| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:CDC42BPA-EPRS |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: CDC42BPA-EPRS | FusionPDB ID: 14836 | FusionGDB2.0 ID: 14836 | Hgene | Tgene | Gene symbol | CDC42BPA | EPRS | Gene ID | 8476 | 2058 |
| Gene name | CDC42 binding protein kinase alpha | glutamyl-prolyl-tRNA synthetase 1 | |
| Synonyms | MRCK|MRCKA|PK428 | EARS|EPRS|GLUPRORS|HLD15|PARS|PIG32|QARS|QPRS | |
| Cytomap | 1q42.13 | 1q41 | |
| Type of gene | protein-coding | protein-coding | |
| Description | serine/threonine-protein kinase MRCK alphaCDC42 binding protein kinase alpha (DMPK-like)CDC42 binding protein kinase betamyotonic dystrophy kinase-related CDC42-binding protein kinase alphamyotonic dystrophy protein kinase-like alphaser-thr protein k | bifunctional glutamate/proline--tRNA ligasebifunctional aminoacyl-tRNA synthetasecell proliferation-inducing gene 32 proteinglutamate tRNA ligaseglutamatyl-prolyl-tRNA synthetaseglutaminyl-tRNA synthetaseproliferation-inducing gene 32 proteinprolif | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q5VT25 | P07814 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000334218, ENST00000366764, ENST00000366765, ENST00000366766, ENST00000366767, ENST00000366769, ENST00000535525, ENST00000488131, | ENST00000468487, ENST00000366923, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 14 X 12 X 9=1512 | 9 X 9 X 4=324 |
| # samples | 15 | 9 | |
| ** MAII score | log2(15/1512*10)=-3.33342373372519 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(9/324*10)=-1.84799690655495 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: CDC42BPA [Title/Abstract] AND EPRS [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | CDC42BPA(227203781)-EPRS(220180680), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | CDC42BPA-EPRS seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. CDC42BPA-EPRS seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. CDC42BPA-EPRS seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. CDC42BPA-EPRS seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CDC42BPA | GO:0006468 | protein phosphorylation | 9092543 |
| Hgene | CDC42BPA | GO:0031532 | actin cytoskeleton reorganization | 9418861 |
| Tgene | EPRS | GO:0006433 | prolyl-tRNA aminoacylation | 24100331 |
| Tgene | EPRS | GO:0017148 | negative regulation of translation | 23071094 |
| Tgene | EPRS | GO:0071346 | cellular response to interferon-gamma | 15479637 |
 Fusion gene breakpoints across CDC42BPA (5'-gene) Fusion gene breakpoints across CDC42BPA (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
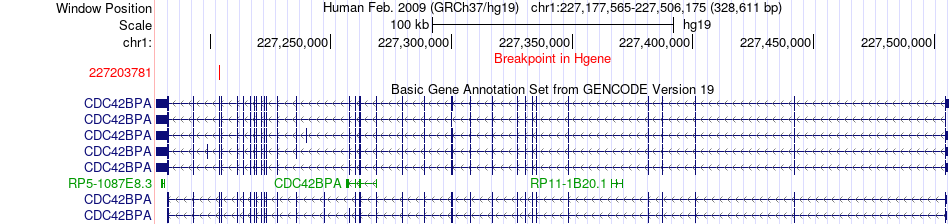 |
 Fusion gene breakpoints across EPRS (3'-gene) Fusion gene breakpoints across EPRS (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
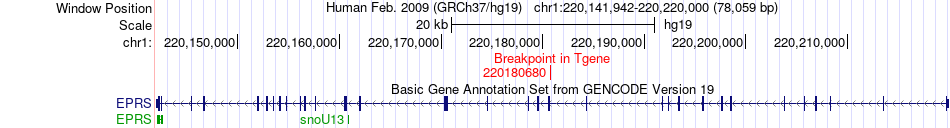 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-E2-A1LK-01A | CDC42BPA | chr1 | 227203781 | - | EPRS | chr1 | 220180680 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000366767 | CDC42BPA | chr1 | 227203781 | - | ENST00000366923 | EPRS | chr1 | 220180680 | - | 7648 | 4509 | 0 | 7442 | 2480 |
| ENST00000366769 | CDC42BPA | chr1 | 227203781 | - | ENST00000366923 | EPRS | chr1 | 220180680 | - | 9183 | 6044 | 1292 | 8977 | 2561 |
| ENST00000366764 | CDC42BPA | chr1 | 227203781 | - | ENST00000366923 | EPRS | chr1 | 220180680 | - | 8750 | 5611 | 943 | 8544 | 2533 |
| ENST00000334218 | CDC42BPA | chr1 | 227203781 | - | ENST00000366923 | EPRS | chr1 | 220180680 | - | 8834 | 5695 | 943 | 8628 | 2561 |
| ENST00000366766 | CDC42BPA | chr1 | 227203781 | - | ENST00000366923 | EPRS | chr1 | 220180680 | - | 8939 | 5800 | 943 | 8733 | 2596 |
| ENST00000535525 | CDC42BPA | chr1 | 227203781 | - | ENST00000366923 | EPRS | chr1 | 220180680 | - | 7858 | 4719 | 27 | 7652 | 2541 |
| ENST00000366765 | CDC42BPA | chr1 | 227203781 | - | ENST00000366923 | EPRS | chr1 | 220180680 | - | 8462 | 5323 | 532 | 8256 | 2574 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000366767 | ENST00000366923 | CDC42BPA | chr1 | 227203781 | - | EPRS | chr1 | 220180680 | - | 0.000452524 | 0.9995474 |
| ENST00000366769 | ENST00000366923 | CDC42BPA | chr1 | 227203781 | - | EPRS | chr1 | 220180680 | - | 0.001576272 | 0.9984237 |
| ENST00000366764 | ENST00000366923 | CDC42BPA | chr1 | 227203781 | - | EPRS | chr1 | 220180680 | - | 0.001100391 | 0.9988996 |
| ENST00000334218 | ENST00000366923 | CDC42BPA | chr1 | 227203781 | - | EPRS | chr1 | 220180680 | - | 0.001078157 | 0.9989219 |
