | 0 | 131.945 | 168.826 | 0 | 447.811 | 1 | 3.5 | 10.574 | 2 | 69.375 | 0 | 0 |
| ZINC000001531009 | 283.114 | 8.624 | 429.179 | 20.113 | 269.828 | 134.694 | 4.543 | 742.511 | 1 | 8.25 | 9.463 | 1 | 26.76 | 0 | 1 |
| ZINC000001531009 | 283.114 | 9.607 | 427.754 | 22.11 | 268.873 | 132.458 | 4.312 | 739.953 | 1 | 8.25 | 9.593 | 1 | 26.859 | 0 | 1 |
| ZINC000001531009 | 283.114 | 7.642 | 425.511 | 19.856 | 269.894 | 133.142 | 2.619 | 746.321 | 1 | 8.25 | 9.496 | 1 | 26.881 | 0 | 1 |
| ZINC000000001706 | 137.138 | 4.434 | 351.185 | 98.444 | 86.892 | 165.848 | 0 | 531.212 | 0 | 3.5 | 10.407 | 3 | 87.307 | 0 | 0 |
| ZINC000006467621 | 274.294 | 9.514 | 495.179 | 0 | 165.642 | 327.077 | 2.46 | 816.857 | 2 | 5.5 | 9.09 | 3 | 70.241 | 0 | 0 |
| ZINC000006467621 | 274.294 | 5.937 | 496.167 | 0 | 165.586 | 328.124 | 2.456 | 817.805 | 2 | 5.5 | 9.121 | 3 | 70.288 | 0 | 0 |
| ZINC000006467621 | 274.294 | 9.57 | 496.894 | 0 | 166.022 | 328.377 | 2.495 | 819.646 | 2 | 5.5 | 8.954 | 3 | 70.264 | 0 | 0 |
| ZINC000000000882 | 135.128 | 2.734 | 303.153 | 0 | 170.481 | 132.672 | 0 | 451.892 | 3 | 4 | 8.538 | 3 | 68.38 | 0 | 0 |
| ZINC000000000882 | 135.128 | 9.438 | 304.278 | 0 | 171.466 | 132.812 | 0 | 454.287 | 3 | 4 | 8.639 | 3 | 68.214 | 0 | 0 |
| ZINC000003831475 | 182.209 | 3.697 | 335.088 | 26 | 184.497 | 0 | 124.591 | 518.524 | 2 | 3 | 10.006 | 2 | 54.049 | 0 | 1 |
| ZINC000003803652 | 272.091 | 6.149 | 414.004 | 25.436 | 272.399 | 109.477 | 6.692 | 698.612 | 1 | 8.75 | 9.817 | 1 | 23.489 | 0 | 1 |
| ZINC000003803652 | 272.091 | 10.346 | 412.626 | 22.493 | 273.968 | 109.284 | 6.881 | 697.133 | 1 | 8.75 | 8.891 | 1 | 23.124 | 0 | 1 |
| ZINC000003803652 | 272.091 | 2.82 | 416.545 | 22.709 | 279.09 | 108.406 | 6.34 | 697.546 | 1 | 8.75 | 9.904 | 1 | 21.992 | 0 | 1 |
| ZINC000003803652 | 272.091 | 11.882 | 412.711 | 24.614 | 274.426 | 108.467 | 5.205 | 696.74 | 1 | 8.75 | 9.026 | 1 | 22.968 | 0 | 1 |
| ZINC000003803652 | 272.091 | 12.418 | 410.051 | 22.071 | 274.507 | 110.827 | 2.646 | 702.503 | 1 | 8.75 | 8.712 | 1 | 23.166 | 0 | 1 |
| ZINC000003803652 | 272.091 | 12.445 | 414.593 | 23.463 | 272.373 | 114.555 | 4.201 | 705.321 | 1 | 8.75 | 8.718 | 1 | 23.722 | 0 | 1 |
| ZINC000008015016 | 176.002 | 3.602 | 323.868 | 34.991 | 281.713 | 0 | 7.164 | 486.492 | 0 | 6 | 9.674 | 1 | 16.795 | 0 | 1 |
| ZINC000008015016 | 176.002 | 13.068 | 326.073 | 33.644 | 285.397 | 0 | 7.032 | 489.653 | 0 | 6 | 9.876 | 1 | 16.139 | 0 | 1 |