| ENST00000366766 | ENST00000366923 | CDC42BPA | chr1 | 227203781 | - | EPRS | chr1 | 220180680 | - | 0.001101106 | 0.99889886 |
| ENST00000535525 | ENST00000366923 | CDC42BPA | chr1 | 227203781 | - | EPRS | chr1 | 220180680 | - | 0.000555753 | 0.9994442 |
| ENST00000366765 | ENST00000366923 | CDC42BPA | chr1 | 227203781 | - | EPRS | chr1 | 220180680 | - | 0.000712139 | 0.9992879 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >14836_14836_1_CDC42BPA-EPRS_CDC42BPA_chr1_227203781_ENST00000334218_EPRS_chr1_220180680_ENST00000366923_length(amino acids)=2561AA_BP=1861 MSGEVRLRQLEQFILDGPAQTNGQCFSVETLLDILICLYDECNNSPLRREKNILEYLEWAKPFTSKVKQMRLHREDFEILKVIGRGAFGE VAVVKLKNADKVFAMKILNKWEMLKRAETACFREERDVLVNGDNKWITTLHYAFQDDNNLYLVMDYYVGGDLLTLLSKFEDRLPEDMARF YLAEMVIAIDSVHQLHYVHRDIKPDNILMDMNGHIRLADFGSCLKLMEDGTVQSSVAVGTPDYISPEILQAMEDGKGRYGPECDWWSLGV CMYEMLYGETPFYAESLVETYGKIMNHKERFQFPAQVTDVSENAKDLIRRLICSREHRLGQNGIEDFKKHPFFSGIDWDNIRNCEAPYIP EVSSPTDTSNFDVDDDCLKNSETMPPPTHTAFSGHHLPFVGFTYTSSCVLSDRSCLRVTAGPTSLDLDVNVQRTLDNNLATEAYERRIKR LEQEKLELSRKLQESTQTVQALQYSTVDGPLTASKDLEIKNLKEEIEKLRKQVTESSHLEQQLEEANAVRQELDDAFRQIKAYEKQIKTL QQEREDLNKELVQASERLKNQSKELKDAHCQRKLAMQEFMEINERLTELHTQKQKLARHVRDKEEEVDLVMQKVESLRQELRRTERAKKE LEVHTEALAAEASKDRKLREQSEHYSKQLENELEGLKQKQISYSPGVCSIEHQQEITKLKTDLEKKSIFYEEELSKREGIHANEIKNLKK ELHDSEGQQLALNKEIMILKDKLEKTRRESQSEREEFESEFKQQYEREKVLLTEENKKLTSELDKLTTLYENLSIHNQQLEEEVKDLADK KESVAHWEAQITEIIQWVSDEKDARGYLQALASKMTEELEALRNSSLGTRATDMPWKMRRFAKLDMSARLELQSALDAEIRAKQAIQEEL NKVKASNIITECKLKDSEKKNLELLSEIEQLIKDTEELRSEKGIEHQDSQHSFLAFLNTPTDALDQFETVDSTPLSVHTPTLRKKGCPGS TGFPPKRKTHQFFVKSFTTPTKCHQCTSLMVGLIRQGCSCEVCGFSCHITCVNKAPTTCPVPPEQTKGPLGIDPQKGIGTAYEGHVRIPK PAGVKKGWQRALAIVCDFKLFLYDIAEGKASQPSVVISQVIDMRDEEFSVSSVLASDVIHASRKDIPCIFRVTASQLSASNNKCSILMLA DTENEKNKWVGVLSELHKILKKNKFRDRSVYVPKEAYDSTLPLIKTTQAAAIIDHERIALGNEEGLFVVHVTKDEIIRVGDNKKIHQIEL IPNDQLVAVISGRNRHVRLFPMSALDGRETDFYKLSETKGCQTVTSGKVRHGALTCLCVAMKRQVLCYELFQSKTRHRKFKEIQVPYNVQ WMAIFSEQLCVGFQSGFLRYPLNGEGNPYSMLHSNDHTLSFIAHQPMDAICAVEISSKEYLLCFNSIGIYTDCQGRRSRQQELMWPANPS SCCYNAPYLSVYSENAVDIFDVNSMEWIQTLPLKKVRPLNNEGSLNLLGLETIRLIYFKNKMAEGDELVVPETSDNSRKQMVRNINNKRR YSFRVPEEERMQQRREMLRDPEMRNKLISNPTNFNHIAHMGPGDGIQILKDLPMNPEVGLKPVWYSPKVFIEGADAETFSEGEMVTFINW GNLNITKIHKNADGKIISLDAKLNLENKDYKKTTKVTWLAETTHALPIPVICVTYEHLITKPVLGKDEDFKQYVNKNSKHEELMLGDPCL KDLKKGDIIQLQRRGFFICDQPYEPVSPYSCKEAPCVLIYIPDGHTKEMPTSGSKEKTKVEATKNETSAPFKERPTPSLNNNCTTSEDSL VLYNRVAVQGDVVRELKAKKAPKEDVDAAVKQLLSLKAEYKEKTGQEYKPGNPPAEIGQNISSNSSASILESKSLYDEVAAQGEVVRKLK AEKSPKAKINEAVECLLSLKAQYKEKTGKEYIPGQPPLSQSSDSSPTRNSEPAGLETPEAKVLFDKVASQGEVVRKLKTEKAPKDQVDIA VQELLQLKAQYKSLIGVEYKPVSATGAEDKDKKKKEKENKSEKQNKPQKQNDGQRKDPSKNQGGGLSSSGAGEGQGPKKQTRLGLEAKKE ENLADWYSQVITKSEMIEYHDISGCYILRPWAYAIWEAIKDFFDAEIKKLGVENCYFPMFVSQSALEKEKTHVADFAPEVAWVTRSGKTE LAEPIAIRPTSETVMYPAYAKWVQSHRDLPIKLNQWCNVVRWEFKHPQPFLRTREFLWQEGHSAFATMEEAAEEVLQILDLYAQVYEELL AIPVVKGRKTEKEKFAGGDYTTTIEAFISASGRAIQGGTSHHLGQNFSKMFEIVFEDPKIPGEKQFAYQNSWGLTTRTIGVMTMVHGDNM GLVLPPRVACVQVVIIPCGITNALSEEDKEALIAKCNDYRRRLLSVNIRVRADLRDNYSPGWKFNHWELKGVPIRLEVGPRDMKSCQFVA VRRDTGEKLTVAENEAETKLQAILEDIQVTLFTRASEDLKTHMVVANTMEDFQKILDSGKIVQIPFCGEIDCEDWIKKTTARDQDLEPGA -------------------------------------------------------------- >14836_14836_2_CDC42BPA-EPRS_CDC42BPA_chr1_227203781_ENST00000366764_EPRS_chr1_220180680_ENST00000366923_length(amino acids)=2533AA_BP=1833 MSGEVRLRQLEQFILDGPAQTNGQCFSVETLLDILICLYDECNNSPLRREKNILEYLEWAKPFTSKVKQMRLHREDFEILKVIGRGAFGE VAVVKLKNADKVFAMKILNKWEMLKRAETACFREERDVLVNGDNKWITTLHYAFQDDNNLYLVMDYYVGGDLLTLLSKFEDRLPEDMARF YLAEMVIAIDSVHQLHYVHRDIKPDNILMDMNGHIRLADFGSCLKLMEDGTVQSSVAVGTPDYISPEILQAMEDGKGRYGPECDWWSLGV CMYEMLYGETPFYAESLVETYGKIMNHKERFQFPAQVTDVSENAKDLIRRLICSREHRLGQNGIEDFKKHPFFSGIDWDNIRNCEAPYIP EVSSPTDTSNFDVDDDCLKNSETMPPPTHTAFSGHHLPFVGFTYTSSCVLSDRSCLRVTAGPTSLDLDVNVQRTLDNNLATEAYERRIKR LEQEKLELSRKLQESTQTVQALQYSTVDGPLTASKDLEIKNLKEEIEKLRKQVTESSHLEQQLEEANAVRQELDDAFRQIKAYEKQIKTL QQEREDLNKELVQASERLKNQSKELKDAHCQRKLAMQEFMEINERLTELHTQKQKLARHVRDKEEEVDLVMQKVESLRQELRRTERAKKE LEVHTEALAAEASKDRKLREQSEHYSKQLENELEGLKQKQISYSPGVCSIEHQQEITKLKTDLEKKSIFYEEELSKREGIHANEIKNLKK ELHDSEGQQLALNKEIMILKDKLEKTRRESQSEREEFESEFKQQYEREKVLLTEENKKLTSELDKLTTLYENLSIHNQQLEEEVKDLADK KESVAHWEAQITEIIQWVSDEKDARGYLQALASKMTEELEALRNSSLGTRATDMPWKMRRFAKLDMSARLELQSALDAEIRAKQAIQEEL NKVKASNIITECKLKDSEKKNLELLSEIEQLIKDTEELRSEKGIEHQDSQHSFLAFLNTPTDALDQFERKTHQFFVKSFTTPTKCHQCTS LMVGLIRQGCSCEVCGFSCHITCVNKAPTTCPVPPEQTKGPLGIDPQKGIGTAYEGHVRIPKPAGVKKGWQRALAIVCDFKLFLYDIAEG KASQPSVVISQVIDMRDEEFSVSSVLASDVIHASRKDIPCIFRVTASQLSASNNKCSILMLADTENEKNKWVGVLSELHKILKKNKFRDR SVYVPKEAYDSTLPLIKTTQAAAIIDHERIALGNEEGLFVVHVTKDEIIRVGDNKKIHQIELIPNDQLVAVISGRNRHVRLFPMSALDGR ETDFYKLSETKGCQTVTSGKVRHGALTCLCVAMKRQVLCYELFQSKTRHRKFKEIQVPYNVQWMAIFSEQLCVGFQSGFLRYPLNGEGNP YSMLHSNDHTLSFIAHQPMDAICAVEISSKEYLLCFNSIGIYTDCQGRRSRQQELMWPANPSSCCYNAPYLSVYSENAVDIFDVNSMEWI QTLPLKKVRPLNNEGSLNLLGLETIRLIYFKNKMAEGDELVVPETSDNSRKQMVRNINNKRRYSFRVPEEERMQQRREMLRDPEMRNKLI SNPTNFNHIAHMGPGDGIQILKDLPMNPEVGLKPVWYSPKVFIEGADAETFSEGEMVTFINWGNLNITKIHKNADGKIISLDAKLNLENK DYKKTTKVTWLAETTHALPIPVICVTYEHLITKPVLGKDEDFKQYVNKNSKHEELMLGDPCLKDLKKGDIIQLQRRGFFICDQPYEPVSP YSCKEAPCVLIYIPDGHTKEMPTSGSKEKTKVEATKNETSAPFKERPTPSLNNNCTTSEDSLVLYNRVAVQGDVVRELKAKKAPKEDVDA AVKQLLSLKAEYKEKTGQEYKPGNPPAEIGQNISSNSSASILESKSLYDEVAAQGEVVRKLKAEKSPKAKINEAVECLLSLKAQYKEKTG KEYIPGQPPLSQSSDSSPTRNSEPAGLETPEAKVLFDKVASQGEVVRKLKTEKAPKDQVDIAVQELLQLKAQYKSLIGVEYKPVSATGAE DKDKKKKEKENKSEKQNKPQKQNDGQRKDPSKNQGGGLSSSGAGEGQGPKKQTRLGLEAKKEENLADWYSQVITKSEMIEYHDISGCYIL RPWAYAIWEAIKDFFDAEIKKLGVENCYFPMFVSQSALEKEKTHVADFAPEVAWVTRSGKTELAEPIAIRPTSETVMYPAYAKWVQSHRD LPIKLNQWCNVVRWEFKHPQPFLRTREFLWQEGHSAFATMEEAAEEVLQILDLYAQVYEELLAIPVVKGRKTEKEKFAGGDYTTTIEAFI SASGRAIQGGTSHHLGQNFSKMFEIVFEDPKIPGEKQFAYQNSWGLTTRTIGVMTMVHGDNMGLVLPPRVACVQVVIIPCGITNALSEED KEALIAKCNDYRRRLLSVNIRVRADLRDNYSPGWKFNHWELKGVPIRLEVGPRDMKSCQFVAVRRDTGEKLTVAENEAETKLQAILEDIQ VTLFTRASEDLKTHMVVANTMEDFQKILDSGKIVQIPFCGEIDCEDWIKKTTARDQDLEPGAPSMGAKSLCIPFKPLCELQPGAKCVCGK -------------------------------------------------------------- >14836_14836_3_CDC42BPA-EPRS_CDC42BPA_chr1_227203781_ENST00000366765_EPRS_chr1_220180680_ENST00000366923_length(amino acids)=2574AA_BP=1874 MSGEVRLRQLEQFILDGPAQTNGQCFSVETLLDILICLYDECNNSPLRREKNILEYLEWAKPFTSKVKQMRLHREDFEILKVIGRGAFGE VAVVKLKNADKVFAMKILNKWEMLKRAETACFREERDVLVNGDNKWITTLHYAFQDDNNLYLVMDYYVGGDLLTLLSKFEDRLPEDMARF YLAEMVIAIDSVHQLHYVHRDIKPDNILMDMNGHIRLADFGSCLKLMEDGTVQSSVAVGTPDYISPEILQAMEDGKGRYGPECDWWSLGV CMYEMLYGETPFYAESLVETYGKIMNHKERFQFPAQVTDVSENAKDLIRRLICSREHRLGQNGIEDFKKHPFFSGIDWDNIRNCEAPYIP EVSSPTDTSNFDVDDDCLKNSETMPPPTHTAFSGHHLPFVGFTYTSSCVLSDRSCLRVTAGPTSLDLDVNVQRTLDNNLATEAYERRIKR LEQEKLELSRKLQESTQTVQALQYSTVDGPLTASKDLEIKNLKEEIEKLRKQVTESSHLEQQLEEANAVRQELDDAFRQIKAYEKQIKTL QQEREDLNKELVQASERLKNQSKELKDAHCQRKLAMQEFMEINERLTELHTQKQKLARHVRDKEEEVDLVMQKVESLRQELRRTERAKKE LEVHTEALAAEASKDRKLREQSEHYSKQLENELEGLKQKQISYSPGVCSIEHQQEITKLKTDLEKKSIFYEEELSKREGIHANEIKNLKK ELHDSEGQQLALNKEIMILKDKLEKTRRESQSEREEFESEFKQQYEREKVLLTEENKKLTSELDKLTTLYENLSIHNQQLEEEVKDLADK KESVAHWEAQITEIIQWVSDEKDARGYLQALASKMTEELEALRNSSLGTRATDMPWKMRRFAKLDMSARLELQSALDAEIRAKQAIQEEL NKVKASNIITECKLKDSEKKNLELLSEIEQLIKDTEELRSEKGIEHQDSQHSFLAFLNTPTDALDQFERSPSCTPASKGRRTVDSTPLSV HTPTLRKKGCPGSTGFPPKRKTHQFFVKSFTTPTKCHQCTSLMVGLIRQGCSCEVCGFSCHITCVNKAPTTCPVPPEQTKGPLGIDPQKG IGTAYEGHVRIPKPAGVKKGWQRALAIVCDFKLFLYDIAEGKASQPSVVISQVIDMRDEEFSVSSVLASDVIHASRKDIPCIFRVTASQL SASNNKCSILMLADTENEKNKWVGVLSELHKILKKNKFRDRSVYVPKEAYDSTLPLIKTTQAAAIIDHERIALGNEEGLFVVHVTKDEII RVGDNKKIHQIELIPNDQLVAVISGRNRHVRLFPMSALDGRETDFYKLSETKGCQTVTSGKVRHGALTCLCVAMKRQVLCYELFQSKTRH RKFKEIQVPYNVQWMAIFSEQLCVGFQSGFLRYPLNGEGNPYSMLHSNDHTLSFIAHQPMDAICAVEISSKEYLLCFNSIGIYTDCQGRR SRQQELMWPANPSSCCYNAPYLSVYSENAVDIFDVNSMEWIQTLPLKKVRPLNNEGSLNLLGLETIRLIYFKNKMAEGDELVVPETSDNS RKQMVRNINNKRRYSFRVPEEERMQQRREMLRDPEMRNKLISNPTNFNHIAHMGPGDGIQILKDLPMNPEVGLKPVWYSPKVFIEGADAE TFSEGEMVTFINWGNLNITKIHKNADGKIISLDAKLNLENKDYKKTTKVTWLAETTHALPIPVICVTYEHLITKPVLGKDEDFKQYVNKN SKHEELMLGDPCLKDLKKGDIIQLQRRGFFICDQPYEPVSPYSCKEAPCVLIYIPDGHTKEMPTSGSKEKTKVEATKNETSAPFKERPTP SLNNNCTTSEDSLVLYNRVAVQGDVVRELKAKKAPKEDVDAAVKQLLSLKAEYKEKTGQEYKPGNPPAEIGQNISSNSSASILESKSLYD EVAAQGEVVRKLKAEKSPKAKINEAVECLLSLKAQYKEKTGKEYIPGQPPLSQSSDSSPTRNSEPAGLETPEAKVLFDKVASQGEVVRKL KTEKAPKDQVDIAVQELLQLKAQYKSLIGVEYKPVSATGAEDKDKKKKEKENKSEKQNKPQKQNDGQRKDPSKNQGGGLSSSGAGEGQGP KKQTRLGLEAKKEENLADWYSQVITKSEMIEYHDISGCYILRPWAYAIWEAIKDFFDAEIKKLGVENCYFPMFVSQSALEKEKTHVADFA PEVAWVTRSGKTELAEPIAIRPTSETVMYPAYAKWVQSHRDLPIKLNQWCNVVRWEFKHPQPFLRTREFLWQEGHSAFATMEEAAEEVLQ ILDLYAQVYEELLAIPVVKGRKTEKEKFAGGDYTTTIEAFISASGRAIQGGTSHHLGQNFSKMFEIVFEDPKIPGEKQFAYQNSWGLTTR TIGVMTMVHGDNMGLVLPPRVACVQVVIIPCGITNALSEEDKEALIAKCNDYRRRLLSVNIRVRADLRDNYSPGWKFNHWELKGVPIRLE VGPRDMKSCQFVAVRRDTGEKLTVAENEAETKLQAILEDIQVTLFTRASEDLKTHMVVANTMEDFQKILDSGKIVQIPFCGEIDCEDWIK -------------------------------------------------------------- >14836_14836_4_CDC42BPA-EPRS_CDC42BPA_chr1_227203781_ENST00000366766_EPRS_chr1_220180680_ENST00000366923_length(amino acids)=2596AA_BP=1896 MSGEVRLRQLEQFILDGPAQTNGQCFSVETLLDILICLYDECNNSPLRREKNILEYLEWAKPFTSKVKQMRLHREDFEILKVIGRGAFGE