| ZINC000003875368 | 238.159 | 4.578 | 425.423 | 58.928 | 250.52 | 115.975 | 0 | 683.161 | 1 | 5 | 9.961 | 2 | 53.722 | 0 | 0 |
| ZINC000003875368 | 238.159 | 4.877 | 423.946 | 59.354 | 248.434 | 116.158 | 0 | 680.796 | 1 | 5 | 9.943 | 2 | 54.077 | 0 | 0 |
| ZINC000013585233 | 241.249 | 7.548 | 455.149 | 158.349 | 273.982 | 22.819 | 0 | 749.387 | 7 | 8.9 | 7.637 | 2 | 28.323 | 1 | 0 |
| ZINC000013585233 | 241.249 | 11.056 | 455.167 | 158.05 | 274.486 | 22.631 | 0 | 750.263 | 7 | 8.9 | 7.777 | 2 | 28.249 | 1 | 0 |
| ZINC000001035331 | 244.207 | 7.612 | 427.913 | 90.998 | 287.486 | 49.429 | 0 | 713.931 | 5 | 12.3 | 9.744 | 2 | 34.344 | 0 | 1 |
| ZINC000038212689 | 130.078 | 5.522 | 259.136 | 0 | 161.594 | 52.299 | 45.242 | 383.043 | 2 | 3.5 | 9.983 | 2 | 65.855 | 0 | 0 |
| ZINC000038212689 | 130.078 | 5.054 | 262.519 | 0 | 162.511 | 54.553 | 45.455 | 386.145 | 2 | 3.5 | 9.862 | 2 | 65.704 | 0 | 0 |
| ZINC000003872277 | 230.668 | 10.522 | 415.073 | 94.247 | 119.686 | 128.269 | 72.87 | 661.918 | 0 | 4.5 | 9.329 | 3 | 82.986 | 0 | 0 |
| ZINC000003872277 | 230.668 | 11.183 | 413.943 | 93.631 | 118.113 | 129.281 | 72.918 | 659.366 | 1 | 5.5 | 9.406 | 3 | 83.218 | 0 | 0 |
| ZINC000008101109 | 126.005 | 6.9 | 264.917 | 0 | 260.554 | 0 | 4.362 | 372.032 | 3 | 7 | 9.757 | 1 | 17.558 | 0 | 1 |
| ZINC000008101109 | 126.005 | 11.927 | 257.279 | 0 | 253.345 | 0 | 3.934 | 359.323 | 3 | 7 | 9.932 | 1 | 18.931 | 0 | 1 |
| ZINC000002015035 | 213.19 | 5.76 | 408.685 | 26.39 | 279.826 | 102.469 | 0 | 664.786 | 5 | 5.2 | 8.908 | 1 | 0 | 1 | 1 |
| ZINC000004640636 | 170.229 | 6.707 | 408.008 | 178.11 | 116.084 | 32.743 | 81.071 | 619.311 | 2 | 3 | 9.191 | 3 | 82.875 | 0 | 0 |
| ZINC000004640636 | 170.229 | 7.083 | 390.596 | 167.972 | 99.585 | 42.266 | 80.773 | 608.72 | 2 | 3 | 8.797 | 3 | 90.531 | 0 | 0 |
| ZINC000003831477 | 182.209 | 7.58 | 330.563 | 25.281 | 178.679 | 0 | 126.603 | 516.577 | 2 | 3 | 10.071 | 2 | 55.288 | 0 | 1 |
Top |
Drug Toxicity Information |
 Toxicity information of individual drugs using eToxPred Toxicity information of individual drugs using eToxPred |
| ZINC ID | Smile | Surface Accessibility | Toxicity |
| ZINC000003927870 | Nc1nc(F)nc2c1ncn2[C@@H]1O[C@H](COP(=O)(O)O)[C@@H](O)[C@@H]1O | 0.050585445 | 0.380605086 |
| ZINC000000388081 | NCCc1cnc[nH]1 | 0.212944784 | 0.352067312 |
| ZINC000003830813 | CC(O)(P(=O)(O)O)P(=O)(O)O | 0.095573527 | 0.535505529 |