VAVVKLKNADKVFAMKILNKWEMLKRAETACFREERDVLVNGDNKWITTLHYAFQDDNNLYLVMDYYVGGDLLTLLSKFEDRLPEDMARF YLAEMVIAIDSVHQLHYVHRDIKPDNILMDMNGHIRLADFGSCLKLMEDGTVQSSVAVGTPDYISPEILQAMEDGKGRYGPECDWWSLGV CMYEMLYGETPFYAESLVETYGKIMNHKERFQFPAQVTDVSENAKDLIRRLICSREHRLGQNGIEDFKKHPFFSGIDWDNIRNCEAPYIP EVSSPTDTSNFDVDDDCLKNSETMPPPTHTAFSGHHLPFVGFTYTSSCVLSDRSCLRVTAGPTSLDLDVNVQRTLDNNLATEAYERRIKR LEQEKLELSRKLQESTQTVQALQYSTVDGPLTASKDLEIKNLKEEIEKLRKQVTESSHLEQQLEEANAVRQELDDAFRQIKAYEKQIKTL QQEREDLNKELVQASERLKNQSKELKDAHCQRKLAMQEFMEINERLTELHTQKQKLARHVRDKEEEVDLVMQKVESLRQELRRTERAKKE LEVHTEALAAEASKDRKLREQSEHYSKQLENELEGLKQKQISYSPGVCSIEHQQEITKLKTDLEKKSIFYEEELSKREGIHANEIKNLKK ELHDSEGQQLALNKEIMILKDKLEKTRRESQSEREEFESEFKQQYEREKVLLTEENKKLTSELDKLTTLYENLSIHNQQLEEEVKDLADK KESVAHWEAQITEIIQWVSDEKDARGYLQALASKMTEELEALRNSSLGTRATDMPWKMRRFAKLDMSARLELQSALDAEIRAKQAIQEEL NKVKASNIITECKLKDSEKKNLELLSEIEQLIKDTEELRSEKGIEHQDSQHSFLAFLNTPTDALDQFETDPVENTYVWNPSVKFHIQSRS TSPSTSSEAEPVKTVDSTPLSVHTPTLRKKGCPGSTGFPPKRKTHQFFVKSFTTPTKCHQCTSLMVGLIRQGCSCEVCGFSCHITCVNKA PTTCPVPPEQTKGPLGIDPQKGIGTAYEGHVRIPKPAGVKKGWQRALAIVCDFKLFLYDIAEGKASQPSVVISQVIDMRDEEFSVSSVLA SDVIHASRKDIPCIFRVTASQLSASNNKCSILMLADTENEKNKWVGVLSELHKILKKNKFRDRSVYVPKEAYDSTLPLIKTTQAAAIIDH ERIALGNEEGLFVVHVTKDEIIRVGDNKKIHQIELIPNDQLVAVISGRNRHVRLFPMSALDGRETDFYKLSETKGCQTVTSGKVRHGALT CLCVAMKRQVLCYELFQSKTRHRKFKEIQVPYNVQWMAIFSEQLCVGFQSGFLRYPLNGEGNPYSMLHSNDHTLSFIAHQPMDAICAVEI SSKEYLLCFNSIGIYTDCQGRRSRQQELMWPANPSSCCYNAPYLSVYSENAVDIFDVNSMEWIQTLPLKKVRPLNNEGSLNLLGLETIRL IYFKNKMAEGDELVVPETSDNSRKQMVRNINNKRRYSFRVPEEERMQQRREMLRDPEMRNKLISNPTNFNHIAHMGPGDGIQILKDLPMN PEVGLKPVWYSPKVFIEGADAETFSEGEMVTFINWGNLNITKIHKNADGKIISLDAKLNLENKDYKKTTKVTWLAETTHALPIPVICVTY EHLITKPVLGKDEDFKQYVNKNSKHEELMLGDPCLKDLKKGDIIQLQRRGFFICDQPYEPVSPYSCKEAPCVLIYIPDGHTKEMPTSGSK EKTKVEATKNETSAPFKERPTPSLNNNCTTSEDSLVLYNRVAVQGDVVRELKAKKAPKEDVDAAVKQLLSLKAEYKEKTGQEYKPGNPPA EIGQNISSNSSASILESKSLYDEVAAQGEVVRKLKAEKSPKAKINEAVECLLSLKAQYKEKTGKEYIPGQPPLSQSSDSSPTRNSEPAGL ETPEAKVLFDKVASQGEVVRKLKTEKAPKDQVDIAVQELLQLKAQYKSLIGVEYKPVSATGAEDKDKKKKEKENKSEKQNKPQKQNDGQR KDPSKNQGGGLSSSGAGEGQGPKKQTRLGLEAKKEENLADWYSQVITKSEMIEYHDISGCYILRPWAYAIWEAIKDFFDAEIKKLGVENC YFPMFVSQSALEKEKTHVADFAPEVAWVTRSGKTELAEPIAIRPTSETVMYPAYAKWVQSHRDLPIKLNQWCNVVRWEFKHPQPFLRTRE FLWQEGHSAFATMEEAAEEVLQILDLYAQVYEELLAIPVVKGRKTEKEKFAGGDYTTTIEAFISASGRAIQGGTSHHLGQNFSKMFEIVF EDPKIPGEKQFAYQNSWGLTTRTIGVMTMVHGDNMGLVLPPRVACVQVVIIPCGITNALSEEDKEALIAKCNDYRRRLLSVNIRVRADLR DNYSPGWKFNHWELKGVPIRLEVGPRDMKSCQFVAVRRDTGEKLTVAENEAETKLQAILEDIQVTLFTRASEDLKTHMVVANTMEDFQKI -------------------------------------------------------------- >14836_14836_5_CDC42BPA-EPRS_CDC42BPA_chr1_227203781_ENST00000366767_EPRS_chr1_220180680_ENST00000366923_length(amino acids)=2480AA_BP=1780 MSGEVRLRQLEQFILDGPAQTNGQCFSVETLLDILICLYDECNNSPLRREKNILEYLEWAKPFTSKVKQMRLHREDFEILKVIGRGAFGE VAVVKLKNADKVFAMKILNKWEMLKRAETACFREERDVLVNGDNKWITTLHYAFQDDNNLYLVMDYYVGGDLLTLLSKFEDRLPEDMARF YLAEMVIAIDSVHQLHYVHRDIKPDNILMDMNGHIRLADFGSCLKLMEDGTVQSSVAVGTPDYISPEILQAMEDGKGRYGPECDWWSLGV CMYEMLYGETPFYAESLVETYGKIMNHKERFQFPAQVTDVSENAKDLIRRLICSREHRLGQNGIEDFKKHPFFSGIDWDNIRNCEAPYIP EVSSPTDTSNFDVDDDCLKNSETMPPPTHTAFSGHHLPFVGFTYTSSCVLSDRSCLRVTAGPTSLDLDVNVQRTLDNNLATEAYERRIKR LEQEKLELSRKLQESTQTVQALQYSTVDGPLTASKDLEIKNLKEEIEKLRKQVTESSHLEQQLEEANAVRQELDDAFRQIKAYEKQIKTL QQEREDLNKLEVHTEALAAEASKDRKLREQSEHYSKQLENELEGLKQKQISYSPGVCSIEHQQEITKLKTDLEKKSIFYEEELSKREGIH ANEIKNLKKELHDSEGQQLALNKEIMILKDKLEKTRRESQSEREEFESEFKQQYEREKVLLTEENKKLTSELDKLTTLYENLSIHNQQLE EEVKDLADKKESVAHWEAQITEIIQWVSDEKDARGYLQALASKMTEELEALRNSSLGTRATDMPWKMRRFAKLDMSARLELQSALDAEIR AKQAIQEELNKVKASNIITECKLKDSEKKNLELLSEIEQLIKDTEELRSEKGIEHQDSQHSFLAFLNTPTDALDQFETVDSTPLSVHTPT LRKKGCPGSTGFPPKRKTHQFFVKSFTTPTKCHQCTSLMVGLIRQGCSCEVCGFSCHITCVNKAPTTCPVPPEQTKGPLGIDPQKGIGTA YEGHVRIPKPAGVKKGWQRALAIVCDFKLFLYDIAEGKASQPSVVISQVIDMRDEEFSVSSVLASDVIHASRKDIPCIFRVTASQLSASN NKCSILMLADTENEKNKWVGVLSELHKILKKNKFRDRSVYVPKEAYDSTLPLIKTTQAAAIIDHERIALGNEEGLFVVHVTKDEIIRVGD NKKIHQIELIPNDQLVAVISGRNRHVRLFPMSALDGRETDFYKLSETKGCQTVTSGKVRHGALTCLCVAMKRQVLCYELFQSKTRHRKFK EIQVPYNVQWMAIFSEQLCVGFQSGFLRYPLNGEGNPYSMLHSNDHTLSFIAHQPMDAICAVEISSKEYLLCFNSIGIYTDCQGRRSRQQ ELMWPANPSSCCYNAPYLSVYSENAVDIFDVNSMEWIQTLPLKKVRPLNNEGSLNLLGLETIRLIYFKNKMAEGDELVVPETSDNSRKQM VRNINNKRRYSFRVPEEERMQQRREMLRDPEMRNKLISNPTNFNHIAHMGPGDGIQILKDLPMNPEVGLKPVWYSPKVFIEGADAETFSE GEMVTFINWGNLNITKIHKNADGKIISLDAKLNLENKDYKKTTKVTWLAETTHALPIPVICVTYEHLITKPVLGKDEDFKQYVNKNSKHE ELMLGDPCLKDLKKGDIIQLQRRGFFICDQPYEPVSPYSCKEAPCVLIYIPDGHTKEMPTSGSKEKTKVEATKNETSAPFKERPTPSLNN NCTTSEDSLVLYNRVAVQGDVVRELKAKKAPKEDVDAAVKQLLSLKAEYKEKTGQEYKPGNPPAEIGQNISSNSSASILESKSLYDEVAA QGEVVRKLKAEKSPKAKINEAVECLLSLKAQYKEKTGKEYIPGQPPLSQSSDSSPTRNSEPAGLETPEAKVLFDKVASQGEVVRKLKTEK APKDQVDIAVQELLQLKAQYKSLIGVEYKPVSATGAEDKDKKKKEKENKSEKQNKPQKQNDGQRKDPSKNQGGGLSSSGAGEGQGPKKQT RLGLEAKKEENLADWYSQVITKSEMIEYHDISGCYILRPWAYAIWEAIKDFFDAEIKKLGVENCYFPMFVSQSALEKEKTHVADFAPEVA WVTRSGKTELAEPIAIRPTSETVMYPAYAKWVQSHRDLPIKLNQWCNVVRWEFKHPQPFLRTREFLWQEGHSAFATMEEAAEEVLQILDL YAQVYEELLAIPVVKGRKTEKEKFAGGDYTTTIEAFISASGRAIQGGTSHHLGQNFSKMFEIVFEDPKIPGEKQFAYQNSWGLTTRTIGV MTMVHGDNMGLVLPPRVACVQVVIIPCGITNALSEEDKEALIAKCNDYRRRLLSVNIRVRADLRDNYSPGWKFNHWELKGVPIRLEVGPR DMKSCQFVAVRRDTGEKLTVAENEAETKLQAILEDIQVTLFTRASEDLKTHMVVANTMEDFQKILDSGKIVQIPFCGEIDCEDWIKKTTA -------------------------------------------------------------- >14836_14836_6_CDC42BPA-EPRS_CDC42BPA_chr1_227203781_ENST00000366769_EPRS_chr1_220180680_ENST00000366923_length(amino acids)=2561AA_BP=1861 MSGEVRLRQLEQFILDGPAQTNGQCFSVETLLDILICLYDECNNSPLRREKNILEYLEWAKPFTSKVKQMRLHREDFEILKVIGRGAFGE VAVVKLKNADKVFAMKILNKWEMLKRAETACFREERDVLVNGDNKWITTLHYAFQDDNNLYLVMDYYVGGDLLTLLSKFEDRLPEDMARF YLAEMVIAIDSVHQLHYVHRDIKPDNILMDMNGHIRLADFGSCLKLMEDGTVQSSVAVGTPDYISPEILQAMEDGKGRYGPECDWWSLGV CMYEMLYGETPFYAESLVETYGKIMNHKERFQFPAQVTDVSENAKDLIRRLICSREHRLGQNGIEDFKKHPFFSGIDWDNIRNCEAPYIP EVSSPTDTSNFDVDDDCLKNSETMPPPTHTAFSGHHLPFVGFTYTSSCVLSDRSCLRVTAGPTSLDLDVNVQRTLDNNLATEAYERRIKR LEQEKLELSRKLQESTQTVQALQYSTVDGPLTASKDLEIKNLKEEIEKLRKQVTESSHLEQQLEEANAVRQELDDAFRQIKAYEKQIKTL QQEREDLNKELVQASERLKNQSKELKDAHCQRKLAMQEFMEINERLTELHTQKQKLARHVRDKEEEVDLVMQKVESLRQELRRTERAKKE LEVHTEALAAEASKDRKLREQSEHYSKQLENELEGLKQKQISYSPGVCSIEHQQEITKLKTDLEKKSIFYEEELSKREGIHANEIKNLKK ELHDSEGQQLALNKEIMILKDKLEKTRRESQSEREEFESEFKQQYEREKVLLTEENKKLTSELDKLTTLYENLSIHNQQLEEEVKDLADK KESVAHWEAQITEIIQWVSDEKDARGYLQALASKMTEELEALRNSSLGTRATDMPWKMRRFAKLDMSARLELQSALDAEIRAKQAIQEEL NKVKASNIITECKLKDSEKKNLELLSEIEQLIKDTEELRSEKGIEHQDSQHSFLAFLNTPTDALDQFETVDSTPLSVHTPTLRKKGCPGS TGFPPKRKTHQFFVKSFTTPTKCHQCTSLMVGLIRQGCSCEVCGFSCHITCVNKAPTTCPVPPEQTKGPLGIDPQKGIGTAYEGHVRIPK PAGVKKGWQRALAIVCDFKLFLYDIAEGKASQPSVVISQVIDMRDEEFSVSSVLASDVIHASRKDIPCIFRVTASQLSASNNKCSILMLA DTENEKNKWVGVLSELHKILKKNKFRDRSVYVPKEAYDSTLPLIKTTQAAAIIDHERIALGNEEGLFVVHVTKDEIIRVGDNKKIHQIEL IPNDQLVAVISGRNRHVRLFPMSALDGRETDFYKLSETKGCQTVTSGKVRHGALTCLCVAMKRQVLCYELFQSKTRHRKFKEIQVPYNVQ WMAIFSEQLCVGFQSGFLRYPLNGEGNPYSMLHSNDHTLSFIAHQPMDAICAVEISSKEYLLCFNSIGIYTDCQGRRSRQQELMWPANPS SCCYNAPYLSVYSENAVDIFDVNSMEWIQTLPLKKVRPLNNEGSLNLLGLETIRLIYFKNKMAEGDELVVPETSDNSRKQMVRNINNKRR YSFRVPEEERMQQRREMLRDPEMRNKLISNPTNFNHIAHMGPGDGIQILKDLPMNPEVGLKPVWYSPKVFIEGADAETFSEGEMVTFINW GNLNITKIHKNADGKIISLDAKLNLENKDYKKTTKVTWLAETTHALPIPVICVTYEHLITKPVLGKDEDFKQYVNKNSKHEELMLGDPCL KDLKKGDIIQLQRRGFFICDQPYEPVSPYSCKEAPCVLIYIPDGHTKEMPTSGSKEKTKVEATKNETSAPFKERPTPSLNNNCTTSEDSL VLYNRVAVQGDVVRELKAKKAPKEDVDAAVKQLLSLKAEYKEKTGQEYKPGNPPAEIGQNISSNSSASILESKSLYDEVAAQGEVVRKLK AEKSPKAKINEAVECLLSLKAQYKEKTGKEYIPGQPPLSQSSDSSPTRNSEPAGLETPEAKVLFDKVASQGEVVRKLKTEKAPKDQVDIA VQELLQLKAQYKSLIGVEYKPVSATGAEDKDKKKKEKENKSEKQNKPQKQNDGQRKDPSKNQGGGLSSSGAGEGQGPKKQTRLGLEAKKE ENLADWYSQVITKSEMIEYHDISGCYILRPWAYAIWEAIKDFFDAEIKKLGVENCYFPMFVSQSALEKEKTHVADFAPEVAWVTRSGKTE LAEPIAIRPTSETVMYPAYAKWVQSHRDLPIKLNQWCNVVRWEFKHPQPFLRTREFLWQEGHSAFATMEEAAEEVLQILDLYAQVYEELL AIPVVKGRKTEKEKFAGGDYTTTIEAFISASGRAIQGGTSHHLGQNFSKMFEIVFEDPKIPGEKQFAYQNSWGLTTRTIGVMTMVHGDNM GLVLPPRVACVQVVIIPCGITNALSEEDKEALIAKCNDYRRRLLSVNIRVRADLRDNYSPGWKFNHWELKGVPIRLEVGPRDMKSCQFVA VRRDTGEKLTVAENEAETKLQAILEDIQVTLFTRASEDLKTHMVVANTMEDFQKILDSGKIVQIPFCGEIDCEDWIKKTTARDQDLEPGA -------------------------------------------------------------- >14836_14836_7_CDC42BPA-EPRS_CDC42BPA_chr1_227203781_ENST00000535525_EPRS_chr1_220180680_ENST00000366923_length(amino acids)=2541AA_BP=1841 MSGEVRLRQLEQFILDGPAQTNGQCFSVETLLDILICLYDECNNSPLRREKNILEYLEWAKPFTSKVKQMRLHREDFEILKVIGRGAFGE VAVVKLKNADKVFAMKILNKWEMLKRAETACFREERDVLVNGDNKWITTLHYAFQDDNNLYLVMDYYVGGDLLTLLSKFEDRLPEDMARF YLAEMVIAIDSVHQLHYVHRDIKPDNILMDMNGHIRLADFGSCLKLMEDGTVQSSVAVGTPDYISPEILQAMEDGKGRYGPECDWWSLGV CMYEMLYGETPFYAESLVETYGKIMNHKERFQFPAQVTDVSENAKDLIRRLICSREHRLGQNGIEDFKKHPFFSGIDWDNIRNCEAPYIP EVSSPTDTSNFDVDDDCLKNSETMPPPTHTAFSGHHLPFVGFTYTSSCVLSDRSCLRVTAGPTSLDLDVNVQRTLDNNLATEAYERRIKR LEQEKLELSRKLQESTQTVQALQYSTVDGPLTASKDLEIKNLKEEIEKLRKQVTESSHLEQQLEEANAVRQELDDAFRQIKAYEKQIKTL QQEREDLNKELVQASERLKNQSKELKDAHCQRKLAMQEFMEINERLTELHTQKQKLARHVRDKEEEVDLVMQKVESLRQELRRTERAKKE LEVHTEALAAEASKDRKLREQSEHYSKQLENELEGLKQKQISYSPGVCSIEHQQEITKLKTDLEKKSIFYEEELSKREGIHANEIKNLKK ELHDSEGQQLALNKEIMILKDKLEKTRRESQSEREEFESEFKQQYEREKVLLTEENKKLTSELDKLTTLYENLSIHNQQLEEEVKDLADK