| ZINC000000001795 | O=C(O)c1cccnc1 | 0.556974396 | 0.499160568 |
| ZINC000001531009 | O=P(O)(O)C(O)(Cc1cccnc1)P(=O)(O)O | 0.151740248 | 0.484505913 |
| ZINC000000001706 | COC(=O)c1cccnc1 | 0.562051569 | 0.439867718 |
| ZINC000006467621 | O=S(=O)(O)c1ccc2[nH]c(-c3ccccc3)nc2c1 | 0.38504104 | 0.405031793 |
| ZINC000000000882 | Nc1ncnc2[nH]cnc12 | 0.174637229 | 0.465480638 |
| ZINC000003831475 | O=C(O)[C@H](S)[C@H](S)C(=O)O | 0.046119564 | 0.585775635 |
| ZINC000003803652 | O=P(O)(O)C(O)(Cn1ccnc1)P(=O)(O)O | 0.100052268 | 0.353941778 |
| ZINC000008015016 | O=P(O)(O)CP(=O)(O)O | 0.097212655 | 0.606313084 |
| ZINC000003875368 | O=C1CN(/N=C/c2ccc([N+](=O)[O-])o2)C(=O)N1 | 0.144023256 | 0.52186584 |
| ZINC000013585233 | C[C@H](O)[C@H](O)[C@H]1CNc2nc(N)[nH]c(=O)c2N1 | 0.031410569 | 0.318425003 |
| ZINC000001035331 | NC(=O)c1ncn([C@@H]2O[C@H](CO)[C@@H](O)[C@H]2O)n1 | 0.05676866 | 0.366258102 |
| ZINC000038212689 | O=c1[nH]cc(F)c(=O)[nH]1 | 0.190312538 | 0.481453644 |
| ZINC000003872277 | CC1=NS(=O)(=O)c2cc(Cl)ccc2N1 | 0.17675056 | 0.347952196 |
| ZINC000008101109 | O=C(O)P(=O)(O)O | 0.151109156 | 0.530839371 |
| ZINC000002015035 | N[C@@H](C(=O)O)[C@@H](O)c1ccc(O)c(O)c1 | 0.133345121 | 0.276427601 |
| ZINC000004640636 | CCCc1cc(=O)[nH]c(=S)[nH]1 | 0.185646757 | 0.507723366 |
| ZINC000003831477 | O=C(O)[C@H](S)[C@@H](S)C(=O)O | 0.046119564 | 0.585775635 |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| CD74 | MIF, CD74, APP, HADHA, LAMTOR5, MAP3K1, TM4SF20, COPG1, COPG2, HLA-DPB1, HLA-DQA1, nef, CD44, AKAP8, ANKRD28, AP3D1, ARCN1, ASCC1, ASCC3, ASNS, CANX, CHAF1B, COPA, COPB1, COPB2, COPE, DDX60, DGKE, DNAJB11, DNAJB12, EIF1, EIF2B3, EIF2B4, EIF2B5, EIF2S1, EIF2S3, EIF3A, EIF3B, EIF3C, EIF3D, EIF3E, EIF3F, EIF3G, EIF3H, EIF3I, EIF3J, EIF3K, EIF3L, EIF3M, EIF4A1, EIF4G1, EIF4G2, FKBP8, GANAB, GEMIN4, GRAMD1A, GTF2I, GTF3C1, HADHB, HMOX2, HUWE1, IPO13, IQGAP2, KPNB1, KRT1, LTN1, MAP1B, MTOR, NCAPD2, NCAPD3, NCAPH, OSBPL8, PCNA, PDAP1, PDCD4, PGRMC1, PGRMC2, PHB, POLR2E, POLR3A, POLR3B, PPP6R1, PPP6R3, PRRC2A, PRRC2B, PRRC2C, PSMC2, PSMD3, PSMD6, RCN1, RHOT2, RWDD1, SGPL1, SNRPE, SRP9, TBRG4, TJP2, TPP2, TUBA1A, TUBA1C, UBE3C, UBR4, UBR5, VDAC1, VWA8, YWHAB, YWHAE, YWHAQ, YWHAZ, IFITM3, FDFT1, POMGNT1, BET1, TMEM97, UPK2, TEX11, SERP1, SLC35B1, SEC22A, PLP1, TMEM254, C14orf1, TMEM243, TMEM60, FXYD6, CMTM7, CLDN19, LPAR3, CLEC7A, EDDM3B, C14orf180, PTCH1, TMEM120B, RTP2, PPAPDC1A, PGA4, SMIM1, CUL7, CCR1, ROPN1L, HLA-DRB3, HLA-DRB1, HLA-DRA, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| CD74 |  |