KESVAHWEAQITEIIQWVSDEKDARGYLQALASKMTEELEALRNSSLGTRATDMPWKMRRFAKLDMSARLELQSALDAEIRAKQAIQEEL NKVKASNIITECKLKDSEKKNLELLSEIEQLIKDTEELRSEKASKGRRTVDSTPLSVHTPTLRKKGCPGSTGFPPKRKTHQFFVKSFTTP TKCHQCTSLMVGLIRQGCSCEVCGFSCHITCVNKAPTTCPVPPEQTKGPLGIDPQKGIGTAYEGHVRIPKPAGVKKGWQRALAIVCDFKL FLYDIAEGKASQPSVVISQVIDMRDEEFSVSSVLASDVIHASRKDIPCIFRVTASQLSASNNKCSILMLADTENEKNKWVGVLSELHKIL KKNKFRDRSVYVPKEAYDSTLPLIKTTQAAAIIDHERIALGNEEGLFVVHVTKDEIIRVGDNKKIHQIELIPNDQLVAVISGRNRHVRLF PMSALDGRETDFYKLSETKGCQTVTSGKVRHGALTCLCVAMKRQVLCYELFQSKTRHRKFKEIQVPYNVQWMAIFSEQLCVGFQSGFLRY PLNGEGNPYSMLHSNDHTLSFIAHQPMDAICAVEISSKEYLLCFNSIGIYTDCQGRRSRQQELMWPANPSSCCYNAPYLSVYSENAVDIF DVNSMEWIQTLPLKKVRPLNNEGSLNLLGLETIRLIYFKNKMAEGDELVVPETSDNSRKQMVRNINNKRRYSFRVPEEERMQQRREMLRD PEMRNKLISNPTNFNHIAHMGPGDGIQILKDLPMNPEVGLKPVWYSPKVFIEGADAETFSEGEMVTFINWGNLNITKIHKNADGKIISLD AKLNLENKDYKKTTKVTWLAETTHALPIPVICVTYEHLITKPVLGKDEDFKQYVNKNSKHEELMLGDPCLKDLKKGDIIQLQRRGFFICD QPYEPVSPYSCKEAPCVLIYIPDGHTKEMPTSGSKEKTKVEATKNETSAPFKERPTPSLNNNCTTSEDSLVLYNRVAVQGDVVRELKAKK APKEDVDAAVKQLLSLKAEYKEKTGQEYKPGNPPAEIGQNISSNSSASILESKSLYDEVAAQGEVVRKLKAEKSPKAKINEAVECLLSLK AQYKEKTGKEYIPGQPPLSQSSDSSPTRNSEPAGLETPEAKVLFDKVASQGEVVRKLKTEKAPKDQVDIAVQELLQLKAQYKSLIGVEYK PVSATGAEDKDKKKKEKENKSEKQNKPQKQNDGQRKDPSKNQGGGLSSSGAGEGQGPKKQTRLGLEAKKEENLADWYSQVITKSEMIEYH DISGCYILRPWAYAIWEAIKDFFDAEIKKLGVENCYFPMFVSQSALEKEKTHVADFAPEVAWVTRSGKTELAEPIAIRPTSETVMYPAYA KWVQSHRDLPIKLNQWCNVVRWEFKHPQPFLRTREFLWQEGHSAFATMEEAAEEVLQILDLYAQVYEELLAIPVVKGRKTEKEKFAGGDY TTTIEAFISASGRAIQGGTSHHLGQNFSKMFEIVFEDPKIPGEKQFAYQNSWGLTTRTIGVMTMVHGDNMGLVLPPRVACVQVVIIPCGI TNALSEEDKEALIAKCNDYRRRLLSVNIRVRADLRDNYSPGWKFNHWELKGVPIRLEVGPRDMKSCQFVAVRRDTGEKLTVAENEAETKL QAILEDIQVTLFTRASEDLKTHMVVANTMEDFQKILDSGKIVQIPFCGEIDCEDWIKKTTARDQDLEPGAPSMGAKSLCIPFKPLCELQP -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:227203781/chr1:220180680) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CDC42BPA | EPRS |
| FUNCTION: Serine/threonine-protein kinase which is an important downstream effector of CDC42 and plays a role in the regulation of cytoskeleton reorganization and cell migration (PubMed:15723050, PubMed:9418861, PubMed:9092543). Regulates actin cytoskeletal reorganization via phosphorylation of PPP1R12C and MYL9/MLC2 (PubMed:21457715). In concert with MYO18A and LURAP1, is involved in modulating lamellar actomyosin retrograde flow that is crucial to cell protrusion and migration (PubMed:18854160). Phosphorylates: PPP1R12A, LIMK1 and LIMK2 (PubMed:11340065, PubMed:11399775). May play a role in TFRC-mediated iron uptake (PubMed:20188707). In concert with FAM89B/LRAP25 mediates the targeting of LIMK1 to the lamellipodium resulting in its activation and subsequent phosphorylation of CFL1 which is important for lamellipodial F-actin regulation (By similarity). Triggers the formation of an extrusion apical actin ring required for epithelial extrusion of apoptotic cells (PubMed:29162624). {ECO:0000250|UniProtKB:Q3UU96, ECO:0000269|PubMed:11340065, ECO:0000269|PubMed:11399775, ECO:0000269|PubMed:15723050, ECO:0000269|PubMed:18854160, ECO:0000269|PubMed:20188707, ECO:0000269|PubMed:21457715, ECO:0000269|PubMed:29162624, ECO:0000269|PubMed:9092543, ECO:0000269|PubMed:9418861}. | FUNCTION: Multifunctional protein which is primarily part of the aminoacyl-tRNA synthetase multienzyme complex, also know as multisynthetase complex, that catalyzes the attachment of the cognate amino acid to the corresponding tRNA in a two-step reaction: the amino acid is first activated by ATP to form a covalent intermediate with AMP and is then transferred to the acceptor end of the cognate tRNA (PubMed:1756734, PubMed:24100331, PubMed:23263184). The phosphorylation of EPRS1, induced by interferon-gamma, dissociates the protein from the aminoacyl-tRNA synthetase multienzyme complex and recruits it to the GAIT complex that binds to stem loop-containing GAIT elements in the 3'-UTR of diverse inflammatory mRNAs (such as ceruplasmin), suppressing their translation. Interferon-gamma can therefore redirect, in specific cells, the EPRS1 function from protein synthesis to translation inhibition (PubMed:15479637, PubMed:23071094). Also functions as an effector of the mTORC1 signaling pathway by promoting, through SLC27A1, the uptake of long-chain fatty acid by adipocytes. Thereby, it also plays a role in fat metabolism and more indirectly influences lifespan (PubMed:28178239). {ECO:0000269|PubMed:15479637, ECO:0000269|PubMed:1756734, ECO:0000269|PubMed:23071094, ECO:0000269|PubMed:23263184, ECO:0000269|PubMed:24100331, ECO:0000269|PubMed:28178239}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000334218 | - | 33 | 37 | 437_820 | 1584.0 | 1782.