| NRG1 |  |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to CD74-NRG1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
| CD74 | NRG1 | Afatinib | PubMed | 29169524 |
Top |
Related Diseases to CD74-NRG1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
| CD74 | NRG1 | Lung Cancer | PubMed | 29169524 |
| CD74 | NRG1 | Lung Adenocarcinoma | MyCancerGenome | |
| CD74 | NRG1 | Breast Invasive Ductal Carcinoma | MyCancerGenome | |
| CD74 | NRG1 | Invasive Breast Carcinoma | MyCancerGenome | |
| CD74 | NRG1 | Adenocarcinoma Of Unknown Primary | MyCancerGenome | |
| CD74 | NRG1 | Bladder Urothelial Carcinoma | MyCancerGenome |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | CD74 | C0006142 | Malignant neoplasm of breast | 1 | CTD_human |
| Hgene | CD74 | C0007131 | Non-Small Cell Lung Carcinoma | 1 | CTD_human |
| Hgene | CD74 | C0023893 | Liver Cirrhosis, Experimental | 1 | CTD_human |
| Hgene | CD74 | C0162557 | Liver Failure, Acute | 1 | CTD_human |
| Hgene | CD74 | C0678222 | Breast Carcinoma | 1 | CTD_human |
| Hgene | CD74 | C1257931 | Mammary Neoplasms, Human | 1 | CTD_human |
| Hgene | CD74 | C1458155 | Mammary Neoplasms | 1 | CTD_human |
| Hgene | CD74 | C4704874 | Mammary Carcinoma, Human | 1 | CTD_human |
| Tgene | NRG1 | C0036341 | Schizophrenia | 7 | CTD_human |
| Tgene | NRG1 | C0005586 | Bipolar Disorder | 5 | PSYGENET |
| Tgene | NRG1 | C0024809 | Marijuana Abuse | 3 | PSYGENET |
| Tgene | NRG1 | C0011570 | Mental Depression | 2 | PSYGENET |
| Tgene | NRG1 | C0011581 | Depressive disorder | 2 | PSYGENET |
| Tgene | NRG1 | C0006142 | Malignant neoplasm of breast | 1 | CTD_human |
| Tgene | NRG1 | C0006870 | Cannabis Dependence | 1 | PSYGENET |
| Tgene | NRG1 | C0007621 | Neoplastic Cell Transformation | 1 | CTD_human |
| Tgene | NRG1 | C0011616 | Contact Dermatitis | 1 | CTD_human |
| Tgene | NRG1 | C0018801 | Heart failure | 1 | CTD_human |
| Tgene | NRG1 | C0018802 | Congestive heart failure | 1 | CTD_human |
| Tgene | NRG1 | C0019569 | Hirschsprung Disease | 1 | CTD_human |
| Tgene | NRG1 | C0023212 | Left-Sided Heart Failure | 1 | CTD_human |
| Tgene | NRG1 | C0023893 | Liver Cirrhosis, Experimental | 1 | CTD_human |
| Tgene | NRG1 | C0024121 | Lung Neoplasms | 1 | CTD_human |