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000334218 | - | 33 | 37 | 880_943 | 1584.0 | 1782.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366764 | - | 32 | 35 | 437_820 | 1556.0 | 1692.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366764 | - | 32 | 35 | 880_943 | 1556.0 | 1692.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366765 | - | 34 | 37 | 437_820 | 1597.0 | 1733.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366765 | - | 34 | 37 | 880_943 | 1597.0 | 1733.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366766 | - | 34 | 37 | 437_820 | 1619.0 | 1755.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366766 | - | 34 | 37 | 880_943 | 1619.0 | 1755.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366767 | - | 32 | 35 | 437_820 | 1503.0 | 1639.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366767 | - | 32 | 35 | 880_943 | 1503.0 | 1639.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366769 | - | 33 | 36 | 437_820 | 1584.0 | 1720.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366769 | - | 33 | 36 | 880_943 | 1584.0 | 1720.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000334218 | - | 33 | 37 | 1082_1201 | 1584.0 | 1782.0 | Domain | PH |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000334218 | - | 33 | 37 | 1227_1499 | 1584.0 | 1782.0 | Domain | CNH |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000334218 | - | 33 | 37 | 1571_1584 | 1584.0 | 1782.0 | Domain | CRIB |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000334218 | - | 33 | 37 | 344_414 | 1584.0 | 1782.0 | Domain | AGC-kinase C-terminal |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000334218 | - | 33 | 37 | 77_343 | 1584.0 | 1782.0 | Domain | Protein kinase |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366764 | - | 32 | 35 | 1082_1201 | 1556.0 | 1692.0 | Domain | PH |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366764 | - | 32 | 35 | 1227_1499 | 1556.0 | 1692.0 | Domain | CNH |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366764 | - | 32 | 35 | 344_414 | 1556.0 | 1692.0 | Domain | AGC-kinase C-terminal |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366764 | - | 32 | 35 | 77_343 | 1556.0 | 1692.0 | Domain | Protein kinase |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366765 | - | 34 | 37 | 1082_1201 | 1597.0 | 1733.0 | Domain | PH |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366765 | - | 34 | 37 | 1227_1499 | 1597.0 | 1733.0 | Domain | CNH |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366765 | - | 34 | 37 | 1571_1584 | 1597.0 | 1733.0 | Domain | CRIB |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366765 | - | 34 | 37 | 344_414 | 1597.0 | 1733.0 | Domain | AGC-kinase C-terminal |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366765 | - | 34 | 37 | 77_343 | 1597.0 | 1733.0 | Domain | Protein kinase |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366766 | - | 34 | 37 | 1082_1201 | 1619.0 | 1755.0 | Domain | PH |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366766 | - | 34 | 37 | 1227_1499 | 1619.0 | 1755.0 | Domain | CNH |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366766 | - | 34 | 37 | 1571_1584 | 1619.0 | 1755.0 | Domain | CRIB |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366766 | - | 34 | 37 | 344_414 | 1619.0 | 1755.0 | Domain | AGC-kinase C-terminal |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366766 | - | 34 | 37 | 77_343 | 1619.0 | 1755.0 | Domain | Protein kinase |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366767 | - | 32 | 35 | 1082_1201 | 1503.0 | 1639.0 | Domain | PH |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366767 | - | 32 | 35 | 1227_1499 | 1503.0 | 1639.0 | Domain | CNH |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366767 | - | 32 | 35 | 344_414 | 1503.0 | 1639.0 | Domain | AGC-kinase C-terminal |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366767 | - | 32 | 35 | 77_343 | 1503.0 | 1639.0 | Domain | Protein kinase |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366769 | - | 33 | 36 | 1082_1201 | 1584.0 | 1720.0 | Domain | PH |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366769 | - | 33 | 36 | 1227_1499 | 1584.0 | 1720.0 | Domain | CNH |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366769 | - | 33 | 36 | 1571_1584 | 1584.0 | 1720.0 | Domain | CRIB |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366769 | - | 33 | 36 | 344_414 | 1584.0 | 1720.0 | Domain | AGC-kinase C-terminal |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366769 | - | 33 | 36 | 77_343 | 1584.0 | 1720.0 | Domain | Protein kinase |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000334218 | - | 33 | 37 | 83_91 | 1584.0 | 1782.0 | Nucleotide binding | ATP |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366764 | - | 32 | 35 | 83_91 | 1556.0 | 1692.0 | Nucleotide binding | ATP |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366765 | - | 34 | 37 | 83_91 | 1597.0 | 1733.0 | Nucleotide binding | ATP |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366766 | - | 34 | 37 | 83_91 | 1619.0 | 1755.0 | Nucleotide binding | ATP |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366767 | - | 32 | 35 | 83_91 | 1503.0 | 1639.0 | Nucleotide binding | ATP |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366769 | - | 33 | 36 | 83_91 | 1584.0 | 1720.0 | Nucleotide binding | ATP |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000334218 | - | 33 | 37 | 1012_1062 | 1584.0 | 1782.0 | Zinc finger | Phorbol-ester/DAG-type |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366764 | - | 32 | 35 | 1012_1062 | 1556.0 | 1692.0 | Zinc finger | Phorbol-ester/DAG-type |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366765 | - | 34 | 37 | 1012_1062 | 1597.0 | 1733.0 | Zinc finger | Phorbol-ester/DAG-type |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366766 | - | 34 | 37 | 1012_1062 | 1619.0 | 1755.0 | Zinc finger | Phorbol-ester/DAG-type |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366767 | - | 32 | 35 | 1012_1062 | 1503.0 | 1639.0 | Zinc finger | Phorbol-ester/DAG-type |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366769 | - | 33 | 36 | 1012_1062 | 1584.0 | 1720.0 | Zinc finger | Phorbol-ester/DAG-type |
| Tgene | EPRS | chr1:227203781 | chr1:220180680 | ENST00000366923 | 12 | 32 | 961_991 | 535.0 | 1513.0 | Compositional bias | Lys-rich | |
| Tgene | EPRS | chr1:227203781 | chr1:220180680 | ENST00000366923 | 12 | 32 | 749_805 | 535.0 | 1513.0 | Domain | Note=WHEP-TRS 1 | |
| Tgene | EPRS | chr1:227203781 | chr1:220180680 | ENST00000366923 | 12 | 32 | 822_878 | 535.0 | 1513.0 | Domain | Note=WHEP-TRS 2 | |
| Tgene | EPRS | chr1:227203781 | chr1:220180680 | ENST00000366923 | 12 | 32 | 900_956 | 535.0 | 1513.0 | Domain | Note=WHEP-TRS 3 | |
| Tgene | EPRS | chr1:227203781 | chr1:220180680 | ENST00000366923 | 12 | 32 | 1152_1154 | 535.0 | 1513.0 | Nucleotide binding | ATP | |
| Tgene | EPRS | chr1:227203781 | chr1:220180680 | ENST00000366923 | 12 | 32 | 1163_1164 | 535.0 | 1513.0 | Nucleotide binding | ATP | |
| Tgene | EPRS | chr1:227203781 | chr1:220180680 | ENST00000366923 | 12 | 32 | 1237_1240 | 535.0 | 1513.0 | Nucleotide binding | ATP | |
| Tgene | EPRS | chr1:227203781 | chr1:220180680 | ENST00000366923 | 12 | 32 | 1007_1512 | 535.0 | 1513.0 | Region | Note=Proline--tRNA ligase | |
| Tgene | EPRS | chr1:227203781 | chr1:220180680 | ENST00000366923 | 12 | 32 | 1121_1123 | 535.0 | 1513.0 | Region | L-proline binding | |
| Tgene | EPRS | chr1:227203781 | chr1:220180680 | ENST00000366923 | 12 | 32 | 760_956 | 535.0 | 1513.0 | Region | Note=3 X 57 AA approximate repeats | |
| Tgene | EPRS | chr1:227203781 | chr1:220180680 | ENST00000366923 | 12 | 32 | 959_991 | 535.0 | 1513.0 | Region | Note=Charged |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366764 | - | 32 | 35 | 1571_1584 | 1556.0 | 1692.0 | Domain | CRIB |
| Hgene | CDC42BPA | chr1:227203781 | chr1:220180680 | ENST00000366767 | - | 32 | 35 | 1571_1584 | 1503.0 | 1639.0 | Domain | CRIB |
| Tgene | EPRS | chr1:227203781 | chr1:220180680 | ENST00000366923 | 12 | 32 | 204_214 | 535.0 | 1513.0 | Motif | Note='HIGH' region | |
| Tgene | EPRS | chr1:227203781 | chr1:220180680 | ENST00000366923 | 12 | 32 | 432_436 | 535.0 | 1513.0 | Motif | Note='KMSKS' region | |
| Tgene | EPRS | chr1:227203781 | chr1:220180680 | ENST00000366923 | 12 | 32 | 164_759 | 535.0 | 1513.0 | Region | Note=Glutamate--tRNA ligase |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| CDC42BPA | |
| EPRS |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to CDC42BPA-EPRS |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to CDC42BPA-EPRS |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