| Tgene | NRG1 | C0026650 | Movement Disorders | 1 | CTD_human |
| Tgene | NRG1 | C0027626 | Neoplasm Invasiveness | 1 | CTD_human |
| Tgene | NRG1 | C0030193 | Pain | 1 | CTD_human |
| Tgene | NRG1 | C0032460 | Polycystic Ovary Syndrome | 1 | CTD_human |
| Tgene | NRG1 | C0038358 | Gastric ulcer | 1 | CTD_human |
| Tgene | NRG1 | C0085758 | Aganglionosis, Colonic | 1 | CTD_human |
| Tgene | NRG1 | C0087031 | Juvenile-Onset Still Disease | 1 | CTD_human |
| Tgene | NRG1 | C0162351 | Contact hypersensitivity | 1 | CTD_human |
| Tgene | NRG1 | C0234230 | Pain, Burning | 1 | CTD_human |
| Tgene | NRG1 | C0234238 | Ache | 1 | CTD_human |
| Tgene | NRG1 | C0234254 | Radiating pain | 1 | CTD_human |
| Tgene | NRG1 | C0235527 | Heart Failure, Right-Sided | 1 | CTD_human |
| Tgene | NRG1 | C0236733 | Amphetamine-Related Disorders | 1 | CTD_human |
| Tgene | NRG1 | C0236804 | Amphetamine Addiction | 1 | CTD_human |
| Tgene | NRG1 | C0236807 | Amphetamine Abuse | 1 | CTD_human |
| Tgene | NRG1 | C0242379 | Malignant neoplasm of lung | 1 | CTD_human |
| Tgene | NRG1 | C0266487 | Etat Marbre | 1 | CTD_human |
| Tgene | NRG1 | C0458257 | Pain, Splitting | 1 | CTD_human |
| Tgene | NRG1 | C0458259 | Pain, Crushing | 1 | CTD_human |
| Tgene | NRG1 | C0678222 | Breast Carcinoma | 1 | CTD_human |
| Tgene | NRG1 | C0751407 | Pain, Migratory | 1 | CTD_human |
| Tgene | NRG1 | C0751408 | Suffering, Physical | 1 | CTD_human |
| Tgene | NRG1 | C0876994 | Cardiotoxicity | 1 | CTD_human |
| Tgene | NRG1 | C1136382 | Sclerocystic Ovaries | 1 | CTD_human |
| Tgene | NRG1 | C1257840 | Aganglionosis, Rectosigmoid Colon | 1 | CTD_human |
| Tgene | NRG1 | C1257931 | Mammary Neoplasms, Human | 1 | CTD_human |
| Tgene | NRG1 | C1458155 | Mammary Neoplasms | 1 | CTD_human |
| Tgene | NRG1 | C1959583 | Myocardial Failure | 1 | CTD_human |
| Tgene | NRG1 | C1961112 | Heart Decompensation | 1 | CTD_human |
| Tgene | NRG1 | C3495559 | Juvenile arthritis | 1 | CTD_human |
| Tgene | NRG1 | C3661523 | Congenital Intestinal Aganglionosis | 1 | CTD_human |
| Tgene | NRG1 | C3714758 | Juvenile psoriatic arthritis | 1 | CTD_human |
| Tgene | NRG1 | C4552091 | Polyarthritis, Juvenile, Rheumatoid Factor Negative | 1 | CTD_human |
| Tgene | NRG1 | C4704862 | Polyarthritis, Juvenile, Rheumatoid Factor Positive | 1 | CTD_human |
| Tgene | NRG1 | C4704874 | Mammary Carcinoma, Human | 1 | CTD_human |